Abstract
The establishment of effective high throughput screening cascades to identify nuclear receptor (NR) ligands that will trigger defined, therapeutically useful sets of NR activities is of considerable importance. Repositioning of existing approved drugs with known side effect profiles can provide advantages because de novo drug design suffers from high developmental failure rates and undesirable side effects which have dramatically increased costs. Ligands that target estrogen receptor β (ERβ) could be useful in a variety of diseases ranging from cancer to neurological to cardiovascular disorders. In this context, it is important to minimize cross-reactivity with ERα, which has been shown to trigger increased rates of several types of cancer. Because of high sequence similarities between the ligand binding domains of ERα and ERβ, preferentially targeting one subtype can prove challenging. Here, we describe a sequential ligand screening approach comprised of complementary in-house assays to identify small molecules that are selective for ERβ. Methods include differential scanning fluorimetry, fluorescence polarization and a GAL4 transactivation assay. We used this strategy to screen several commercially-available chemical libraries, identifying thirty ERβ binders that were examined for their selectivity for ERβ versus ERα, and tested the effects of selected ligands in a prostate cancer cell proliferation assay. We suggest that this approach could be used to rapidly identify candidates for drug repurposing.
Keywords: Screening, gene expression, drugs, nuclear receptors, chemical libraries, gene regulation
Introduction
Nuclear receptors (NRs) regulate a variety of biological processes and are critically important in the emergence, prevention, and treatment of an array of diseases. NRs are regulated by small, exchangeable, lipophilic molecules, which make them optimal targets for drug discovery [1–3]. While NRs are considered useful therapeutic targets for prevention and/or treatment of diseases such as cancer [4], metabolic [5], and neurodegenerative diseases [6], efforts to develop new NR-based therapeutics have often been stalled or curtailed due to unexpected side effects that can arise from undesired cross-reactivity with other NRs and off-target effects. It is important to develop approaches to selectively target therapeutically useful subsets of NR activities in order to obtain effective NR drugs with improved side effect profiles.
One rapid route towards development of new safe treatments to target NRs could involve repurposing of existing drugs. Drug repurposing includes several advantages such as safety, financial, market potential, return on investment, out-licensing potential, and time to market [7]. During clinical trials, safety accounts for approximately 30% of drug failure rates [8]. Since repurposed drugs already meet established regulatory safety requirements, they possess a significant advantage over competing drugs in development, leaving drug effectiveness for the particular indication as the primary concern. While it may cost more than USD 800 million to develop a drug de novo, repurposing averages approximately USD 8.4 million [9, 10]. The financial savings therefore create a significant economic incentive for this avenue of drug development. What may be most significant to the end user, however, is time to market. It is estimated to take 10 to 17 years to develop a drug de novo, whereas a repositioned drug could be moved directly to phase II clinical trials [9]. For these purposes, it is essential to develop reliable methods to identify useful candidates among large numbers of approved candidate drugs.
Estrogen receptor β (ERβ) is an attractive target for drug development. While original models of estrogen action suggested that only a single ER gene (which encodes a protein that is now termed ERα) was responsible for transducing signals of estradiol and other ligands [11], the discovery in 1996 of a second ER gene, encoding ERβ, prompted a reevaluation of the estrogen signaling system. It is now known that ERα and ERβ play different roles in gene regulation [12] and that ERα and ERβ have overlapping but distinctive tissue distributions and non-redundant roles [13]. These considerations have led to the suggestion that ERβ could be an attractive therapeutic target for the development of selective agonists to treat and prevent neurodegenerative diseases [14] and other diseases, including autoimmune diseases, endometriosis, depression, hypertension, and colon, breast, prostate, lung, and skin cancer [15]. It is important that such ligands should not cross-react with ERα, which triggers classical estrogenic side effects, such as breast or uterine stimulation, thereby increasing a woman’s chance of developing breast or uterine cancer, and gynecomastia and decreased libido in men [16]. Presently, natural and synthetic estrogens for ERβ are being studied in colon cancer, breast cancer, lung cancer, schizophrenia, and metabolic syndrome [17]. While the agonist ERB-041 failed to demonstrate efficacy in a Phase II double-blind clincial trial for rheumatoid arthritis, further studies are warranted to examine ERβ-selective efficacy in other inflammatory disorders [18]. With many other clinical trials still in progress awaiting completion of the study followed by publication of the findings, it is still too early to make any definitive conclusions about these drugs and their effects. The discovery of potentially beneficial effects of selective ERβ ligands on prostate cancer proliferation and apoptosis in the absence of full ERα or ERβ agonism has raised hopes that applications for new safe selective ERβ modulator ligands could emerge in the context of this disease [19–22].
Although ERα and ERβ are highly homologous in their ligand-binding domains (LBDs), differences exist in ligand binding affinity and/or specificity between the ER subtypes. Further, the structural diversity of estrogenic chemicals is very broad [23]. Environmental chemicals (such as polychlorinated hydroxybiphenyls, dichlorodiphenyltrichloroethane (DDT) and derivatives, alkylphenols, bisphenol A, methoxychlor and chlordecone) and phytoestrogens (such as genistein, coumestrol and zearalenone) show estrogenic activity and bind both ER subtypes, with some ligands showing stronger binding to ERβ [24]. Additionally, synthetic estrogen agonists and antagonists have been developed with diverse chemical structures, such as diethylstilbestrol, moxestrol, and tamoxifen. Fink et al. [25] used a combinatorial synthetic approach to modify an azole core structure to generate binding, while others, such as Manas et al. [26], employed molecular modeling for selectivity enhancement. To date, however, highly selective antagonists have not been designed [27].
We hypothesize that it may be possible to identify novel and potentially useful ER ligands among libraries of existing approved drugs. As mentioned, the ERs recognize a broad range of non-steroidal ligand structures, exemplified by their recognition of the hydroxystilbene backbone of diethylstilbestrol and 4-hydroxytamoxifen [28] and by the fact that many compounds act as estrogenic endocrine disruptors. Non-steroidal ligands have gained attention as potential ER therapeutics since they are thought to have diminished cross-reactivity with other NRs, which can eliminate side effects, as well as altered physicochemical properties, which can result in unique and potentially useful tissue distributions.
To identify novel ER modulators, it is important to devise rapid and reliable methods to detect ER ligands and to define subtype selectivity. Here, we report a screening strategy for ERβ ligands that relies on application of sequential orthogonal assays. We present evidence that our overall screening strategy is effective at identifying selective ERβ modulators from a large collection of compound libraries and test effects of representative ligands in an ERβ-dependent prostate cancer model system.
