Abstract
• Background and Aims Thymus loscosii (Lamiaceae) is a tetraploid perennial species endemic to the Ebro river basin (north‐eastern Spain), which is included in the National Catalogue of Endangered Species. It is a tetraploid species (2n = 54), presumably an autotetraploid originated by the duplication of a 2n = 28 genome and the subsequent loss of two chromosomes. Allozyme electrophoresis was conducted to survey the levels and distribution of genetic diversity and to test the previous autopolyploid hypothesis for its origin. In addition, both in situ and ex situ conservation measures are proposed.
• Methods Eight populations were sampled for analysis by standard methods of starch gel electrophoresis, and six putative enzymatic loci were resolved (five consistently and one only partially).
• Key Results Banding patterns exhibited no evidence of fixed heterozygosity and showed both balanced and unbalanced heterozygotes. In addition, most individuals showed a pattern consistent with the presence of three or four alleles at a single locus. High levels of genetic variability were found at population level (P = 85 %, A = 3·0, He = 0·422), in addition to a trend of an excess of heterozygotes.
• Conclusions Allozyme data support the hypothesis that T. loscosii is an autotetraploid, and the high number of alleles at some loci may be due to repeated polyploidization events. The high values of genetic variation found in this species agree with those expected for tetraploids. The excess of heterozygotes may be due to some barriers to inbreeding (e.g. occurrence of gynodioecy) and/or selection for heterozygosity.
Key words: Allozyme electrophoresis, genetic diversity, conservation, endemic species, tetraploidy, Lamiaceae, Thymus loscosii
INTRODUCTION
Polyploidy is considered a significant mode of species formation and an important source of evolution in higher plants (Stebbins, 1980). In fact, estimates of the frequency of polyploidy in angiosperms vary from 30 % to 80 % (Otto and Whitton, 2000; Soltis and Soltis, 2000). The success of polyploidy can be explained by several factors such as increased heterozygosity and genetic diversity, enzyme multiplicity and increased biochemical diversity (Levin, 1983; Soltis and Soltis, 2000) and their establishment and commonness in nature may be favoured by vegetative (asexual) multiplication and a perennial habit (Otto and Whitton, 2000; Soltis and Soltis, 2000). Moreover, polyploids have a broader ecological amplitude than the parental diploids and a better colonizing ability, which allows for an increased availability of new ecological niches (Ramsey and Schemske, 1998; Soltis and Soltis, 2000).
Two major types of polyploidy are recognized in nature, allopolyploidy and autopolyploidy (Stebbins, 1947; Crawford, 1989; Soltis and Soltis, 2000), although these two terms represent the ends of a spectrum of variation. Allopolyploidy is thought to be the result of an interspecific hybridization process and subsequent chromosome duplication, while autopolyploidy is probably the consequence of polyploidization of conspecific individuals, generally by fusion of non‐reduced gametes. Nevertheless, both allo‐ and autopolyploids may also be formed via a triploid bridge, in which there takes place a union of a reduced and an unreduced gamete (Ramsey and Schemske, 1998). Unfortunately, because allopolyploidy and autopolyploidy are often equated with interspecific and intraspecific polyploidy, respectively, classification of polyploids may also depend on species circumscription (Ramsey and Schemske, 1998).
Allopolyploids are characterized by disomic inheritance, bivalent formation at meiosis and fixed heterozygosity (non‐segregating) due to the combination of two divergent parental genomes. In contrast, autopolyploids are expected to express polysomic inheritance (tetrasomic in autotetraploids) and may also show multivalent formation at meiosis. Two types of heterozygotes may be formed: balanced and unbalanced.
Thymus loscosii Willk. (Lamiaceae) is endemic to the Ebro river basin in north‐eastern Spain. The classification of Thymus has long been considered difficult (Jalas, 1972; Morales, 1986), and T. loscosii has been included in three different sections, namely Serpyllum (Miller) Bentham, Hyphodromi (A. Kerner) Hálacsy and Thymus. The inclusion in the latter is the most widely recognized today (Greuter et al., 1986; Morales, 1986). Thymus loscosii is a tetraploid (2n = 54; Morales, 1986), presumably an autotetraploid, which probably originated by the duplication of a 2n = 28 genome and the subsequent loss of two chromosomes, i.e. dysploidy (2n = 4× – 2) (Morales, 1986). Morales (1986, 2002) has suggested a basic number in the genus Thymus of x = 7, which probably gave rise to the secondary basic numbers x = 14 and x = 15. Several studies report different ploidy levels in the same species, which indicates that polyploidization may occur frequently in this genus. Perhaps one of the most interesting cases is that of Thymus herba‐barona Loisel., with 2n = 28, 56 and 84 (Morales, 2002). In section Thymus, two ploidy levels (diploid and tetraploid) and five different numbers, 2n = 28, 30, 54, 56 and 58, are found. According to Morales (1986), the tetraploid numbers (54, 56 and 58) have different origins: 2n = 56 is probably derived from a duplication of a 2n = 28 genome (i.e. autopolyploidy); 2n = 58 may have originated from a hybridation of two taxa with n = 14 and n = 15 and a subsequent doubling of the chromosome number; and 2n = 54 is probably derived from a 2n = 56 plant which has lost two chromosomes (i.e. autopolyploidy followed by dysploidy). The taxonomically closest species to T. loscosii is T. zygis Loefl. ex L. (Morales, 1986), which has three subspecies in the Iberian peninsula, two diploid (subsp. zygis and subsp. gracilis, both with 2n = 28) and one probably autotetraploid (subsp. sylvestris, with 2n = 56). Morales (1986) studied the meiosis of T. carnosus Boiss. (2n = 56), from the same section, and reported some tetravalents, which supports the hypothesis that the number 2n = 56 is derived by autopolyploidy. Judging from its morphology, it is difficult to believe that Thymus loscosii should have arisen by hybridization and, accordingly, an allopolyploid origin of the species is not supported by morphological data (R. Morales, pers. comm.).
Thymus loscosii is a perennial woody plant, 9–10 (15) cm tall, with abundant stoloniferous branches. The inflorescences are composed of whorls of small, whitish, zygomorphic flowers, with seeds disposed in up to four nutlets of about 0·4 × 1·1 mm. The pollination in T. loscosii is entomophilous, as in most Thymus species (Morales, 1986). The main pollinators are Apis mellifera and some species of Bombylius. It is gynodioecious, as for most species of Thymus (Morales, 1986; Manicacci et al., 1998). Despite being a self‐compatible species, self‐pollination is rare. The germination rate is low: 0 % reported by Morales (1986) and Sainz et al. (1996), 2 % by our group (Bosch et al., 2002). However, other researchers found higher rates under different temperature treatments, although with remarkable variation among populations (Albert et al., 2002). There is no active fruit dispersal; the nutlets are probably dispersed by gravity and/or surface water movements. Thymus loscosii might have a combined strategy of sexual and asexual reproduction, since vegetative propagation of stolons has been observed in the field. Thymus loscosii grows at 130–1010 m, in open sites in alkaline or saline loam substrates.
