Abstract
• Aims Receptors for plant hormones are becoming identified with increasing rapidity, although a frustrating number remain unknown. There have also been many more hormone‐binding proteins described than receptors. This Botanical Briefing summarizes what has been discovered about hormone binding sites, their discovery and descriptions, and will not dwell on receptor functions or activities except where these are relevant to understand binding.
• Scope Of those receptors identified, each falls into recognized protein superfamilies. Ethylene and cytokinin receptors have intracellular histidine kinase phosphorelay domains, but the ligand‐binding sites are distinct, one being buried within membrane‐spanning helices, the other in an extracellular loop domain. Brassinosteroid and phytosulfokine receptors are members of the leucine‐rich repeat receptor‐like protein superfamily and for these the ligand binding sites are likely to be in one of the loops of the extracellular leucine‐rich domain. For auxin, the auxin‐binding protein ABP1 is a member of the cupin superfamily and the binding site is in a hydrophobic pocket at the head of which is a zinc ion to coordinate the acid group of the ligand. Receptors for other plant hormones have still to be identified.
• Conclusions Plant hormone receptors have been identified through the application of many different techniques; no one technique is likely to prove more successful than any other for discovering new receptors. At present there is structural detail only for auxin binding, although a good model exists for the amino acid residues needed for Cu(I) and ethylene binding. In this respect plant biology is very poor and effort needs to be put into receptor discovery and molecular characterizetion. The information accumulated by such work will undoubtedly indicate many new ways in which plant growth and development can be manipulated, but knowledge‐led design of new ligands or of altered sensitivities is still some way off.
Key words: Phytohormone, receptor, protein structure
INTRODUCTION
The discovery of each plant hormone has led, in turn, to the desire to find and understand their receptors. Once a receptor gene and protein have been described, questions about how the protein works become accessible. Two leading questions are: (1) How does the receptor recognize the ligand? (2) How does the receptor signal that its ligand‐binding site is occupied? An understanding of binding provides an entry for molecular modelling and the possibility of knowledge‐led design of new and modified ligands, as well as ways of altering sensitivity to ligands. Knowledge of the signalling functions of the receptor will help identify downstream components in the signalling cascade and offer possibilities of manipulating physiological responses to the hormone. This Briefing will look only at binding because for some receptor candidates the signalling pathway is unknown; for others, the signalling domain is a kinase about which much has been written, although in plants the substrates for phosphorylation are mostly unrecognized.
For a few plant hormones there are still no convincing receptor candidates, a situation that is in striking contrast to the wealth of information known about animal and microbial hormone‐receptors. Progress towards receptor identification will be noted, but even for verified plant hormone receptors, there is still a very great deal to be learnt. There have also been numerous descriptions of plant hormone binding proteins, only some of which have turned out to be bona fide receptors. Some mention of these has been included where specific information about the binding site was gained, or if it helps illustrate the approaches used to identify receptors.
Binding sites and structure–activity relationships
For each of the plant hormones a considerable literature exists cataloguing the exacting experiments done to determine the structure–activity relationships between compound libraries and hormone‐specific responses. Summary or example data sets can be found for auxin (Veldstra, 1944; Thimann, 1963; Katekar, 1979), for absciscic acid (ABA) (Hill et al., 1995), for gibberellins (GAs) (Hoad, 1983; Takahaski et al., 1986), for cytokinins (Matsubara, 1990) and for brassinosteroids (Baron et al., 1998; Bajguz and Tretyn, 2003). Almost without exception, this experimentation was done using bioassays and such work goes on to this day for compounds likely to prove of agronomic importance. For example, such testing continues for auxins due to their importance as agrochemicals, particularly as selective herbicides. Clearly, the ability to introduce screening in silico could accelerate compound discovery. For this reason, even before crystal structures for hormone receptors started becoming available there were attempts to model hormone binding sites. Published accounts exist only for auxin, probably because the complexity of the problem in three‐dimensional space is too great for larger ligands. Even so, no published record exists for novel compounds selected using binding site models.
It is also interesting to note that a related approach, chemical genetics, is being used to help uncover receptors and their signalling networks (Stockwell, 2000). Synthetic organic compounds are screened for modulations of plant responses, in the same way that mutant populations are scored in forward genetic screening. If the compound is a variant or mimetic of a plant hormone, or even if its action is allosteric, hormone‐specific responses can be scored. The protein target of an active compound is then sought by screening mutant populations for resistance to the compound and cloning the mutated gene. Such experimentation has identified both sirtinol as an activator of many auxin pathways, and its target SIR1 (Zhao et al., 2003).
The rest of this Briefing describes how close plant scientists are to definitive, high‐resolution molecular detail for the binding domain of each class of hormone.