Materials and Methods
Reagents
Reagents were obtained from the following manufacturers: LB Broth, Ampicillin Sodium Salt, HisPur Cobalt Resin (Fisher Scientific, Waltham, MA); IPTG (RPI Corp., Mt. Prospect, IL); Complete EDTA-Free Protease Inhibitor Cocktail Tablets, Lightcycler 480 II RT-PCR (Roche Applied Science, Indianapolis, IN); SYPRO® Orange Protein Gel Stain 5000x Concentrate in DMSO (Life Technologies, Grand Island, NY); 2-Mercaptoethanol, Electrophoresis ≥98%, (Fisher BioReagents, Waltham, MA); DMSO (Sigma Aldrich, St. Louis, MO); Amicon Ultra-15 Centrifugal Filter Units (EMD Millipore, Billerica, MA); MultiTron Incubated Shaker (INFORS HT, Bottmingen, Switzerland); Ultrasonic Liquid Processor (Misonix, Inc., Farmingdale, NY); ÄKTA purifier FPLC, Nanodrop Spectrophotometer ND-1000 (GE Healthcare Life Sciences, Piscataway, NJ); Envision 2104 multilabel reader (Perkin Elmer, Waltham, MA); Prestwick Chemical Library® (Prestwick Chemical, Illkirch, France); NIH Clinical Collection 1 and 2 (Evotec, San Francisco, CA); Custom Clinical Collection (provided by Cliff Stephan at the GCC); National Cancer Institute Diversity, Natural Products, Mechanistic, and Challenge Sets (NCI/NIH Developmental Therapeutics Program, Bethesda, MD); BL21 Star™ (DE3), Competent E. coli, PolarScreen™ ER-β Competitor Assay, Green and PolarScreen™ ER-α Competitor Assay, Green (Life Technologies, Carlsbad, CA).
Production and purification of ERβ
BL21 E. Coli cells were transformed with an expression vector containing a sequence encoding His6-hERβ LBD (261-530). Freshly transformed cells were grown at 22°C at 160 rpm in 1 L LB Broth Miller supplemented with 1 mL of 100 mg/mL ampicillin per flask with a MultiTron Incubated Shaker for approximately 24 hours until reaching mid-log phase of growth. The culture was cooled to 16°C, induced with 1 mL of 100 mM IPTG per 1 L culture, and allowed to continue to grow while shaking at 160 rpm for another 24 hours. Cells were pelleted at 4000 x g at 4°C for 20 minutes and resuspended in 40 mL of lysis buffer supplemented with 0.1% BME (300 mM NaCl, 20 mM Tris, 20 mM Imidazole, 10% glycerol @ pH 8.0) per 1 L culture. Cells and/or protein were kept on ice or at 4°C throughout the purification. Cells were pelleted again for 10 minutes at 3400 x g, the supernatant was removed, and stored at -80°C until use. One tablet of EDTA-free protease inhibitor cocktail was added per 1 L culture, and the cells were resuspended in lysis buffer and lysed by sonication using an Ultrasonic Liquid Processor. The lysate was clarified by centrifugation at 34500 x g for 40 minutes. The supernatant was added to HisPur Cobalt Resin (4 mL beads per culture) and placed on a shaker in the cold room for between 1 and 1.5 hours and pelleted at 215 x g for 5 minutes. The supernatant was discarded and the beads were resuspended in lysis buffer supplemented with 0.1% BME. The beads and buffer were poured through a gravity-flow column in the cold room. After allowing the lysate to flow-through, the column was washed with 20 mL of elution buffer supplemented with 0.1% BME (300 mM NaCl, 20 mM Tris, 300 mM Imidazole, 10% glycerol @ pH 8.0). The eluted protein was then concentrated by centrifuging with 10 kDa centrifugal filter units at 2600 x g until the volume was less than 5 mL and injected into an ÄKTApurifier FPLC and purified by gel filtration using a HiLoad 16/600 Superdex 75 in gel filtration buffer (150 mM NaCl, 20 mM Tris, 10 mM DTT, 10% glycerol, @ pH 8.0). Fractions under the peak spanning at a retention volume of around 60 mL were pooled and concentrated by centrifuging with 10 kDa centrifugal filter units at 2600 x g.
Protein melting curves
The Roche Lightcycler 480 II RT-PCR machine was programmed to equilibrate samples at 25°C for 10 seconds and then increase temperature to 95°C at a rate of 0.05°C/second, taking 11 acquisitions per °C. The melting point of the protein was obtained as the lowest point of first derivative plot, as calculated by the software included with the RT-PCR machine.
Optimal concentrations of ERβ LBD and SYPRO orange dye were determined by performing differential scanning fluorimetry (DSF) on varying concentrations of protein and SYPRO orange in screening buffer (0.15 M NaCl, 0.01 M Tris, 0.001 M DTT @ pH 7.4). For ERβ, the optimal conditions were 0.02 mg/mL protein (as determined by the absorbance at 280nm from a Nanodrop Spectrophotometer), screening buffer and 2X SYPRO orange (diluted from 5000X concentrate) within the reaction well.
20 μL reactions were conducted in a single well of a 384-well white PCR plate by combining 19 μL of protein solution (0.02 mg/mL in screening buffer) with 2X Sypro orange (diluted from a 500X substock in DMSO) and 1μL of solution containing either 10 mM ligand in DMSO or DMSO (as vehicle control). Approximately 3,000 compounds evaluated from several compound library sources, including The Prestwick Chemical Library® (100% FDA approved drugs), the NIH Clinical Collection (small molecules that have a history of use in human clinical trials), the Custom Clinical Collection (of which 57% of the compounds are currently used in the clinic for the treatment of various forms of cancer and 37% of the compounds are in clinical trials), and the NCI Diversity (compounds identified using the program Chem-X), Natural Products (compounds selected from an open repository of 140,000 compounds), Mechanistic (compounds that have been tested in the NCI human tumor 60 cell line screen), and Challenge (compounds of novel structural types) Sets were screened. All library test compounds were performed in duplicate wells in 384-well white Roche qPCR microplates. The plates were sealed and centrifuged and then loaded on a Roche Light Cycler 480II.
Fluorescence polarization
A secondary fluorescence polarization (FP) assay was used to quantitatively determine receptor subtype preference and affinity (β versus α). The FP assay was performed using a Polarscreen™ ERβ Competitor Assay, Green from Life Technologies. Compounds were serially diluted in DMSO, and transferred into ERβ Green Screening Buffer. Black, multiwell plates were used and the assay was incubated for two hours in the dark before reading on a Perkin Elmer EnVision® 2104 Multilabel Reader capable of reading fluorescence polarization.
Transactivation assays
A tertiary cell-based GAL4 transactivation assay was used to qualitatively determine ligand-dependent transactivation of ERβ to distinguish agonism from partial agonism or antagonism. Human embryonic kidney (HEK) 293T cells were transiently transfected with GAL4 DBD-ERβ LBD and a GAL4 response element linked to a luciferase reporter gene (luc2P/9XGAL4UAS/Hygro) reporter. Cells were plated and transfected at a density of 1.2x105 cells per well of a 24-well plate using FuGENE® HD Transfection Reagent (Promega) and were maintained in DMEM without phenol-red supplemented with charcoal stripped serum and 1x Pen/Strep antibiotics (500 μL/well). Either vehicle (DMSO) or increasing doses of the test compounds were added to the cells 24 hours after transfection, and cells were assayed for luciferase activity after overnight treatment. Luciferase values were normalized to a Renilla control. Luminescence and Renilla luciferase activity were measured on a Tecan Safire2™ Microplate Reader. The data was fit using Prism software. Similar assays that employed expression vectors for full length ERα or ERβ were used to define effects of selected ligands on both receptor subtypes. HeLa cells were transiently transfected with full length ERα or ERβ cloned into the pSG5 expression vector (Promega) and a ERE response element linked to a luciferase reporter gene in pGL4 vector as previously described [29]. Cells were plated at a density of 1.0x105 cells per well of a 12-well plate and maintained in DMEM without phenol-red supplemented with charcoal stripped serum and 1x Pen/Strep antibiotics (1000 μL/well). The co-transfections were performed using the TransFectin Lipid reagent (Bio-Rad) with approximately 10 ng/well of full length ERα or ERβ, 10 ng/well of renilla luciferase gene (Promega) for internal control and 200 ng/well of ERE-Luc. Approximately, 5 h after transfections, cells were treated with compounds of interest (1 μM final concentration), or solvent (DMSO). After overnight incubation, luciferase activities were assessed using Dual Luciferase assay reagent (Promega). Luciferase activities were calculated by normalizing firefly luciferase to renilla luciferase signal. Normalized luciferase activities were then represented relative to control (DMSO-treated cells).
