Abstract
Cks is an evolutionarily conserved protein that regulates cyclin-dependent kinase (Cdk) activity. Clarifying the underlying mechanisms and cellular contexts of Cks function is critical, as Cks is essential for proper cell growth, and its overexpression has been linked to cancer. We observe that budding yeast Cks associates with select phosphorylated sequences in cell cycle regulatory proteins. We characterize the molecular interactions responsible for this specificity and demonstrate that Cks enhances Cdk activity in response to specific priming phosphosites. Identification of the binding consensus sequence allows us to identify putative Cks-directed Cdk substrates and binding partners. We characterize novel Cks binding sites in the mitotic regulator Wee1 and discover a novel role for Cks in regulating Cdk activity at mitotic entry. Together our results portray Cks as a multifunctional phosphoadaptor that serves as a specificity factor for Cdk activity.
Protein kinases must recognize their proper target substrates and regulators among a large number of proteins in the cell1. How specificity is achieved is a critical question considering the prominent role of phosphorylation in signal transduction and the misregulation of kinase activity in disease2. In the cell cycle, cyclin-dependent kinases (Cdks) process signals that lead to cell division3. Hundreds of Cdk substrates have been identified in proteomic screens, and Cdk phosphorylation alters the location, interactions, stability, and activity of these target proteins4,5. A deregulated cell cycle is a hallmark of cancer, emphasizing the need for tight coordination of Cdk activity6. Still, many questions remain regarding how regulatory proteins recognize Cdks and how Cdks discriminate among substrates to phosphorylate them in the appropriate order and at the appropriate times in the cell cycle.
The Cdk complex is composed of the kinase subunit, the cyclin subunit, and Cks. The active site of the kinase recognizes a minimum consensus sequence of (S/T)P and an optimal consensus of (S/T)PX(R/K)7. In addition to activating the kinase domain, the cyclin subunit binds docking sequences present in some substrates and confers specificity8,9. Although Cks is essential for viability and its deregulated expression correlates with tumorigenesis and poor cancer prognosis, its particular molecular functions have been less clear10–16. Genetic analysis has shown Cks genes regulate cell growth and division10,11,14. In addition to binding Cdk and influencing kinase function, Cks has been implicated in other cellular processes such as transcription and the degradation of the Cdk inhibitor p2717–20.
Several studies have suggested that Cks associates with phosphorylated cell cycle regulator proteins and plays a role in Cdk multisite phosphorylation. Multisite phosphorylation is critical for producing proper signaling output21–26, as it influences properties such as sensitivity and switch-like behavior and permits integration of a large number of inputs to produce diverse outputs27–30. Entry into mitosis, for example, is a switch-like transition, and multisite phosphorylation of the mitotic regulators Wee1 and Cdc25 is critical for this behavior22,23,25,31,32. Similarly, multisite phosphorylation of Sic1 in budding yeast generates an ultrasensitive response for S phase entry21,24. Different signaling behaviors are generated by differences in enzyme mechanism such as degree of cooperativity and processivity of substrate processing29. Therefore, uncovering mechanistic details of how Cdk acts on multisite substrates is important for understanding the molecular origins of highly complex, vastly tunable signaling through phosphorylation.
A role for Cks in binding phosphorylated substrates was postulated after structures of Cks revealed a conserved cationic pocket that weakly binds free phosphate and other anions33,34. When Cks is bound to Cdk, structural modeling suggests this cationic pocket is part of a continuous surface including the Cdk active site and cyclin35. In the specific context of the Skp2-Skp1-Cullin ubiquitin ligase, human Cks1 binds phosphorylated p27 to stimulate its ubiquitylation and degradation36. From these structural insights, it has been proposed that Cks binds Cdk substrates primed by initial phosphorylation and facilitates further phosphorylation of the primed substrate. This hypothesis is supported by experiments that show phosphorylation of cell cycle regulatory proteins is reduced when Cks is immuno-depleted from Xenopus egg extracts37. We found that semiprocessive phosphorylation of the G1/S regulator Sic1 depends on an intact Cks cationic pocket24. However, it has not been clarified whether the stimulatory role of Cks on Cdk activity relies on specific priming sites or whether any site can prime the multisite phosphorylation reaction.
We demonstrate here that Cks recognizes specific phosphorylated sequences in Cdk substrates, uncovering a novel mechanism for Cdk substrate targeting, Cdk interactions with its regulators, and signaling kinetics. We identify a Cks-binding consensus sequence and characterize the complex with a phosphopeptide-Cks crystal structure. Cks interaction with the consensus sequence is required for efficient multisite phosphorylation of a subset of Cdk substrates and for budding yeast viability. Using the Cks consensus as a search motif, we identify novel Cks-binding proteins and demonstrate a critical role for Cks in facilitating Wee1 regulation of Cdk at mitotic entry.
Results
Cks binds to specific phosphorylated sites in Cdk substrates
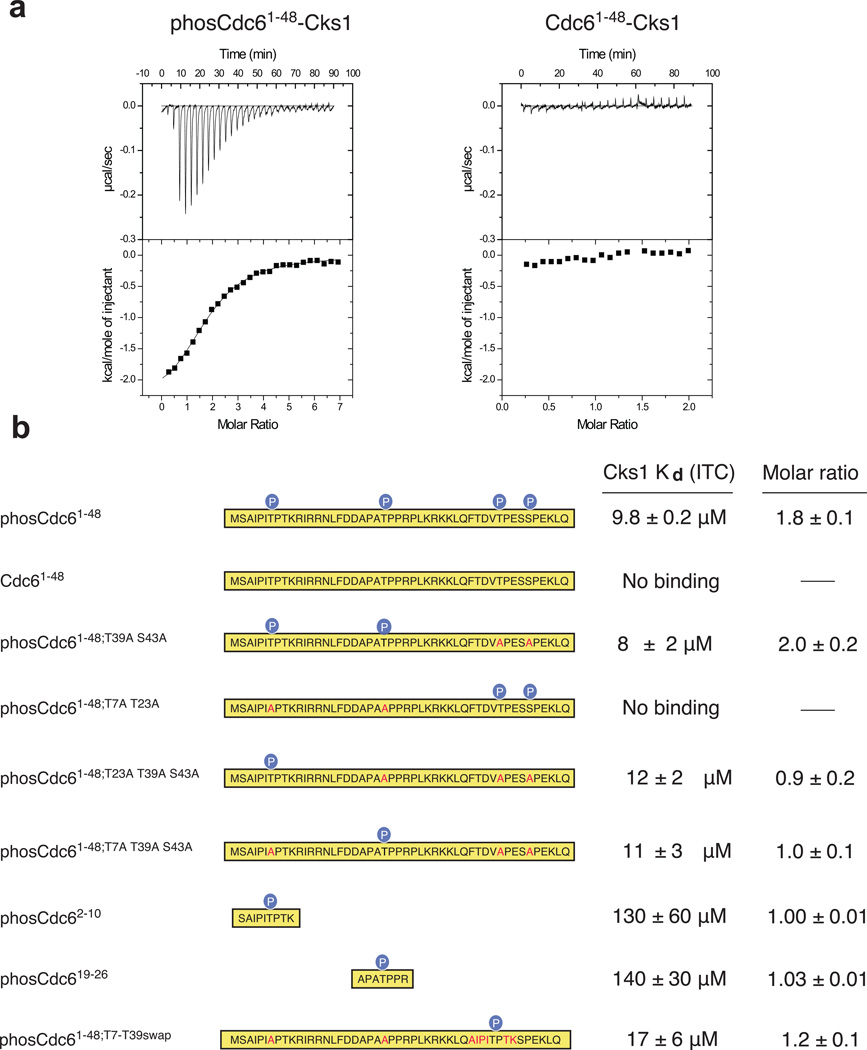
A number of phosphorylation-dependent Cdk-substrate complexes have been reported22,38, and we and others have demonstrated that Cks binds to phosphorylated sequences in intrinsically disordered domains of several Cdk substrates24,39. To test whether Cks binds specific phosphorylated sites, we examined the affinity of S. cerevisiae Cks (Cks1) for the N-terminal domain of the replication-licensing factor Cdc6. As determined by isothermal titration calorimetry (ITC), Cks1 binds to phosphorylated Cdc61–48 with Kd = 9.8 ± 0.2 µM, while no detectable heat was measured for the unphosphorylated protein (Figure 1a). The presence of two distinct and independent Cks1 binding sites in phosCdc61–48 is supported by the stoichiometry parameter; the data fit to a one site model with n ≈ 2, indicating that two molecules of Cks1 bind noncooperatively to each phosphorylated Cdc6 molecule.
