Abstract
• Background Phosphorus (P) is an essential macronutrient for plants. Plants take up P as phosphate (Pi) from the soil solution. Since little Pi is available in most soils, P fertilizers are applied to crops. However, the use of P fertilizers is unsustainable and may cause pollution. Consequently, there is a need to develop more P-use-efficient (PUE) crops and precise methods to monitor crop P-status.
• Scope Manipulating the expression of genes to improve the PUE of crops could reduce their P fertilizer requirement. This has stimulated research towards the identification of genes and signalling cascades involved in plant responses to P deficiency. Genes that respond to P deficiency can be grouped into ‘early’ genes that respond rapidly and often non-specifically to P deficiency, or ‘late’ genes that impact on the morphology, physiology or metabolism of plants upon prolonged P deficiency.
• Summary The use of micro-array technology has allowed researchers to catalogue the genetic responses of plants to P deficiency. Genes whose expression is altered by P deficiency include various transcription factors, which are thought to coordinate plant responses to P deficiency, and other genes involved in P acquisition and tissue P economy. Several common cis-regulatory elements have been identified in the promoters of these genes, suggesting that their expression might be coordinated. It is suggested that knowledge of the genes whose expression changes in response to P deficiency might allow the development of crops with improved PUE, and could be used in diagnostic techniques to monitor P deficiency in crops either directly using ‘smart’ indicator plants or indirectly through transcript profiling. The development of crops with improved PUE and the adoption of diagnostic technology could reduce production costs, minimize the use of a non-renewable resource, reduce pollution and enhance biodiversity.
Key words: Arabidopsis thaliana, phosphate, phosphorus, micro-arrays, roots, cis-regulatory elements, gene expression
INTRODUCTION
Phosphorus (P) is one of six essential macronutrients (N, P, K, Ca, Mg and S) required by plants. Their roots acquire P as phosphate (Pi), primarily in the form of H2PO4−, from the soil solution (Vance et al., 2003). The concentration of Pi in the soil solution is often low (2 to 10 µm; Raghothama, 1999) and, consequently, the supply of Pi to the root surface by diffusion is slow (Fitter and Hay, 2002). Hence, P is one of the most unavailable and inaccessible macronutrients in the soil (Vance et al., 2003) and frequently limits plant growth. For this reason, crops are supplied with inorganic P fertilizers. However, the non-renewable nature of inorganic P fertilizers means that cheap sources of P, such as phosphate rocks, will be exhausted within the next 60–90 years (Runge-Metzger, 1995). In addition, excess P added to crops can pollute local watercourses, contributing to the process of eutrophication (Withers et al., 2001). Therefore, P fertilization should be minimized. This might be achieved by developing crops that either acquire P or use P more efficiently, so that less P fertilizer is required, or developing more precise methods to monitor crop P status, such that P fertilization can be managed efficiently. This briefing focuses on the genetics of P-deficiency in an agricultural context. In an ecological context, plants may grow in P-deficient soils from germination to senescence and, consequently, may express a different set of genes from those discussed here in order to cope with a perpetually low availability of P.
GENETIC RESPONSES TO P DEFICIENCY
Plants have developed many physiological strategies to cope with low P (Fig. 1). It is thought that manipulating the expression of genes enabling growth in low-P environments could improve the P-use-efficiency of plants and reduce the P-fertilizer requirement of crops (López-Bucio et al., 2000b; Vance et al., 2003). This has stimulated research to identify genes and signalling cascades involved in plant responses to P deficiency. Several studies have identified individual genes that respond to P deficiency and, more recently, micro-array technology has been used to catalogue the effects of P deficiency on the expression of thousands of genes simultaneously (Wang et al., 2002; Hammond et al., 2003; Uhde-Stone et al., 2003; Wasaki et al., 2003b; Wu et al., 2003). It is thought that the expression of these genes is coordinated by both general stress-related and low-P-specific signalling cascades (Fig. 1; Hammond et al., 2003; Vance et al., 2003; Franco-Zorrilla et al., 2004). The genes identified as responding to P deficiency can be grouped into ‘early’ genes that respond rapidly and often non-specifically to P deficiency, and ‘late’ genes that alter the morphology, physiology or metabolism of plants upon prolonged P deficiency (Fig. 1). These ‘late’ genes generally improve the acquisition of P or promote the efficient use of P within the plant (Vance et al., 2003).
Fig. 1.
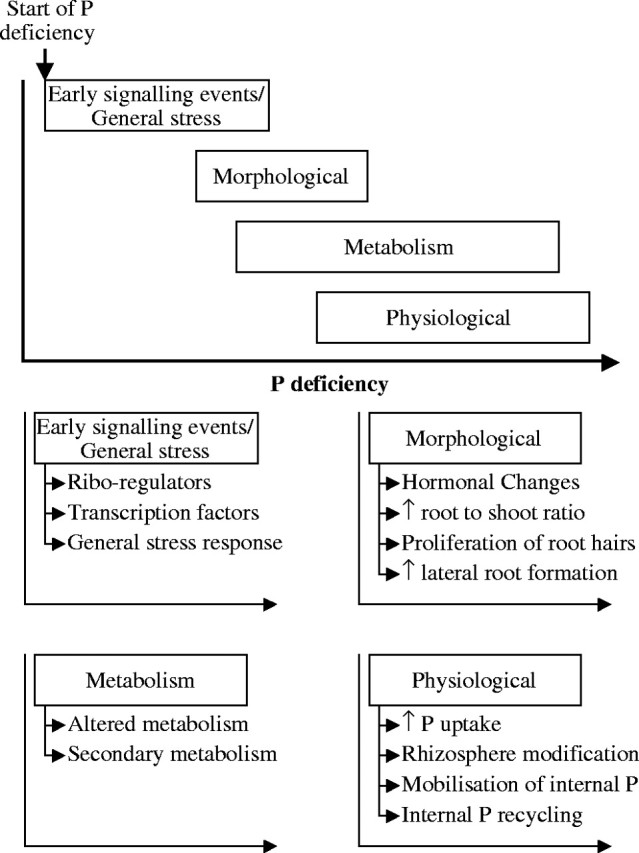
A hypothetical description of changes in gene expression in response to P deficiency in which genes are grouped by function into early (both specific and non-specific), morphological, physiological and metabolic processes. These groups have been arranged against an arbitrary scale of P deficiency going from slight P deficiency to extreme P deficiency.