Cell proliferation
Cell proliferation of LNCaP cells (ATCC® CRL-1740™) was assessed using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega). 50,000 cells per well were plated using TrypLE™ Select Enzyme (1X), no phenol red (Gibco®) and phenol-free RPMI (Mediatech) supplemented with charcoal stripped serum and 1x Pen/Strep antibiotics. Cells were centrifuged, redispersed in complete media, and seeded in Corning black 96-well flat clear bottom microplates. Wells were filled with either 100 μL of complete media without cells (to obtain a value for background luminescence) or media with cells. After 24 h, cells were treated with either 1 μL of DMSO or test compound (1 μM final concentration). After an additional 24 h, the plate and its contents were equilibrated to room temperature for approximately 30 minutes and an equal volume (100 μL) of CellTiter-Glo® Reagent was added. Plates were shaken for 2 minutes on an orbital shaker to induce cell lysis and allowed to incubate at room temperature for 10 minutes to stabilize luminescent signal. Luminescence was recorded using an integration time of 1 second per well.
Results
We expressed his-tagged ERβ LBD in E. coli as described in methods and purified the material using a standard affinity column. Upon further purification over a gel filtration column, we noted that the chromatogram yielded two distinct 280 nm absorbance peaks at retention volumes of ~45 mL and ~61 mL, respectively (Figure 1a). Once pooled and concentrated, SDS-PAGE (Figure 1b) indicated that the second protein eluted from the column has a mass estimate of 30 kDa and should correspond to our protein of interest, His6-hERβ LBD (30864 Da). Mass spectrometry analysis indicated that the first peak contained mostly transcription termination factor Rho OS from E. coli (strain K12) and confirmed that the second peak is His6-hERβ LBD. The ERβ peptides identified are shown in Supplementary File 1.
Figure 1. ÄKTA chromatogram and SDS-PAGE for the purification of ERβ LBD.
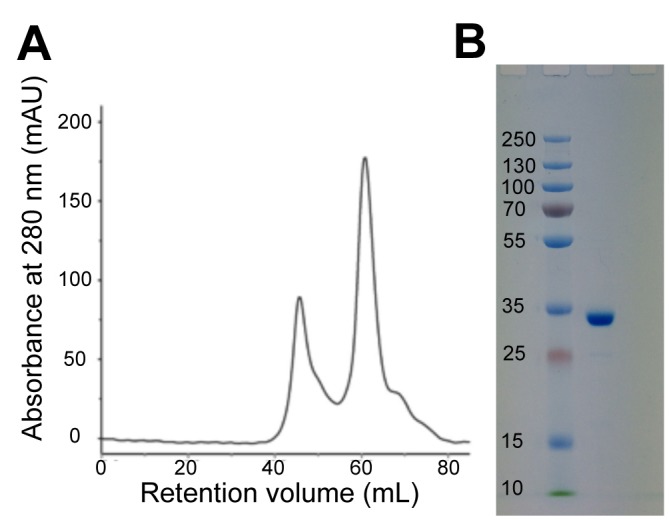
Representative size-exclusion chromatogram from the ÄKTA purifier FPLC for the purification of ERβ LBD (A) and Coomassie stained SDS-PAGE gel after SEC of the pooled and concentrated eluted fractions at ~61 mL corresponding to ERβ LBD (B).
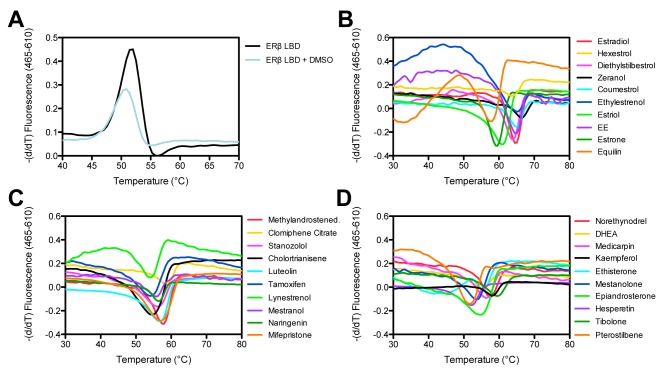
We chose to apply DSF, also known as the thermofluor-binding assay, as our first approach towards ERβ ligand identification. This technique relies upon the fact that protein stability is commonly enhanced upon ligand binding. Fundamentally, as the increasing temperature forces protein unfold, normally buried residues of a protein's hydrophobic core are exposed increasing the fluorescence of a dye with an affinity for hydrophobic surface. The assay has been adapted for a conventional real-time PCR instrument in our laboratory and can be performed with 96 or 384-well plate formats [30–32]. We used doubly purified ERβ LBD as we reasoned that use of partially pure material that retained the RhoOS contaminant in DSF could result in detection of multiple melting point (Tm) transitions and potentially confusing results. Purified unliganded His6-hERβ LBD produced a denaturation curve with a Tm of ~51 ºC when tested in DSF (Figure 2a). While slightly depressed in intensity, no significant shift in the melting temperature of ERβ LBD was recorded following treatment of the purified protein with DMSO (Figure 2a).
Figure 2. DSF assay to determine the binding of ligands from compound libraries to ERβ LBD.
Representative DSF output from the RT-PCR machine as graphs displaying the first derivative of fluorescence versus temperature for (A) unliganded ERβ LBD and unliganded ERβ LBD with DMSO and (B-D) ERβ LBD with the addition of ligand (final concentration 500 μM). Ethinyl estradiol (EE) is represented by the purple trace in B.
We used DSF to evaluate binding of approximately 3,000 test compounds derived from several sources: The Prestwick Chemical Library® (100% FDA approved drugs), the NIH Clinical Collection (small molecules with a history of use in human clinical trials), the Custom Clinical Collection (57% of compounds are currently used in the clinic for the treatment of various forms of cancer and 37% of compounds are in clinical trials), NCI Diversity (compounds identified using the program Chem-X, which uses defined centers and defined distance intervals to create a particular finite set of pharmacophores) [33], Natural Products (selected from an open repository of 140,000 compounds), Mechanistic (compounds tested in the NCI human tumor 60 cell line screen), and Challenge Sets (compounds of novel structural types). We were able to eliminate a large number of false positives by manual curation. Several compounds which scored as hits were eliminated because of large volume and complex chemical structure which suggested that they should not bind inside the ERβ pocket (which is approximately 450 cubic angstroms and twice the molecular volume occupied by the estradiol molecule itself) [24, 34, 35] in a conventional manner. After this step, we were left with 60 potential hit compounds that elicited a noticeable and significant change in the melting temperature of the purified ERβ LBD protein.
