Abstract
• Background and Aims Genetic structure and variability were examined in the only three extant populations of the narrow‐endemic tree Antirhea aromatica (Rubiaceae, Guettardeae), an endangered species of the tropical forest of eastern Mexico. Patterns of genetic diversity within and among populations for adult plants and seedlings were obtained.
• Methods Allozyme electrophoresis of 15 loci was conducted and the data analysed with statistical approximation for obtaining genetic diversity, structure and gene flow.
• Key Results The mean expected heterozygosity (He) in the adult and seedling populations was 0·18 ± 0·08 and 0·20 ± 0·09, respectively. The genetic variation explained by differences among populations was 51 and 35 %, for adult and seedling populations, respectively. On average, gene flow between paired adult populations was low (Nm = 0·26 ± 0·09), compared with other trees from the tropical forest.
• Conclusions The results indicated that the populations evaluated have high genetic variability, compared with other endemic and geographically narrowly distributed plant species, in areas with high levels of environmental heterogeneity (e.g. tropical forests). The conservation implications of the results are discussed, and in this regard it is proposed that A. aromatica should be considered as an indicator species with economic potential. It is suggested that sustainable management practices should be implemented and that the areas where the species is distributed should be declared a natural reserve to ensure the species conservation.
Key words: Antirhea aromatica, endemic, genetic variability, conservation biology, tropical forest, Mexico
Introduction
The genetic structure of populations refers to the distribution of genetic variation within and among populations, and is affected by demographic factors (Antonovics and Via, 1987; Loveless and Hamrick, 1984) as well as evolutionary processes (Wright, 1951). The genetic variation within a population is considered to represent its evolutionary potential (Wright, 1978), and the range of geographical distribution is one of the major factors correlated with the genetic variability of plant populations (Hamrick and Godt 1996a, b; Savolainen and Kuittien, 2000). Thus, genetic variation has implications for conservation at the species level (Holsinger et al., 1999; Lande, 1999), and the assessment of genetic variability is the first step in evaluating the long‐term conservation status of species in natural conditions. This is particularly important in plant species with low population sizes exposed to the effects of inbreeding and genetic drift (Barrett and Kohn 1991; Frankham, 1995).
Plant species with restricted geographical distributions tend to have lower levels of genetic variation than their more widespread congeners (Gitzendanner and Soltis, 2000). However, high gene diversity has been reported for the rare ferns Adenophorus periens (Ranker, 1994), and Polystichum otomasui (Maki and Asada, 1998), the endangered tree Caesalpinia echinata (Cardoso et al., 1998), an endangered pine Pinus rzedowskii (Delgado et al., 1999), the rare Mexican pinyon pine Pinus maximartinezii (Ledig et al., 1999), the endemic Agave victoriae‐reginae (Martínez‐Palacios et al., 1999), three endemic plants from Florida (Eryngium cuneifolium, Hypericum cumulicola and Liatris ohlingerae; Dolan et al., 1999), the annual endemic Warea carteri (Evans et al., 2000), the endemics Iris cristata and I. lacustris (Hannan and Orick, 2000), the narrow and endemic species Antirrhinum charidemi and A. valentinum (Mateu‐Andrés and Segarra‐Moragues, 2000), the endemic monoecious shrub Brongniartia vazquezii, of tropical dry forests of Central Mexico (González‐Astorga and Núñez‐Farfán, 2001), Viola palmensis endemic of Canary Islands (Batista and Sosa, 2002), and the cycad Dioon edule of eastern Mexico (González‐Astorga et al., 2003).
Conservation programs for long‐lived tropical trees must take into account the ecological and genetic relevance of environmental conditions fluctuating over large periods of time (Alvarez‐Buylla et al., 1996b; Lande, 1999; Hedrick, 2001).
Antirhea aromatica (Rubiaceae, Guettardeae) is a monoecious tree, of 6–15 m height and a diameter at breast height of 10–30 cm. It takes 7–10 years for the species to reach maturity, and the plant’s lifespan is approx. 150 years. Local people of the region use the fruits and the bark of A. aromatica as a natural remedy for dental diseases (Castillo‐Campos, 1995). Active components with antiseptic, antioxidant and antibiotic principles have been isolated from a congener, A. acutata (Lee et al., 2001). The species A. aromatica is endemic to central Veracruz, with a highly restricted geographical distribution. The species has been registered only in the type locality area, in Jalcomulco and Apazapan, Veracruz, from 350–500 m a.s.l. (Castillo‐Campos and Lorence, 1985). Antirhea aromatica inhabits remnant patches of tropical lowland rainforest (sensu Miranda and Hernández, 1963) with a population density of approx. 200 individual adults ha–1, and these populations cover an area of approximately 20 ha (Castillo‐Campos, 1995). This species blooms from July to September (43·4 ± 7·2 flowers per plant) during the summer. Its white aromatic flowers (6·4 ± 0·3 cm long) are visited by moths, bumblebees (Bombus sp.) and bats; the seeds are dispersed by bats. The aim of the present study was to determine the patterns of genetic variation and differentiation among the three existing populations of A. aromatica. Additionally, both adults and seedlings were analyzed and compared. Finally, the population size of the species and its fragmented distribution offers the opportunity to determine the relationship between the genetic structure, population size and geographic isolation. It is hypothesized that genetic diversity will be low and genetic differentiation high, with a subsequent decrease in gene flow of the extant populations.