Although several genes or groups of genes have been identified that consistently respond to P deficiency in all plant species, the precise time at which these genes change their expression during P deficiency is still unclear. This is a consequence of the different techniques used to impose P deficiency and the contrasting ontogenies of the plant material used. Consequently, constructing a precise response pathway of the genes involved in plant responses to P deficiency is speculative. However, a hypothetical description of changes in gene expression in response to P deficiency can be proposed (Fig. 1) in which genes are grouped by function into early (both low-P-specific and non-specific), morphological, physiological and metabolic processes. These groups can be arranged against an arbitrary scale of P deficiency going from slight P deficiency to extreme P deficiency.
Regulating the early genetic responses to P deficiency
Analysis of the promoters of the PHO regulon genes in yeast revealed two consensus cis-regulatory elements, GCACGTGGG and GCACGTTTT, for PHO2 and PHO4 binding and control of gene expression in response to P deficiency (Oshima et al., 1996). A cis-regulatory element, similar to the yeast PHO4 binding site (CACGT(G/T)), was subsequently identified in two plant P-responsive genes, TPSI1 from tomato and Mt4 from Medicago truncatula. These genes represented a novel type of P-responsive gene thought to be involved in regulating plant responses to P deficiency (Liu et al., 1997; Burleigh and Harrison, 1999), and orthologous genes in arabidopsis (At4 and AtIPS1) and rice (OsPI1) have been identified (Martín et al., 2000; Wasaki et al., 2003a). These genes encode short, non-conserved open reading frames, respond rapidly and specifically to P deficiency, and are likely to function as riboregulators controlling the function of other molecules such as RNA, DNA or proteins (Martín et al., 2000; Wasaki et al., 2003a).
Analysis of Chlamydomonas reinhardtii and arabidopsis mutants defective in a number of P responses revealed mutations in the CrPSR1 and AtPHR1 genes, respectively (Wykoff et al., 1999; Rubio et al., 2001). These genes encode proteins that contain MYB DNA-binding domains and coiled-coil (CC) domains for protein-protein interactions. Further analysis of the AtPHR1 protein demonstrated that it bound to an imperfect-palindromic sequence (GNATATNC) as a dimer (Rubio et al., 2001). This sequence is present in the promoter regions of several P-responsive genes (Rubio et al., 2001; Hammond et al., 2003; Table 1).
Table 1.
Percentage of genes differentially expressed by P deficiency containing cis-regulatory elements in their promoter regions compared with the arabidopsis genome
|
Hammond et al. (2003) |
Wu et al. (2003) |
|||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) Data set | ||||||||||||||||||||||||||||||||||||||||||||
| Tissuea | L | L | L | L | L | L | L | L | L | L | L | L | R | R | R | R | R | R | R | R | Affymetrix | |||||||||||||||||||||||
| Time after P withdrawalb (h) | 4 | 4 | 28 | 100 | 100 | 6 | 24 | 24 | 48 | 48 | 72 | 72 | 6 | 6 | 24 | 24 | 48 | 48 | 72 | 72 | ATH1 | |||||||||||||||||||||||
| Up- or down-regulated genesc | + | − | + | + | − | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | GeneChip | |||||||||||||||||||||||
| Number of genes in group | 56 | 4 | 4 | 44 | 12 | 4 | 150 | 114 | 495 | 418 | 47 | 27 | 16 | 8 | 62 | 39 | 244 | 294 | 36 | 31 | 22746 | |||||||||||||||||||||||
| (B) P response elements | ||||||||||||||||||||||||||||||||||||||||||||
| PHO-liked | C(G/T/A)(C/T/A)GTGG | 41·1 | 25·0 | 0·0 | 11·4 | 25·0 | 25·0 | 12·0 | 16·7 | 14·5 | 18·9 | 10·6 | 25·9 | 12·5 | 25·0 | 14·5 | 17·9 | 11·9 | 17·7 | 13·9 | 16·1 | 16·93 | ||||||||||||||||||||||
| TATA box liked | TATAAATA | 39·3 | 0·0 | 25·0 | 20·5 | 8·3 | 25·0 | 14·0 | 21·1 | 16·2 | 16·3 | 25·5 | 33·3 | 18·8 | 12·5 | 16·1 | 15·4 | 12·7 | 20·1 | 13·9 | 19·4 | 15·28 | ||||||||||||||||||||||
| TC element | TCTCTCT | 17·9 | 50·0 | 0·0 | 31·8 | 41·7 | 75·0 | 51·3 | 45·6 | 48·7 | 36·4 | 40·4 | 29·6 | 31·3 | 37·5 | 41·9 | 43·6 | 41·0 | 39·5 | 55·6 | 41·9 | 36·07 | ||||||||||||||||||||||
| NIT 2-like | AAATATCT | 8·9 | 25·0 | 50·0 | 13·6 | 25·0 | 25·0 | 8·0 | 26·3 | 10·1 | 13·2 | 6·4 | 18·5 | 12·5 | 12·5 | 12·9 | 23·1 | 10·7 | 11·9 | 11·1 | 12·9 | 8·20 | ||||||||||||||||||||||
| PHR1 elemente | GNATATNC | 26·8 | 0·0 | 50·0 | 13·6 | 25·0 | 50·0 | 14·7 | 15·8 | 16·2 | 17·0 | 23·4 | 14·8 | 6·3 | 0·0 | 16·1 | 15·4 | 19·3 | 15·3 | 22·2 | 16·1 | 18·61 | ||||||||||||||||||||||
| PHO elementf | CACGT(G/C) | 30·4 | 0·0 | 25·0 | 29·5 | 8·3 | 25·0 | 23·3 | 27·2 | 23·2 | 25·8 | 25·5 | 25·9 | 12·5 | 37·5 | 22·6 | 28·2 | 28·3 | 27·6 | 25·0 | 29·0 | 22·70 | ||||||||||||||||||||||
| P responsiveg | ATGCCAT | 1·8 | 25·0 | 0·0 | 2·3 | 0·0 | 0·0 | 2·7 | 5·3 | 2·4 | 5·5 | 0·0 | 3·7 | 0·0 | 0·0 | 3·2 | 7·7 | 4·5 | 2·4 | 8·3 | 3·2 | 4·10 | ||||||||||||||||||||||
