Abstract
• Background and Aims Quercus petraea colonized Ireland after the last glaciation from refugia on mainland Europe. Deforestation, however, beginning in Neolithic times, has resulted in small, scattered forest fragments, now covering less than 12 000 ha.
• Methods Plastid (three fragments) and microsatellite variation (13 loci) were characterized in seven Irish populations sampled along a north–south gradient. Using Bayesian approaches and Wright’s F‐statistics, the effects of colonization and fragmentation on the genetic structure and mating patterns of extant oak populations were investigated.
• Key Results All populations possessed cytotypes common to the Iberian Peninsula. Despite the distance from the refugial core and the extensive deforestation in Ireland, nuclear genetic variation was high and comparable to mainland Europe. Low population differentiation was observed within Ireland and populations showed no evidence for isolation by distance. As expected of a marker with an effective population size of one‐quarter relative to the nuclear genome, plastid variation indicated higher differentiation. Individual inbreeding coefficients indicated high levels of outcrossing.
• Conclusions Consistent with a large effective population size in the historical migrant gene pool and/or with high gene flow among populations, high within‐population diversity and low population differentiation was observed within Ireland. It is proposed that native Q. petraea populations in Ireland share a common phylogeographic history and that the present genetic structure does not reflect founder effects.
Key words: Quercus petraea, microsatellites, plastid DNA, population differentiation, outbreeding
INTRODUCTION
Remnants of the formerly extensive Atlantic woodland that covered up to 80 % of the island of Ireland are now fragmented, covering less than 0·2 % (Mitchell, 1995; Rackham, 1995). Pinus sylvestris L., formerly an abundant taxon in this woodland, is thought to have declined to extinction by a combination of climatic change and, more recently, exploitation (Mitchell, 2003). One of the predominant species in scattered woodland remnants is Quercus petraea (Matt.) Liebl., now covering less than 12 000 ha (Keogh, 1987). The history of woodland exploitation in Ireland has parallels with the decline of Caledonian native pinewoods in Scotland. Both have been exploited for industrial purposes, particularly during the last 400 years, whereas only more recently has attention turned to their management and conservation. Surveys in Caledonian pinewoods indicate that despite the reduction in size of some stands to only hundreds of individuals, genetic variation remains high (Provan et al., 1998). In addition, increased selfing rates have not been detected as a result of the reduction in tree density (Helgason and Ennos, 1991).
Counts of radiocarbon‐dated pollen indicate that Ireland was colonized by Q. petraea from south to north (Mitchell, 2003). Maternally inherited plastid markers support this migration route and indicate that Irish populations of Q. petraea were derived from glacial refugia located on the Iberian Peninsula (Dumolin‐Lapègue et al., 1997; Petit et al., 2002). Nevertheless, sample sizes in these studies were small in Ireland, and Irish oaks have thus not been well characterized with molecular markers compared with British or mainland European populations.
Radiocarbon‐dated pollen also indicates a reduction in forest cover from a maximum of 80 % (approx. 7000 years ago), with a decline across all woody taxa beginning in Neolithic times (Mitchell, 1988). While climate influenced the postglacial dynamics of colonization at this time as well, the impact of human activity was sufficiently intense to register on the pollen record by the Bronze Age, approx. 3000 years ago (Mitchell, 1995). By the 1600s, census data indicate that woodland cover had declined to 2·1 % (Rackham, 1995). The effects of this exploitation on levels of genetic variation are unclear, but the value of these remnants as part of the natural heritage of Ireland is high.
Remnants of Irish oak represent populations at the north‐western margin of the range of Q. petraea in Europe. Irish populations are far from the refugial core and may have been exposed to (multiple) founding events. Latterly, fragmentation and reduced population sizes may also have led to a reduction in gene flow among previously connected populations and to an increase in mating between closely related individuals. To evaluate the impact of genetic drift, populations were sampled along a north–south gradient in close geographical proximity, as well as at more distant locations. If drift was a significant force then isolation by distance would be expected under colonization from south to north. Bayesian statistics and a model‐based clustering algorithm were also used to estimate the most probable population genetic structure in Ireland. Using a common European haplotype nomenclature (Dumolin‐Lapègue et al., 1997), plastid markers were used to establish if Irish oaks share a common postglacial colonization history with British oaks (Cottrell et al., 2002). In addition, nuclear microsatellites were used to compare the level of genetic diversity in Ireland with mainland European populations.
Genetic bottlenecks can lead to an increase in mating between closely related individuals within populations and to purging of deleterious alleles. In turn, this may lead to a change in the mating system (Vogl et al., 2002). In common with other trees, Q. petraea is a highly outcrossing species and maintains a high genetic load (Ledig, 1986). Given the contraction of ancient woodland in Ireland to current levels of less than 0·2 % and the persistence of only small and disjunct populations, past mating between closely related individuals may have influenced the mating system of Q. petraea. To infer the degree of reproductive isolation among remnant individuals, a recently developed Bayesian approach was used to place posterior confidence intervals on individual inbreeding coefficients. Rather than estimate a population average that may mask differences among individuals, individual inbreeding coefficients were estimated based on microsatellites (13 loci) to provide a higher resolution of mating patterns.