Figure 1.
Cks1 binds specific phosphorylated Cdk sites in the N-terminal domain of Cdc6. (a) Purified Cdc61–48 was phosphorylated with recombinant Cdk and affinity for Cks1 was measured by isothermal titration calorimetry. phosCdc61–48 binds Cks1 with Kd = 9.8 ± 0.2 µM (fit of data on left), while titration of unphosphorylated Cdc61–48 does not yield any binding heat (right). (b) ITC measurements with phosphorylated Cdc61–48 constructs and synthetic peptides demonstrate that two sites, T7 and T23, are each sufficient for Cks binding. Sequence mutations are highlighted in red within each construct. ITC data curves not shown in part (a) are shown in Supplementary Figure 1A–1G.
We then mutated different combinations of phosphoacceptor sites and found that Cks1 binds to Cdc61–48 phosphorylated only at T7 and T23 (Cdc61–48;T39A S43A) with a similar affinity and stoichiometry as fully phosphorylated Cdc61–48 (Figure 1b and Supplementary Figure 1). In contrast, no detectable binding signal was observed with Cdc61–48 phosphorylated only at T39 and S43. Cdc6 constructs in which only T7 (Cdc61–48;T23A T39A S43A) or T23 (Cdc61–48;T7A T39A S43A) are phosphorylated bind with similar affinity as the wild-type protein but have stoichiometries n ≈ 1. We also tested binding of Cks1 to synthetic peptides, phosCdc2–10 and phosCdc619–26, which only contain phosphorylated T7 or T23 respectively. Each bound to Cks1 with an affinity about 10-fold less than phosphorylated Cdc1–48. A Cdc61–48 construct in which T7 and T23 are mutated to alanine and the sequence surrounding T39 is replaced with the T7 sequence (Cdc61–48;T7-T39swap) binds Cks1 with wild-type affinity. We conclude that the majority of the cohesive interactions are contained within short sequences including the phosphothreonines and suggest that the peptides bind with slightly lower affinity due to a greater entropy penalty. Together these data demonstrate that T7 or T23 phosphorylation is necessary and sufficient for Cks1 association and that Cks1 has binding requirements beyond the minimum phosphorylated (S/T)P Cdk site.
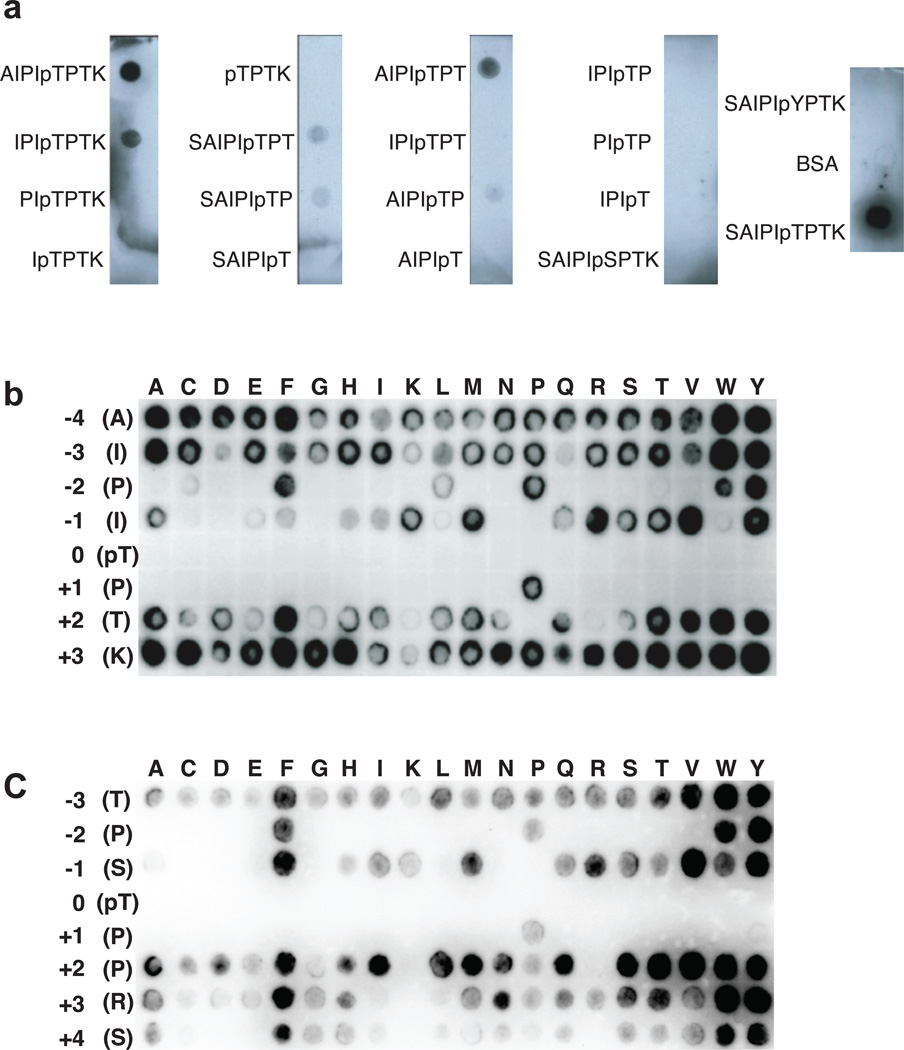
We tested the minimum sequence around T7 required for binding Cks1 using a dot-blot assay. Synthetic phosphopeptides were cross-linked to BSA, spotted onto a PVDF membrane, and the membrane was then incubated with His-tagged Cks1 and probed with an anti-His antibody (Figure 2a). We found that the 7-mers AIPIpTPT and IPIpTPTK peptide were sufficient for Cks1 binding. ITC measurements confirmed that AIPI(pT)P, SAIPI(pT)P, and AIPI(pT)PT bind Cks1 with similar affinity as phosCdc62–10 (Table 1). We substituted phosphoserine and phosphotyrosine for phosphothreonine and observed no binding to Cks1 in both the dot-blot and ITC assays.