| Helix–loop–helixh | CA(T/G)(A/C)TG | 42·9 | 75·0 | 50·0 | 40·9 | 16·7 | 0·0 | 46·7 | 47·4 | 42·4 | 50·0 | 34·0 | 51·9 | 31·3 | 50·0 | 38·7 | 43·6 | 48·4 | 47·3 | 47·2 | 48·4 | 48·20 | ||||||||||||||||||||||
| NIT 2i | TATC(A/T)(A/T) | 87·5 | 75·0 | 75·0 | 75·0 | 75·0 | 75·0 | 88·0 | 86·0 | 86·1 | 85·2 | 89·4 | 85·2 | 81·3 | 87·5 | 90·3 | 94·9 | 85·7 | 91·8 | 86·1 | 87·1 | 85·60 | ||||||||||||||||||||||
| (C) Other stress-related elements | ||||||||||||||||||||||||||||||||||||||||||||
| GCC–boxj | GCCGCC | 10·7 | 0·0 | 25·0 | 4·5 | 0·0 | 0·0 | 5·3 | 4·4 | 6·9 | 5·7 | 8·5 | 3·7 | 12·5 | 12·5 | 1·6 | 2·6 | 4·9 | 5·4 | 2·8 | 22·6 | 5·16 | ||||||||||||||||||||||
| DRE-likej | (A/G/T)(A/G)CCGACN(A/T) | 8·9 | 0·0 | 0·0 | 6·8 | 16·7 | 0·0 | 5·3 | 8·8 | 5·7 | 6·9 | 2·1 | 18·5 | 12·5 | 12·5 | 6·5 | 12·8 | 4·5 | 4·8 | 5·6 | 9·7 | 6·61 | ||||||||||||||||||||||
| AtMyb1k | (A/C)TCC(A/T)ACC | 0·0 | 0·0 | 0·0 | 4·5 | 0·0 | 0·0 | 5·3 | 4·4 | 3·8 | 3·1 | 6·4 | 3·7 | 12·5 | 0·0 | 6·5 | 5·1 | 7·8 | 4·1 | 5·6 | 3·2 | 3·56 | ||||||||||||||||||||||
| AtMyb2k | TAAC(G/C)GTT | 0·0 | 0·0 | 0·0 | 0·0 | 0·0 | 25·0 | 1·3 | 1·8 | 3·4 | 3·1 | 0·0 | 7·4 | 0·0 | 0·0 | 1·6 | 7·7 | 2·9 | 4·8 | 2·8 | 6·5 | 2·58 | ||||||||||||||||||||||
| AtMyb3k | TACCTACC | 1·8 | 0·0 | 0·0 | 6·8 | 0·0 | 0·0 | 0·7 | 0·9 | 0·6 | 1·0 | 0·0 | 3·7 | 0·0 | 0·0 | 1·6 | 7·7 | 0·4 | 1·7 | 2·8 | 6·5 | 0·60 | ||||||||||||||||||||||
| AtMyb4k | A(A/C)C(A/T)A(A/C)C | 64·3 | 75·0 | 75·0 | 56·8 | 58·3 | 50·0 | 56·0 | 48·2 | 53·1 | 49·8 | 61·7 | 66·7 | 56·3 | 62·5 | 64·5 | 48·7 | 48·4 | 54·1 | 69·4 | 41·9 | 49·90 | ||||||||||||||||||||||
| WRKYl | TTGAC(C/T) | 44·6 | 0·0 | 50·0 | 50·0 | 25·0 | 50·0 | 42·7 | 42·1 | 40·0 | 41·4 | 40·4 | 44·4 | 37·5 | 37·5 | 46·8 | 38·5 | 39·3 | 43·5 | 33·3 | 38·7 | 43·60 | ||||||||||||||||||||||
| ABREj | ACGTG(T/G)C | 8·9 | 0·0 | 25·0 | 4·5 | 8·3 | 25·0 | 12·7 | 17·5 | 12·3 | 10·5 | 12·8 | 14·8 | 12·5 | 25·0 | 14·5 | 12·8 | 15·2 | 10·5 | 11·1 | 19·4 | 9·23 | ||||||||||||||||||||||
The occurrence of cis-regulatory elements in the promoters of genes whose expression changes in shoots and roots in response to P withdrawal in comparison to the promoters of all genes on the Affymetrix ATH1 GeneChip.
Values in parts B and C are the percentage of each group that contain at least one copy of the cis-element between 10 and 1000 bases upstream of the ATG start codon identified using the ‘Find regulatory sequences’ function in GeneSpring (Silicon Genetics, CA, USA).
Values in bold are present significantly (P < 0·05) more often than would be expected based on the occurrence in the promoter regions of all genes on the ATH1 GeneChip.
Tissue from which RNA was isolated to challenge micro-array, L = leaves, R = roots.
Time after P was removed from the nutrient solution.
Genes whose expression increased or decreased in response to P deficiency as described by Hammond et al. (2003) and Wu et al. (2003).
Other potential cis-regulatory elements have been identified in the promoter sequences of P-responsive genes (Mukatira et al., 2001; Rubio et al., 2001; Tang et al., 2001; Hammond et al., 2003; Table 1). Cataloguing genes differentially expressed in response to P deficiency through micro-array technology has facilitated their identification. Hammond et al. (2003) reported the occurrence of putative PHO-like (CGCGTGGG) and TATA box-like (TATAAATA) cis-regulatory elements in the promoter regions of arabidopsis genes up-regulated 4 h after the withdrawal of P. The positive regulator of the yeast PHO regulon, PHO4, contains a basic helix–loop–helix (bHLH) DNA binding domain. Although no orthologues of the yeast PHO4 gene exist in arabidopsis, another bHLH transcription factor might bind to plant PHO-like sequences. Alternatively, since the core sequences of the arabidopsis PHO-like elements (CACGTG) are also similar to ABRE-like and G-box elements, proteins from the bZIP class of transcription factors might bind to them (Schindler et al., 1992). Previously, G-box and ABRE-like elements were shown to occur more often in the promoter regions of genes responding to cold stress (Chen et al., 2002), which, amongst other things, affects P metabolism (Stitt and Hurry, 2002). Analysis of the promoter regions from genes described as differentially regulated under P deficiency by Wu et al. (2003) identified two more novel putative cis-regulatory elements that occur more frequently in the promoters of genes up-regulated in leaves 48 h after withdrawing P (TCTCTCT) and down-regulated in leaves 24 h after withdrawing P (AAAATATC) than in the promoter regions of all genes present on the Affymetrix ATH1 GeneChip (Table 1). Repetitive TC elements have also been identified in genes differentially expressed under cation stress (Maathuis et al., 2003) and low oxygen stress (Klock et al., 2002) and could therefore represent a general stress response element or be an artefact of identifying these elements in silico.