Detection of 60 putative binders from the initial screen corresponded to a 2% primary hit rate, which appeared slightly high. It is not unusual for promiscuous compounds to invade compound screening and appear active in many assays; therefore, substructure filters have been developed to help reduce pan assay interference [36]. We systematically and manually filtered and removed interference compounds and reduced the list to 40 potential putative ERβ binding compounds (yielding a 1.3% hit rate, which appeared genuine). Our remaining ligand set produced denaturation curves with discernable transitions when compared to unliganded ERβ LBD. Figure 2 shows the negative derivative of the melting traces (derivative curves) obtained with several small molecules screened that were identified as potential ERβ ligands. These compounds yielded ideal traces with a symmetric, sharp, and upward-shifted peak. For all these ERβ putative binders, the derivative curves invert in intensity and shifted upward representing conditions in which the protein is the most stable (Figure 2b-d). The compounds fall in various chemical and structural classes such as nonsteroidal (ex: pterostilbene) and steroidal (ex: ethylestrenol), triphenylethylene antiestrogens (ex: clomiphene citrate, tamoxifen), diphenyl derivatives (ex: hexestrol), natural estrogens (ex: estrone, equilin, dehydroepiandrosterone (DHEA)), phytoestrogens (ex: zeranol, coumestrol, naringenin), as well as synthetic (ex: stanozolol) and semisynthetic (ex: ethinyl estradiol). It is not surprising that such structurally distinct ligands with full agonistic, SERMs, or antagonistic properties all show affinity for this receptor and are identified in the screening cascade.
We also determined whether DSF could verify whether a ligand introduced during protein expression and purification was incorporated into the LBD. This is important because purification of a protein does not guarantee that it remains folded in its native state and addition of ligand during bacterial expression can increase yields of soluble correctly folded protein. When the small molecule DHEA was added to the bacterial culture at 1 μM in DMSO prior to induction with IPTG and maintained at 1 μM in the gel filtration buffer throughout purification, the His6-hERβ LBD purification product produced an inverted derivative curve with a Tm of ~55 ºC, increased over Tm values obtained with purified unliganded ERβ LBD (see Supplementary File 2). These data provide independent verification that DHEA scored as an ERβ binder in DSF assays and confirmed that the addition of ligand during purification steps allows for the ligand to be incorporated into the LBD and maintain proper tertiary structure.
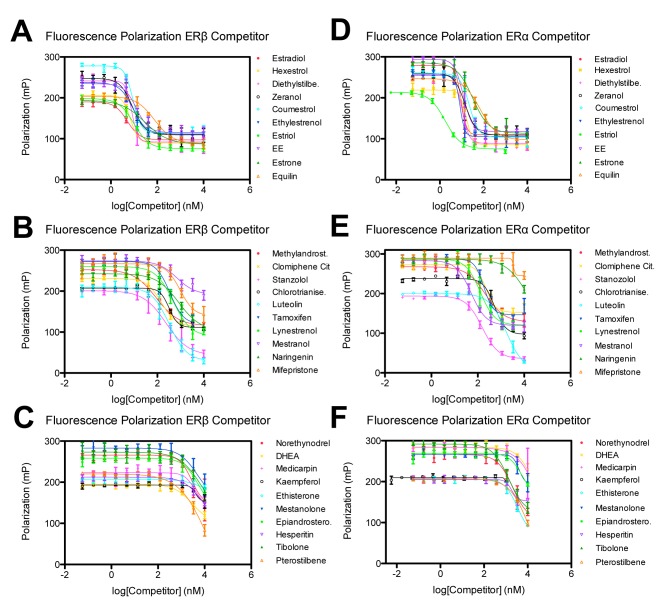
While DSF offers a simple platform for rapid identification of interacting ligands, the assay is essentially qualitative and does not assess binding affinity. To confirm initial hits and more accurately rank the ligands based on affinity, we applied a secondary screening assay based on a commercially available fluorescence polarization (FP) assay to define ligand displacement (Figure 3). Here, test compounds were analyzed for their ability to displace a fluorescently labeled ligand (in this case fluorescein labeled E2) from ERs. The reduced size of the free molecule can be discerned by increased mobility in solution, which is detected by FP. The observed FP value depends on free and bound fractions of labeled molecules. We utilized paired kits to measure the ability of test compounds to displace ligands from both ERβ and ERα to obtain initial estimates of selectivity.
Figure 3. FP assay to verify ligand hits from compound libraries and determine selectivity for ERβ and ERα.
Fluorescence Polarization versus concentration with (A-C) ERβ and (D-F) ERα for ligands identified as ERβ binders from the primary DSF assay. Curves shifted the furthest to the left represent those compounds with the greatest affinity for the protein. Error bars represent one standard deviation from the mean of triplicate reaction wells.
The initial 40 hit compounds identified from the primary assay were reduced to 30 hit compounds (see Supplementary File 3) after the secondary FP assay, indicating that these 30 compounds can effectively compete with E2 for binding to either ERβ or ERα at physiological concentrations. The data are fit with a non linear regression curve fit using SigmaPlot® software. The relative affinities of the 30 test compounds for ERβ and ERα are indicated by the IC50 values listed in Supplementary File 3, in which compounds are listed by their affinity for ERβ. Selectivity was determined by dividing the IC50 values obtained with ERβ by ERα. It should be noted that the ranking of the ligands based on change in ERβ melting temperature correlated very well with the affinity determined from the FP assay. These data also fit with previous analyses in which coumestrol, zeranol, narigenin, and kaempferol were all found to compete more effectively with E2 for binding to ERβ than to ERα, and in which coumestrol and zeranol had higher ranking in affinity than narigenin and kaempferol for both ER subtypes [24]. It was surprising that an unambiguous fit for kaempferol with ERα could not be obtained since it displays ERα binding affinity in a radioligand-competition assay, however the authors of that study did note that complete displacement of the radioligand from the ERα protein could not be obtained for this ligand [24]. Another surprising finding was the order of competition for estriol and estrone, such that estriol displayed greater relative affinity than estrone for both ERβ and ERα. These two physiological estrogens were ranked in the reverse order by Kuiper et al. [37]. Otherwise, the Kuiper et al. [37] radioligand binding competition assay results show high similarities to our data confirming that estradiol, hexestrol, diethylstilbestrol, coumestrol, zeranol, estrone, estriol, clomiphene, tamoxifen and DHEA were binders for both ERα and ERβ.