Populations of T. loscosii are located in several autonomous territories of Spain (Aragon, Castilla‐León, Catalonia, Euskadi, La Rioja and Navarra), comprising 52 UTM 10 × 10 km squares (Blanché et al., 2000). The total number of individuals is probably greater than one million and the extent of occurrence is approx. 28 000 km2. The species was included in the Catálogo Nacional de Especies Amenazadas (National Catalogue of Endangered Species) as endangered (Boletín Oficial del Estado, 1990), and at regional level is listed as rare in Aragon (Sainz et al., 1996), vulnerable in Navarra (Boletín Oficial de Navarra, 1997), and of special interest in Euskadi (Nekazal Ikerketa Eta Teknologia, 2002). However, the increasing number of new localities found in recent years has led to the exclusion of this species from the recent Lista Roja de la Flora Vascular Española (Red List of the Vascular Spanish Flora) (Aizpuru et al., 2000).
Allozyme electrophoresis was used to address the following issues: (a) whether there is evidence of auto polyploidy in T. loscosii, and (b) to describe the levels and distribution of genetic diversity. In addition, inferences are made about the status of conservation of this species together with some suggestions for strategies for its preservation.
MATERIALS AND METHODS
Sampling strategy
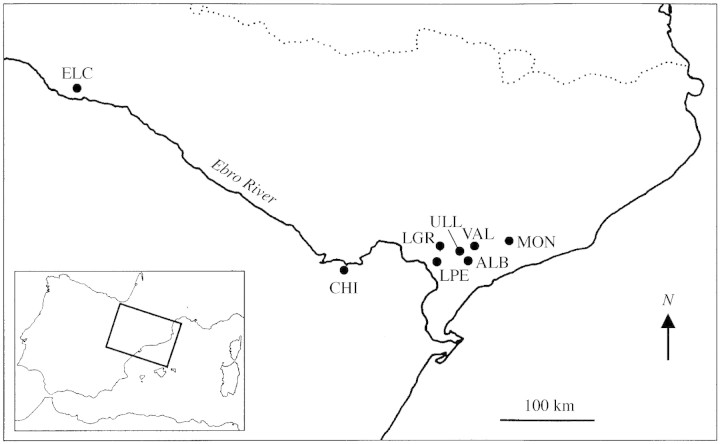
The study was focused in Catalonia (where six populations were sampled), but two additional populations were also sampled (one each from Aragon and Euskadi) to cover the entire geographic range of Thymus loscosii (Table 1 and Fig. 1). All populations were sampled in January–March 2001, and about 30 samples per population were collected. Sampling was conducted along a linear transect within each population and samples were collected about 50–100 cm apart to avoid collecting ramets from the same genet. Samples consisted of small fragments of branches which were placed into envelopes, transported to the laboratory, and stored at 4 °C until extraction 1 or 2 d later. Collection of samples was done carefully to minimize the potential damage to populations.
Table 1.
Populations of Thymus loscosii studied
| Population code | Location | Population size | Sample size |
| ULL | Ulldemolins (Catalonia, Spain), 31TCF2075 | ∼100 | 32 |
| ALB | Albarca (Catalonia, Spain), 31TCF2075 | 300–500 | 33 |
| LGR | La Granadella (Catalonia, Spain), 31TCF0380 | ∼200 | 33 |
| VAL | Vallclara (Catalonia, Spain), 31TCF3283 | >500 | 32 |
| MON | Montagut (Catalonia, Spain), 31TCF6885 | ∼100 | 32 |
| LPE | La Palma d’Ebre (Catalonia, Spain), 31TCF0172 | 50–100 | 30 |
| CHI | Chiprana (Aragon, Spain), 30TYL3868 | ∼100 | 32 |
| ELC | Elciego (Basque Country, Spain), 30TWN3205 | ∼100 | 33 |
Location of populations is detailed by UTM 1 × 1 km squares.
Fig. 1. Sampled populations of Thymus loscosii. ALB, Albarca, CHI, Chiprana, ELC, Elciego, LGR, La Granadella, LPE, La Palma d’Ebre, MON, Montagut, ULL, Ulldemolins, and VAL, Vallclara.
Electrophoresis
Genetic data were obtained through standard methods for starch gel electrophoresis of allozymes (Soltis et al., 1983; Wendel and Weeden, 1989). Leaves were detached from branches, and homogenized on refrigerated porcelain plates using a cold extraction buffer consisting of 0·011 m Tris–HCl, pH 7·6, 4 % sodium thioglycolate, 2 % polyethy leneglycol, and 8 % PVP‐40 (polyvinyl‐pyrrolidone). Analyses with an alternative extraction buffer (the composition is detailed in López‐Pujol et al., 2001) were also performed but with poorer results. Extracts were absorbed onto 3 mm Whatman filter paper, and either analysed immediately or stored at –20 °C until analysis 1 or 2 d later. Using 12·5 % starch gels, 19 enzyme systems were assayed. Aspartate aminotransferase (AAT; EC 2.6.1.1), diaphorase (DIA; EC 1.6.99.) and phosphoglucoisomerase (PGI; EC 5.3.1.9) were satisfactorily resolved on a tris–citrate/lithium–borate buffer at pH 8·2; aconitate hydratase (ACO; EC 4.2.1.3) and phosphogluconate dehydrogenase (6‐PGD; EC 1.1.1.44) were resolved on a morpholine buffer at pH 6·1 and phosphoglucomutase (PGM; EC 5.4.2.2) was resolved on histidine–citrate buffer pH 5·7.