All the putative cis-regulatory elements identified in groups of genes that respond rapidly to P deficiency could act as binding sites for proteins controlling the initial responses of plants to P deficiency. Further analysis is now required to identify the factors that bind to these regulatory elements. At present, the spatial and temporal expression of transcription factors is poorly understood, and some are thought to perform house-keeping functions under certain conditions and show overlapping roles with several response pathways (Chen et al., 2002; Venter and Botha, 2004).
Several studies have shown that general stress response genes are differentially regulated in response to P deficiency, although the temporal patterns and magnitude of these changes vary (Hammond et al., 2003; Uhde-Stone et al., 2003; Wasaki et al., 2003b; Wu et al., 2003). This could imply that plants either respond generically to the withdrawal of P before initiating specific responses to P deficiency, or that the regulatory signal cascades that control plant responses to P deficiency have redundancy with other stress response pathways. For example, the involvement of Pi in the acclimation of plants to cold, suggests that P and cold stress might share common regulatory signal cascades (Stitt and Hurry, 2002), and the differential regulation of many genes previously identified as being responsive to cold treatments during P deficiency (Hammond et al., 2003), may reflect commonality or redundancy in signalling these two stresses.
LATE RESPONSES TO P DEFICIENCY: INCREASING THE ACQUISITION OF P
Changes in root morphology
When subjected to P deficiency, plants maximize the volume of soil exploited and, thereby, the amount of Pi available, by increasing their effective root surface area. Plants responding to P deficiency increased lateral root formation and elongation, and reduce primary root elongation (Lynch and Brown, 2001; Williamson et al., 2001; Linkohr et al., 2002; Hodge, 2004); they also form more associations with mycorrhizal fungi. In addition, the number and length of root hairs increases in response to P deficiency (Jungk, 2001; Ma et al., 2001). Recently, the response of 19 root hair mutants to P deficiency was characterized and a genetic model for root hair development under these conditions was suggested (Müller and Schmidt, 2004).
A central role has been proposed for plant growth regulators (PGRs) in controlling the development of the root system in response to P deficiency or localized patches of nutrient (Casson and Lindsey, 2003). Changes in root morphology and growth are proportional to the concentration of PGRs, in particular auxins, ethylene and cytokinins (Casson and Lindsey, 2003). Under optimal conditions (P-replete) there is evidence that auxin is involved in lateral root development (Casimiro et al., 2003), root hair elongation (Bates and Lynch, 1996) and modulating root hair density (Ma et al., 2001). The involvement of ethylene in lateral root development is less clear, but it has been speculated that, since auxin is known to stimulate ethylene biosynthesis, ethylene may be involved together with auxin in controlling root elongation (Casson and Lindsey, 2003). Ethylene is better known for its role in root epidermal cell patterning and as a positive regulator of root hair development (Michael, 2001). An increase in cytokinins is normally associated with stimulating shoot growth and inhibiting root growth (Martín et al., 2000).
It is not known how the concentration of endogenous auxins responds to P-deficient conditions. The determination of changes in endogenous auxin levels in P-deficient plants would clarify the role auxin plays in modifying root morphology in response to P deficiency. Ethylene production is stimulated in plant roots by P deficiency (Borch et al., 1999), and might be responsible for the increased root hair formation in P-deficient plants (Michael, 2001). Cytokinin levels decrease in the roots of P-deficient plants (Kuiper et al., 1988). It is possible that this decrease in root cytokinin levels in response to P deficiency may release the inhibition of root growth and act as a negative regulator for P-induced root growth (Martín et al., 2000).
The application of exogenous PGRs has been used to rescue or simulate P-deficient phenotypes, and the use of mutants which over-express, or are insensitive or resistant to various PGRs, have been employed to investigate the influence of PGRs in modulating root morphology in response to P deficiency, with mixed results. Application of auxins to P-replete plants stimulates lateral root development similar to that observed in P-deficient plants, suggesting a role for auxin in changing root morphology in response to P deficiency (López-Bucio et al., 2002; Al-Ghazi et al., 2003). However, the growth of auxin-resistant or auxin-insensitive mutants under P deficiency has revealed similar changes in root-system architecture to wild-type plants (Schmidt and Schikora, 2001; Williamson et al., 2001; Al-Ghazi et al., 2003).
The use of ethylene inhibitors and precursors on arabidopsis grown at high and low P suggest a role for ethylene in root elongation but not in lateral root formation under P deficiency (López-Bucio et al., 2002; Ma et al., 2003). Treatment of roots with ethylene-synthesis inhibitors restricts root hair formation, whilst treatment of roots with an ethylene precursor promotes the formation of root hairs (Tanimoto et al., 1995). However, treating P-deficient plant roots with an ethylene-biosynthesis inhibitor or precursor (and an auxin-transport inhibitor) did not alter root hair density relative to control plants, suggesting that other signal transduction pathways may be responsible for altering root morphology under P deficiency (Ma et al., 2001). This is supported by the observation that ethylene insensitive or resistant mutants produce a similar number of root hairs as wild-type plants in response to P deficiency (Schmidt and Schikora, 2001). Exogenous cytokinins suppress lateral root initiation in P-deficient arabidopsis (López-Bucio et al., 2002), which fits with their perceived role as a negative regulator of root growth.