BINDING SITES FOR PLANT HORMONES
Ethylene
The first unequivocal identification of a plant hormone receptor was for ethylene (Chang et al., 1993; Schaller and Bleecker, 1995) and this was based on cloning the ETR1 gene from arabidopsis. As soon as the sequence was available, it became clear that ETR1 contained domains with similarity to a large family of signal transducing proteins known as two component regulators. The paradigms were bacterial environmental sensor receptors. The homologous domains lie in the C‐terminal, cytoplasmic part of ETR1 and include a histidine kinase domain and a response regulator receiver domain (Fig. 1). In the bacterial two component regulators, the response regulator is the second component and comprises receiver and output domains. In ETR1 the receiver is part of a single polypeptide and the output domain is absent. Ethylene binding leads to autophosphorylation of the histidine kinase domain followed by a phosphorelay to the receiver domain. In bacterial two component receptors the phosphorelay passes to the output domain and onto a kinase cascade. In plants, it appears that the constitutive triple response protein (CTR1) is part of an ethylene receptor–signalling complex that sits on the membranes of the endoplasmic reticulum (Gao et al., 2003). CTR1 is similar to Raf‐type MAP kinase, kinase, kinases and a MAP kinase cascade has recently been shown to be part of the ethylene signal transduction pathway (Ouaked et al., 2003).
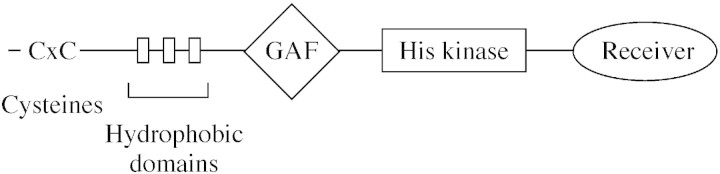
Fig. 1. Representation of ETR1. In arabidopsis and many other species there are just three residues N‐terminal to the C × C motif. The GAF domain is present in phytochromes and cyclic GMP‐specific phosphodiesterases. The other domains are discussed in the text.
Since the cloning of ETR1, a family of ethylene receptor genes has been identified in arabidopsis; there are five members: ETR1, ERS1, ERS2, ETR2 and EIN4. All contain the C‐terminal histidine kinase domain and this domain has been studied in detail using site‐directed mutagenesis (Gamble et al., 2002) and by crystallography (Müller‐Dieckmann et al., 1999). In the N‐terminal half of the ethylene receptor there are three hydrophobic membrane‐spanning motifs. It is in this hydrophobic domain that the ethylene binding site is buried. At the N‐terminus and located in the apoplast there is a short section carrying two cysteine residues and these are involved in disulfide bonding of receptor monomers to give dimers.
The identification of ETR1 (and family members) as an ethylene receptor was done by phenotypic selection of mutants defective in their responses to ethylene. Confirmation that ETR1 bound the hormone was achieved by expression of the protein in yeast (Schaller and Bleecker, 1995). The calculated dissociation constant was 2·4 × 10–9 m ethylene with a dissociation half‐time of 12·5 h. Binding was inhibited by known ethylene competitors. A series of experiments using truncated versions of ETR1 showed that ethylene‐binding activity was contained in the N‐terminal hydrophobic domain and analysis of the mutation in etr1‐1 suggested that Cys65 in the second transmembrane helix was an important residue (Schaller and Bleecker, 1995).
Other mutations that abolish ethylene binding also lie in the hydrophobic domain (Hall et al., 2000). There had been suggestions in earlier literature that the ethylene receptor would contain a Cu(I) ion as cofactor, based on the specificities of alkenes binding to this transition metal (Burg and Burg, 1967). Metal coordination by proteins often involves Cys, His or Met residues and so the finding that Cys65 was critical was consistent with the likelihood that this was the Cu(I) coordination site. Comparison of the hydrophobic domains of homologues from the blue‐green alga Synechocystis (Rodríguez et al., 1999) and the Never‐Ripe gene of tomato (Hua et al., 1995) identified conserved residues and targets for site‐directed mutagenesis. A two‐dimensional model for the structure of the ethylene‐binding domain of ETR1 nominated Cys65 and His19 as the coordination residues for the Cu(I) ion (Rodríguez et al., 1999). The structural determination of this domain by 3‐D crystallography is awaited with interest. Crystallography of membrane‐bound proteins presents methodological challenges, but examples of such determinations are becoming available.
Cytokinins
The first cytokinin‐binding protein (CBP) was affinity purified from wheat extracts using a conjugate of 6‐benzyladenine (Erion and Fox, 1981). Rapid progress led to identification of the protein as a vicilin, a large family of storage proteins. Some progress was made in modelling the cytokinin site using crystallographic data from other vicilins, but a full definition was not possible because crystals of this CBP were not obtained (Fox, 1992). Progress was remarkably good, although the vicilin CBP is not considered a cytokinin receptor because it is unlikely that storage proteins have such dual functionality and because no response was determined that resulted from cytokinin binding.
Similar cytokinin affinity chromatography using purine‐based cytokinins has succeeded in purifications of CBPs from other cereals and dicots (Kulaeva et al., 2000), but no sequence or structural data are available. Affinity purifications using a phenylurea‐type conjugate pulled out a number of proteins, including a high‐affinity CBP from mung bean seedlings with homology to PR‐10 pathogenesis‐related proteins (Nagata et al., 1993). Expression in Escherichia coli of the PR‐10 protein yielded both pure protein and crystal data (Bujacz et al., 2003). The phase problem has not yet been solved and so the protein structure remains unknown. Resolution of the cytokinin‐binding site is awaited with interest. However, the authors note that PR‐10 proteins contain a highly conserved region that is likely to form a nucleoside‐binding loop, the likely (non‐cytokinin‐specific) binding site for free cytokinins.