MATERIALS AND METHODS
Plant material
To obtain a latitudinal representation of the genetic variation present in Irish oakwoods, Q. petraea individuals (n = 25–33; see Table 3) were sampled from each of seven woods on a north–south gradient (see Fig. 1). All samples came from mature, even‐aged woods (125–175 years old) that form disjunct populations and have a density of approx. 300–500 stems ha–1. The two woods from the Killarney Valley (Tomies and Derrycunihy), however, were separated by only 6 km. Leaves were sampled from: Breen, Co. Antrim 55°7′N 6°20′W, Correl Glen, Co. Fermanagh 54°25′N 7°47′W, Garranon, Co. Clare 52°41′N 8°38′W, Pollnaknockaun, Co. Clare 53°3′N 8°24′W, Tomies, Co. Kerry 52°1′N 9°30′W, Derrycunihy, Co. Kerry 51°58′N 9°29′W and Glengarriff, Co. Cork 51°45′N 9°33′W. Latitudinal and longitudinal co‐ordinates were obtained from http://www.getty.edu/research/tools/vocabulary/tgn/index.html. Derrycunihy belongs to a cluster of woods in the Killarney Valley, thought to represent the largest ancient woodlands in Ireland (Kelly, 1981; Mitchell, 1988). Counts of radiocarbon‐dated pollen from Derrycunihy indicate a continuous Quercus pollen record for at least the last 5000 years and, most probably, direct continuity with post‐glacial primary forest (Mitchell, 1988). Nevertheless, historical records indicate that Tomies wood, also in the Killarney Valley, was felled and replanted at the beginning of the 19th century (Mitchell, 1988). It is likely that regrowth of this wood occurred via natural regeneration or locally sourced seeds. However, forest management in these woods cannot be discounted. Oakwoods are referred to as populations in the text.
Table 3.
Diversity indices by population across 13 microsatellite loci in Quercus petraea
| No. | Population | H O | H E | V | A | n |
| 1 | Breen | 0·70 | 0·81 | 20 | 7 | 25 |
| 2 | Correl Glen | 0·66 | 0·79 | 20 | 7 | 33 |
| 3 | Pollnaknockaun | 0·68 | 0·79 | 24 | 7 | 29 |
| 4 | Garranon | 0·67 | 0·80 | 21 | 7 | 29 |
| 5 | Tomies | 0·68 | 0·79 | 17 | 7 | 32 |
| 6 | Derrycunihy | 0·71 | 0·79 | 19 | 7 | 29 |
| 7 | Glengarriff | 0·72 | 0·81 | 24 | 7 | 32 |
| Eastern France | 0·64 | 0·82 | 21 | 7 | 13 | |
| SW France | 0·66 | 0·77 | 16 | 6 | 10 | |
| Spain | 0·61 | 0·76 | 19 | 6 | 16 |
No., population numbers in Fig. 1; HO, observed heterozygosity; HE, expected heterozygosity; V, variance in microsatellite repeat number; A, allelic richness after rarefaction to 16 chromosomes; and n, number of analysed individuals.
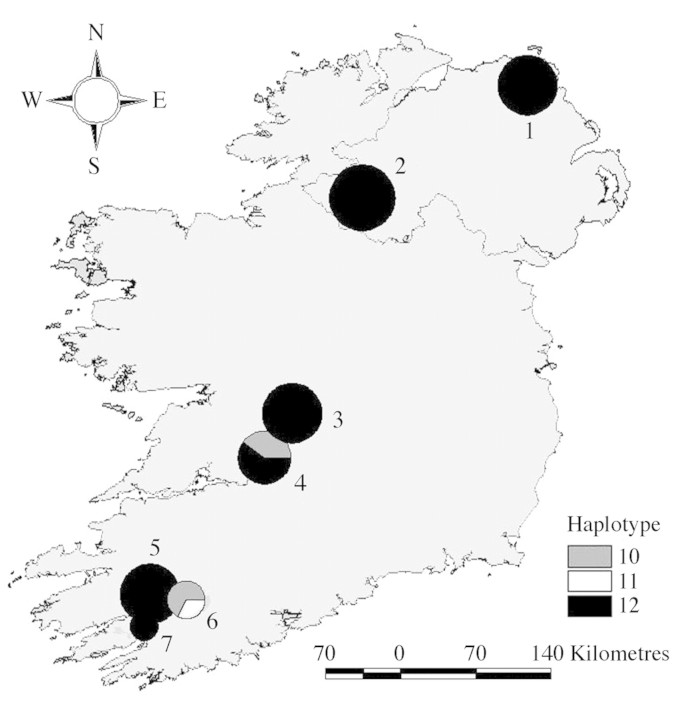
Fig. 1. Frequency of three plastid haplotypes detected in seven Irish populations of Quercus petraea. The size of the circle is proportional to the number of individuals sampled (n = 2–6). Population numbers correspond to those given in Table 3. Haplotype nomenclature follows Dumolin‐Lapègue et al. (1997).