How these PGRs coordinate genetic responses to P deficiency is still unclear, but it is likely to result from a fine balance between PGRs, both spatially and temporally, and significant cross-talk between many partially redundant PGR signalling pathways (Forde and Lorenzo, 2001). Several genes regulated by PGRs are differentially regulated in response to P withdrawal. These include increased expression of auxin responsive genes, including AIR1, AIR3, AIR9, AIR12, HRGP and LRP1, which control lateral root development (Al-Ghazi et al., 2003; Casimiro et al., 2003; Uhde-Stone et al., 2003). The expression of several genes involved in ethylene biosynthesis (ACC oxidase, methioninesynthase and S-adenosyl methionine synthetase) and signalling (ethylene responsive element binding factor 2; EREB2), which could mediate the transcription of ethylene-responsive genes, increased their expression in the proteiod roots of white lupin (Uhde-Stone et al., 2003) and P-deficient arabidopsis roots (Wu et al., 2002). Genes encoding cytokinin oxidases, which may be involved in the breakdown of cytokinin in the roots and, thereby, release the negative control cytokinins have on root development, were observed to increase their expression during P deficiency in the proteiod roots of white lupin (Uhde-Stone et al., 2003) and in arabidopsis roots (J. P. Hammond, unpubl. res.). Changes in the expression of genes known to be regulated by, or involved in the regulation of PGRs, implies the coordinated involvement of ethylene, auxin and cytokinin in controlling root development in P-deficient plants (Martín et al., 2000; López-Bucio et al., 2002; Al-Ghazi et al., 2003).
Increasing the acquisition of P
Plants also respond to P deficiency by increasing the Pi transport capacity of root cells (Lee, 1993). Regulation of plant Pi transporters in response to P deficiency is thought to occur at the level of transcription (Smith et al., 2003). However, the over-expression of Pi transporters does not always increase P uptake (Rae et al., 2004). High affinity Pi transporters are primarily expressed in root hairs, which are ideally suited to P acquisition (Mudge et al., 2002; Schünmann et al., 2004). Transcripts from many members of the Pht1 subfamily of Pi transporters increase in response to P deficiency. With the exception of Pht1;6, the expression of all Pht1 subfamily members increased in the roots of arabidopsis in response to P deficiency (Mudge et al., 2002; Rausch and Bucher, 2002). In contrast, the expression of Pht2;1 from arabidopsis does not respond to P deficiency and is expressed mainly in leaves (Daram et al., 1999; Versaw and Harrison, 2002). Using a Pht2;1::GFP fusion protein, this Pi transporter was shown to be localized to the chloroplast and its expression varied during the photoperiod, suggesting a role in photosynthesis (Versaw and Harrison, 2002). Changes in the expression of the mitochondrial Pi transporters (Pht3 subfamily) in response to P deficiency have not yet been determined.
Plants also have the ability to modify their rhizosphere and improve the availability of Pi in the soil solution. An increased exudation of organic acids has been observed in several species in response to P deficiency, and serves to release Pi from insoluble inorganic salts (López-Bucio et al., 2000a). Organic acids are derived from the TCA cycle. The activity of TCA cycle enzymes increases in P-deficient plants, but little evidence is available as to whether this is effected at the level of transcription. The expression of PEPCase, which feeds carbon skeletons into the TCA cycle, but not citrate synthase or malate dehydrogenase (MDH) increases in P-deficient white lupin and tobacco (Toyota et al., 2003; Uhde-Stone et al., 2003). In contrast, micro-array analysis of P-deficient arabidopsis roots revealed a significant down-regulation of both PEPCase and MDH by P deficiency (Wu et al., 2003). Interestingly, transgenic tobacco engineered to overproduce citrate had improved PUE and produced up to 50 % more dry weight under P-limiting conditions than wild-type plants (López-Bucio et al., 2000b), although this does not appear to be easily reproducible (Delhaize et al., 2001).
Plants also release enzymes into the rhizosphere or present them on extracellular membranes to increase the availability of Pi from organic compounds containing P (Miller et al., 2001). The transcription and activity of secreted acid phosphatases are increased by P deficiency (Haran et al., 2000; Coello, 2002; Li et al., 2002) and can serve to release Pi from extracellular organic compounds. A gene encoding a secreted acid phosphatase, AtsAPase, was shown to be strongly induced by P deficiency in arabidopsis (Haran et al., 2000) and the expression of purple acid phosphatase genes AtPAP11 and AtPAP12 increased in P-deficient arabidopsis suspension cells (Li et al., 2002).
The acquisition of P from phytate could potentially improve crop PUE by providing plants with an alternative P source. However, transgenic arabidopsis expressing an extracellular phytase from Aspergillus niger under the control of a Pi transporter promoter or a constitutive promoter, only showed enhanced growth and P nutrition compared with control plants when phytate was the sole source of P (Richardson et al., 2001; Mudge et al., 2003). Presumably, this is a manifestation of the diverse strategies plants use to acquire P, which show considerable environmental dependence.
LATE RESPONSES TO P DEFICIENCY: IMPROVING THE INTERNAL USE OF P
Changes in metabolism to conserve P
The increased activity of proteins involved in alternative respiratory and photosynthetic pathways has been observed during P deficiency (Plaxton and Carswell, 1999; Fig. 2). However, it is unclear if this increase in activity reflects de novo protein synthesis or the activation of existing proteins.
Fig. 2.

Alternative metabolic processes for cytosolic glycolysis, mitochondrial electron transport and tonoplast H+ pumping (bold arrows) that may enable plants to survive under P limiting conditions. The enzymes that catalyse these reactions are (1) invertase, (2) sucrose synthase, (3) hexokinase, (4) fructokinase, (5) UDP-glucose pyrophosphorylase, (6) nucleoside diphosphate kinase, (7) phosphoglucomutase, (8) phosphoglucose isomerase, (9) phosphofructokinase (ATP-dependent), (10) phosphofructokinase (PPi-dependent), (11) NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (G3PDH; phosphorylating), (12) 3-phosphoglycerate (PGA) kinase, (13) NADP-dependent G3PDH (non-phosphorylating), (14) pyruvate kinase, (15) phosphoenolpyruvate (PEP) phosphatase, (16) PEP carboxylase, (17) malate dehydrogenase, (18) malic enzyme, (19) 3-deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) synthase, (20) 3-dehydroquinate (DHQ) dehydratase, (21) 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, (22) chorismate (CHA) synthase, (23) tonoplast H+-ATPase and (24) tonoplast H+-PPiase. Abbreviations for compounds are in the text or as follows; Glu-1-P, glucose 1-phosphate; Glu-6-P, glucose 6-phosphate; Fru-6-P, fructose 6-phosphate; Fru-1,6-P2, fructose 1,6-bisphosphate; G3P, glyceraldehyde-3-phosphate; 1,3-DPGA, 1,3-dephosphoglycerate; OAA, oxaloacetate; E4P, erythrose 4-phosphate; S3P, shikimate-3-phosphate. Redrawn from Plaxton and Carswell (1999).