After all the work using affinity ligands, molecular genetic screens have recently identified a bona fide receptor named CRE1 (Inoue et al., 2001). Affinity determinations suggest that a range of active cytokinins bind strongly with dissociation constants in the range of 0·5–5 × 10–9 m (Yamada et al., 2001). A similar protein named CKI1 is also likely to be involved, but its cytokinin‐binding activity is uncertain (Kakimoto, 1996). Like the ethylene receptors above (Fig. 1), CRE1 and CKI1 are members of the two component histidine kinase family (Fig. 2) with cytoplasmic histidine kinase and phosphorelay receiver domains. The N‐terminus of CRE1 (but not CKI1) is also likely to be intracellular and the protein is anchored in the membrane (probably the plasma membrane) by two trans‐membrane helices flanking an extracellular domain. The CKI1 protein is more similar to ETR1.
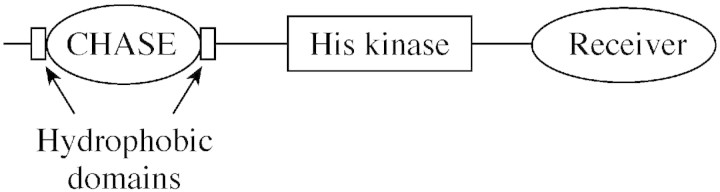
Fig. 2. Representation of CRE1.
The long extracellular domain of CRE1 has been shown to be similar to an equivalent loop on two slime mould receptor histidine kinases and a number of bacterial receptor‐like proteins. The domain has been named the CHASE domain (cyclases/histidine kinases associated sensory extracellular) because of its presence in many diverse receptor‐like proteins with histidine kinase domains and nucleotide cyclase domains (Anantharaman and Aravind, 2001). The CHASE domain is frequently associated with binding low molecular weight ligands. A BLAST search using the domain identified three other arabidopsis genes with high homology, which are interesting targets for hormone researchers. Characterizetions of the domain have failed to predict a structure so far. The alignments of all recognized CHASE sequences do identify conserved residues, but these are likely to be crucial folding determinants rather than ligand binding residues. As soon as there is crystal structure for a CHASE domain, modelling will rapidly allow identification of the cytokinin binding site, and, it is hoped, how the ligand‐bound signal is transduced across the membrane to the phosphorelay domains.
The downstream events from cytokinin perception are mapped in some detail and involve phosphorylation of proteins, known as AHP1 and AHP2, which shuttle into the nucleus to derepress transcription of cytokinin‐activated genes (Hwang and Sheen, 2001).
Brassinosteroids, phytosulfokine and other receptor‐like kinases
Receptors for brassinosteroids and phytosulfokine have been identified as members of a quite different superfamily of proteins, those carrying extracellular leucine‐rich repeat domains (LRR domains) coupled to intracellular serine‐threonine kinase domains (Fig. 3). Included in this superfamily are a number of receptors that bind peptides, such as CLV1, the meristem identity protein thought to bind a small peptide encoded by CLV3 (Clark et al., 1997) and FLS2, which binds flagellin (Gomez‐Gomez et al., 2001). The arabidopsis genome contains over 200 of these LRR receptor‐like kinases, some of which appear to contribute to cross‐talk between signalling pathways (Godiard et al., 2003). These LRR receptor‐like kinases are, in turn, part of a much bigger family of receptor‐like kinases, all of which have intracellular serine/threonine kinase domains, a transmembrane domain and an extracellular domain which might be a LRR, or a lectin‐like domain or a number of other domains known to interact with particular ligands (Shiu and Bleecker, 2001, 2003). Although many contribute to the control of plant growth and development, only the LRR kinases shown to recognize plant hormones will be considered further here, although it will be seen that the brassinosteroid receptor is also the target for the peptide systemin.
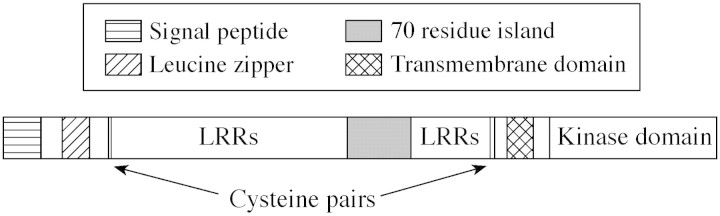
Fig. 3. Representation of BRI1.
The brassinosteroid receptor BRI1, CLV1 and most of the named receptor‐like kinases above were identified from mutant screens. In contrast, the receptors for the peptide derivatives systemin and phytosulfokine were identified using photoaffinity labelling and affinity chromatography. The ligands for very few of the LRR receptor‐like kinases are known, but where they are known, as in these examples, each binds its ligand with nanomolar affinity.
The BRI1 protein (Fig. 3) has a single transmembrane domain splitting the intracellular, C‐terminal kinase domain from the rest of the polypeptide which comprises a large set of extracellular LRRs interrupted by an island of around 70 residues important for ligand binding. Identification of BRI1 as the brassinosteroid receptor has been questioned because of initial concerns that the steroid receptor site should be intracellular. Most other ligands for LRR receptors are also peptides or proteins, not steroids. However, a series of experiments has supported the annotation of BRI1, including characterization of the initial set of loss‐of‐function mutants (Li and Chory, 1997), brassinosteroid responsiveness of a hybrid protein in yeast carrying the extracellular LRR domain (He et al., 2000) and brassinsteroid‐specific binding by the BRI1 complex after immunoprecipitation (Wang et al., 2001).