Genetic analyses
Loci were numbered consecutively, and alleles at each locus were labelled alphabetically, beginning from the most anodal form. Isozyme phenotypes were interpreted genetically according to standard principles (Wendel and Weeden, 1989), but special attention was paid to phenotypes characteristic of tetraploids and to their interpretation (Gottlieb, 1981). To describe the levels of genetic diversity, the following statistics were calculated: P, the percentage of polymorphic loci at which the most common allele had a frequency of <0·95; A, the mean number of alleles per locus; Ho, the observed heterozygosity; and He, the expected panmictic heterozygosity. In autotetraploids, two types of He can be computed; He (Ce), the expected heterozygosity assuming random chromosomal segregation, and He (Cd), the expected heterozygosity assuming some level of chromatid segregation. Chromatid segregation is produced if ‘double reduction’ takes place, i.e sister chromatids segregate into the same gamete, a phenomenon specific to autopolyploids and which is dependent on the amount of tetravalent formation and the proximity of the locus to the centromere (see Bever and Felber, 1992; Ronfort et al., 1998). It is not known whether double reduction takes place in T. loscosii, and it was assumed that only chromosomal segregation occurs. This consideration allows a more conservative data processing method to be used regarding the type of polyploid origin for this species (auto‐ or allopolyploid), since double reduction increases the production of homozygous gametes as compared with what is expected under random chromosomal segregation in diploids and allopolyploids (Ronfort et al., 1998). The mean fixation index (F) was also calculated for all variable loci in each population, to compare genotype proportions with those expected under the Hardy–Weinberg equilibrium. We used a chi‐square (χ2) test to evaluate deviations of F from zero. The partitioning of genetic diversity within and between populations was analysed using Nei’s gene diversity statistics (Nei, 1973): total genetic diversity (HT), mean genetic diversity within populations (HS), genetic diversity between populations (DST), and proportion of total genetic diversity between populations (GST) were calculated for all populations. Gene flow (Nm) was estimated by Wright’s equation (Wright, 1951): Nm = 1– GST/4 GST. Nei’s genetic identity (I) (Nei, 1978) was also calculated between pairs of populations. A program for analysis of autotetraploid genotypic data, AUTOTET (Thrall and Young, 2000), was used for calculations of Ho, He and F. Employing the appropriate options for tetraploids, BIOSYS‐1 (Swofford and Selander, 1989) was used for the calculation of P, while GeneStat version 3·31 (Whitkus, 1988) was used to calculate HT, HS, DST and GST. The mean number of alleles per locus (A) was calculated by hand because data was included for the partially resolved locus Aco‐1.
RESULTS
Interpretation of enzyme banding patterns
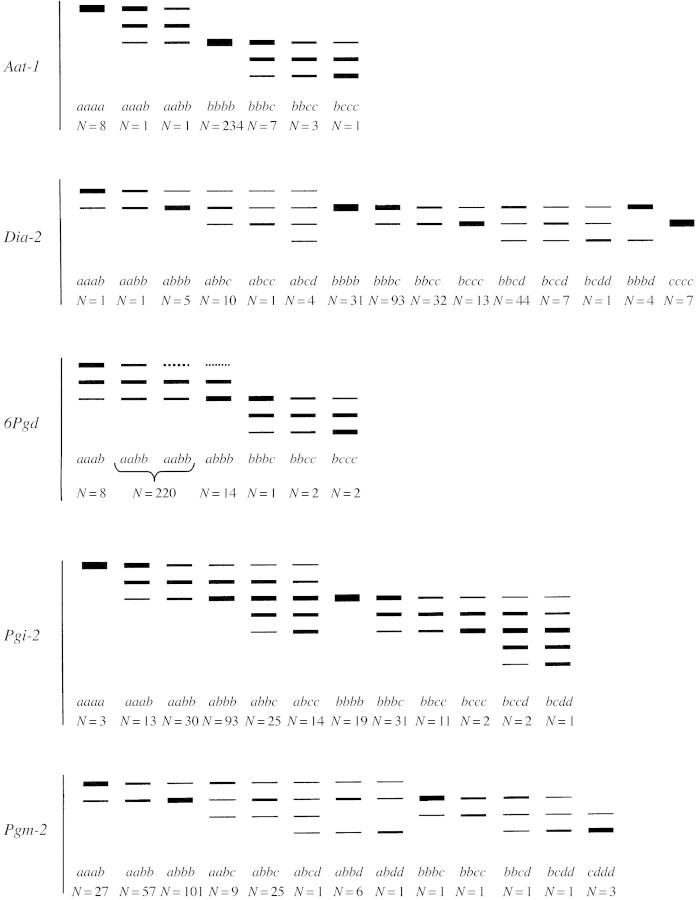
There were considerable difficulties in resolving enzymes in T. loscosii, due to interference during the extraction from polyphenolic substances and volatile oils. Up to 100 preliminary experiments were needed to obtain acceptable resolution. Of the 19 enzymes assayed, five were satisfactorily resolved, each with one interpretable locus: Aat‐1, Dia‐2, 6Pgd, Pgi‐2 and Pgm‐2 (Fig. 2). Aco‐1 was poorly stained on gels and allelic dosage in heterozygote phenotypes could not be determined, but the locus was used for calculation of A.
Fig. 2. Schematic banding patterns obtained for the five resolved loci in Thymus loscosii. All the inferred genotypes and the number of individuals showing each genotype are given below each phenotype.
Banding patterns in AAT, 6‐PGD and PGI were consistent with dimeric enzymes, and in ACO, DIA and PGM with monomeric enzymes. AAT displayed two regions of activity in T. loscosii, encoded by two independent putative loci: Aat‐1 and Aat‐2. Aat‐1 was highly monomorphic, with the majority of individuals being homozygous for Aat‐1b allele. Alleles Aat‐1a and Aat‐1c, although less frequent, were also present. The locus Aat‐2 was too faint to be interpretable, but appeared to be variable.
ACO presented two zones of activity, encoded by two different putative loci (Aco‐1 and Aco‐2). Aco‐1 was too poorly stained to interpret the allelic dosage, but four different alleles were detected at this locus, Aco‐1b and Aco‐1c being the most common. Some analysed individuals showed patterns of three and four bands, which could be interpreted as possessing three and four different alleles for this enzyme. The locus Aco‐2 could not be interpreted due to the poor staining of the bands.
Two different regions of activity were identified for DIA, from which only the most cathodal was consistently interpreted (Dia‐2). Four alleles were detected, with Dia‐2b displaying the highest frequency. Single‐banded homozygotes and several types of heterozygotes were observed at this locus. Heterozygous phenotypes were distinguished by the number of bands and/or the relative intensity of band staining. Thus, it was possible to observe two‐banded heterozygotes, with balanced and unbalanced staining activity, three‐banded phenotypes with unbalanced staining activity, and four‐banded phenotypes with equally strong bands.
6‐PGD exhibited a single zone of activity, encoded by only one putative locus (6Pgd). All studied individuals showed either a two‐banded or a three‐banded phenotype, which is not consistent with the dimeric structure of this enzyme. This pattern can be attributed to the inactivity of the dimer aa in some populations, since this band appeared in other populations but was poorly stained. Three‐banded patterns are consistent with both balanced phenotypes (aabb), which were the majority of observed genotypes, and unbalanced heterozygotes (aaab, abbb), which were less frequent. In the Vallclara (VAL) population we also observed heterozygous phenotypes with the 6Pgdc allele.