In order to designate ligands as agonists, partial agonists or antagonists, we next evaluated the ability of these compounds to modulate the transcriptional activity of ERβ. The transcriptional response of the ligands was measured using a well-defined system comprised of transfected receptor LBD linked to a GAL4 DNA binding function and a GAL4 responsive reporter in an ER-negative mammalian cell line. Results are expressed as normalized luciferase activity (normalized to Renilla for transfection efficiency). As observed in Figure 4, many compounds induced transactivation of ERβ in a dose-dependent manner. Compared with E2, the other dose-response curves are shifted to the right, reflecting the differences in affinity of these compounds for ERβ. The system also demonstrated different ligand classifications, namely, agonists, partial agonists, and/or antagonists. For example, when examined in a dose response manner in antagonist mode with 5 nM E2, clomiphene citrate (Figure 4b, yellow x) displays an IC50 of 221 nM. With increasing concentration of clomiphene citrate, a dose-dependent decrease in luciferase activity is found in the presence of E2, demonstrating the compound’s ability to block the estrogen response. Calculated EC50/IC50 values are listed in Supplementary File 3.
Figure 4. Cell-based transactivation assay to evaluate the ability of ligands to modulate the transcriptional activity of ERβ.
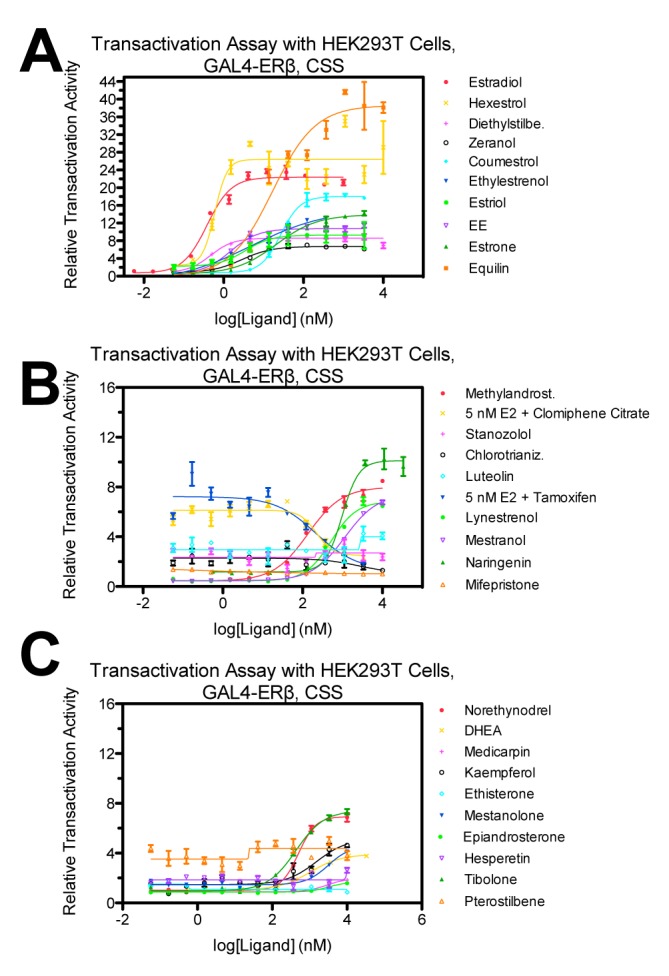
Relative transactivation activity for the ligands identified as ERβ binders from the primary and secondary assays using HEK293T cells transiently transfected with GALDBD-ERβLBD. Data are means ± SE of triplicate wells and normalized to Dimethyl Sulfoxide (DMSO). Charcoal stripped serum was used.
Those ligands which appeared to be coming through the cascade as ERβ selective are listed in Supplementary File 4, where β-selectivity (fold) was greater than 2 and calculated by dividing the compound’s half maximal inhibitory concentration (IC50) for ERα by that for ERβ as determined from FP. In this screening cascade we had set out to look for β-selectivity and discovered nine leads compounds based solely on the data from fluorescence polarization assays with the DSF assay lending additional confirmation of ligand binding. These nine lead ligands (Supplementary File 4) include hexestrol, coumestrol, methylandrostenediol, luteolin, naringenin, mifepristone, DHEA, medicarpin, and kaempferol.
We next examined effects of selected ligands that were identified in the cascade and displayed different activities and ER subtype selectivity in transfections with full length ERs versus cells treated with E2 control (Figure 5A). E2 (full agonist) activated both receptors, with ERα activity higher than that of ERβ. By contrast, one of the ligands described in Supplementary File 4, DHEA, displayed strong ERβ selectivity, with greater capacity for ERβ activation than the selective partial agonist 27-hydroxycholesterol [38–40]. Finally, clomifene failed to activate either ER subtype, consistent with antagonist activity.
Figure 5. Cell-based transactivation assay to evaluate the ability of ligands to modulate the transcriptional activity of ERβ.
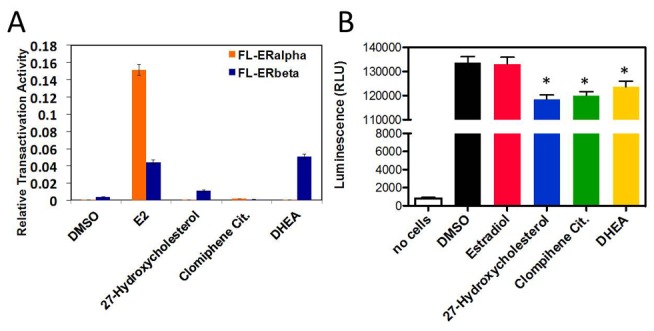
(A) Relative transactivation activity for selected ligands (final concentration 1 μM) using HeLa cells transiently transfected with full length ERα and full length ERβ. (B) Cell viability assay measured in LNCaP cells after 24 h treatment with selected ligands at a final concentration 1 μM. Average of three wells each for three independent experiments.
We determined effects of the same ligands on ERβ-dependent suppression of prostate cancer cell proliferation. In accordance with previous results [20, 21], the full agonist E2 did not affect LNCaP cell proliferation (Figure 5B). Suppression of proliferation was obtained with DHEA, 27-hydroxycholesterol, and clomifene. Accordingly, our findings support the concept that certain ERβ ligands induce anti-proliferative activities in prostate cancer cells that are not shared with the full agonist E2. We propose that our screening cascade has the capacity to identify compounds with these potentially desirable activities.
Discussion
Although DSF is a commonly used technique for primary screens, it is also known to generate false positives, and we observed this problem in the current study. For example, dactinomycin (NIH Clinical Collection), goserelin acetate (NIH Clinical Collection), antimycin A (Prestwick Library), lithocholic acid (Prestwick Library) and nystatin (Prestwick Library) were omitted but also elicited changes in the protein melting temperature. These antibiotics, antifungals, and amphiphiles act as nonspecific effectors by either adsorbing to plastics, creating micelles and interacting with the dye to form aggregates that may destabilize the protein; or by weakly electrostatically interacting with the protein, as opposed to binding inside the ligand-binding pocket. Nystatin, for example, has been shown to perturb ligand binding to receptors [41] and dactinomycin, also known as actinomycin D, has been reported to inhibit the nuclear processing of estrogen receptors in MCF-7 cells [42]. Additionally, some compounds, such as felodipine (Prestwick Library) and dactinomycin (NIH Clinical Collection) were colored, another common cause of interference with the optical detection of fluorescence in DSF. While these compounds also showed up as hits in the primary assay, they were omitted from Supplementary File 3 based on their complicated chemical structures. For ERα LBD when the data was fit using Graphpad Prism with a nonlinear regression curve fit for the compounds mifepristone and kaempferol (Figure 3) the IC50 values yielded were ambiguous and therefore excluded. The remaining compounds, however, displayed good correlation between the different assays, where the melting temperature of the protein with the addition of test compound from DSF can be compared with the IC50 determined from the fluorescence polarization assay. Generally, as the melting temperature identified using DSF decreased, the IC50 of the corresponding ligand, identified using FP, increased (Supplementary File 3).