As a mainland European reference, Q. petraea individuals (n = 10–16; see Table 3) were genotyped from three locations in Spain and France using DNA kindly provided by C. Bodénès and A. Kremer. These locations were in northern Spain 41°07′N 3°30′W, south‐western France 43°11′N 0°40′E and eastern France 48°18′N 4°28′E.
DNA extraction and PCR application
Genomic DNA was extracted from leaves using the Nucleon Phytopure Kit (Amersham) according to the manufacturer’s instructions. PCR reaction, amplification and electrophoresis conditions were as described in Schlötterer (1998). In total, ten populations (248 individuals) were genotyped using 13 microsatellite loci: 1/5, 1/2, 7, 9, 15, 110, 119 (Steinkellner et al., 1997) and 7, 87, 96, 101, 108, 112 (Kampfer et al., 1998). To establish the affinity of Irish oaks with plastid lineages from the European mainland, we determined the cytotypes for approx. five individuals from each of the seven Irish populations (n = 40). The restriction fragment length polymorphism analysis of plastid PCR was as described in Cottrell et al., (2002) and scoring of these fragments followed a widely used nomenclature (Dumolin‐Lapègue et al., 1997).
Data analysis
Microsatellite analyses.
Diversity indices and general statistics were calculated using MICROSATELLITE ANALYZER (MSA; Dieringer and Schlötterer, 2003). Estimates of allelic richness were corrected using the rarefaction option in FSTAT (Goudet, 2001). The MSA program was also used to determine Θ, an unbiased estimate of Wright’s Fixation Index (Weir and Cockerham, 1984), to characterize the extent of population differentiation. The significance of pairwise Θ values (referred to as FST in the text) was tested by performing permutations of genotypes among groups. This method of permutation does not rely on Hardy–Weinberg assumptions (Goudet et al., 1996). A Bonferroni correction was applied to account for multiple testing.
To test for a significant correlation between genetic and geographic distances, we compared an FST/(1 – FST) matrix with a geographical distance matrix (ln km) using a Mantel test (10 000 permutations) in GENEPOP (Raymond and Rousset, 1995). A web‐based distance calculator was used to determine the surface distance between two sampling points (http://www.wcrl.ars.usda.gov/cec/java/lat‐long.htm).
A model‐based clustering method implemented in the program STRUCTURE (Pritchard et al., 2000) was used to assign individuals to homogeneous clusters (populations) without consideration of sampling localities. Estimated posterior probabilities were calculated assuming a uniform prior for K, where 1 ≤ K ≤ 4. To minimize the effect of the starting configuration during the Monte Carlo simulation we simulated 50 × 103 updates of the Markov chain before data for the parameter estimation were collected from another 106 iterations. Three independent runs of the Markov chain, each of at least 106 updates were performed to assure convergence of the chain and homogeneity between runs for each prior of K. The posterior probabilities of K were then calculated using Bayes’ rule. The program was run without population identifiers (USEPOPINFO = 0) and in the admixture mode (NOADMIX = 0).
Inbreeding coefficients were estimated for the 209 Irish individuals using MUSTATS (Vogl et al., 2002). The program uses Gibbs sampling to estimate the posterior distribution of inbreeding coefficients by cyclically updating the conditional distribution of three sets of key variables. These are: (1) a set of variables that indicates identity by descent for each individual–locus combination; (2) the inbreeding coefficient for each individual; and (3) the population allelic proportion for each locus. From the marginal posterior distribution, the mean inbreeding coefficient and the lower 0·05 and upper 0·95 posterior interval were then calculated for each individual.
Plastid analyses.
Restriction fragment polymorphisms were scored as unordered binary characters. Total diversity (hT) and total differentiation as measured by GST were estimated using HAPLODIV (Pons and Petit 1995). Glengarriff was omitted from the latter because of the low number of individuals assayed (n = 2).
RESULTS
Colonization and population structure
All 40 individuals analysed for plastid DNA variation possessed one of three cytotypes, 10 (10 %), 11 (5 %) and 12 (85 %), all three belong to the west European lineage B (Dumolin‐Lapègue et al., 1997). Hence, in common with British oaks, native populations of Q. petraea in Ireland represent lineal descendants of refugial ancestors from the Iberian Peninsula (Cottrell et al., 2002). Five of the seven populations were monomorphic and fixed for cytotype 12. The remainder, located in the south (Fig. 1), were poly morphic, possessing at least two cytotypes. GST was relatively high among Irish populations (0·501) but hT was considerably lower (0·384) than published estimates for Q. petraea in Britain (hT = 0·569; Cottrell et al., 2002), and the Iberian Peninsula (hT = 0·804; Olalde et al., 2002).