Figure 2.
Sequence requirements for Cks1 binding. (a) Dot-blot peptide experiment to determine the minimum length required for association with Cks1. The indicated peptides were conjugated to BSA, spotted onto PVDF membrane, incubated with His-tagged Cks1, and probed with an anti-His antibody. The data demonstrate the requirement of a 7-mer containing a phosphothreonine. The full blot for this experiment can be seen in Supplemental Figure 7a. (b) Scanning mutagenesis SPOT peptide array based on the Cdc63–10 AIPIpTPTK peptide. Peptides containing the indicated single-amino acid substitution were synthesized onto the membrane and probed for Cks1 binding as in (a). (c) Similar to (b), except the array is constructed from a phosphorylated Sic12–9 peptide.
Table 1.
Binding affinities of Cks1 to phosphorylated Cdk substrate peptides
| Peptide | Sequence | Cks1 Kd (µM) | Supplemental Figure |
|---|---|---|---|
| phosCdc62–10 | SAIPI(pT)PTK | 130 ± 60 | 1E |
| phosCdc619–26 | APAp(T)PPR | 140 ± 30 | 1F |
| phosCdc63–8 | AIPI(pT)P | 81 ± 6 | 1H |
| phosCdc62–8 | SAIPI(pT)P | 62 ± 2 | 1I |
| phosCdc63–9 | AIPI(pT)PT | 61 ± 9 | 1J |
| phosCdc62–10; pT7pS | SAIPI(pS)PTK | no heat | 1K |
| phosCdc62–10; pT7pY | SAIPI(pY)PTK | no heat | 1L |
| phosCdc62–10; P8K | AIPI(pT)KTK | no heat | 1M |
| phosCdc62–10; P5Y | AIYI(pT)PTK | 190 ± 1 | 1N |
| phosCdc62–10; P5F | AIFI(pT)PTK | 50 ± 30 | 1O |
| phosCdc62–10; P5K | AIKI(pT)PTK | no heat | 1P |
| phosCdc62–10; T9P | AIPI(pT)PPK | 129 ± 8 | 1Q |
| phosCdc62–10; T9K | AIPI(pT)PKK | 158 ± 4 | 1R |
| phosSic12–9 | TPS(pT)PPRS | 95 ± 8 | 1S |
| phosSic12–9; pT5pS | TPS(pS)PPRS | no heat | 1T |
| phosSic12–9; P6K | TPS(pT)KPRS | no heat | 1U |
| phosSic12–9; P3Y | TYS(pT)PPRS | 90 ± 50 | 1V |
| phosSic12–9; P3K | TKS(pT)PPRS | no heat | 1W |
| phosSic12–9; P7T | TPS(pT)PTRS | 70 ± 20 | 1X |
| phosSic12–9; P7K | TPS(pT)PKRS | 120 ± 20 | 1Y |
| phosSwe141–48 | IGGS(pT)PTN | no heat | 1EE |
| phosSwe1117–124 | ESVT(pT)PIT | 170 ± 90 | 1FF |
| phosSwe1192–199 | RIPE(pT)PVK | 60 ± 10 | 1GG |
| phosSwe1369–376 | EEIS(pT)PTR | 160 ± 70 | 1HH |
In order to identify the Cdc6 sequence determinants of Cks affinity, we used SPOT arrays with peptides directly synthesized on a membrane (Figure 2b). Every spot contains a version of the phosCdc63–10 sequence in which a single substitution is made at each position with each of the twenty amino acids. The array was probed as described above, except Cks1 was alkylated to prevent the formation of disulfide linkages (Supplementary Figure 2). We observed that no residue is tolerated at the pT position (0 position), including the phosphomimetics glutamate and aspartate, and that proline is required in the +1 position. This result indicates that all Cks1 binding sequences contain the phosphorylated Cdk consensus site TP. Cks1 also displays a marked propensity for binding to peptides with a bulky hydrophobic residue in the −2 position. The array demonstrates that any residue is tolerated in the −4, −3, +2, and +3 positions, although a basic residue or proline seems to be disfavored in the +2 position. There appears to be a varied preference in the −1 position, however there is no clear correlation between the chemical properties of the tolerated sidechains.
In order to corroborate that these observations are general for Cks1 binding and are not influenced by subtle effects of the specific Cdc63–10 sequence context, we conducted the array based on a sequence in Sic1 (Figure 2c). Sic1 T5 is a preferred Cdk phosphorylation site that serves as a priming site for Cks-dependent phosphorylation24. Probing a phosSic12–9 array for Cks1 binding, we again found a requirement for phosphothreonine and proline in the 0 and +1 positions respectively, a preference for a bulky hydrophobic residue in the −2 position, and a disfavoring of a positive residue in the +2 position.
We performed ITC with Cks1 and phosCdc62–10 peptides to confirm and quantify the spot array results (Table 1). Replacing the phosphothreonine for a different phosphorylated sidechain (pT7pS and pT7pY) or substitution of the +1 proline (P8K) results in loss of detectable heat. Substitutions of aromatic residues for the proline in the −2 position have little effect or slightly increase binding affinity relative to wild-type, whereas substitution of a charged lysine at the −2 position (P5K) results in no detectable heat. The +2 position in the SPOT array disfavored a positive charge or proline. However, by ITC, we found that substitution of a proline (T9P) or a lysine (T9K) for the +2 threonine results in a binding affinity similar to wild-type peptide.
Three Cdk sites (T5, T33, and T45) in the Sic N-terminal domain (Sic11–215, “Sic1ΔC”) facilitate Cks binding, and these sites are known to promote priming-dependent phosphorylation24. T5 and T45 contain a proline in the −2 position and their phosphorylation promotes higher affinity binding to Cks than T33, which contains a glutamate in the −2 position (data not shown). We measured affinity for wild-type and substituted phosSic12–9 peptides (Table 1 and Supplementary Figure 1). PhosSic12–9 binds with a slightly weaker affinity (Kd = 95 ± 8 µM) than the entire phosphorylated Sic1 N-terminus and phosSic1ΔC that only contains T5 as a phosphoacceptor site (Kd = 11 ± 3 µM and Kd = 20 ± 10 µM respectively24). As in the case of Cdc6, this observation suggests an interaction between the immediate sequence surrounding T5 and Cks1. The affinities of phosSic12–9 peptides with single amino acid substitutions show the same trend as Cdc6 peptides (Table 1). The ITC measurements are also consistent with the SPOT array data, with the exception that a positive lysine is tolerated in the +2 position in the ITC measurements.