PGI showed two different regions of activity, encoded by two putative independent loci (Pgi‐1 and Pgi‐2), of which only Pgi‐2 could be interpreted. Four different alleles were detected, Pgi‐2b being the most common in all populations. Complex banding patterns with up to five bands were observed in some individuals, which are consistent with the expression of three different alleles. Five (not six) bands were observed due to the co‐migration of a homodimeric band encoded by the ‘intermediate’ allele with the heterodimeric band formed between the ‘fastest’ and the ‘slowest’ allele. Both balanced and unbalanced phenotypes with three or five bands were observed, but homozygous phenotypes were also present.
PGM presented two zones of activity. Only the most cathodal region was interpretable, controlled by the putative locus Pgm‐2. Analysed individuals showed four different alleles at this locus, Pgm‐2a and Pgm‐2b being the most common. Some phenotypes of three and four bands were found, which correspond to individuals possessing three and four different alleles at this locus.
Levels and distribution of genetic diversity
Among the six interpretable loci (including Aco‐1) 22 alleles were detected, the frequencies of which are given in Table 2. The richest populations were ALB, VAL and ELC (all displaying 16 alleles); the poorest populations were ULL, LGR and LPE (all with 13 alleles). Only one population‐specific allele was found in T. loscosii, at VAL (6Pgd‐c). In contrast, rare alleles (alleles with a frequency of <0·05) were common to all populations, representing 8 % (LGR) to 31 % (ALB) of the total number of alleles. It was found that 37·2 % of all plants possessed three or four alleles for at least one of the loci examined, with a maximum percentage of 58 % at Pgm‐2 in the MON population. Genetic diversity was high in T. loscosii, with the mean values over all eight populations of P = 85 %, A = 3·0, and He = 0·422 (Table 3). The most variable population was LGR (He = 0·479), while the least diversity was found in CHI (He = 0·399); however, values of the standard deviation (Table 3) showed that differences between populations were not statistically significant.
Table 2.
Allele frequencies for five loci in eight populations of Thymus loscosii
| Locus | Allele | ULL | ALB | LGR | VAL | MON | LPE | CHI | ELC |
| Aat‐1 | a | 0·000 | 0·000 | 0·235 | 0·000 | 0·000 | 0·000 | 0·000 | 0·045 |
| b | 1·000 | 0·962 | 0·727 | 0·976 | 1·000 | 1·000 | 1·000 | 0·932 | |
| c | 0·000 | 0·038 | 0·038 | 0·023 | 0·000 | 0·000 | 0·000 | 0·023 | |
| Dia‐2 | a | 0·000 | 0·016 | 0·000 | 0·016 | 0·039 | 0·008 | 0·039 | 0·078 |
| b | 0·609 | 0·672 | 0·656 | 0·766 | 0·297 | 0·675 | 0·656 | 0·641 | |
| c | 0·266 | 0·250 | 0·258 | 0·180 | 0·609 | 0·292 | 0·297 | 0·203 | |
| d | 0·125 | 0·062 | 0·086 | 0·039 | 0·055 | 0·025 | 0·008 | 0·078 | |
| 6Pgd | a | 0·509 | 0·507 | 0·485 | 0·427 | 0·492 | 0·475 | 0·500 | 0·476 |
| b | 0·491 | 0·492 | 0·515 | 0·484 | 0·508 | 0·525 | 0·500 | 0·523 | |
| c | 0·000 | 0·000 | 0·000 | 0·089 | 0·000 | 0·000 | 0·000 | 0·000 | |
| Pgi‐2 | a | 0·274 | 0·288 | 0·280 | 0·336 | 0·298 | 0·339 | 0·089 | 0·069 |
| b | 0·653 | 0·614 | 0·636 | 0·594 | 0·645 | 0·536 | 0·661 | 0·698 | |
| c | 0·072 | 0·091 | 0·083 | 0·070 | 0·056 | 0·125 | 0·234 | 0·224 | |
| d | 0·000 | 0·007 | 0·000 | 0·000 | 0·000 | 0·000 | 0·016 | 0·009 | |
| Pgm‐2 | a | 0·342 | 0·281 | 0·393 | 0·208 | 0·298 | 0·400 | 0·577 | 0·537 |
| b | 0·639 | 0·687 | 0·607 | 0·617 | 0·540 | 0·583 | 0·388 | 0·361 | |
| c | 0·009 | 0·016 | 0·000 | 0·050 | 0·137 | 0·017 | 0·034 | 0·102 | |
| d | 0·009 | 0·016 | 0·000 | 0·125 | 0·024 | 0·000 | 0·000 | 0·000 |
The most frequent allele for each locus and population is indicated in bold.
Table 3.
Summary of genetic variation for five loci in eight populations of Thymus loscosii
| Population | N | P | A* | H o | H e | F |
| ULL | 30·2 | 80·0 | 2·83 | 0·491 (0·279) | 0·402 (0·226) | –0·222 |
| ALB | 32·6 | 80·0 | 3·33 | 0·497 (0·249) | 0·407 (0·189) | –0·221 |
| LGR | 29·0 | 100 | 2·83 | 0·469 (0·239) | 0·479 (0·038) | 0·022 |
| VAL | 31·4 | 80·0 | 3·17 | 0·439 (0·246) | 0·418 (0·222) | –0·051 |
| MON | 31·4 | 80·0 | 3·00 | 0·470 (0·281) | 0·425 (0·242) | –0·104 |
| LPE | 29·6 | 80·0 | 2·50 | 0·479 (0·270) | 0·408 (0·232) | –0·175 |
| CHI | 31·2 | 80·0 | 3·00 | 0·452 (0·266) | 0·399 (0·223) | –0·132 |
| ELC | 30·6 | 100 | 3·33 | 0·476 (0·246) | 0·438 (0·178) | –0·086 |
| Mean | 30·7 | 85·0 | 3·00 | 0·472 | 0·422 | – |
| s.d. | 9·2 | 0·28 | 0·019 | 0·026 | – |
N, sample size; P, percentage of polymorphic loci; A, mean number of alleles per locus; Ho, observed heterozygosity; He, expected panmictic heterozygosity; F, mean fixation index.
Standard deviation in parentheses.
* Locus Aco‐1 is included in its calculation.
Values of observed heterozygosity were higher than those of expected heterozygosity in all populations except LGR, the only population with a positive value of F (fixation index) (Table 3). At loci level, 15 F‐values were in accordance with Hardy–Weinberg expectations (P ≥ 0·05), while 21 F‐values differed significantly from zero (P < 0·05). Of these 21 values, four were positive and 17 were negative (Table 4). Positive values indicate deficiency of heterozygotes; negative values indicate excess.
Table 4.