Low population substructure both in Ireland and mainland Europe was observed for microsatellites by FST‐analysis (Table 1). There was no statistically significant difference between average pairwise FST‐values in Ireland (F̄ST = 0·008) and mainland Europe (F̄ST = 0·020), respectively (Mann–Whitney U‐test, P = 0·0795). With two exceptions (from a total of 21), values for pairwise comparisons within Ireland were not statistically significant.
Table 1.
Genetic differentiation among populations of Quercus petraea measured by pairwise FST (below diagonal): significance values based on the permutation of microsatellite genotypes among populations (above diagonal)
| North | Central | South | ||||||||
| Breen | Correl Glen | Pollnaknockaun | Garranon | Tomies | Derrycunihy | Glengarriff | Eastern France | SW France | Spain | |
| Breen | ns | ns | ns | ns | ns | ns | * | ns | ns | |
| Correl Glen | 0·013 | ** | ** | ns | ns | ns | ** | ns | ** | |
| Pollnaknockaun | 0·008 | 0·020 | ns | ns | ns | ns | ** | ns | ns | |
| Garranon | 0·011 | 0·024 | 0·004 | ns | ns | ns | ** | ns | ns | |
| Tomies | 0·002 | 0·006 | 0·001 | 0·006 | ns | ns | ** | ns | ns | |
| Derrycunihy | 0·006 | 0·012 | 0·004 | 0·008 | 0·002 | ns | ** | ns | ns | |
| Glengarriff | 0·007 | 0·019 | 0·003 | 0·008 | 0·004 | 0·004 | ** | ns | ns | |
| Eastern France | 0·028 | 0·036 | 0·033 | 0·029 | 0·031 | 0·027 | 0·033 | ns | ns | |
| SW France | 0·005 | 0·016 | 0·018 | –0·001 | 0·007 | 0·008 | 0·010 | 0·010 | ns | |
| Spain | 0·014 | 0·032 | 0·015 | 0·010 | 0·017 | 0·016 | 0·009 | 0·040 | 0·011 | |
P‐values determined using 10 000 replicates. **, P ≤ 0·01, *, P ≤ 0·05,
ns, not significant (after Bonferroni correction); SW, south‐western.
Values for the highest pairwise FST comparisons within Ireland were confined to the two most northerly populations. One of these populations (Correl Glen) was significantly different from both populations located in central Ireland (Pollnaknockaun, Garranon). Nevertheless, consistent with the single west European cytotype (12) detected in this population and a shared colonization history, Correl Glen was not significantly different from its northern neighbour (Breen).
In the model‐based clustering approach, individuals from seven locations in Ireland were assigned to populations (K) on the basis of their microsatellite genotype, ignoring their actual geographic origins within Ireland. This mimics a scenario in which there is no a priori information as to the geographic origin of individuals. Most of the posterior probability was on K = 1 (Table 2). Hence, despite the substructure observed in Correl Glen based on the FST‐analysis, Irish Q. petraea as a whole can be regarded as one genetic cluster.
Table 2.
Estimated posterior probabilities of K assuming four genetic clusters
| K* | ln P(X|K) | P(K|X)** |
| 1 | –11172·8 | ∼1 |
| 2 | –11214·2 | ∼0 |
| 3 | –11290·4 | ∼0 |
| 4 | –11289·3 | ∼0 |
Calculations based on seven locations (209 individuals of Q. petraea from Ireland).
* number of clusters assumed using STRUCTURE; ** assuming a uniform prior for K, where 1 ≤ K ≤ 4 (for explanation, see text).
As the level of genetic differentiation depends on gene flow and genetic drift, the analyses above suggest high rates of historical gene flow between populations and/or large effective population sizes. If genetic drift and low gene flow were significant forces, then adjacent populations along the north–south gradient in Ireland should show more genetic similarity than distantly located ones. However, the Mantel test detected no significant correlation between pairwise genetic distance and geographic distance in Irish Q. petraea.
Levels of genetic variation
We observed substantial levels of microsatellite variation (H̄E > 0·7 and V̄ > 20, Table 3), comparable with those obtained using the same 13 loci (plus seven) in a larger survey of 20 microsatellite loci, scored in five populations of Q. petraea from mainland Europe (G. Muir and C. Schlötterer, ITG, Vienna, Austria, unpubl. res.). Hence, observed levels of variation in Ireland cannot be an artefact of a higher sampling variance associated with fewer loci. Irish populations also showed comparable levels of variation to putative source populations in mainland Europe. We compared diversity indices (in Table 3) for each of the 13 loci surveyed among the two groups: Ireland and mainland Europe. With the exception of allelic richness (A), there was no statistically significant difference among the diversity indices (Table 3) between populations from mainland Europe and Ireland (Wilcoxon sign test).