Structural basis of Cks binding specificity
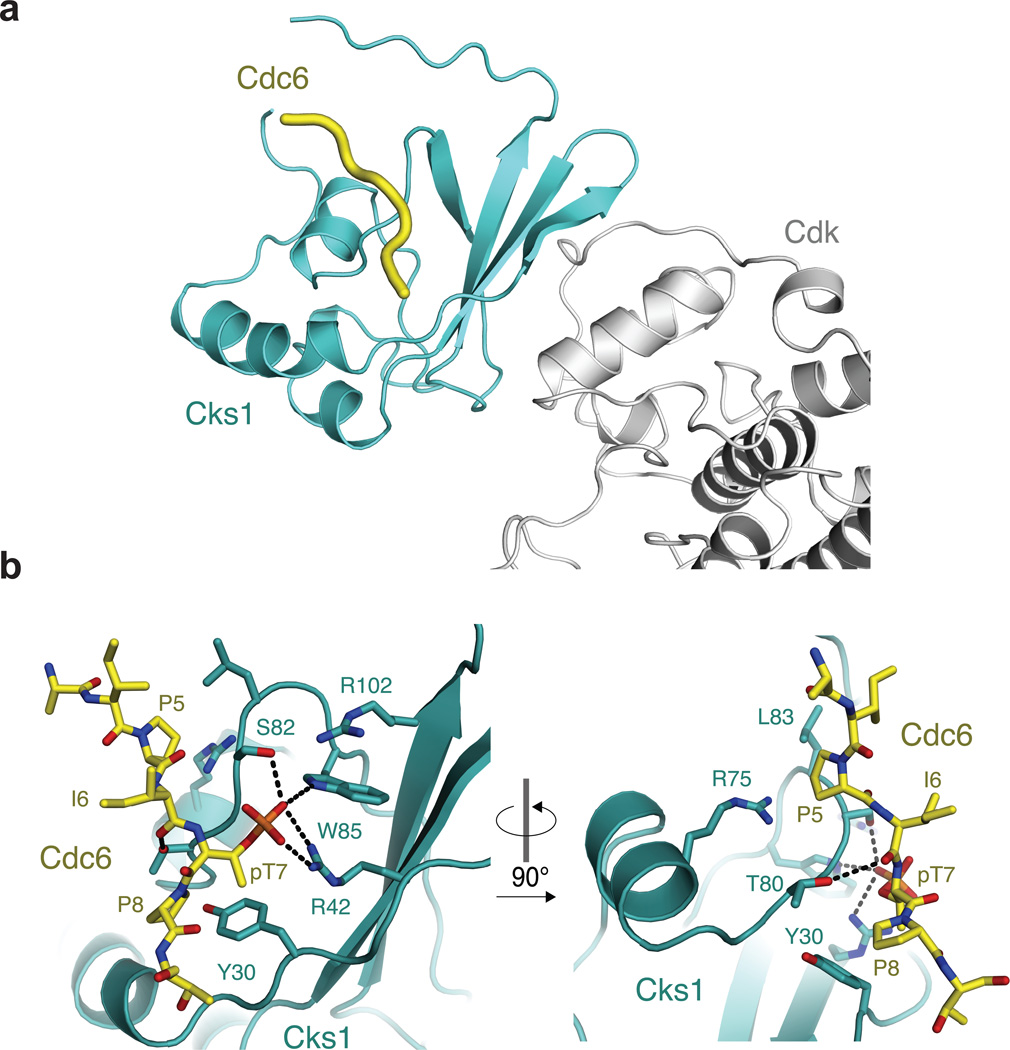
We next determined the 2.9 Å crystal structure of a Cks1-phosCdc6 complex to understand the molecular determinants of the specificity observed in our binding experiments (Table 2 and Figure 3). Crystals were grown using a fusion protein construct, phosphorylated after purification, in which the Cdc63–9 sequence was appended to the C-terminus of Cks11–112 (Supplementary Figure 3). The Cdc6 sequence binds across a surface of Cks1 that is distal to the Cdk binding site and made up of one face of the Cks1 β-sheet and helix 2 (Figure 3a). The phosphate on the Cdc6 T7 sidechain is bound to the previously described cationic pocket (Figure 3b)33,34. The Cdc6 T7 γ-methyl makes van der Waals contacts with Y30 and R42 in Cks1, which explains the specificity for phosphothreonine over phosphoserine. Cdc6 P8, which is in the +1 position, fits into a pocket formed by both Y30 and T80 in Cks1. The fact that no other hydrophobic residues are tolerated in this +1 position suggests that geometry constraints in the Cdc6 backbone are important for proline specificity. The dihedral angles adopted by the +1 proline (phi = ~−80, psi = ~−160) to contact Y30 and T80 are relatively disfavored by other residues. The loss of affinity upon proline substitution likely arises from the compromise of maintaining the sidechain-Cks1 contacts and adopting such strained torsion angles. The γ-hydroxyl of Cks1 T80 acts as a hydrogen bond donor to the backbone carbonyl of I6 (−1 position) in Cdc6. Although there is no clear correlation among the chemical properties of the tolerated amino acids at the −1 position in the array data (Figure2b and 2c), this hydrogen bond may select for particular backbone dihedral preferences that best facilitate its formation.
Table 2.
Data collection and refinement statistics
| phosCdc63–9 -Cks11–112 | |
|---|---|
| Data collection | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 84.7, 84.7, 239.9 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 60.0–2.9 (3.06–2.90) |
| Rmerge | 0.0463 (0.2598) |
| I / σI | 12.65 (3.57) |
| Completeness (%) | 99.79 (99.90) |
| Redundancy | 10.0 (9.9) |
| Refinement | |
| Resolution (Å) | 58.1–2.9 |
| No. reflections | 20,140 |
| Rwork / Rfree | 24.5% / 28.1% |
| No. atoms | 3852 |
| Protein | 3800 |
| Ligand/ion | 0 |
| Water | 52 |
| B factors | 56.5 Å2 |
| Protein | 47.60 |
| Ligand/ion | n/a |
| Water | 52.30 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.39 |
One crystal was used.
Values in parentheses are for highest-resolution shell.
Figure 3.
Crystal structure of a phosCdc63–9 -Cks11–112 complex. (a) Ternary complex including Cdk shows that the phosphorylated substrate binding site in Cks is distal from Cdk. The model was created by aligning Cks1 from the structure solved here and hsCks1 in the hsCks1-Cdk2 complex (PDB code: 1buh35). (b) Close-up views of the phosCdc63–9 binding site in Cks1.
A second pocket formed by Cks L83 and R75 fits Cdc6 P5 (the −2 position). The distances to the −2 proline sidechain explain the preference for a large hydrophobic residue in this position. The presence of R75, which is highly conserved in Cks proteins, is also consistent with the observed preference for aromatic residues in the −2 position of the binding consensus (Figure2b and 2c), as the guanidinium group is capable of making cation-pi interactions with aromatic sidechains40. There are few interactions in the crystal structure between Cks1 and residues in the +2, +3, and −3 positions in the Cdc6 peptide, which is consistent with our observation that any amino acid type can substitute there.
The importance of the molecular interactions observed in the crystal structure of the Cks1-phosCdc6 complex were verified with single amino acid mutations to Cks1. Y30E, R75A, R75K, T80D, and L83D Cks1 mutants were purified and tested for binding to phosCdc6 by ITC. The structure shows that Y30 and T80 are important for binding the +1 proline, while R75 and L83 form the pocket to accommodate the −2 proline. All the mutants were folded properly, as determined by circular dichroism (Supplementary Figure 4a). As predicted by the structure, mutations at these positions inhibited binding to the phosCdc62–10 peptide in the calorimetry assay; in each case, no heat was detected (Supplementary Figure 1Z–1DD).