Having toiled to gather evidence for the activity of BRI1, a good number of mutant alleles were available to help assign key functional residues (He et al., 2000). Loss‐of‐function mutations cluster to the kinase domain and the 70 amino acid island between the LRRs, suggesting that ligand specificity is conferred within this island. Further evidence arose from identification of a receptor for the peptide systemin. Interestingly, BRI1 is more closely related by sequence to the systemin receptor of tomato than BRI1 is to other LRRs in arabidopsis, yet systemin is an 18‐residue peptide found only in members of the Solanaceae.
The systemin receptor (SR160) was purified and identified from tomato extracts (Scheer and Ryan, 2002) in a classical photoaffinity labelling protocol combined with sequence identification from mass spectrometry. The receptor binds its ligand with an affinity of 0·2 × 10–9 m. It has been argued that SR160 also binds brassinosteroids given the similarity of the two sequences, a possibility supported by the cloning of the tomato curl3 mutant which is systemin‐insensitive and yet found to be the tomato homologue of BRI1 (Montoya et al., 2002). Comparison of the sequences across the 70‐residue island of arabidopsis BRI1 and SR160 (68 residues) shows that 51 residues are conserved. A series of BRI1 loss‐of‐function mutant alleles all map to conserved glycine residues. Unfortunately, it is likely that these residues are important for functional folding rather than being directly responsible for ligand binding.
Phytosulfokine is a sulphated five‐residue peptide that binds to its receptor with a high affinity (around 1 × 10–9 m). The phytosulfokine receptor was purified from carrot cell suspension cultures using a photoaffinity labelled probe and affinity chromatography (Matsubayashi et al., 2002). The receptor is highly glycosylated (as in SR160). Once sequenced, the receptor was seen to fall into the LRR receptor‐like kinase family (Fig. 3) and, in this case, the island amongst the LRRs is 36 residues in length. Structural determination of the extracellular domains will be required for a detailed analysis of ligand binding in each case, but the data so far have narrowed down the receptor binding sites to a small part of each protein and structural data for all these LRR receptor domains, with ligands bound, should become a high priority for further research.
Auxin
Affinity labelling, affinity purification and genetic screens have all been used to try to identify candidate auxin receptors. Affinity labelling has yielded a long list of proteins that clearly do bind auxin but, for most, their principal function is not as a receptor (Venis and Napier, 1995; Napier et al., 2002). Photoactive IAA appears to bind with differing, but generally low affinity to lipophilic binding sites. For some there are preferences for active auxins, but in only one case has auxin binding led to auxin‐mediated responses, this being the auxin‐binding protein known as ABP1. Similarly, affinity chromatography using immobilized auxins (and tryptophan) has added to the list of auxin‐binding proteins. However, auxin specificity data have not been fully consistent with the characteristics anticipated for an auxin receptor. These proteins bind auxin, but information about their binding sites is unlikely to be useful for understanding receptor affinities. However, as for photoaffinity labelling, one of the proteins to have been purified by auxin affinity chromatography is ABP1.
The one set of auxin‐receptor searches that has not pulled out ABP1 remains genetic screens. This is not for lack of attempts and many other proteins necessary for auxin action and transport have been identified from screens for auxin insensitivity and constitutive auxin‐like action but, to date, no auxin receptor has been found. For a review of the many auxin‐related mutants and the proteins and pathways encoded by mutated genes see Leyser (2002).
The protein ABP1 was first studied and purified using its capacity to bind radiolabelled auxin 1‐NAA (Hertel et al., 1972; Löbler and Klämbt, 1985). APB1 has an affinity for 1‐NAA of between 0·5 and 2 × 10–7 m. The case that ABP1 is a functional auxin receptor has been built up using diverse experimental approaches over many years (Jones, 1994; Napier et al., 2002). Recently, this has included overexpression in tobacco plants and cell cultures (Jones et al., 1998; Bauly et al., 2000) and the description of an embryo‐lethal phenotype in a homozygous null line of arabidopsis (Chen et al., 2001). All the experiments suggest that ABP1 perceives auxin to induce an increase in cell expansion. Any dependence of auxin‐mediated cell division or auxin‐regulated gene expression on ABP1 remains unproven. Similarly, the signalling pathways through which an ABP1 response would pass to elevate H+ efflux, K+ influx and membrane hyperpolarization remain undiscovered, even though all these processes have been shown to be modulated by ABP1.