The use of highly purified material in initial screens is critical. Our initial trials with partially purifed protein identified more than one melting transition, and we speculate that one of the two melting transitions observed by DeSantis et al. [43] for their human ERα LBD (likely the ligand independent transition at 41 ºC) is due to the presence of a contaminant such as a chaperone or ribosomal protein that yielded its own melting transition. The use of additional purification steps such as a gel filtration column, can help yield a more pure product and eliminate additional unrelated melting transitions that might otherwise confound the experimental system and results.
The trial screen used here identified nine ERβ selective ligands from around 3000 compounds. In agreement with previous radioligand binding assay findings from Kuiper et al. [37], the Ki for the competitior compounds DHEA and coumestrol were both lower for ERβ than ERα, indicating ERβ selectivity, while the Ki for hexestrol was the same for both isoforms. In fact, coumestrol is widely considered an ERβ-selective agonist ligand [44, 45]. The literature is not as definitive for the other phytoestrogens identified here, likely because the ability of a compound to selectively bind to a particular ER subtype can be species dependent [46]. In a radioligand binding assay-based investigation of the ligand binding profiles of both ER subtypes from human, rat, and mouse, Harris et al. reported a lower IC50 for the interaction of kaempferol with human ERβ than with ERα, while for narigenin, an IC50 value was obtained for human ERβ only [46]. Additionally, both narigenin and kaempferol were found to act as weaker agonists (as compared to coumestrol and others) in a study evaluating transcriptional activation of MCF-7 cells transiently transfected with either ERα or ERβ and an ERE-reporter plasmid. Moreover, a sigmoidal dose-response curve model was fit and an EC50 determined for ERβ transfected cells, the EC50 for both narigenin and kaempferol for ERα was not determined due to a lack of full dose response to these ligands [45]. Luteolin has also been studied for its ability to activate ERα or ERβ in transiently transfected MCF-7 cells, and was found to have a very slight effect on ERβ but no effect on ERα [47]. While direct binding measurements of medicarpin to ERs have not been reported, selective knockdown of both ERα and ERβ in osteoblasts demonstrated that the osteogenic action of medicarpin is ERβ-dependent [48]. Although the selectivity of methyandrostenediol (methandriol) for ERα or ERβ has also not been reported, its parent compound 5-androstenediol activates ERs [49] and is ERβ selective [50], suggesting that similar properties for methyandrostenediol would not be unexpected. Moreover, Mifepristone (RU-486) has been found to be ERβ selective in a screen of the ToxCast library, although no affinity values were reported [51]. Our screen has identified a mix of known ERβ selective ligands, as well as novel ligands with possible ERβ selective activities.
It should be mentioned that some compounds and plant extracts, such as MF101, have been shown to have equal binding to both ERα and ERβ subtypes, but display ERβ specific proliferative gene activation [52, 53]. Additionally the compound 3,3’-diindolylmethane did not bind selectively to ERβ but selectively activated multiple endogenous genes by recruiting ERβ and coactivators to target genes [54]. Therefore, it is clear that binding preference, activation, and gene targeting create a complex issue in determining ligand effectiveness.
Our screen relies primarily upon detection of ERβ selective binding and examining large numbers of compounds in gene activation and cell proliferation assays requires further studies. We did, however, assess capacity of our screen to identify ligands with selective ERβ modulator activities during prostate cancer proliferation (Figure 5). Previous studies showed that certain ERβ ligands inhibit prostate cancer cell proliferation and increase apoptosis and that this capacity is distinct from full agonist activity [20, 21]. Accordingly, whereas the full agonist estradiol does not alter LNCaP proliferation, DHEA (identified from our screening cascade), 27-hydroxycholesterol (a known SERM with ERβ activity [38]), and the antagonist clomifene do. Of these, DHEA displays striking selectivity for ERβ versus ERα. Accordingly, we were able to identify ligands with potentially useful effects on prostate cancer proliferation, and further screening of this ligand subset could identify other compounds with useful selective modulator activities.
While the LBD specificity of some ligands for ERα and ERβ has been assessed, and selective estrogen receptor modulators (SERMs) for ERα have been ranked in different species, such as human, dog, and cat [55], the identification of differential binding of compounds substantiates the prospect of developing ER-selective drugs. Our approach also helps make evident the structural features that are important for ER binding and contributes to the accumulated knowledge of ligand structure activity relationship (SAR) for this molecule.
Conclusions
Herein we have demonstrated the use of a screening cascade consisting of a primary thermofluor assay to qualitatively monitor changes in protein melting upon ligand binding and allow for the rapid elimination of non-binders from our small molecule collection. We included a secondary FP assay to quantitatively determine receptor subtype preference and affinity (β versus α). This assay was performed to eliminate any potential false positives initially obtained and determine relative binding affinity. The tertiary cell-based GAL4 transactivation assay offers the ability to qualitatively determine ligand dependent transactivation of ERβ. Transactivation activity information is highly valuable to generate SAR and further identifies which compounds elicit an ERβ mediated response. The overall screening strategy provides a method to rank affinity for lead-optimization and reduces the uncertainty of the potential of a compound to bind to the protein of interest. Compounds with overlapping activity in binding, functional, and cell-based assays provide the best screening hits and lead compounds to be further explored for drug repurposing as they indicate activity regardless of the assay employed. We showed that several selected ligands, including a known SERM (27-hydroxycholesterol), a ERβ selective ligand identified from the cascade (DHEA), and an antagonist identified from the cascade (clomiphene), demonstrated effects on cell proliferation using a prostate cancer cell line as our disease model. As we have shown, several ERβ-selective SERMs were identified from a large collection of compound libraries, including known endogenous ligands as well as synthetic compounds that are now off patent. By beginning with libraries containing a pool of safe, off patent compounds, chemicals that are selective for ERβ can be further evaluated for drug repurposing.
Acknowledgements
PW, JÅG, CSF, PSG, AS, and ALB were supported by the Texas Emerging Technology Fund (TETF) State of Texas Superiority grant 300-9-1958 for the Texas International Center for Cell Signaling and Nuclear Receptors. The authors would like to thank and acknowledge The NCI/DTP Open Chemical Repository (http://dtp.cancer.gov) for providing the source of several materials, including agents: NSC 22842, NSC 180973, and NSC 350085. We also thank Dr. David Engler and Dr. Rise Matsunami for the Mass Spectrometry experiments and analyses and Dr. Margaret Warner, Dr. Kevin Phillips, Dr. Douglas Sieglaff, and Dr. Sena Rajagopalan for helpful discussions.
Supplementary Material
Supplementary Material (.zip)
- ER
estrogen receptor
- DSF
differential scanning fluorimetry
- DHEA
dehydroepiandrosterone
- E2
estradiol
- FP
fluorescence polarization
- Gen
genistein
- LBD
ligand binding domain
- NR
nuclear receptor
Footnotes
Competing interests: The authors declare no competing financial interests.