Priming-dependent Cdk phosphorylation requires Cks-substrate binding
We next explored the implications of Cks-substrate docking for its function in stimulating Cdk activity towards cell cycle substrates. We previously found that priming phosphorylation of Sic1 facilitates semiprocessive phosphorylation of critical phosphodegrons, and the requirement for an intact Cks1 cationic pocket demonstrated the importance of Cks-dependent kinetics for Cdk signaling24. The structural details of the Cks-substrate complex observed here allowed us to test specifically the effect of disrupting this complex on Cdk kinetics. We tested whether Cks1 mutations that abolish interactions with substrate residues inhibit multisite phosphorylation (Figure 4). As previously described24, recombinant Cks1 was added to Cdk1-Clb5 purified from budding yeast, and the enzyme was used to phosphorylate the non-inhibitory truncation Sic1ΔC in a kinase assay. In the reaction with wild-type Cks1, hyperphosphorylated species are present within 8 minutes and are the dominant product (Figure 4a). Previous substrate competition experiments established that the rapid accumulation of these hyperphosphorylated forms is due to a semiprocessive mechanism24. In contrast, hyperphosphorylated Sic1ΔC does not appear in the same reaction time upon addition of Cks1 with mutations to the phosphate-binding pocket (“+pocket”, R33E S82E R102A) or a Y30E mutation. Stronger bands corresponding to hypophosphorylated forms are instead observed. Use of R75A, T80D, and L83D mutants also results in considerable loss of hyperphosphorylated Sic1ΔC, and intermediate forms containing fewer phosphates are the dominant product. All the mutants are capable of forming a complex with Cdk (Supplementary Figure 4b). In reactions with a Sic1ΔC containing a single phosphorylation site (all sites mutated to alanine except T5), the mutants behave similarly to wild-type Cks1 (Figure 4a). This control reaction demonstrates that Cks1 mutations that disrupt substrate docking have no effect on the catalytic activity of the kinase towards a single phosphoacceptor site, but instead result in defects specifically in the mechanism of multisite phosphorylation.
Figure 4.
Cks association with phosphorylated Cdk substrates Cdc6 and Sic1 enhances the kinetics of their multisite phosphorylation. (a) The indicated recombinant Cks1 was mixed with purified Cdk1-Clb5 and reacted with the N-terminal domain of Sic1 (Sic1ΔC), and phosphorylated products were separated with a PhosTag gel (left). Mutants that show defects in binding phosphorylated Cks consensus peptides also show defects in hyperphosphorylated Sic1, while the activity of all proteins towards a substrate with a single phosphorylation site (right) is similar. The full gels for this experiment can be seen in Supplemental Figure 7b. (b) Steady-state kinetics of Cdk1-Clb5 mixed with wild-type and mutant Cks1 proteins. The substrate is a synthetic Cdc6 peptide that contains a priming Cks consensus phosphate (phosT7) and an additional phosphoacceptor site (T23). (c) As in (b) except that wild-type Cks1 is used in each experiment with the indicated Cdc6 peptide mutation. (d) Cks1 mutants defective in consensus binding cannot rescue temperature-sensitive Cks1-mutant budding yeast. Temperature sensitive (TS) and wild-type (WT) strains were transformed with a CEN vector containing either wild-type yeast Cks1 or a Cks1 point mutant, grown in selective media to saturation, plated at 1:5 serial dilutions, and grown again at room temperature or 34°C.
To demonstrate directly the function of Cks in priming-dependent phosphorylation, we assayed kinase activity on primed substrates (Figure4b and 4c). A Cdc62–29 peptide was synthesized with T7 as a phosphothreonine. We then measured the steady-state kinetics of phosphate incorporation at T23 in the presence of the phosphorylated T7. The KM for the kinase reaction using Cdk1-Clb5 and wild-type Cks1 is less than that measured for a generic single-site substrate or Sic1 with a single T5 phosphoacceptor site available (Supplementary Figure 5). Conversely, the absolute kcat value for the wild-type reaction, calculated in these experiments by comparing Vmax to the published absolute kcat for the H1 peptide9, is similar to the kcat for reactions with single-site substrates. We tested in similar reactions Cks1 that contains mutations in the consensus sequence binding residues (Figure 4b). These reactions all proceed with KM values greater than reactions with wild-type protein. As observed in the Sic1ΔC multisite phosphorylation assay, mutations to the cationic pocket or Y30 have the largest effect on KM, while the T80D, L83D, and R75A mutations all show clear but more modest effects. These data demonstrate that Cks1 residues critical for binding phosphorylated substrates are required for the KM decrease that results from priming in a multisite kinase reaction.
We also assayed the importance of the Cks-binding consensus in the substrate. Cdc6 peptides with mutations in the phosphoacceptor site or surrounding consensus residues were synthesized and used in kinase reactions with wild-type Cks1 (Figure 4c). Mutation of the priming phosphothreonine to an alanine (T7A) leads to a KM increase for the reaction measuring phosphate incorporation at T23. Consistent with our binding measurements, mutation of the proline in the −2 position of the consensus to a tyrosine (P5Y) does not change the KM from the wild-type reaction, whereas mutation to a lysine (P5K) increases the KM by 3-fold. Mutation of the +1 proline (P8A) results in a 6-fold increase in KM. These results together further establish the importance of the Cks-substrate association for enhancing Cdk multisite kinetics and demonstrate that a binding consensus, and not solely a phosphate, directs the priming-dependent reaction.
To demonstrate the importance of Cks1 association with specific priming site sequences in vivo, we examined the viability of Cks1 mutants that are defective in consensus binding in budding yeast (Figure 4d). We identified a mutation (L98S) in Cks1 that has temperature sensitivity at 34°C. At the restrictive temperature, growth is impaired as described previously11 but is rescued by wild-type Cks1 expressed from a CEN vector and under control of the alcohol dehydrogenase promoter. In contrast, expression of a phosphate-binding pocket mutant or the consensus-binding mutants fails to rescue the temperature sensitivity.
The Cks-binding consensus is present in a subset of Cdk substrates
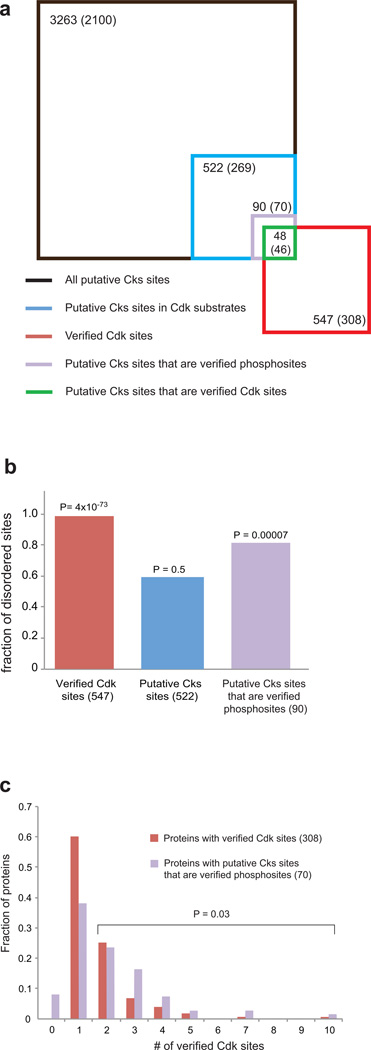
To find Cdk substrates that associate with Cks, we considered the sequence (F/I/L/P/V/W/Y)XTP, which contains the minimum requirements for binding Cks1 based on our data. We input this sequence into the Yeast Genome Database Pattern Matching tool (http://www.yeastgenome.org/cgi-bin/PATMATCH/nph-patmatch). 3263 sequences match the consensus; these sequences are found in 2100 different ORFs (Figure 5a). 269 of the 2100 ORFS are known Cdk substrates, as determined in at least one of the two global Cdk-substrate identification studies4,5. There are 522 putative Cks-binding sequences within these 269 ORFs, which we list in Supplementary Table 1.