Mating system
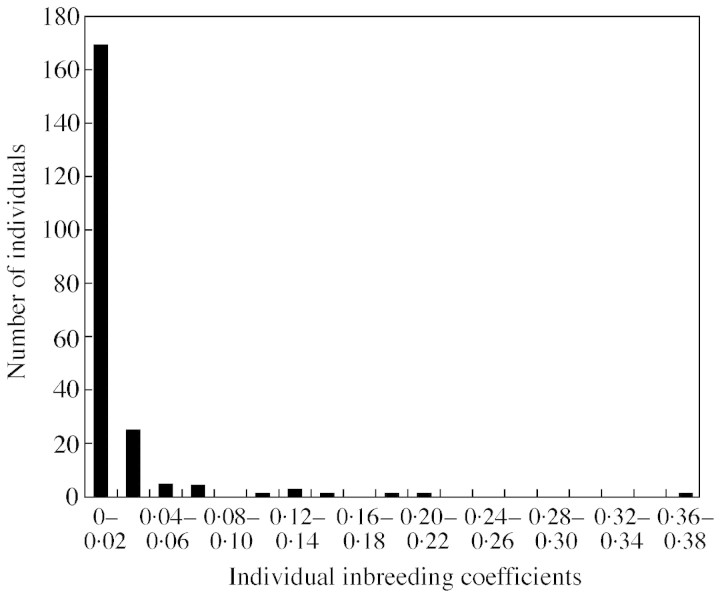
The majority of individual inbreeding coefficients were close to zero with average inbreeding coefficients per population between 0·010 and 0·031 (Table 4). On average, only one individual per population possessed coefficients above 0·12, and no individual in the whole data set possessed coefficients above 0.38 (Fig. 2; see also supplementary material). Hence, there is little evidence for mature selfed individuals, or for a change in mating system as a result of population fragmentation in Ireland.
Table 4.
Estimates of average inbreeding coefficients, E(F̄I), and lower (0·05) and upper (0·95) posterior intervals for seven Irish Quercus petraea populations (estimated using 13 microsatellite loci)
| Population | E(F̄I) | 0·05 | 0·95 |
| Breen | 0·031 | 0·005 | 0·060 |
| Correl Glen | 0·018 | 0·001 | 0·046 |
| Pollnaknockaun | 0·014 | 0 | 0·042 |
| Garranon | 0·021 | 0·001 | 0·045 |
| Tomies | 0·024 | 0·003 | 0·053 |
| Derrycunihy | 0·020 | 0·001 | 0·046 |
| Glengarriff | 0·010 | 0 | 0·026 |
Fig. 2. Frequency distribution of inbreeding coefficients estimated for 209 Quercus petraea individuals from seven Irish populations. Corresponding posterior intervals can be found at http://i122server.vu‐wien.ac.at.
DISCUSSION
Low genetic differentiation among populations
Approximately 100 generations separate the current cohort of trees from the first post‐refugial colonizers to Ireland. Assuming a mutation rate of 10–4 per generation for plant microsatellites (Thuillet et al., 2002), the number of mutations that have arisen in this time‐frame can be considered negligible. Hence, new mutations are unlikely to influence the genetic variation observed at microsatellite loci between Ireland and the mainland or within and among Irish populations. As such, the present population structure in Ireland either reflects historical levels of differentiation or, as an alternative, high gene flow among populations.
The FST analysis indicated that population substructure within Ireland was limited to Correl Glen. Nevertheless, the model‐based clustering algorithm indicated that Irish Q. petraea as a whole can be regarded as one genetic cluster. The difference in the detection level between these analyses may stem from the a priori groupings of individuals made for the FST analysis. The clustering approach uses the genotypes of single individuals rather than a priori groupings to infer population substructure. Together with a relatively small number of loci, this may have provided insufficient power to detect the population substructure in Correl Glen.
Pairwise FST comparisons of Irish populations with those in mainland Europe indicated no statistical difference between most Irish populations and those from Spain and south‐western France. All Irish populations, however, were statistically different from the population in eastern France (Table 1); a result which is consistent with a colonization route along the north‐western seaboard of Spain and France (Olalde et al., 2002; Petit et al., 2002). Surveys of additional populations would be required to support this claim.
High levels of genetic variation within populations
Despite the peripheral location and the large reduction in forest cover, Q. petraea populations in Ireland have similar allelic proportions and comparable levels of microsatellite variation with those in mainland Europe. This pattern contrasts with the decrease in variation expected with distance from a refugial source, but is supported by other comparisons of diversity indices from the margins and core distribution of the species (e.g. Cottrell et al., 2003). The lack of genetic differentiation between the above two regions may reflect admixture (at mid‐latitudes) of populations derived from independent glacial refugia (Petit et al., 2003).