References
- 1.Sladek FM: Nuclear Receptors as Drug Targets: New Developments in Coregulators, Orphan Receptors and Major Therapeutic Areas. Expert Opin Ther Targets 2003, 7:679–684. 10.1517/14728222.7.5.679 [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM: The nuclear receptor superfamily: the second decade. Cell 1995, 83:835–839. 10.1016/0092-8674(95)90199-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov 2006, 5:993–996. 10.1038/nrd2199 [DOI] [PubMed] [Google Scholar]
- 4.Ariazi EA, Ariazi JL, Cordera F, Jordan VC: Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem 2006, 6:181–202. 10.2174/156802606776173483 [DOI] [PubMed] [Google Scholar]
- 5.Schulman IG: Nuclear receptors as drug targets for metabolic disease. Adv Drug Deliv Rev 2010, 62:1307–1315. 10.1016/j.addr.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang P, Chandra V, Rastinejad F: Structural Overview of the Nuclear Receptor Superfamily: Insights into Physiology and Therapeutics. Annu Rev Physiol 2010, 72:247–272. 10.1146/annurev-physiol-021909-135917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Persidis A: The benefits of drug repositioning. Drug Discov World 2011. [Google Scholar]
- 8.Kola I, Landis J: Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 2004, 3:711–715. 10.1038/nrd1470 [DOI] [PubMed] [Google Scholar]
- 9.Tobinick EL: The value of drug repositioning in the current pharmaceutical market. Drug News Perspect 2009, 22:119–125. 10.1358/dnp.2009.22.2.1343228 [DOI] [PubMed] [Google Scholar]
- 10.Slade O: Drug Repurposing the Most Affordable Relaunch Option for Pharma Companies. PR Newswire 2013. [Google Scholar]
- 11.Couse JF, Lindzey J, Grandien K, Gustafsson JA, Korach KS: Tissue distribution and quantitative analysis of estrogen receptor-alpha (ERalpha) and estrogen receptor-beta (ERbeta) messenger ribonucleic acid in the wild-type and ERalpha-knockout mouse. Endocrinology 1997, 138:4613–4621. [DOI] [PubMed] [Google Scholar]
- 12.Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS: Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science 1997, 277:1508–1510. 10.1126/science.277.5331.1508 [DOI] [PubMed] [Google Scholar]
- 13.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, Gustafsson J-A: Estrogen receptors: how do they signal and what are their targets. Physiol Rev 2007, 87:905–931. 10.1152/physrev.00026.2006 [DOI] [PubMed] [Google Scholar]
- 14.Enmark E, Gustafsson J-A: Estrogen receptor beta - a novel receptor opens up new possibilities for cancer diagnosis and treatment. Endocr Relat Cancer 1998, 5:213–222. 10.1677/erc.0.0050213 [DOI] [Google Scholar]
- 15.Warner M, Gustafsson J-A: The role of estrogen receptor beta (ERbeta) in malignant diseases--a new potential target for antiproliferative drugs in prevention and treatment of cancer. Biochem Biophys Res Commun 2010, 396:63–66. 10.1016/j.bbrc.2010.02.144 [DOI] [PubMed] [Google Scholar]
- 16.Deroo BJ: Estrogen receptors and human disease. J Clin Invest 2006, 116:561–570. 10.1172/JCI27987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. www.clinicaltrials.gov
- 18.Roman-Blas JA, Castañeda S, Cutolo M, Herrero-Beaumont G: Efficacy and safety of a selective estrogen receptor β agonist, ERB-041, in patients with rheumatoid arthritis: A 12-week, randomized, placebo-controlled, phase II study. Arthritis Care Res 2010, 62:1588–1593. 10.1002/acr.20275 [DOI] [PubMed] [Google Scholar]
- 19.Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P: Loss of ER expression as a common step in estrogen-dependent tumor progression. Endocr Relat Cancer 2004, 11:537–551. 10.1677/erc.1.00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey P, Ström A, Gustafsson J-Å: Estrogen receptor β upregulates FOXO3a and causes induction of apoptosis through PUMA in prostate cancer. Oncogene 2014,. 33:4213–25. 10.1038/onc.2013.384 [DOI] [PubMed] [Google Scholar]
- 21.Dey P, Jonsson P, Hartman J, Williams C, Ström A, Gustafsson J-Å: Estrogen receptors β1 and β2 have opposing roles in regulating proliferation and bone metastasis genes in the prostate cancer cell line PC3. Mol Endocrinol 2012, 26:1991–2003. 10.1210/me.2012.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng J, Lee EJ, Madison LD, Lazennec G: Expression of estrogen receptor β in prostate carcinoma cells inhibits invasion and proliferation and triggers apoptosis. FEBS Lett 2004, 566:169–172. 10.1016/j.febslet.2004.04.025 [DOI] [PubMed] [Google Scholar]
- 23.Blair RM: The Estrogen Receptor Relative Binding Affinities of 188 Natural and Xenochemicals: Structural Diversity of Ligands. Toxicol Sci 2000, 54:138–153. 10.1093/toxsci/54.1.138 [DOI] [PubMed] [Google Scholar]
- 24.Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, Gustafsson JA: Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology 1998, 139:4252–4263. [DOI] [PubMed] [Google Scholar]
- 25.Fink BE, Mortensen DS, Stauffer SR, Aron ZD, Katzenellenbogen JA: Novel structural templates for estrogen-receptor ligands and prospects for combinatorial synthesis of estrogens. Chem Biol 1999, 6:205–219. 10.1016/S1074-5521(99)80037-4 [DOI] [PubMed] [Google Scholar]
- 26.Manas ES, Unwalla RJ, Xu ZB, Malamas MS, Miller CP, Harris HA, Hsiao C, Akopian T, Hum W-T, Malakian K, Wolfrom S, Bapat A, Bhat RA, Stahl ML, Somers WS, Alvarez JC: Structure-based design of estrogen receptor-beta selective ligands. J Am Chem Soc 2004, 126:15106–15119. 10.1021/ja047633o [DOI] [PubMed] [Google Scholar]
- 27.Harris HA: Preclinical Characterization of Selective Estrogen Receptor β Agonists: New Insights into Their Therapeutic Potential. In Tissue-Specif Estrogen Action. Volume 2006/1. Edited by Korach KS, Wintermantel T. Berlin, Heidelberg: Springer Berlin Heidelberg; 2007:149–162. [Google Scholar]
- 28.Weatherman RV, Fletterick RJ, Scanlan TS: Nuclear-receptor ligands and ligand-binding domains. Annu Rev Biochem 1999, 68:559–581. 10.1146/annurev.biochem.68.1.559 [DOI] [PubMed] [Google Scholar]
- 29.Webb P, Lopez GN, Uht RM, Kushner PJ: Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol 1995, 9:443–456. [DOI] [PubMed] [Google Scholar]
- 30.Lo M-C, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G: Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem 2004, 332:153–159. 10.1016/j.ab.2004.04.031 [DOI] [PubMed] [Google Scholar]