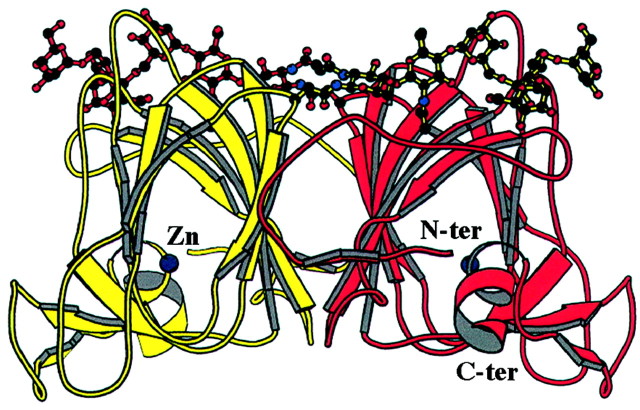
The auxin‐binding site of ABP1 has been analysed in some detail since its crystal structure was solved at high resolution (1·9 Å) with 1‐NAA both absent and bound (Woo et al., 2002). Sequence homologues for ABP1 had previously suggested that it was a member of a protein superfamily known as the cupins, which includes representatives from microbes and animals as well as plants (Dunwell et al., 2000; Warwicker, 2001). Amongst the plant homologues are oxalate oxidase (germin) and the vicilins. The structure confirmed the relationships of ABP1 and germin. Amongst the features conserved are the protein fold, a β‐jellyroll barrel formed by two antiparallel β‐sheets, and a metal ion‐binding site (Fig. 4). For germin, the metal is manganese and the ion is likely to be a redox centre involved in the enzyme activity of that protein, oxidation of oxalate. For ABP1, the predominant metal ion was found to be zinc. No enzymic activity of any sort has been reported for ABP1. The zinc is coordinated by a cluster of histidine residues and one glutamic acid residue, leaving one coordination axis free. In the auxin‐unbound form this is filled by a water molecule. Auxin binding displaces the water and the auxinic carboxylic acid group makes a bidentate contact with the zinc ion, orientating the auxin deep inside an otherwise hydrophobic pocket.
Fig. 4. Ribbon diagram showing the structure of an ABP1 dimer. The β‐sheets are shown as broad arrows. ABP1 is N‐glycosylated and some of the sugar residues are shown at the top of each monomer. Three C‐terminal residues were not resolved and would extend the α‐helices at the foot of the molecules. The zinc ion is shown in green. Reproduced from The EMBO Journal, Vol. 21 No. 12, pp. 2877–2885, 2002, with permission from Woo et al. (2002), Oxford University Press.
The naphthalene ring system of 1‐NAA is orientated by an end‐to‐face interaction with a tryptophan residue (Fig. 5). This tryptophan fulfils the role of the hydrophobic platform that has been an integral part of all models of the auxin binding site (Napier, 2001). Other hydrophobic residues also line the binding pocket. Several different auxins have been resolved bound in ABP1 crystals and each sits at a slightly different pitch in the binding pocket, which will relate to relative binding affinities. However, in each case the ring system sits in exactly the same plane and the carboxylate coordinates the zinc ion (R. Napier, O. Opaleye, J. Marshall and R. W. Pickersgill, unpublished).
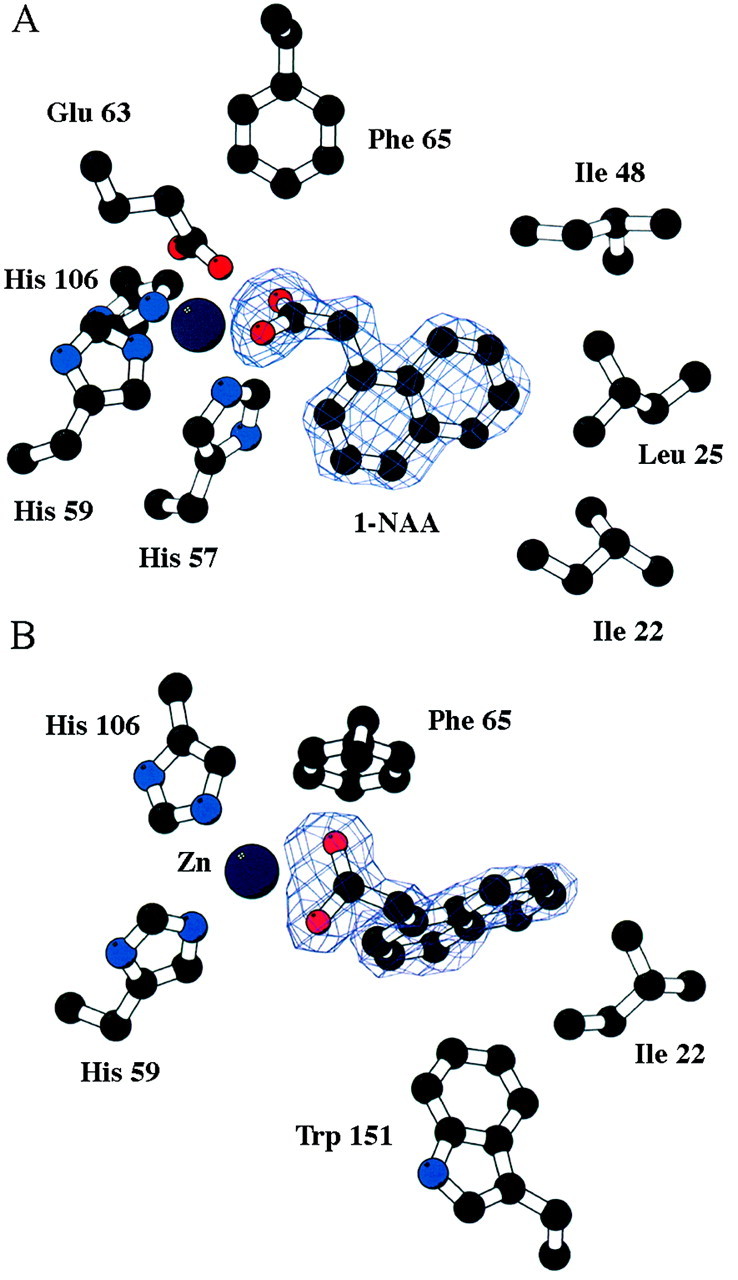
Fig. 5. An electron density map showing 1‐NAA bound in ABP1. The zinc ion and some of the residues lining the binding site are identified, including Trp151, the hydrophobic platform. The view in (B) is rotated approx. 60° around the x‐axis compared with the view in (A), and His57, Glu63, Leu25 and Ile48 are omitted from this view for clarity. Reproduced from The EMBO Journal, Vol. 21 No. 12, pp. 2877–2885, 2002, with permission from Woo et al. (2002).