Additional information on the demography of oaks can be obtained from the maternally inherited plastid genome (Dumolin et al. 1995). With an effective population size of one‐quarter relative to nuclear markers under neutrality, plastid DNA is likely to reveal higher levels of differentiation among populations. Cytotype differentiation and diversity were higher in the south of Ireland, as has been observed within Britain (Cottrell et al., (2002). Whereas variability of cytotypes in Ireland was reduced compared with mainland Europe (see also Cottrell et al., 2002), this was not indicative of a severe genetic bottleneck. Hence, the observed difference in differentiation among nuclear and cytoplasmic markers can be explained by the differences in effective population size of the two genomes. In the remainder of this paper, several scenarios that may account for the level of genetic variation observed in Ireland are discussed.
High rates of gene flow
Founder events during the colonization of Ireland may have resulted in stochastic loss of genetic variation and divergence among populations. On the other hand, gene flow may have compensated for these processes. Populations may formerly have been continuously connected by gene flow (via pollen and seed). This is supported by the analysis of populations sampled along the north–south gradient in Ireland. With nuclear markers, we detected low and (mostly) statistically insignificant population differentiation and an absence of a distance effect (no isolation by distance) expected under colonization from south to north. Additionally, no evidence for population substructure was detected using the model‐based clustering algorithm. These observations all suggest extensive (at least historical) gene flow among populations or colonization in large numbers or both.
Recent exploitation
Extensive pollen flow may have maintained historically large effective population sizes despite recent negative demographic effects. However, levels of forest cover in Ireland are documented only for the last 400 years, when pressures were intense. Since this period corresponds to less than five oak generations, the effects of exploitation may not yet be detectable. Nevertheless, deforestation did occur prior to this period, at least since the Bronze Age (approx. 30 generations ago). To consider the potential for drift within populations, we assumed a standard model of population divergence with migration, and estimated the evolutionary divergence of subpopulations within Ireland. Given a single panmictic population split t generations ago into subpopulations of size Ne, population differentiation between subpopulations over time can be estimated by FST = 1 – e–t/Ne (Crow and Kimura, 1970). Thus, the change in effective population size expected over five generations (assuming an effective population size of 1000) is 0·005. Over 30 generations, the expected change is 0·03. Hence, given a relatively small number of generations exposed to drift and a large effective population size (maintained by gene flow and/or many colonizers), it is possible that more recent demographic effects have not yet manifested themselves.
The ameliorating effects of a large colonization pool and/or gene flow notwithstanding, observed levels of genetic differentiation in Ireland are also consistent with theoretical predictions that show an absence of founder effects when the long juvenile phase in woody plants is incorporated into migration models (Austerlitz et al., 2000; Austerlitz and Garnier‐Géré, 2003).
Admixture
Irish populations of Q. petraea may have gained additional genetic variation by admixture from the closely related pedunculate oak (Q. robur). The sharing of cytotypes within the natural distribution of these species has been interpreted as evidence for hybridization between the two taxa within glacial refugia (Ferris et al., 1993; Dumolin‐Lapègue et al., 1997). Cytotypes observed in Q. petraea in this study were also observed in one Q. robur population from central Ireland (results not shown). Hence, the two species may share common colonization routes. Both species share substantial amounts of genetic variation (Bodénès et al., 1997; Muir et al., 2000; Gömöry et al., 2001; Coart et al., 2002). This may result from shared ancestral polymorphism rather than recurrent interspecific gene flow. But even with low rates of introgression, the introduction of ‘new’ genetic variants may result in the rapid spread of advantageous alleles in heterospecific genetic backgrounds (Barton, 2001). Hence, interspecific hybridization cannot be discounted as a source of additional genetic variation.
CONCLUSIONS
Consistent with a large effective population size in the historical migrant gene pool and/or with high gene flow among populations, low population differentiation and high levels of genetic variation were observed in Ireland. Extensive pollen flow has been documented in Q. petraea, e.g. by Streiff et al. (1999). Deforestation at the level recorded in Ireland, however, can affect the number of migrants and can result in a decline in the number of effective mating partners. Studies on the effects of deforestation among insect‐pollinated trees indicate an inverse correlation between pollen flow and tree density (Dick, 2001; White et al., 2002). However, among wind‐pollinated trees, such as oak, the effective number of pollen donors, as defined by Smouse et al. (2001), can vary positively with tree density (Sork et al., 2002). Hence pollen may travel further on average among fragmented forest‐trees pollinated by insects. Nevertheless, Sork et al. (2002) infer a decline in the effective number of pollen donors in Quercus lobata (Californian valley oak) over a relatively short (55‐year) period.
Our results do not indicate historical genetic isolation of native oaks in Ireland. However, they continue to be threatened because the present tree structure is even‐aged and natural regeneration is unprotected from browsing. Recognition of an ageing demography has resulted in the initiation of restoration programs. As seed collected from native/autochthonous populations is unlikely to be inbred, as shown by the present study, this material may be used to expand the present resource. Nevertheless, the transfer of genetic material for planting should attempt to match differences in adaptive genetic variation (Ennos et al., 1998).