Values of fixation index (F) for all polymorphic loci in eight populations of Thymus loscosii
| Locus | ULL | ALB | LGR | VAL | MON | LPE | CHI | ELC |
| Aat‐1 | – | 0·238** | 0·854*** | 0·204ns | – | – | – | 0·492ns |
| Dia‐2 | –0·248** | –0·254*** | –0·230** | 0·013ns | 0·057** | –0·199ns | 0·077ns | –0·001ns |
| 6Pgd | –0·299*** | –0·324*** | –0·314*** | –0·084*** | –0·301*** | –0·303*** | –0·303*** | –0·305*** |
| Pgi‐2 | –0·113* | –0·129* | 0·078ns | –0·150ns | 0·027ns | –0·043* | –0·065ns | 0·007ns |
| Pgm‐2 | –0·223*** | –0·256** | –0·148ns | 0·015*** | –0·192* | –0·179ns | –0·217ns | –0·179ns |
Conformance to Hardy–Weinberg equilibrium was tested using chi‐square analysis: ns, P ≥ 0·05; * P < 0·05; **, P < 0·01;***, P < 0·001.
Genetic diversity in T. loscosii was distributed mainly within populations (HS = 0·429) rather than between them (DST = 0·015), i.e. genetic diversity attributable to interpopulation differentiation was low (GST = 0·033) (Table 5). Gene flow was high (Nm = 7·32), indicating a substantial interchange of genes between populations. Values for Nei’s genetic identity (I) (Nei, 1978) were very high between pairs of populations (mean = 0·973, range 0·929–1·000; Table 6).
Table 5.
Gene diversity statistics (Nei, 1973) for five loci in eight populations of Thymus loscosii
| Locus | H T | H S | D ST | G ST |
| Aat‐1 | 0·099 | 0·084 | 0·014 | 0·146 |
| Dia‐2 | 0·528 | 0·497 | 0·031 | 0·059 |
| 6Pgd | 0·511 | 0·517 | 0·000 | 0·000 |
| Pgi‐2 | 0·531 | 0·520 | 0·010 | 0·019 |
| Pgm‐2 | 0·552 | 0·528 | 0·024 | 0·043 |
| Mean | 0·444 | 0·429 | 0·015 | 0·033 |
| s.d. | 0·193 | 0·193 | 0·012 | 0·056 |
HT, Total genetic diversity; HS, genetic diversity within populations; DST, genetic diversity between populations; GST, proportion of total genetic diversity between populations.
Table 6.
Matrix of Nei’s genetic identity (Nei, 1978) between populations of Thymus loscosii
| Populations | ULL | ALB | LGR | VAL | MON | LPE | CHI | ELC |
| ULL | – | |||||||
| ALB | 1·000 | – | ||||||
| LGR | 0·991 | 0·992 | – | |||||
| VAL | 0·994 | 1·000 | 0·976 | – | ||||
| MON | 0·968 | 0·957 | 0·938 | 0·932 | – | |||
| LPE | 1·000 | 1·000 | 0·988 | 0·995 | 0·961 | – | ||
| CHI | 0·978 | 0·971 | 0·967 | 0·953 | 0·941 | 0·985 | – | |
| ELC | 0·972 | 0·965 | 0·968 | 0·950 | 0·929 | 0·975 | 1·000 | – |
DISCUSSION
Autopolyploidy in Thymus loscosii
Morales (1986) hypothesized that Thymus loscosii is an autotetraploid. This hypothesis was based on chromosome numbers (as presented in the Introduction), and phylogenetic studies. Furthermore, according to R. Morales (pers. comm.) it may be inferred from its morphology that T. loscosii did not arise from hybridization of the potential diploid parentals, e.g. T. zygis or T. vulgaris.
To verify the hypothesis of an autopolyploid origin, extensive cytological (i.e. karyological meiotic analyses) and inheritance studies (allozyme progeny tests) would be needed. However, cytological analyses are technically extremely difficult in Thymus, due to the very small size of the chromosomes (cf. Morales, 1986), and inheritance studies would also be difficult due to low germination rates.
Allozyme data are extremely useful in distinguishing between autopolyploids and allopolyploids (Soltis and Rieseberg, 1986; Crawford, 1989). Tetrasomic inheritance in autotetraploids results in the formation of unbalanced as well as balanced heterozygotes in all possible combinations. In contrast, allotetraploids are expected to display fixed heterozygosity, at least at some loci at which loci on homoeologous chromosomes are fixed for different alleles. For example, when two alleles (a and b) are present at a locus in an autotetraploid, three types of heterozygotes can be produced, one balanced (aabb) and two unbalanced (aaab and abbb), which can be differentiated on zymograms by the intensity of band staining. Such a pattern would only be possible in an allopolyploid if the locus was heterozygous for a and b on both pairs of homoelogous chromosomes.
Allozyme data support the hypothesis that T. loscosii is an autotetraploid. No evidence of fixed heterozygosity was found in T. loscosii. Except for 6Pgd and Pgm‐2, all loci showed both homozygotes and heterozygotes (balanced and unbalanced). The 6Pgd and the Pgm‐2 loci displayed only heterozygous phenotypes, but these were both balanced and unbalanced. Support for autopolyploidy was also given by genetic diversity data, as discussed below.
Levels of diversity in autopolyploids
The high levels of heterozygosity found in autotetraploids may also be a consequence of tetrasomic inheritance (Soltis and Soltis, 2000), because this mode of inheritance would reduce the effects of population bottlenecks and genetic drift. The literature provides us with several examples of higher levels of heterozygosity in autotetraploid plants than in their diploid counterparts: Tolmiea menziesii Torr. & Gray (Ho = 0·237 in tetraploid populations and Ho = 0·070 in diploid ones; Soltis and Soltis, 1989), Heuchera micrantha Dougl. (Ho = 0·150 in tetraploid and Ho = 0·075 in diploid; Ness et al., 1989), Heuchera grossulariifolia Rydb. (Ho = 0·159 in tetraploid and Ho = 0·058 in diploid; Wolf et al., 1990), Dactylis glomerata L. (Ho = 0·43 in tetraploid and Ho = 0·17 in diploid; Soltis and Soltis, 1993), Rutidosis leptorrhinchoides F. Muell. (Ho = 0·34 in tetraploid and Ho = 0·22 in diploid; Brown and Young, 2000), Vaccinium oxycoccos L. (Ho = 0·213 in tetraploid and Ho = 0·067 in diploid; Mahy et al., 2000) and Centaurea jacea L. (Ho = 0·54 in tetraploid and Ho = 0·29 in diploid; Hardy and Vekemans, 2001), among others. Judging from allozyme banding patterns, no population in the examined material of T. loscosii appeared to be diploid. Accordingly, only its levels of heterozygosity (Ho = 0·472) can be compared, although cautiously, with the closely related diploid species T. vulgaris (Ho = 0·295; Tarayre and Thompson, 1997).