Figure 5.
Sequence context of putative Cks binding sites. (a) Venn diagram showing in each set the number of sequences that match the minimal Cks consensus (F/I/L/P/V/W/Y-X-T-P) and the number of proteins containing those sequences (in parentheses). The set of all verified Cdk sites is taken from a mass spectrometry based screen5. Square areas are proportional to site number. (b) The fraction of sites found in disordered protein sequences. P values are calculated using the Chi-squared test and assuming an expected distribution equivalent to that found for all (S/T)P sites in the yeast proteome (60.6% disordered)5. (c) Multiplicity of Cdk sites in Cdk and putative Cks-directed substrates. The bars show the number of Cdk sites verified in the mass spectrometry screen5 for all Cdk substrates found in that screen (red) and for substrates that contain putative Cks sequences that are also verified phosphorylation sites (purple). The substrates that contain zero verified Cdk sites in the mass spectrometry screen contain Cks sequences that are phosphorylated according to the PhosphoGRID database. The P value is calculated for a distribution between either zero or one Cdk sites or more than one Cdk site. The Chi-squared test was used assuming an expected distribution for the Cks-containing proteins that is equivalent to the distribution for all verified Cdk sites in the mass spectrometry screen.
In order to filter and characterize better the properties of Cks targets, we further analyzed the 522 putative Cks-binding sites found in Cdk substrates (Figure 5). We first searched for evidence validating that the sites are phosphorylated by Cdk or other kinases. 48 of the 522 sites were identified as Cdk phosphorylation sites in a mass spectrometry-based proteomics screen to identify Cdk substrates (Figure 5a)5, and 42 other sites were identified as phosphorylation sites in the PhosphoGRID database (www.phosphogrid.org). These 90 Cks consensus sites that correspond to validated phosphorylation sites (highlighted in Supplementary Table 1) are found in 70 different ORFS.
Cdk sites tend to be found in regions of proteins that lack structure5,41. We analyzed whether the Cks consensus sites are in sequences that are likely structured or disordered using a web-based disorder prediction tool42. We found that 309 of the 522 putative Cks sites (59%) are in regions of likely disorder, while 73 of the 90 (81%) Cks sites that are verified phosphorylation sites are found in likely disordered regions (Figure 5b). When all putative Cks sites are considered, there is no significant preference for disordered sequences; however, the statistics for the subset of Cks sites corresponding to verified phosphorylation sites do suggest that Cks preferentially binds unstructured sequences. The disorder frequency of Cks sites is less than the disorder frequency (99%) of all verified Cdk sites in the mass spectrometry screen5. It may be that Cks sites are more likely to be found in predicted structured regions because of the hydrophobic amino acid in the −2 position.
Considering the role of Cks in stimulating multisite phosphorylation kinetics, we predicted and found that Cks sites preferentially occur in substrates that contain multiple Cdk sites. Putative Cks sites that are verified phosphorylation sites occur in substrates with more than one Cdk site at a frequency that is greater than expected from the multiplicity distribution of all Cdk substrates found in the mass spectrometry screen (Figure 5c)5.
Cks mediates Cdk1-directed Wee1 activity at mitotic entry
In examining the list of putative Cks binding partners, we found the potential docking association of Cks1 with budding yeast Wee1 (called Swe1) of particular interest, as Swe1 is known to form a phosphorylation-dependent complex with Cdk1 during mitotic entry. This complex is required for efficient inhibitory phosphorylation of Cdk1 by Swe1 on Y1922,31,32. Formation of the complex requires phosphorylation of 8 consensus sites in Wee1 by Cdk132. Similar mechanisms are likely to act in vertebrate cells31,43,44. We hypothesized that Cks docking to specific phosphorylated consensus sites mediates the Cdk1-Wee1 complex, and we tested this idea with mutational analysis of Swe1 in budding yeast.
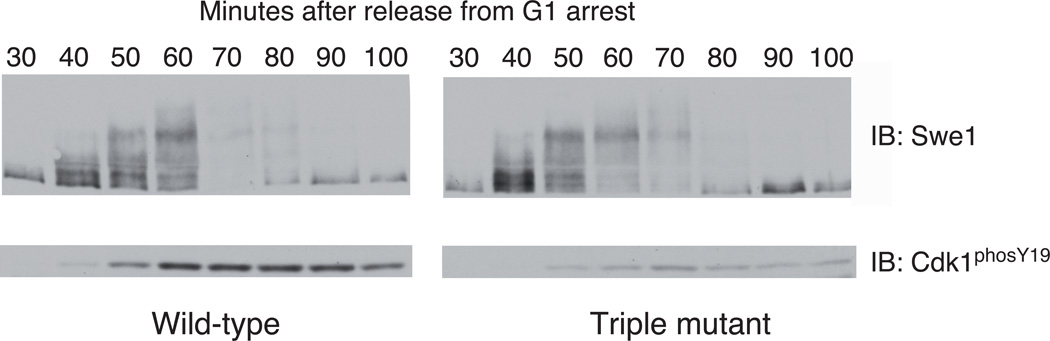
Of the eight consensus Cdk1 phosphosites in Swe1, there are four phosphothreonines: T45, T121, T196, and T373. All except T45 possess a bulky hydrophobic residue in the −2 position, and T196 and T373 were previously found to play an important role in Cdk1 complex formation in budding yeast32. Although neither the precise sequences surrounding the sites nor their location in the primary sequence are conserved in higher eukaryotes, most Swe1 orthologs contain 3 or 4 putative Cks binding sites. We tested Cks1 binding to each of these phosphopeptides by ITC and found as predicted that all except T45 bound with an affinity similar to phosCdc62–10 (Table 1). To assay the importance of the Cks-binding sites in vivo, a yeast strain was constructed in which the three consensus threonines in Swe1 were mutated to serines (T121S, T196S, T373S; “triple mutant”), which preserves Cdk phosphorylation of Swe1 but inhibits Cks association. We observed no considerable difference in the phosphorylation pattern of mutant and wild-type Swe1 (Figure 6) following release from G1 arrest, suggesting that the Cks consensus sites do not play a role in Swe1 hyperphosphorylation in vivo. The triple mutant cells do show a defect in phosphorylation of Cdk1 on Y19 (Figure 6). The decreased Cdk1 phosphorylation is not due to reduced Swe1, as the total amount of Swe1 and its phosphorylation state are consistent between mutant and wild-type cells (Figure 6). We conclude that the Cks consensus sites are critical for Swe1 activity towards Cdk1. The fact that hyperphosphorylation of Swe1 appears to be normal in the mutant suggests that Cks1 binding does not influence multisite phosphorylation in this context. Rather, Cks1 mediates formation of the complex in response to Cks1-consensus site phosphorylation, which in turn allows Swe1 to phosphorylate Cdk1 efficiently on its inhibitory site.