ACKNOWLEDGEMENTS
We are grateful to R. Munro and S. Cavers for technical assistance and to C. Lexer, F. Mitchell, C. Schlötterer and one anonymous reviewer for critical comments on the manuscript. Thanks also to D. Thompson and J. Fennessy (Coillte Teoranta) and to P. Hunter‐Blair (Northern Ireland Forest Service) for information on current forest practice, policy and research. Financial support was provided by the European Science Foundation and the Austrian Science Fund to G.M., the European Union and the Natural Environmental Research Council to A.J.L. and the Department of Education and Department of Environment (Northern Ireland) to C.C.F.
Supplementary Material
Received: 10 September 2003; Returned for revision: 14 October 2003; Accepted: 19 February 2004; Published electronically: 21 April 2004
References
- AusterlitzF, Mariette S, Machon N, Gouyon P‐H, Godelle B.2000. Effects of colonization processes on genetic diversity: differences between annual plants and tree species. Genetics 154: 1309–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AusterlitzF, Garnier‐Géré PH.2003. Modelling the impact of colonisation on genetic diversity and differentiation of forest trees: interaction of life cycle, pollen flow and seed long‐distance dispersal. Heredity 90: 282–290. [DOI] [PubMed] [Google Scholar]
- BartonNH2001. The role of hybridization in evolution. Molecular Ecology 10: 551–568. [DOI] [PubMed] [Google Scholar]
- BodénèsC,Joandet S, Laigret F, Kremer A.1997. Detection of genomic regions differentiating two closely related oak species Quercus petraea (Matt.) Liebl. and Quercus robur L. Heredity 78: 433–444. [Google Scholar]
- CoartE,Lamote V, De Loose M, VanBockstaele E, Lootens P, Roldán‐Ruiz I.2002. AFLP markers demonstrate local genetic differentiation between two indigenous oak species (Quercus robur L. and Quercus petraea (Matt.) Liebl.) in Flemish populations. Theoretical and Applied Genetics 105: 431–439. [DOI] [PubMed] [Google Scholar]
- CottrellJE, Munro RC, Tabbener HE, Gillies ACM, Forrest GI, Deans JD, Lowe AJ.2002. Distribution of chloroplast variation in British oaks (Quercus robur and Q petraea): the influence of postglacial recolonization and human management. Forest Ecology and Management 156: 181–195. [Google Scholar]
- CottrellJE, Munro RC, Tabbener HE, Milner AD, Forrest GI, Lowe AJ.2003. Comparison of fine‐scale genetic structure using nuclear microsatellites within two British oakwoods differing in population history. Forest Ecology and Management 176: 287–303. [Google Scholar]
- CrowJF, Kimura M.1970.An introduction to population genetics theory. New York: Harper and Row. [Google Scholar]
- DickCW.2001. Genetic rescue of remnant tropical trees by an alien pollinator. Proceedings of the Royal Society of London B 268: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DieringerD, Schlötterer C.2003. MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite datasets. Molecular Ecology Notes 3: 167–169. [Google Scholar]
- DumolinS, Demesure B, Petit RJ.1995. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 91: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Dumolin‐LapègueS, Demesure B, Fineschi S, Le Corre V, Petit RJ.1997. Phylogeographic structure of white oaks throughout the European continent. Genetics 146: 1475–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EnnosRA, Worrell R, Malcolm DC.1998. The genetic management of native species in Scotland. Forestry 71: 1–23. [Google Scholar]
- FerrisC, Oliver RP, Davy AJ, Hewitt GM.1993. Native oak chloroplasts reveal an ancient divide across Europe. Molecular Ecology 2: 337–344. [DOI] [PubMed] [Google Scholar]
- GömöryD,Yakovlev I, Zhelev P, Jedináková J, Paule L.2001. Genetic differentiation of oak populations within the Quercus robur/Quercus petraea complex in Central and Eastern Europe. Heredity 86: 557–563. [DOI] [PubMed] [Google Scholar]
- GoudetJ2001. FSTAT, a program to estimate and test gene diversities and fixation indices. www.unil.ch/izea/softwares/fstat.html. [Google Scholar]
- GoudetJ, Raymond M, de Meeüs T, Rousset F.1996. Testing differentiation in diploid populations. Genetics 144: 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HelgasonT, Ennos RA.1991. The outcrossing rate and gene frequencies of a native Scots pinewood population determined using isozyme markers. Scottish Forestry 45: 111–119. [Google Scholar]
- KampferS, Lexer C, Glössl J, Steinkellner H.1998. Characterization of (GA)n microsatellite loci from Quercus robur Hereditas 129: 183–186. [Google Scholar]