Of the features described for the structure of ABP1, perhaps the most surprising was the absence of a change in conformation between auxin‐bound and ‐unbound forms (Woo et al., 2002). Of all the plant hormone binding sites, the auxin‐binding site of ABP1 (from a monocot) has been described in the greatest detail. Molecular models of dicot ABP1s are likely to follow as auxinic herbicide susceptibility and tolerance are investigated. It will be interesting to see if these detailed analyses prove useful to those pursuing novel auxins for agriculture.
ABA and gibberellins
There has been no lack of effort put into tracing receptors for these two phytohormones but, in each case, there are no clear candidates yet. For ABA, large numbers of ABA‐sensitive mutants have been characterized and genes cloned (Gazzarrini and McCourt, 2003), but so far all appear to be signalling intermediates or transcription factors. Affinity labelling contributed a lead back in the 1980s (Hornberg and Weiler, 1984), but this has gone no further. The most recent affinity chromatography describes identification of a 42 kD protein from bean leaves with an affinity for ABA of 21 × 10–9 m (Zhang et al., 2002). Treatment of leaves with an antibody raised to the protein decreased the activity of an ABA‐induced enzyme and so the cloning of this candidate ABA receptor is awaited with interest.
For gibberellins there have been still fewer candidates for a receptor. A good deal is known about GA signalling, particularly of GA‐mediated gene repression (Harberd, 2003). A LRR receptor‐like transmembrane kinase (named OsTMK) has been found to be transcriptionally upregulated in response to GA, but no GA binding has been reported (van der Knaap et al., 1999). Consequently, although it is tempting to recall BRI1 and other receptors of this protein superfamily, there are no indications yet that OsTMK is a candidate GA receptor.
Progress towards understanding GA binding has been made using an anti‐GA monoclonal antibody. This has binding specificities for active GAs similar to those expected of a receptor (Murata et al., 2002). The Fab fragment of the antibody was crystallized and its structure determined at 2·8 Å resolution in the presence of bound GA4. These experiments identified all the hydrogen bonds and van der Waals contacts between key groups in the hormone molecule and antibody recognition site, but how useful such information might be for understanding hormone action at a receptor will only be determined once there is a receptor site for comparison.
SUMMARY
Considerable progress has been made in both the identification and, in some cases, detailed analysis of plant hormone binding sites and receptors. This progress has accelerated, but there are still hormones for which no receptor candidates have been described. It is clear that no one technique or approach is more likely to identify novel receptor candidates than any other. Novel analogues of many of the major classes of plant hormone continue to be synthesized (e.g. Hill et al., 1995; Baron et al., 1998) and conjugates of any one of these might help identify new binding proteins from either genetic or biochemical screens. More diverse families of chemicals might also throw up novel receptor candidates using chemical genetics.
Of the receptors characterized, all have high affinities for their ligands, generally in the nanomolar range, and this is likely to have aided their identification. For a number there is some knowledge of the amino acids involved in ligand binding but, so far, there is structural information only for ABP1. It is unlikely that plant biology will remain so poorly informed about receptor functionalities for much longer and detailed crystallographic determinations of each confirmed receptor must become high priorities for researchers. The information accumulated by such work will undoubtedly indicate new ways in which plant growth and development might be advantageously manipulated, but knowledge‐led design of new ligands or of altered sensitivities is still some way off.