Figure 6.
Cks consensus sites in Wee1 are required for Cdk1 inhibitory phosphorylation. Yeast expressing wild-type and mutant (T121S, T196S, T373S) Wee1 (Swe1) were released from a G1 arrest. Cells at the indicated time points were lysed and Swe1 (a) and phosphorylation on Cdk1 Y19 (b) were probed. Exposures of the full gels for this experiment can be seen in Supplemental Figure 7c.
Discussion
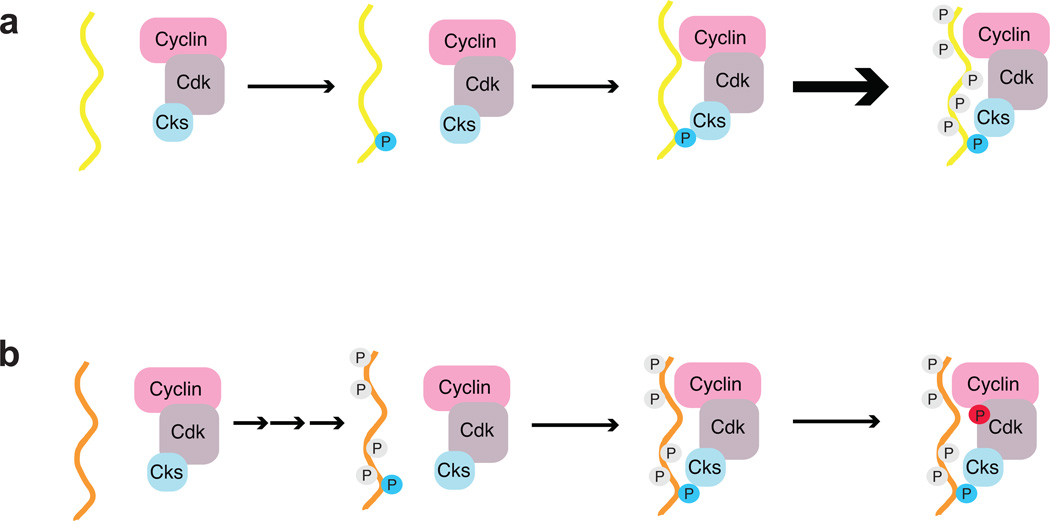
Although it has been postulated that Cks stimulates Cdk activity by binding primed substrates3,12, little evidence has been provided for this model, and the association of Cks with phosphorylated substrates has not been well characterized. We demonstrate here that Cks binds phosphorylated Cdk substrates in a sequence dependent manner and find that efficient hyperphosphorylation of substrates depends on recognition of sequence elements that surround the phosphate. Our results support a model in which Cks binds specific priming sites to facilitate phosphorylation at other sites (Figure 7a). Other data suggest that Cks1 orients substrates for Cdk phosphorylation at sites that are C-terminal to the Cks1 binding site (M.K. and M.L. unpublished). Characterization of the Cks-binding consensus sequence has permitted identification of a large set of Cdk substrates (269 proteins containing 522 Cks sites) that may utilize the Cks-dependent hyperphosphorylation mechanism. Considering that a smaller subset of putative Cks-binding sites contains sites for which phosphorylation has been directly observed (90 sites in 70 proteins), we anticipate that the actual number of Cks-interacting substrates in the cell is more modest. It remains an open question how many real Cks-directed Cdk substrates exist, and it will be interesting to explore the different cell cycle contexts in which Cks contributes to the desired signaling response.
Figure 7.
Cks is a specificity factor that mediates Cdk-substrate association for multiple functions. a) Cks binds substrates phosphorylated at specific consensus sites (blue phosphate), and the Cks-Cdk-substrate complex stimulates further phosphorylation. The stimulatory effect requires that the Cks-consensus site be phosphorylated early in the reaction, as is true in Sic1. b) Cks acts as an adaptor that targets Cdk to its regulators. In the case of Wee1, association with Cks consensus sites induces inhibitory phosphorylation in Cdk (red phosphate).
The observation that only a subset of Cdk sites have the required surrounding sequence for Cks binding has important implications for our understanding of multisite phosphorylation in cell cycle regulation. The data presented here demonstrate that Cdk phosphoacceptor sites differentially influence the kinetics of phosphate incorporation at other sites. This regulatory mechanism can be used to control whether and when sites with critical outputs are phosphorylated. For example, in Sic1, phosphorylation at specific sites required for ubiquitylation and degradation requires priming phosphorylation at the Cks consensus site (M.K. and M.L. unpublished). It has also been shown recently that different phosphorylation events in the Retinoblastoma protein induce distinct conformational changes26, which motivates the need for unique control of phosphorylation at distinct sites. This notion that specific phosphorylation sites play distinct roles in tuning signaling outputs differs from other models for the mechanism of multisite phosphorylation in the cell cycle21,23, in which phosphorylation sites play generic roles.
Our data suggest that the influence of Cks on Cdk substrate phosphorylation kinetics depends on whether Cks-binding consensus sites are phosphorylated early or late in the multisite reaction. In the case that a Cks-binding site is phosphorylated early, phosphorylation triggers Cks binding to enhance phosphorylation of remaining sites (Figure 7a). The consensus for optimal Cks1 affinity contains a Cdk acceptor site (TP), which is consistent with a self-priming mechanism. We note that other kinases could phosphorylate the Cks consensus, including MAP kinases, which also recognize TP45. The scenario depicted in Figure 7a is true for Sic1. Two optimal Cdk sites that act as Cks-dependent priming (T5 and T33) sites are required for semiprocessive phosphorylation of Sic1 and its degradation24. Cdc25 phosphorylation also uses the priming mechanism; mutation of Cks consensus sites abrogates cooperativity in the phosphorylation kinetics25. Alternatively, if Cks consensus sites are phosphorylated late in the multisite reaction, Cdk phosphorylation proceeds without or with limited Cks-induced stimulation for that particular substrate (Figure 7b). Swe1/Wee1 may be an example of such a substrate. We observe no effect on Swe1 hyperphosphorylation in vivo when the Cks consensus is disrupted, and it has been proposed that the best Cks consensus sites (T196 and T373) are phosphorylated late33.
We found that the Cks consensus in budding yeast Wee1 is required for its ability to phosphorylate Cdk on the inhibitory Y19 site. This observation demonstrates a novel role for Cks in regulation of Cdk activity through influencing posttranslational modifications and explains one mechanism by which Cks regulates mitotic entry. It also emphasizes that the cellular role of Cks extends beyond its capacity to stimulate Cdk kinetics. Cks mediates phosphorylation-dependent interactions between Cdk and its own regulators (Figure 7b). The Wee1-Cdk1 complex is a second example of this function in addition to hsCks1 binding phosphorylated p27 (phosp27) within a ubiquitin ligase18–19.