- KellyDL.1981. The native forest vegetation of Killarney, southwest Ireland: an ecological account. Journal of Ecology 69: 437–472. [Google Scholar]
- KeoghRM.1987. Statistics on broadleaved forest cover in Ireland. Irish Forestry 44: 146–147. [Google Scholar]
- LedigFT.1986. Heterozygosity, heterosis, and fitness in outbreeding plants. In: Soulé ME, ed. Conservation biology: the science of scarcity and diversity Sunderland MA: Sinauer, 77–104. [Google Scholar]
- MitchellFJG.1988. The vegetational history of the Killarney oakwoods, SW Ireland: evidence from fine spatial resolution pollen analysis. Journal of Ecology 76: 415–436. [Google Scholar]
- MitchellFJG.1995. The dynamics of Irish post‐glacial forests. In: Mac an tSaoir SS, ed. Wood, trees and forests in Ireland Dublin: Royal Irish Academy, 13–22. [Google Scholar]
- MitchellFJG.2003. Natural invaders: the postglacial tree colonisation of Ireland. In: Murray DA, ed. Biological invaders: the impact of exotic species Dublin: Royal Irish Academy, 2–13. [Google Scholar]
- MuirG,Fleming CC, SchlöttererC.2000. Species status of hybridizing oaks Nature 405: 1016. [DOI] [PubMed] [Google Scholar]
- OlaldeM, Herrán A, Espinel S, Goicoechea PG.2002. White oaks phylogeography in the Iberian Peninsula. Forest Ecology and Management 156: 89–102. [Google Scholar]
- PetitRJ, Aguinagalde I, de Beaulieu J‐L, Bittkau C, Brewer S, Cheddadi Ret al.2003. Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300: 1563–1565. [DOI] [PubMed] [Google Scholar]
- PetitRJ, Brewer S, Bordács S, Burg K, Cheddadi R, Coart Eet al.2002. Identification of refugia and post‐glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. Forest Ecology and Management 156: 49–74. [Google Scholar]
- PonsO, Petit RJ.1995. Estimation, variance and optimal sampling of gene diversity. I. Haploid locus. Theoretical and Applied Genetics 90: 462–470. [DOI] [PubMed] [Google Scholar]
- PritchardJK, Stephens M, Donnelly PJ.2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ProvanJ, Soranzo N, Wilson NJ, McNicol JW, Forrest GI, Cottrell J, Powell W.1998. Gene‐pool variation in Caledonian and European Scots pine (Pinus sylvestris L.) revealed by chloroplast simple‐sequence repeats. Proceedings of the Royal Society of London B 265: 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RackhamO.1995. Looking for ancient woodland in Ireland. In: Mac an tSaoir SS, ed. Wood, trees and forests in Ireland Dublin: Royal Irish Academy, 1–12. [Google Scholar]
- RaymondM, Rousset F.1995. GENEPOP (version 3.3): population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. [Google Scholar]
- SchlöttererC.1998. Microsatellites. In: Hoelzel RA, ed. Molecular genetic analysis of populations: a practical approach Oxford: Oxford University Press, 237–261. [Google Scholar]
- SmousePE, Dyer RJ, Westfall RD, Sork VL.2001. Two generation analysis of pollen flow across a landscape. I. Male heterogeneity among females. Evolution 55: 260–271. [DOI] [PubMed] [Google Scholar]
- SorkVL, Davis FW, Smouse PE, Apsit VJ, Dyer RJ, Fernandez‐M JF, Kuhn B.2002. Pollen movement in declining populations of California Valley oak, Quercus lobata: where have all the fathers gone? Molecular Ecology 11: 1657–1668. [DOI] [PubMed] [Google Scholar]
- SteinkellnerH, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A, Burg K, Glössl J.1997. Identification and characterization of (GA/CT)n – microsatellite loci from Quercus petraea Plant Molecular Biology 33: 1093–1096. [DOI] [PubMed] [Google Scholar]
- StreiffR, Ducousso A, Lexer C, Steinkellner H, Glössl J, Kremer A.1999. Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Q petraea (Matt.) Liebl. Molecular Ecology 8: 831–841. [Google Scholar]
- ThuilletA‐C, Bru D, David J, Roumet P, Santoni S, Sourdille P, Bataillon T.2002. Direct estimation of mutation rate for 10 microsatellite loci in durum wheat, Triticum turgidum (L.) Thell. ssp. durum Desf. Molecular Biology and Evolution 19: 122–125. [DOI] [PubMed] [Google Scholar]
- VoglC, Karhu A, Moran G, Savolainen O.2002. High resolution analysis of mating systems: inbreeding in natural populations of Pinus radiata Journal of Evolutionary Biology 15: 433–439. [Google Scholar]
- WeirBS, Cockerham CC.1984. Estimating F‐statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- WhiteGM, Boshier DH, Powell W.2002. Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proceedings of the National Academy of Sciences of the USA 99: 2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.