During P deficiency, the conversion of sucrose to hexose-P can proceed via a pyrophosphate (PPi) dependent pathway requiring UDP-glucose pyrophosphorylase, the expression of which increases in P-deficient arabidopsis (Ciereszko et al., 2001). The activity of PEPCase appears central to regulating many metabolic adaptations to P deficiency (Fig. 2). An increase in the expression of PEPCase occurred in P-deficient tobacco (Toyota et al., 2003) and white lupin (Uhde-Stone et al., 2003). In contrast, the expression of PEPCase was repressed in P-deficient arabidopsis (Wu et al., 2003). Wu et al. (2003) also reported the down-regulation of many photosystem subunits and small subunits of RuBisCo in P-deficient arabidopsis. The expression of the alternative oxidase 1a precursor in the shoots of arabidopsis was significantly up-regulated following the withdrawal of P (Hammond et al., 2003). This may serve to reduce the production of reactive oxygen species by increasing electron transport through the alternative oxidase pathway and maintain energy production in P-deficient plants. The differential regulation of genes involved in primary metabolism demonstrates the activation of genes involved in bypassing the ATP- and Pi-dependent enzymes, and the changing metabolism required to generate energy and carbon skeletons during P deficiency (Plaxton and Carswell, 1999).
Genes involved in secondary metabolism are also differentially regulated in P-deficient plants, including genes involved in anthocyanin biosynthesis (Hammond et al., 2003; Uhde-Stone et al., 2003; Wu et al., 2003). The accumulation of anthocyanins in the aerial tissues is a characteristic response of P-deficient plants, which is thought to protect nucleic acids from UV damage and chloroplasts from photoinhibitory damage caused by P-limited photosynthesis (Hoch et al., 2001).
Changes in physiology to conserve P
To optimize the internal use of P, plants recycle P from old to new tissues. Under P-limiting conditions plants also re-mobilize P from non-essential uses to essential uses. The activities of phosphatases and nucleases re-mobilize Pi from cellular metabolites under P deficiency (Duff et al., 1991; Bariola et al., 1994). However, phosphatases also play an important role in protein regulatory cascades, and their increased expression in P-deficient plants might also indicate the initiation of P-specific signalling cascades (Luan, 2003). Increases in the activities of intra-cellular acid phosphatases and nucleases correspond with increased expression of their genes, indicating control at the level of transcription in response to P deficiency. The expression of AtACP5 increased in response to P deficiency and is thought to function in the internal remobilization of P (del Pozo et al., 1999). Changes in the expression of genes encoding nucleases in response to P deficiency are less well documented. The expression of three ribonucleases, RNS1, RNS2 and RNS3, all increase in P-starved arabidopsis, suggesting that ribonucleases may play a role in the internal remobilization of P under P deficiency (Bariola et al., 1994; Hammond et al., 2003).
The substitution and recycling of phospholipids during P deficiency can release Pi from phospholipid head groups. Under P deficiency, the proportion of mono- and di-galactolipids and sulpholipids increases in the thylakoid membranes, allowing photosynthesis to continue despite a reduction in phospholipid content (Yu et al., 2002; Andersson et al., 2003). The genes SQD1 and SQD2 encode proteins involved in the biosynthesis of sulpholipids, and their expression increases in P-deficient plants (Essigmann et al., 1998; Yu et al., 2002; Hammond et al., 2003; Wasaki et al., 2003b). Expression of glycerophosphodiesterases also increases under P deficiency (van der Rest et al., 2003). The activity of glycerophosphocholine phosphodiesterase, which breaks down glycerophosphocholine into glycerol-3-phosphate and choline, increased under P deficiency in carrot, arabidopsis and sycamore cell cultures (van der Rest et al., 2003). Glycerol-3-phosphate is thought to be converted to glycerol and Pi by acid phosphatases increasing the availability of Pi under P-limiting conditions.
SUMMARY AND FUTURE DIRECTIONS
It is clear that gene expression changes in both roots and shoots in response to P deficiency (Fig. 1; Raghothama, 1999; Hammond et al., 2003; Vance et al., 2003; Franco-Zorrilla et al., 2004). These changes in gene expression are likely to be coordinated by regulatory cascades involving transcription factors and their cognate cis-regulatory elements. Transcriptional profiling has identified several transcription factors that are increased under P deficiency (Hammond et al., 2003; Wasaki et al., 2003b; Wu et al., 2003) and several putative cis-regulatory elements have been identified in the promoters of genes whose expression changes in response to P deficiency (Table 1). These are likely to be involved in controlling general stress responses as well as responses specific to P deficiency (Fig. 1). Coordinated changes in the concentration of PGRs appear to effect changes in root structure in response to P deficiency and also impact on the expression of apparently unrelated genes (Forde and Lorenzo, 2001; Al-Ghazi et al., 2003; Vance et al., 2003; Fig. 1). The increase in the size of the root system, which will increase the soil volume exploited by plants, is accompanied by an increased expression of genes encoding Pi transporters of the Pht1 subfamily (Smith et al., 2003) and the release of enzymes and organic acids into the rhizosphere (Haran et al., 2000; López-Bucio et al., 2000a; Coello, 2002; Fig. 1). Thus, the plant increases its ability to release and take up Pi from the soil. Transcriptional cascades will also control the expression of genes encoding enzymes involved in primary and secondary metabolism (Fig. 2). These changes in metabolism, although not always observed, serve to conserve P internally by replacing P in metabolites and structural compounds (Plaxton and Carswell, 1999; Yu et al., 2002).
The identification of transcription factors, regulatory signalling cascades, and genes involved in plant responses to P deficiency could contribute to the development of crops with improved PUE. However, since many transcriptional studies have assayed gene expression in plants grown under laboratory conditions, conclusions from these studies also need to be confirmed in an agronomic context. Furthermore, since transgenic approaches to crop improvement have often been unsuccessful, such an approach should be followed with caution (Flowers, 2004; Sinclair et al., 2004). For example, the constitutive expression of genes to enhance crop PUE may be effective under P-limiting conditions, but could be detrimental under other environmental conditions. The use of promoter regions or transcription factors that regulate gene expression solely in response to P deficiency might improve PUE in crop plants, without a yield penalty in P-replete conditions.