The structure of hsCks1 bound to phosp27 in the context of the Skp2-Skp1-Cullin ligase was previously solved36, and comparison with the phosCdc6-Cks1 structure here indicates several similarities (Supplementary Figure 6). The location of the phosphothreonine phosphate and γ-methyl groups and contacts with the +1 proline are equivalent. One notable difference is the presence of a glutamate in the −2 position in phosp27, which is not preferred in the Cks1 binding consensus. In the phosp27-hsCks1-ligase structure, this phos27 E185 hydrogen bonds with R294 and Y346 of Skp2 and Q52 of hsCks1. In Cks1, L83 is in the equivalent position as Q52. This difference explains why glutamate in the −2 position facilitates the phosp27-hsCks1 interaction in the context of ubiquitylation, whereas a hydrophobic residue is required for general Cks1 binding to phosphorylated targets. HsCks2 has a leucine at the hsCks1-Q52 position, which is similar to Cks1, the fission yeast ortholog suc1, and the frog ortholog p9. Our observations suggest that hsCks2, like its more distant orthologs, has a general role in Cdk binding to its substrates and regulators, while Cks1 has evolved to gain its specific function as an adaptor for the Skp2 SCF ligase. Further dissection of the functional differences between the human Cks paralogs will be important for understanding the role of Cks in tumorigenesis and for its use as a cancer diagnostic.
Supplementary Material
Acknowledgements
The authors acknowledge R. Cook of the MIT Biopolymers group for synthesis of and E. van Veen for providing valuable advice regarding peptide arrays. The authors thank E. Chen for assistance with CD experimental design and analysis. This research was supported by funding to S.M.R. from the American Cancer Society (RSG-12-131-01-CCG).
Footnotes
Accession code
Coordinates and structure factors for the Cks11–112-phosCdc63–9 fusion protein have been deposited in the Protein Data Bank under accession codes 4LPA.
Author contributions
D.A.M., E.R.M.B., M.K., R.L., D.R.K., M.L., and S.M.R. designed the study. D.A.M., E.R.M.B., M.K., R.L., M.V.M., and A.H. performed experiments. All authors analyzed data. D.A.M., E.R.M.B., and S.M.R. wrote the manuscript.
References
- 1.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 2.Brognard J, Hunter T. Protein kinase signaling networks in cancer. Curr Opin Genet Dev. 2011;21:4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan DO. The cell cycle : principles of control. xxvii. LondonSunderland, MA: Publishers; 2007. p. 297. Published by New Science Press in association with Oxford University Press; Distributed inside North America by Sinauer Associates. [Google Scholar]
- 4.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 5.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 7.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 8.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koivomagi M, et al. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayles J, Aves S, Nurse P. suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. Embo J. 1986;5:3373–3379. doi: 10.1002/j.1460-2075.1986.tb04653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Reed SI. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 12.Pines J. Cell cycle: reaching for a role for the Cks proteins. Curr Biol. 1996;6:1399–1402. doi: 10.1016/s0960-9822(96)00741-5. [DOI] [PubMed] [Google Scholar]
- 13.Shapira M, et al. Alterations in the expression of the cell cycle regulatory protein cyclin kinase subunit 1 in colorectal carcinoma. Cancer. 2004;100:1615–1621. doi: 10.1002/cncr.20172. [DOI] [PubMed] [Google Scholar]
- 14.Martinsson-Ahlzen HS, et al. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28:5698–5709. doi: 10.1128/MCB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan Y, et al. Aberrant expression of Cks1 and Cks2 contributes to prostate tumorigenesis by promoting proliferation and inhibiting programmed cell death. Int J Cancer. 2008;123:543–551. doi: 10.1002/ijc.23548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westbrook L, et al. High Cks1 expression in transgenic and carcinogen-initiated mammary tumors is not always accompanied by reduction in p27Kip1. Int J Oncol. 2009;34:1425–1431. [PubMed] [Google Scholar]
- 17.Ganoth D, et al. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- 18.Spruck C, et al. A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- 19.Morris MC, et al. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature. 2003;423:1009–1013. doi: 10.1038/nature01720. [DOI] [PubMed] [Google Scholar]
- 20.Yu VP, Baskerville C, Grunenfelder B, Reed SI. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol Cell. 2005;17:145–151. doi: 10.1016/j.molcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 22.Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 23.Kim SY, Ferrell JE., Jr Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 24.Koivomagi M, et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trunnell NB, Poon AC, Kim SY, Ferrell JE., Jr Ultrasensitivity in the Regulation of Cdc25C by Cdk1. Mol Cell. 2011;41:263–274. doi: 10.1016/j.molcel.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burke JR, Hura GL, Rubin SM. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev. 2012;26:1156–1166. doi: 10.1101/gad.189837.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldbeter A, Koshland DE., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci U S A. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrell JE., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 29.Salazar C, Hofer T. Multisite protein phosphorylation--from molecular mechanisms to kinetic models. Febs J. 2009;276:3177–3198. doi: 10.1111/j.1742-4658.2009.07027.x. [DOI] [PubMed] [Google Scholar]
- 30.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deibler RW, Kirschner MW. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell. 2010;37:753–767. doi: 10.1016/j.molcel.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvey SL, et al. A phosphatase threshold sets the level of Cdk1 activity in early mitosis in budding yeast. Mol Biol Cell. 2011;22:3595–3608. doi: 10.1091/mbc.E11-04-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arvai AS, Bourne Y, Hickey MJ, Tainer JA. Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain. J Mol Biol. 1995;249:835–842. doi: 10.1006/jmbi.1995.0341. [DOI] [PubMed] [Google Scholar]
- 34.Bourne Y, et al. Crystal structure of the cell cycle-regulatory protein suc1 reveals a beta-hinge conformational switch. Proc Natl Acad Sci U S A. 1995;92:10232–10236. doi: 10.1073/pnas.92.22.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourne Y, et al. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84:863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 36.Hao B, et al. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Patra D, Wang SX, Kumagai A, Dunphy WG. The xenopus Suc1/Cks protein promotes the phosphorylation of G(2)/M regulators. J Biol Chem. 1999;274:36839–36842. doi: 10.1074/jbc.274.52.36839. [DOI] [PubMed] [Google Scholar]
- 38.Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–1123. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- 39.Odaert B, et al. Solution NMR study of the monomeric form of p13suc1 protein sheds light on the hinge region determining the affinity for a phosphorylated substrate. J Biol Chem. 2002;277:12375–12381. doi: 10.1074/jbc.M111741200. [DOI] [PubMed] [Google Scholar]
- 40.Crowley PB, Golovin A. Cation-pi interactions in protein-protein interfaces. Proteins. 2005;59:231–239. doi: 10.1002/prot.20417. [DOI] [PubMed] [Google Scholar]
- 41.Tyanova S, Cox J, Olsen J, Mann M, Frishman D. Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS Comput Biol. 2013;9:e1002842. doi: 10.1371/journal.pcbi.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, Coleman TR, Dunphy WG. Two distinct mechanisms for negative regulation of the Wee1 protein kinase. EMBO J. 1993;12:3427–3436. doi: 10.1002/j.1460-2075.1993.tb06017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukhopadhyay NK, et al. An array of insulin-activated, proline-directed serine/threonine protein kinases phosphorylate the p70 S6 kinase. J Biol Chem. 1992;267:3325–3335. [PubMed] [Google Scholar]
- 46.Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem. 2010;285:16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynard GJ, Reynolds W, Verma R, Deshaies RJ. Cks1 is required for G(1) cyclin-cyclin-dependent kinase activity in budding yeast. Mol Cell Biol. 2000;20:5858–5864. doi: 10.1128/mcb.20.16.5858-5864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.