Received: 2 September 2003;; Returned for revision: 24 October 2003; Accepted: 4 December 2003
References
- AnantharamanV, Aravind L.2001. The CHASE domain: a predicted ligand‐binding module in plant cyokinin receptors and other eukaryotic and bacterial receptors. Trends in Biochemical Sciences 26: 579–582. [DOI] [PubMed] [Google Scholar]
- BajguzA, Tretyn A.2003. The chemical characteristics and distribution of brassinosteroids in plants. Phytochemistry 62: 1027–1046. [DOI] [PubMed] [Google Scholar]
- BaronDL, Luo W, Janzen L, Pharis RP, Back TG.1998. Structure‐activity studies of brassinolide B‐ring analogues. Phytochemistry 49: 1849–1858. [Google Scholar]
- BaulyJM, Sealy IM, Macdonald H, Brearley J, Droge S, Hilmer S, Robinson DG, Venis MA, Blatt MR, Lazarus CM, Napier RM.2000. Overexpression of ABPl heightens the sensitivity of guard cells to auxin. Plant Physiology 124: 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BujaczGD, Pasternak O, Fujimoto Y, Hashimoto Y, Sikorski MM, Jaskolski M.2003. Crystallization and preliminary crystallographic studies of mung bean cytokinin‐specific binding protein. Acta Crystallographica Section D‐Biological Crystallography 59: 522–525. [DOI] [PubMed] [Google Scholar]
- BurgSP, Burg EA.1967. Molecular requirements for the biological activity of ethylene. Plant Physiology 42: 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChangC, Kwok SF, Bleecker AB, Meyerowitz EM.1993. Arabidopsis ethylene response gene ETR1: similarity of product to two‐component regulators. Science 262: 539–544. [DOI] [PubMed] [Google Scholar]
- ChenJ‐G, Ullah H, Young JC, Sussman MR, Jones AM. 200l. ABPl is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes and Development 15: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClarkSE, Williams RW, Meyerowitz EM.1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- DunwellJM, Khuri S, Gane PJ.2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiology Molecular Biology Reviews 64: 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ErionJL, Fox JE.1981. Purification and properties of a protein which binds cytokinin‐active 6‐substituted purines. Plant Physiology 67: 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FoxJE.1992. Molecular modelling of cytokinins and the CBF‐1 receptor. In: Kaminek M, Mok, DWS, Zazimalova, E, eds. Physiology and biochemistry of cytokinin in plants: developed from a symposium held at Liblice, Czechoslovakia, 10–14 September 1990. SPB Academic Publishing, Czech Republic, 128–132. [Google Scholar]
- GambleRL, Qu X, Schaller GE.2002. Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiology 128: 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GaoZY, Chen YF, Randlett MD, Zhao XC, Findell JL, Keiber JJ, Schaller GE.2003. Localization of the Raf‐like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signalling complexes. Journal of Biological Chemistry 278: 34725–34732. [DOI] [PubMed] [Google Scholar]
- GazzarriniS, McCourt P.2003. Cross‐talk in plant hormone signalling: what arabidopsis mutants are telling us. Annals of Botany 91: 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GodiardL, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y.2003. ERECTA, an LRR receptor‐like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant Journal 36: 353–365. [DOI] [PubMed] [Google Scholar]
- Gomez‐GomezL, Bauer Z, Boller T.2001. Both the extracellular leucine‐rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- HallAE, Findell JL, Schaller GE, Sisler EC, Bleecker AB.2000. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiology 123: 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HarberdNP.2003. Relieving DELLA restraint. Science 299: 1853–1854. [DOI] [PubMed] [Google Scholar]
- HeZ, Want Z‐Y, Li J, Zhu Q, Lamb C, Ronald P, Chory J.2000. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363. [DOI] [PubMed] [Google Scholar]
- HertelR, Thomson K, Russo VEA.1972. In‐vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta 107: 325–340. [DOI] [PubMed] [Google Scholar]
- HillRD, Liu JH, Durnin D, Lamb N, Shaw A, Abrams SR.1995. Abscisic‐acid structure‐activity‐relationships in barley aleurone layers and protoplasts – biological‐activity of optically‐active, oxygenated abscisic‐acid analogs. Plant Physiology 108: 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoad,1983. Gibberellin bioassays and structure‐activity relationships. In: Crozier, ed. The biochemistry and physiology of gibberellins, Vol. 2 New York: Praeger, 57–94. [Google Scholar]
- HornbergC, Weiler EW.1984. High‐affinity binding sites for abscisic acid on the plasmalemma of Vicia faba guard cells. Nature 310: 321–324. [Google Scholar]
- HuaJ, Chang C, Sun Q, Meyerowitz EM.1995. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714. [DOI] [PubMed] [Google Scholar]
- HwangI, Sheen J.2001. Two‐component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389. [DOI] [PubMed] [Google Scholar]
- InoueT, Kiguchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T.2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis Nature 409: 1060–1063. [DOI] [PubMed] [Google Scholar]
- JonesAM.1994. Auxin‐binding proteins. Annual Review of Plant Physiology and Plant Molecular Biology 45: 393–420. [Google Scholar]
- JonesAM, Im K‐H, Savka MA, Wu M‐J, DeWitt G, Shillito R, Bunns AN.1998. Auxin‐dependent cell expansion mediated by overexpressed auxin‐binding protein 1. Science 282: 1114–1117. [DOI] [PubMed] [Google Scholar]
- KakimotoT.1996. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985. [DOI] [PubMed] [Google Scholar]
- KatekarGF.1979. Auxins: on the nature of the receptor site and molecular requirements for receptor activity. Phytochemistry 18: 223–233. [Google Scholar]