To manage P fertilizer applications more efficiently, there is a need for more precise methods to monitor crop P status. In the field, P availability, P movement through the soil and plant P requirements may differ throughout the day, the season and between crops. The identification of diagnostic genes, whose changes in expression allow the physiological P status of a particular crop to be defined accurately, will enable robust methods to monitor P deficiency in crops either directly, using smart plant technology, or indirectly, through diagnostic micro-arrays (Hammond et al., 2003). Coupled with simulation models that can inform the quantity and temporal application of P fertilizers to crop plants (Greenwood et al., 2001), application of these diagnostics could lower production costs, minimize the use of a non-renewable resource, reduce pollution and enhance biodiversity.
To realize the ambition of crops with improved PUE and molecular diagnostics for P deficiency, further research is needed. Whole genome arrays for A. thaliana and other species are now available, and should be used to complete the transcriptional profiles of plant responses to P deficiency under both controlled and field conditions. Cis-regulatory elements, identified through in silico searches from transcriptional profiling of P-deficient plants, need to be confirmed using promoter dissection techniques before the signal transduction cascades during P deficiency can be established.
The development of transcriptional profiles for crop species will facilitate the development of universal transcript profiling assays for P deficiency. Establishing a characteristic transcriptional pattern for incipient P deficiency would then inform remedial P fertilization of any crop. The adoption of such knowledge and tools may ultimately contribute to a reduction in the use of P fertilizers.
Acknowledgments
We thank Malcolm Bennett, Helen Bowen, Dan Eastwood, Sean May, Clive Rahn, Ranjan Swarup and Kath Woolaway for their contributions to our work on P diagnostics. This work was supported by the Department for Environment, Food and Rural Affairs (UK; project nos HH3504SPO, HH3502SFV and HH3501SFV), by the Biotechnology and Biological Sciences Research Council (UK), and by a HRI Gordon Browning Studentship (to J.P.H.).
LITERATURE CITED
- Al-Ghazi Y, Muller B, Pinloche S, Tranbarger TG, Nacry P, Rossignol M, Tardieu F, Doumas P. 2003. Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signalling. Plant, Cell and Environment 26: 1053–1066. [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AE. 2003. Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyliacylglycerol. FEBS Letters 537: 128–132. [DOI] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. 1994. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant Journal 6: 673–685. [DOI] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. 1996. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant, Cell and Environment 19: 529–538. [Google Scholar]
- Blackwell TK, Weintraub H. 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250: 1104–1110. [DOI] [PubMed] [Google Scholar]
- Borch K, Bouma TJ, Lynch JP, Brown KM. 1999. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant, Cell and Environment 22: 425–431. [Google Scholar]
- Burleigh SH, Harrison MJ. 1999. The down regulation of Mt4-like genes by phosphate fertilization occurs systematically and involves phosphate translocation to the shoots. Plant Physiology 119: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. 2003. Dissecting Arabidopsis lateral root development. Trends in Plant Science 8: 165–171. [DOI] [PubMed] [Google Scholar]
- Casson SA, Lindsey K. 2003. Genes and signalling in root development. New Phytologist 158: 11–38. [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al. 2002. Expression profile matrix of arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Hurry V, Kleczkowski LA. 2001. Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis Planta 212: 598–605. [DOI] [PubMed] [Google Scholar]
- Coello P. 2002. Purification and characterisation of secreted acid phosphatase in phosphorus-deficient Arabidopsis thaliana Physiologia Plantarum 116: 293–298. [Google Scholar]
- Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M. 1999.Pht2;1 encodes a low affinity phosphate transporter from arabidopsis. Plant Cell 11: 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Hebb DM, Ryan PR. 2001. Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiology 125: 2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. 1999. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant Journal 19: 579–589. [DOI] [PubMed] [Google Scholar]
- Duff SMG, Plaxton WC, Lefebvre DD. 1991. Phosphate-starvation response in plant cells: De novo synthesis and degradation of acid phosphatases. Proceedings of the National Academy of Sciences of the USA 88: 9538–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C. 1998. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana Proceedings of the National Academy of Sciences of the USA 95: 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Fitter AH, Hay RKM. 2002.Environmental physiology of plants. London: Academic Press. [Google Scholar]
- Flowers TJ. 2004. Improving crop salt tolerance. Journal of Experimental Botany 55: 307–319. [DOI] [PubMed] [Google Scholar]
- Forde B, Lorenzo H. 2001. The nutritional control of root development. Plant and Soil 232: 51–68. [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. 2004. The transcriptional control of plant responses to phosphate limitation. Journal of Experimental Botany 55: 285–293. [DOI] [PubMed] [Google Scholar]
- Fu YH, Marzluf GA. 1990. NIT-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proceedings of the National Academy of Sciences of the USA 87: 5331–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood DJ, Karpinets TV, Stone DA. 2001. Dynamic model for the effects of soil P and fertilizer P on crop growth, P uptake and soil P in arable cropping: Model description. Annals of Botany 88: 279–291. [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. 2003. Changes in gene expression in arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology 132: 578–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran S, Logendra S, Seskar M, Bratanova M, Raskin I. 2000. Characterization of arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiology 124: 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Zeldin EL, McCown BH. 2001. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiology 21: 1–8. [DOI] [PubMed] [Google Scholar]
- Hodge A. 2004. The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytologist 162: 9–24. [Google Scholar]
- Jungk A. 2001. Root hairs and the acquisition of plant nutrients from soil. Journal of Plant Nutrition and Soil Science 164: 121–129. [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES. 2002. Expression profile analysis of the low-oxygen response in arabidopsis root cultures. Plant Cell 14: 2481–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper D, Schuit J, Kuiper PJC. 1988. Effects of internal and external cytokinin concentrations on root growth and shoot to root ratio of Plantago major ssp pleiosperma at different nutrient conditions. Plant and Soil 111: 231–236. [Google Scholar]