The low number of loci resolved in T. loscosii illustrates the problems raised when allozyme studies are addressed to polyploid, woody and aromatic species, which are traditionally considered to be difficult (P. Arús, pers. comm.). To exemplify this, only four loci were interpretable in the analysis of the population structure of the closely related Thymus vulgaris L. (Tarayre and Thompson, 1997).
Genetic diversity in Thymus loscosii
Tetraploidy allows for the presence of three or four alleles at a single locus since loci are duplicated (Soltis and Rieseberg, 1986; Soltis and Soltis, 1989; Mahy et al., 2000). Compared with diploids, more enzyme variants and an increased biochemical diversity may contribute to the success of polyploids in nature (Levin, 1983). In T. loscosii, 37·2 % of all plants possessed three or four alleles for at least one of the examined loci, a value close to that obtained for an autotetraploid cytotype of Tolmiea menziesii (39 %; Soltis and Soltis, 1989), and higher than that reported for Vaccinium oxycoccos (12·1 %; Mahy et al., 2000). It is possible that the high number of alleles at some loci may be due to repeated polyploidization events, as discussed, e.g. for the grass Dactylis glomerata (Lumaret, 1988).
The expected tetrasomic inheritance leads to high levels of genetic diversity in autotetraploids, as described by values of A, P and heterozygosity (Soltis and Soltis, 1989). High values of genetic variation in T. loscosii are within the range of other autotetraploids (Table 7). As observed in the field, T. loscosii can undergo vegetative propagation through stolons, which can maintain genetic variation, once produced, within populations. This phenomenon has already been reported in other species, such as Populus tremuloides Michx. (Cheliak and Dancik, 1982), Erythronium albidum Nutt. and E. propullans A. Gray (Pleasants and Wendel, 1989), Iris cristata Aiton (Hannan and Orick, 2000) and Delphinium montanum DC. (Simon et al., 2001).
Table 7.
Genetic diversity in Thymus loscosii and in other autotetraploid species
| Taxa | P | A | H o | H e | Reference |
| Aster kantoensis Kitam. | 36·9 | 1·53 | – | 0·142 | Maki et al., 1996 |
| Centaurea jacea L. | – | 3·54 | 0·54 | 0·38 | Hardy and Vekemans, 2001 |
| Dactylis glomerata L. | 80·0 | 2·36 | 0·43 | – | Soltis and Soltis, 1993 |
| Heuchera grossulariifolia Rydb. | 31·0 | 1·55 | 0·159 | – | Wolf et al., 1990 |
| Heuchera micrantha Dougl. | 38·33 | 1·64 | 0·150 | – | Ness et al., 1989 |
| Rutidosis leptorrhynchoides F. Muell. | 98·0 | 3·2 | 0·34 | 0·36 | Brown and Young, 2000 |
| Swainsona recta A.T. Lee | – | 4·3 | 0·24 | 0·42 | Buza et al., 2000 |
| Thymus loscosii Willk. | 85·0 | 3·0 | 0·472 | 0·422 | This study |
| Tolmiea menziesii Torr. & Gray | 40·8 | 1·5 | 0·237 | – | Soltis and Soltis, 1989 |
| Turnera ulmifolia var. elegans Urb. | 65·3 | 2·03 | 0·42 | 0·27 | Shore, 1991 |
| Turnera ulmifolia var. Intermedia Urb. | 20·1 | 1·20 | 0·07 | 0·04 | Shore, 1991 |
| Vaccinium oxycoccos L. | 38·9 | 1·66 | 0·213 | – | Mahy et al., 2000 |
If a species has both diploid and tetraploid populations, values of diversity given here are only for tetraploid populations.
All values given here are population means.
P, Percentage of polymorphic loci; A, mean number of alleles per locus; Ho, observed heterozygosity; He, expected panmictic heterozygosity.
In populations of autotetraploid species, equilibrium frequencies under random mating are reached after several generations, not in a single generation as for randomly mating diploid species (Bever and Felber, 1992). This can theoretically result in deviations from Hardy–Weinberg equilibrium (Mahy et al., 2000). Negative values for the fixation index were found at most loci and most populations of T. loscosii, indicating an excess of heterozygotes compared with Hardy–Weinberg expectations. The 6Pgd and the Pgm‐2 loci displayed only heterozygous phenotypes in all examined populations. A negative fixation index could be explained by: (a) selection against homozygote survival promoting high heterozygosity; (b) random stochastic events; (c) barriers to inbreeding; and (d) sampling error. The excess of heterozygotes detected in T. loscosii might be explained by some barriers to inbreeding, e.g. ocurrence of gynodioecy, together with a marked protandry and low rates of selfing (Bosch et al., 2002). However, other reasons cannot be discarded, as for example selection for heterozygosity. Alternatively, vegetative propagation may have contributed to the maintenance of the heterozygote excess, but the extent to which vegetative reproduction occurs in T. loscosii needs further study.
Distribution patterns of genetic diversity within and among populations are primarily determined by genetic flow, genetic drift and natural selection. Only a minimal fraction of allozyme variation in T. loscosii (around 3 %) is due to differences between populations. This pattern is also expressed as a high value of gene flow (7·32), which is theoretically high enough to prevent divergence by genetic drift (Wright, 1951). However, judging from other species of Thymus, which are characterized by small pollen and seed dispersal distances (Tarayre and Thompson, 1997; Eriksson, 1998), gene flow should also be restricted in T. loscosii. Instead, the very high genetic identities found between populations and the high value of Nm may be explained by recent and rapid fragmentation from a wide, continuous area resulting in genetically similar populations. Chorological research in recent years has revealed the existence of many additional populations, which may indicate that T. loscosii formerly had a continuous distribution along the Ebro river basin. This area has been extensively replanted with forest during the last decades (Blanco et al., 1997), and open sites at which the species could grow have been lost. In addition, this area is currently experiencing a fragmentation process caused by change of land use, e.g. the extensive plantation of new vineyards in the westernmost part of the distribution area (Nekazal Ikerketa Eta Teknologia, 2002, and field observations by J. López‐Pujol). Fragmentation may result in genetic impoverishment (Barrett and Kohn, 1991; Ellstrand and Elam, 1993), but polyploids may be less sensitive since genetic diversity is maintained by genome duplications.