- KulaevaON, Karavaiko NN, Selivankina SY, Kusnetsov VV, Zemlyachenki YA, Cherepneva GN, Maslova GG, Lukevich TV, Smith AR, Hall MA.2000. Nuclear and chloroplast cytokinin‐binding proteins from barley leaves participating in transcription regulation. Plant Growth Regulation 32: 329–335. [Google Scholar]
- LeyserO.2002. Molecular genetics of auxin signaling. Annual Review of Plant Biology 53: 377–398. [DOI] [PubMed] [Google Scholar]
- LiJM, Chory J.1997. A putative leucine‐rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- LöblerM, Klämbt D.1985. Auxin‐binding protein from coleoptile membranes of corn (Zea mays L.): purification by immunological methods and characterization. Journal of Biological Chemistry 260: 9848–9853. [PubMed] [Google Scholar]
- MatsubaraS.1990. Structure‐activity relationships of cytokinins. Critical Review of Plant Sciences 9: 17–57. [Google Scholar]
- MatsubayashiY, Ogawa M, Morita A, Sakagami Y.2002. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472. [DOI] [PubMed] [Google Scholar]
- MontoyaT, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ.2002. Cloning the tomato Curl3 gene highlights the putative dual role of the leucine‐rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signalling. Plant Cell 14: 3163–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller‐DieckmannHJ, Grantz AA, Kim SH.1999. The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure with Folding and Design 7: 1547–1556. [DOI] [PubMed] [Google Scholar]
- MurataT, Fushinobu S, Nakajima M, Asami O, Sassa T, Wakagi T, Yamaguchi I.2002. Crystal structure of the liganded anti‐gibberellin A4 antibody 4‐B8(8)/E9 fragment. Biochemical and Biophysical Research Communications 293: 489–496. [DOI] [PubMed] [Google Scholar]
- NagataR, Kawachi E, Hashimoto Y, Shudo K.1993. Cytokinin‐specific binding‐protein in etiolated mung bean seedlings. Biochemical and Biophysical Research Communications 191: 543–549. [DOI] [PubMed] [Google Scholar]
- NapierRM.2001. Models of auxin binding. Journal of Plant Growth Regulation 20: 244–254. [Google Scholar]
- NapierRM, David KM, Perrot‐Rechenmann CP.2002. A short history of auxin‐binding proteins. Plant Molecular Biology 49: 339–348. [PubMed] [Google Scholar]
- OuakedF, Rozhon W, Lecourieux D, Hirt H.2003. A MAPK pathway mediated ethylene signaling in plants. EMBO Journal 22: 1282–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RodríguezFI, Esch JJ, Hall AE, Binder BM, Shaller GE, Bleecker AB.1999. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis Science 283: 996–998. [DOI] [PubMed] [Google Scholar]
- SchallerGE, Bleecker AB.1995. Ethylene‐binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811. [DOI] [PubMed] [Google Scholar]
- ScheerJM, Ryan CA Jr.2002. The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proceedings of the National Academy of Sciences of the USA 99: 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ShiuS‐H, Bleecker AB.2001. Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences of the USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ShiuS‐H, Bleecker AB.2003. Expansion of the receptor‐like kinase/Pelle gene family and receptor‐like proteins in Arabidopsis. Plant Physiology 132: 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StockwellBR.2000. Chemical genetics: ligand‐based discovery of gene function. Nature reviews of Genetics 1: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TakahashiN.1986. Gibberellins. In: Takahashi, N, ed. Chemistry of plant hormones. Boca Raton, FL: CRC Press. [Google Scholar]
- ThimannKV,1963. Plant growth substances; past, present and future. Annual Reviews of Plant Physiology 14: 1–18. [Google Scholar]
- van der KnaapE, Song WY, Ruan DL, Sauter PC, Kende H.1999. Expression of a gibberellin‐induced leusine‐rich repeat receptor‐like kinase in deepwater rice and its interaction with kinase‐associated protein phosphatase. Plant Physiology 120: 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VeldstraH.1944. Researches on plant growth substances, IV and V. Relation between chemical structure and physiological activity. Enzymologia II: 97–136, 137–163. [Google Scholar]
- VenisMA, Napier RM.1995. Auxin receptors and auxin‐binding proteins. Critical Reviews in Plant Science 14: 27–47. [Google Scholar]
- WangZY, Seto H, Fujioka S, Yoshida S, Chory J.2001. BRI1 is a critical component of a plasma‐membrane receptor for plant steroids. Nature 410: 380–383. [DOI] [PubMed] [Google Scholar]
- WarwickerJ.2001. Modelling of auxin‐binding protein 1 suggests that its C‐terminus and auxin could compete for a binding site that incorporates a metal ion and tryptophan residue 44. Planta 212: 343–347. [DOI] [PubMed] [Google Scholar]
- WooE‐J, Marshall J, Bauly J, Chen J‐G, Venis M, Napier RM, Pickersgill RW.2002. Crystal structure of auxin‐binding protein 1 in complex with auxin. EMBO Journal 21: 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YamadaH, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Tamashino T, Mizuno T.2001. The Arabidopsis AHK4 histidine kinase is a cytokinin‐binding receptor that transduces cytokinin signals across the membrane. Plant and Cell Physiology 42: 1017–1023. [DOI] [PubMed] [Google Scholar]
- ZhangDP, Wu ZY, Li XY, Zhao ZX.2002. Purification and identification of a 42‐kilodalton abscisic acid‐specific‐binding protein from epidermis of broad bean leaves. Plant Physiology 128: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhaoY, Dai X, Blackwell HE, Schreiber SL, Chory J.2003. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110. [DOI] [PubMed] [Google Scholar]