- Lee RB. 1993. Control of net uptake of nutrients by regulation of influx in barley plants recovering from nutrient deficiency. Annals of Botany 72: 223–230. [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D. 2002. Purple acid phosphatases of Arabidopsis thaliana Journal of Biological Chemistry 277: 27772–27781. [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. 2002. Nitrate and phosphate availability and distribution effects on root system architecture of Arabidopsis Plant Journal 29: 751–760. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Raghothama KG. 1997. Differential expression of TPSI1, a phosphate starvation-induced gene in tomato. Plant Molecular Biology 33: 867–874. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez V, Herrera-Estrella L. 2000. Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Science 160: 1–13. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, de la Vega OM, Guevara-García A, Herrera-Estrella L. 2000. Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nature Biotechnology 18: 450–453. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. 2002. Phosphate availability alters architecture and causes changes in hormone sensitivity in the arabidopsis root system. Plant Physiology 129: 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. 2003. Protein phosphatases in plants. Annual Review of Plant Biology 54: 63–92. [DOI] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2001. Topsoil foraging – an architectural adaptation of plants to low phosphorus availability. Plant and Soil 237: 225–237. [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. 2001. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana Plant, Cell and Environment 24: 459–467. [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. 2003. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiology 131: 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM, Filatov V, Herzyk P, Krijger GC, Axelsen KB, Chen SX, Green BJ, Li Y, Madagan KL, Sánchez-Fernández R, et al. 2003. Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant Journal 35: 675–692. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. 1997. MYB transcription factors in plants. Trends in Genetics 13: 67–73. [DOI] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Peña A, Leyva A, Paz-Ares J. 2000. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis Plant Journal 24: 559–567. [DOI] [PubMed] [Google Scholar]
- Michael G. 2001. The control of root hair formation: suggested mechanisms. Journal of Plant Nutrition and Soil Science 164: 111–119. [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP. 2001. Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiology 127: 594–606. [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. 2002. Expression analysis suggests novel roles for members of Pht1 family of phosphate transporters in Arabidopsis Plant Journal 31: 341–353. [DOI] [PubMed] [Google Scholar]
- Mudge SR, Smith FW, Richardson AE. 2003. Root-specific and phosphate-regulated expression of phytase under the control of a phosphate transporter promoter enables Arabidopsis to grow on phytate as a sole P source. Plant Science 165: 871–878. [Google Scholar]
- Mukatira UT, Liu C, Varadarajan DK, Raghothama KG. 2001. Negative regulation of phosphate starvation-inducible genes. Plant Physiology 127: 1854–1862. [PMC free article] [PubMed] [Google Scholar]
- Müller M, Schmidt W. 2004. Environmentally induced plasticity of root hair development in arabidopsis. Plant Physiology 134: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Ogawa N, Harashima S. 1996. Regulation of phosphatase synthesis in Saccharomyces cerevisiae – a review. Gene 179: 171–177. [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Carswell MC. 1999. Metabolic aspects of phosphate starvation in plants. In: HR Lerner, ed. Plant responses to environmental stresses: from phytohormones to genome reorganisation. New York: Dekker, 349–372. [Google Scholar]
- Rae AL, Jarmey JM, Mudge SR, Smith FW. 2004. Over-expression of a high-affinity phosphate transporter in transgenic barley plants does not enhance phosphate uptake rates. Functional Plant Biology 31: 141–148. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology 50: 665–693. [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M. 2002. Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Hodobas PA, Hayes JE. 2001. Extracellular secretion of Aspergillus phytate from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant Journal 25: 641–649. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor in phosphate starvation signalling both in vascular plants and in unicellular algae. Genes and Development 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Metzger A. 1995. Closing the cycle: obstacles to efficient P management for improved global security. In: H Tiessen, ed. Phosphorus in the global environment: transfers, cycles and management. Chichester: John Wiley and Sons, 27–42. [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. 1992. TGA1 and G-box binding factors: two distinct classes of arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4: 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Schikora A. 2001. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiology 125: 2078–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PHD, Richardson AE, Smith FW, Delhaize E. 2004. Characterisation of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). Journal of Experimental Botany 55: 855–865. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2000. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signalling pathways. Current Opinion in Plant Biology 3: 217–223. [PubMed] [Google Scholar]
- Sinclair TR, Purcell LC, Sneller CH. 2004. Crop transformation and the challenge to increase yield potential. Trends in Plant Science 9: 70–75. [DOI] [PubMed] [Google Scholar]
- Smith FW, Mudge SR, Rae AL, Glassop D. 2003. Phosphate transport in plants. Plant and Soil 248: 71–83. [Google Scholar]
- Stitt M, Hurry V. 2002. A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis Current Opinion in Plant Biology 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Tang Z, Sadka A, Morrishige DT, Mullet JE. 2001. Homeodomain leucine zipper proteins bind to the phosphate response domain of the soybean VspB tripartite promoter. Plant Physiology 125: 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto M, Roberts K, Dolan L. 1995. Ethylene is a positive regulator of root hair development in Arabidopsis thaliana Plant Journal 8: 943–948. [DOI] [PubMed] [Google Scholar]
- Toyota K, Koizumi N, Sato F. 2003. Transcriptional activation of phosphoenolpyruvate carboxylase by phosphorus deficiency in tobacco. Journal of Experimental Botany 54: 961–969. [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Zinn KE, Ramirez-Yáñez M, Li A, Vance CP, Allan DL. 2003. Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorous deficiency. Plant Physiology 131: 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist 157: 423–447. [DOI] [PubMed] [Google Scholar]
- van der Rest B, Boisson A-M, Gout E, Bligny R, Douce R. 2003. Glycerophosphocholine metabolism in higher plant cells. Evidence of a new glyceryl-phosphodiester phosphodiesterase. Plant Physiology 130: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M, Botha FC. 2004. Promoter analysis and transcriptional profiling: Integration of genetic data enhances understanding of gene expression. Physiologia Plantarum 120: 74–83. [DOI] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ. 2002. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H, Garvin DF, Kochian LV. 2002. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiology 130: 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Shinano T, Kai M, Osaki M. 2003. Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytologist 158: 239–248. [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, et al. 2003. Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant, Cell and Environment 26: 1515–1523. [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Leyser HMO. 2001. Phosphate availability regulates root system architecture in arabidopsis. Plant Physiology 126: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers PJA, Edwards AC, Foy RH. 2001. Phosphorus cycling in UK agriculture and implications for phosphorus loss from soil. Soil Use and Management 17: 139–149. [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. 2003. Phosphate starvation triggers distinct alterations of genome expression in arabidopsis roots and leaves. Plant Physiology 132: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. 1999. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proceedings of the National Academy of Sciences of the USA 96: 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C. 2002.Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth.Proceedings of the National Academy of Sciences of the USA 99: 5732–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]