- 31.Phillips K, de la Peña AH: The combined use of the Thermofluor assay and ThermoQ analytical software for the determination of protein stability and buffer optimization as an aid in protein crystallization. Curr Protoc Mol Biol Ed Frederick M Ausubel Al 2011, Chapter 10:Unit10.28. [DOI] [PubMed] [Google Scholar]
- 32.Niesen FH, Berglund H, Vedadi M: The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2007, 2:2212–2221. 10.1038/nprot.2007.321 [DOI] [PubMed] [Google Scholar]
- 33.First Diversity Set Information [http://dtp.nci.nih.gov/branches/dscb/diversity_explanation.html]
- 34.Bogan AA, Cohen FE, Scanlan TS: Natural ligands of nuclear receptors have conserved volumes. Nat Struct Biol 1998, 5:679–681. 10.1038/1372 [DOI] [PubMed] [Google Scholar]
- 35.Mason JI: Genetics of Steroid Biosynthesis and Function. London; New York: Taylor & Francis; 2002. [Google Scholar]
- 36.Baell JB, Holloway GA: New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 2010, 53:2719–2740. 10.1021/jm901137j [DOI] [PubMed] [Google Scholar]
- 37.Kuiper GG, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, Gustafsson JA: Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology 1997, 138:863–870. [DOI] [PubMed] [Google Scholar]
- 38.DuSell CD, Umetani M, Shaul PW, Mangelsdorf DJ, McDonnell DP: 27-Hydroxycholesterol Is an Endogenous Selective Estrogen Receptor Modulator. Mol Endocrinol 2008, 22:65–77. 10.1210/me.2007-0383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DuSell CD, Nelson ER, Wang X, Abdo J, Mödder UI, Umetani M, Gesty-Palmer D, Javitt NB, Khosla S, McDonnell DP: The Endogenous Selective Estrogen Receptor Modulator 27-Hydroxycholesterol Is a Negative Regulator of Bone Homeostasis. Endocrinology 2010, 151:3675–3685. 10.1210/en.2010-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umetani M, Domoto H, Gormley AK, Yuhanna IS, Cummins CL, Javitt NB, Korach KS, Shaul PW, Mangelsdorf DJ: 27-Hydroxycholesterol is an endogenous SERM that inhibits the cardiovascular effects of estrogen. Nat Med 2007, 13:1185–1192. 10.1038/nm1641 [DOI] [PubMed] [Google Scholar]
- 41.Pucadyil TJ, Shrivastava S, Chattopadhyay A: The sterol-binding antibiotic nystatin differentially modulates ligand binding of the bovine hippocampal serotonin1A receptor. Biochem Biophys Res Commun 2004, 320:557–562. 10.1016/j.bbrc.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 42.Horwitz KB, McGuire WL: Actinomycin D prevents nuclear processing of estrogen receptor. J Biol Chem 1978, 253:6319–6322. [PubMed] [Google Scholar]
- 43.DeSantis K, Reed A, Rahhal R, Reinking J: Use of differential scanning fluorimetry as a high-throughput assay to identify nuclear receptor ligands. Nucl Recept Signal 2012, 10:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katzenellenbogen JA, Muthyala R, Katzenellenbogen BS: Nature of the ligand-binding pocket of estrogen receptor α and β: The search for subtypeselective ligands and implications for the prediction of estrogenic activity. Pure Appl Chem 2009, 75:2397–2403. [Google Scholar]
- 45.Harris DM, Besselink E, Henning SM, Go VLW, Heber D: Phytoestrogens induce differential estrogen receptor alpha- or Beta-mediated responses in transfected breast cancer cells. Exp Biol Med 2005, 230:558–568. [DOI] [PubMed] [Google Scholar]
- 46.Harris HA, Bapat AR, Gonder DS, Frail DE: The ligand binding profiles of estrogen receptors alpha and beta are species dependent. Steroids 2002, 67:379–384. 10.1016/S0039-128X(01)00194-5 [DOI] [PubMed] [Google Scholar]
- 47.Innocenti G, Vegeto E, Dall’Acqua S, Ciana P, Giorgetti M, Agradi E, Sozzi A, Fico G, Tomè F: In vitro estrogenic activity of Achillea millefolium L. Phytomedicine 2007, 14:147–152. 10.1016/j.phymed.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 48.Bhargavan B, Singh D, Gautam AK, Mishra JS, Kumar A, Goel A, Dixit M, Pandey R, Manickavasagam L, Dwivedi SD, Chakravarti B, Jain GK, Ramachandran R, Maurya R, Trivedi A, Chattopadhyay N, Sanyal S: Medicarpin, a legume phytoalexin, stimulates osteoblast differentiation and promotes peak bone mass achievement in rats: evidence for estrogen receptor β-mediated osteogenic action of medicarpin. J Nutr Biochem 2012, 23:27–38. 10.1016/j.jnutbio.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 49.Purohit A, Woo LW., Chander S., Newman S., Ireson C, Ho Y, Grasso A, Leese M., Potter BV., Reed M.: Steroid sulphatase inhibitors for breast cancer therapy. J Steroid Biochem Mol Biol 2003, 86:423–432. 10.1016/S0960-0760(03)00353-4 [DOI] [PubMed] [Google Scholar]
- 50.Blizzard TA, Gude C, Morgan JD, Chan W, Birzin ET, Mojena M, Tudela C, Chen F, Knecht K, Su Q, Kraker B, Mosley RT, Holmes MA, Sharma N, Fitzgerald PMD, Rohrer SP, Hammond ML: Androstenediol analogs as ER-β-selective SERMs. Bioorg Med Chem Lett 2006, 16:834–838. 10.1016/j.bmcl.2005.11.014 [DOI] [PubMed] [Google Scholar]
- 51.Filer D, Judson R, Rotroff D, Reif D, Richard A, Houck K, Martin M: Protein-fragment complementation assays detect and describe nuclear receptor activity across the ToxCast chemical library. 2014. http://epa.gov/research/sot/2014/posters/protein-fragment-complementation.pdf
- 52.Cvoro A, Paruthiyil S, Jones JO, Tzagarakis-Foster C, Clegg NJ, Tatomer D, Medina RT, Tagliaferri M, Schaufele F, Scanlan TS, Diamond MI, Cohen I, Leitman DC: Selective activation of estrogen receptor-beta transcriptional pathways by an herbal extract. Endocrinology 2007, 148:538–547. 10.1210/en.2006-0803 [DOI] [PubMed] [Google Scholar]
- 53.Mersereau JE, Levy N, Staub RE, Baggett S, Zogovic T, Zogric T, Chow S, Ricke WA, Tagliaferri M, Cohen I, Bjeldanes LF, Leitman DC: Liquiritigenin is a plant-derived highly selective estrogen receptor beta agonist. Mol Cell Endocrinol 2008, 283:49–57. 10.1016/j.mce.2007.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vivar OI, Saunier EF, Leitman DC, Firestone GL, Bjeldanes LF: Selective activation of estrogen receptor-beta target genes by 3,3’-diindolylmethane. Endocrinology 2010, 151:1662–1667. 10.1210/en.2009-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toniti W, Suthiyotha N, Puchadapirom P, Jenwitheesuk E: Binding capacity of ER-α ligands and SERMs: comparison of the human, dog and cat. Asian Pac J Cancer Prev 2011, 12:2875–2879. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material (.zip)