Conservation
The optimal measures for species preservation are those in situ. The conservation of a species in its habitat allows for the maintenance of interactions between the species and its ecosystem without detaining evolutionary processes (Falk and Holsinger, 1991). Protection of habitat is generally limited to the protection of selected populations of species. Using the formula proposed by Ceska et al. (1997), 1 – (GST)n (where n is the number of populations), conservation of only two populations allows us to preserve 99·9 % of genetic diversity found in T. loscosii, due to the high genetic similarity between populations. Nevertheless, given the variation of GST among loci, this may be an underestimate of genetic diversity detected in this species. Moreover, when choosing the populations to preserve, other considerations must also be taken into account, for instance large populations and populations with more intermediate allele frequencies must be favoured in order to reduce the probability of loss of alleles (Martínez‐Palacios et al., 1999). For example, the combination of VAL and ALB populations contains 21 of the 22 detected alleles, and they are the largest sampled populations. However, if population LGR is also preserved (i.e. the three largest populations), then all alleles would be conserved. Collection of seeds for maintenance in germplasm banks as an ex situ strategy does not seem to be an appropriate alternative to conservation policy because of the low germination rates.
Apart from genetic criteria, two additional types of polymorphism may be considered: (1) chemical diversity and (2) the frequency of females within populations. Chemical diversity may play a role in adaptation to environment, as suggested in T. vulgaris (Thompson et al., 1998) and, if this also applies to T. loscosii, all chemotypes must be conserved. At present, only a single population of T. loscosii has been surveyed for phytochemical diversity (Molero and Rovira, 1983).
ACKNOWLEDGEMENTS
We thank J. Thompson for kindly providing the extraction buffer recipe; J. Molero and L. Sáez for their assistance in locating populations; R. Morales, D. Crawford and an anonymous reviewer for their skilled scientific comments; and J. Chaves, N. Álvarez, and N. Cerrillo for their collaboration in the experimental work. This work was partially subsidized by grants REN00‐0829GLO (DGI del MCyT, Spain) and FBG 301022 (DMA, Generalitat de Catalunya) and Research Fellowships FPI (MECD) to J.L.‐P. and Programa Ramón y Cajal (MCyT) to M.B.
Received: 18 July 2003;; Returned for revision: 7 October 2003; Accepted: 25 November 2003, Published electronically: 23 January 2004
References
- AizpuruIet al.2000. Lista Roja de Flora Vascular Española (valoración según categorías UICN). Conservación Vegetal 6: 11–38. [Google Scholar]
- AlbertMJ, Iriondo JM, Pérez‐García F.2002. Effects of temperature and pretreatments on seed germination of nine semiarid species from NE Spain. Israel Journal of Plant Sciences 50: 103–112. [Google Scholar]
- BarrettSCH, Kohn JR.1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE, eds. Genetics and conservation of rare plants New York: Oxford University Press, 3–30. [Google Scholar]
- BeverJD, Felber F.1992. The theoretical population genetics of autopolyploidy. In: Antonovics J, Futuyma D, eds. Oxford surveys in evolutionary biology Oxford: Oxford University Press, 185–217. [Google Scholar]
- BlanchéC, Molero J, Rovira A, Simon J, Bosch M, López‐Pujol J.2000.Estudi de l’estructura i dinàmica de les poblacions de l’espècie de la flora vascular: Thymus loscosii. Memòria del Projecte 2. Departament de Medi Ambient, Generalitat de Catalunya. [Google Scholar]
- BlancoEet al.1997.Los bosques ibéricos. Una interpretación geobotánica. Barcelona: Editorial Planeta. [Google Scholar]
- Boletín Oficial del Estado.1990. Real Decreto 439/1990 por el que se establece el Catálogo Nacional de Especies Amenazadas. Boletín Oficial del Estado 82: 9468–9471. Available at: http://www.mma.es/Naturalia/naturalia_hispanica/catalogo_especies_2000/incluser/ListadoFlora.html [Google Scholar]
- Boletín Oficial de Navarra.1997. Decreto foral 94/1997, de 7 de abril, por el que se crea el Catálogo de la Flora Amenazada de Navarra y se adoptan medidas para la conservación de la flora silvestre catalogada. Available at: http://www.cfnavarra.es/BON/974/97418004.htm [Google Scholar]
- BoschM, Orellana MR, Molero J, Rovira AM.2002. Biología de la reproducción de la especie endémica ginodioica Thymus loscosii Willk. (Labiatae). 1er Congreso de biología de la Conservación de Plantas. Abstracts book: 65. Jardí Botànic de la Universitat de València, Spain. [Google Scholar]
- BrownAHD, Young AG.2000. Genetic diversity in tetraploid populations of the endangered daisy Rutidosis leptorrhynchoides and implications for its conservation. Heredity 85: 122–129. [DOI] [PubMed] [Google Scholar]
- BuzaL, Young A, Thrall P.2000. Genetic erosion, inbreeding and reduced fitness in fragmented populations of the endangered tetraploid pea Swainsona recta Biological Conservation 93: 177–186. [Google Scholar]
- CeskaJF, Affolter JM, Hamrick JL.1997. Developing a sampling strategy for Baptisia arachnifera based on allozyme diversity. Conservation Biology 11: 1133–1139. [Google Scholar]
- CheliakWM, Dancik BP.1982. Genetic diversity of natural populations of a clone‐forming tree Populus tremuloides Canadian Journal of Genetics and Cytology 24: 611–616. [Google Scholar]
- CrawfordDJ.1989. Enzyme electrophoresis and plant systematics. In: Soltis DE, Soltis PS, eds. Isozymes in plant biology Portland: Dioscorides Press, 146–164. [Google Scholar]
- EllstrandNC, Elam DR.1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- ErikssonA.1998. Regional distribution of Thymus serpyllum: management history and dispersal limitation. Ecogeography 21: 35–43. [Google Scholar]
- FalkDA, Holsinger KE.1991.Genetics and conservation of rare plants. New York: Oxford University Press. [Google Scholar]
- GottliebLD.1981. Electrophoretic evidence and plant populations. Progress in Phytochemistry 7: 1–46. [Google Scholar]
- GreuterW, Burdet HM, Long G (eds).1986.Med‐Checklist. Vol. 3 (Convolvulaceae‐Labiatae). Conservatoire Botanique de Genève. [Google Scholar]
- HannanGL, Orick MW.2000. Isozyme diversity in Iris cristata and the threatened glacial endemic I. lacustris (Iridaceae). American Journal of Botany 87: 293–301. [PubMed] [Google Scholar]
- HardyOJ, Vekemans X.2001. Patterns of allozyme variation in diploid and tetraploid Centaurea jacea at different spatial scales. Evolution 55: 943–954. [DOI] [PubMed] [Google Scholar]
- 22.Nekazal Ikerketa Eta Teknologia.2002. Especies amenazadas de flora en el País Vasco. Available at: http://www.nekanet.net/Naturaleza/especies.htm [Google Scholar]
- JalasJ.1972.Thymus L. In: Tutin et al. eds. Flora Europaea, Vol. 3 (Diapensiaceae–Myoporaceae). Cambridge: Cambridge University Press. [Google Scholar]
- LevinDA.1983. Polyploidy and novelty in flowering plants. American Naturalist 122: 1–25. [Google Scholar]
- López‐PujolJ, Bosch M, Simon J, Blanché C.2001. Allozyme diversity of the two endemic Petrocoptis: P. montsicciana and its close relation P. pardoi (Caryophyllaceae). Canadian Journal of Botany 79: 1379–1389. [Google Scholar]
- LumaretR.1988. Cytology, genetics and evolution in the genus Dactylis Critical Reviews in Plant Sciences 7: 55–91. [Google Scholar]
- MahyG, Bruederle LP, Connors B, Van Hofwegen M, Vorsa N.2000. Allozyme evidence for genetic autopolyploidy and high genetic diversity in tetraploid cranberry, Vaccinium oxycoccos (Ericaceae). American Journal of Botany 87: 1882–1889. [PubMed] [Google Scholar]
- MakiM, Masuda M, Inoue K.1996. Genetic diversity and hierarchical population structure of a rare autotetraploid plant, Aster kantoensis (Asteraceae). American Journal of Botany 83: 296–303. [Google Scholar]
- ManicacciD, Atlan A, Elena‐Rosselló JA, Couvet D.1998. Ginodioecy and reproductive trait variation in three Thymus species (Lamiaceae). International Journal of Plant Sciences 159: 948–957. [Google Scholar]
- Martínez‐PalaciosA, Eguiarte LE, Furnier GR.1999. Genetic diversity of the endangered endemic Agave victoriae‐reginae (Agavaceae) in the Chihuahuan Desert. American Journal of Botany 86: 1093–1098. [PubMed] [Google Scholar]
- MoleroJ, Rovira A.1983. Contribución al estudio biotaxonómico de Thymus loscosii Willk. y Thymus fontqueri (Jalas) Molero & Rovira, stat. nov. Anales del Jardín Botánico de Madrid 39: 279–296. [Google Scholar]
- MoralesR.1986. Taxonomía de los géneros Thymus (excluída la sección Serpyllum) y Thymbra en la Península Ibérica. Ruizia 2: 1–324. Monografías Del Real Jardín Botánico, CSIC. [Google Scholar]
- MoralesR.2002. The history, botany and taxonomy of the genus Thymus In: Stahl‐Biskup E, Sáez F, eds. Thyme. The genus Thymus. London: Taylor & Francis, 1–43. [Google Scholar]
- NeiM.1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NeiM.1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NessBD, Soltis DE, Soltis PS.1989. Autopolyploidy in Heuchera micrantha (Saxifragaceae). American Journal of Botany 76: 614–626. [DOI] [PubMed] [Google Scholar]
- OttoSP, Whitton J.2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- PleasantsJM, Wendel JF.1989. Genetic diversity in a clonal narrow endemic, Erythronium propullans, and in its widespread progenitor, Erythronium albidum American Journal of Botany 76: 1136–1151. [Google Scholar]
- RamseyJ, Schemske DW.1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29: 467–501. [Google Scholar]
- RonfortJ, Jenczewski E, Bataillon T, Rousset F.1998. Analysis of population structure in autotetraploid species. Genetics 150: 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SainzH, Franco F, Arias J.1996.Estrategias para la Conservación de la Flora Amenazada de Aragón. Zaragoza: Diputación General de Aragón. [Google Scholar]
- ShoreJS.1991. Tetrasomic inheritance and isozyme variation in Turnera ulmifolia vars. elegans Urb. and intermedia Urb. (Turneraceae). Heredity 66: 305–312. [Google Scholar]
- SimonJ, Bosch M, Molero J, Blanché C.2001. Conservation biology of the Pyrenean larkspur (Delphinium montanum): a case of conflict of plant versus animal conservation? Biological Conservation 98: 305–314. [Google Scholar]
- SoltisDE, Rieseberg LH.1986. Autopolyploidy in Tolmiea menziesii (Saxifragaceae): genetic insights from enzyme electrophoresis. American Journal of Botany 73: 310–318. [Google Scholar]
- SoltisDE, Soltis PS.1989. Genetic consequences of autopolyploidy in Tolmiea (Saxifragaceae). Evolution 43: 586–594. [DOI] [PubMed] [Google Scholar]
- SoltisDE, Soltis PS.1993. Molecular data and the dynamic nature of polyploidy. Critical Reviews in Plant Sciences 12: 243–273. [Google Scholar]
- SoltisPS, Soltis DE.2000. The role of genetic and genomic attributes in the success of polyploids. National Academy of Sciences Colloquium Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins 27–29 January 2000, Irvine, California. Available at: http://www.nap.edu/openbook/0309070791/html [Google Scholar]
- SoltisDE, Haufler H, Darrow D, Gastony J.1983. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers and staining schedules. American Fern Journal 73: 9–27. [Google Scholar]
- StebbinsGL.1947. Types of polyploids: their classification and significance. Advances in Genetics 1: 403–429. [DOI] [PubMed] [Google Scholar]
- StebbinsGL.1980. Polyploidy in plants: unresolved problems and prospects. In: Lewis WH, ed. Polyploidy New York: Plenum Press, 495–520. [Google Scholar]
- SwoffordDL, Selander RB.1989.BIOSYS‐1: Release 1·7 edition. A computer program for the analysis of allelic variation in genetics. User’s manual. Urbana‐Champaign: University of Illinois, Department of Genetics and Development. [Google Scholar]
- TarayreM, Thompson JD.1997. Population genetic structure of gynodioecious Thymus vulgaris L. (Labiatae) in southern France. Journal of Evolutionary Biology 10: 157–174. [Google Scholar]
- ThompsonJD, Manicacci D, Tarayre M.1998. Thirty‐five years of thyme: a tale of two polymorphisms. Why so many females? Why so many chemotypes? Bioscience 48: 805–814. [Google Scholar]
- ThrallPH, Young A.2000. AUTOTET: a program for the analysis of autotetraploid genotypic data. Journal of Heredity 91: 348–349. [DOI] [PubMed] [Google Scholar]
- WendelF, Weeden NF.1989. Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS, eds. Isozymes in plant biology Portland: Dioscorides Press, 5–45. [Google Scholar]
- WhitkusR.1988. Modified version of GENESTAT: a program for computing genetic statistics from allele frequency. Plant Genetics Newsletter 4: 10. [Google Scholar]
- WolfPG, Soltis DE, Soltis PS.1990. Chloroplast‐DNA and allozymic variation in diploid and autotetraploid Heuchera grossulariifolia (Saxifragaceae). American Journal of Botany 77: 232–244. [DOI] [PubMed] [Google Scholar]
- WrightS.1951. The genetic structure of populations. Annals of Eurogenics 15: 323–354. [DOI] [PubMed] [Google Scholar]