Methods
Study sites
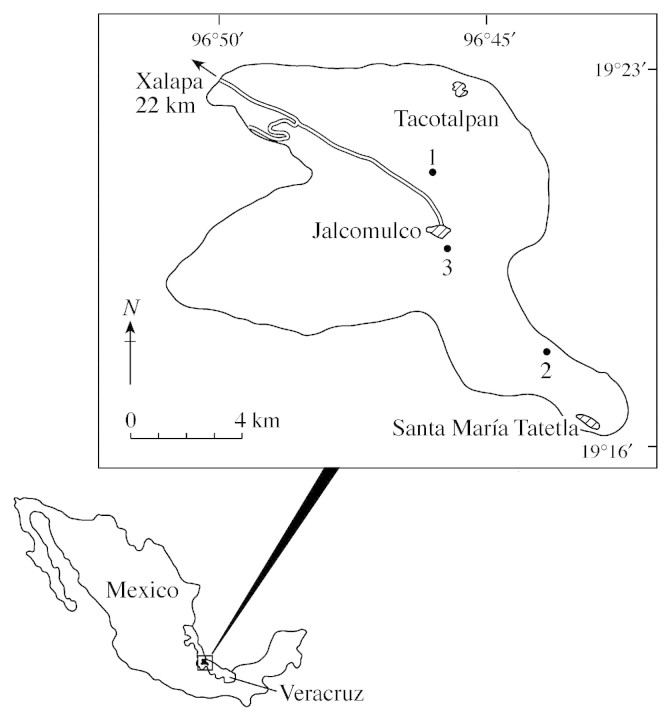
The study was conducted during 1998–99, in the localities of Jalcomulco and Apazapan (96°41′–99°08′W; 19°18′–19°27′N) in central Veracruz, in Eastern Mexico (Fig. 1) (Castillo‐Campos, 1995). The study area is a fragmented landscape of lowland tropical rainforest (sensu Miranda and Hernández, 1963) surrounded by roads, cultivated fields and pasturelands. The climate is semi‐warm humid [(A)C(m)] following the classification of Köeppen (1948), the mean annual precipitation ranges from 1200–1500 mm, and the mean annual temperature ranges from 22–24 °C (García, 1988).
Fig. 1. Geographical distribution of populations examined of Antirhea aromatica in Veracruz, Mexico.
Sample collection
Tissue sampling was done in the three extant populations of Antirhea aromatica Castillo‐Campos & Lorence in Jalcomulco, Veracruz, Mexico. Populations are separated by distances that range from 2·7–15·5 km. The sampled populations of A. aromatica encompassed the total geographic range of the species (Fig. 1). Fully expanded young leaves were collected from 40 reproductive individuals in each population. Simultaneously, leaves of 40 seedlings of each population were collected. This tissue was transported in ice‐filled containers, and then stored in a freezer at –70 °C until extraction for electrophoretic analysis.
Electrophoresis
Multilocus genotypes of 40 mature individuals and 40 seedlings from each population were determined through horizontal starch gel electrophoresis (12 % w/v). Allozymic variation was scored at 15 loci for each individual plant, nine of which were polymorphic: malate‐dehydrogenase (E.C. 1.1.1.37, loci Mdh1 and Mdh2), esterase (E.C. 3.1.1, loci Est1, Est2 and Est3), phosphoglucoisomerase (E.C. 5.3.1.9, loci Pgi1 and Pgi2), glutamate oxaloacetate transaminase (E.C. 2.6.1.1, locus Got), and phosphoglucomutase (E.C. 5.2.2, locus Pgm). The remaining six were monomorphic: 6‐phosphogluconate dehydrogenase (E.C. 1.1.1.44, locus 6Pgd), diaphorase (E.C. 1.6.99.‐, loci Dia1 and Dia2), isocitrate dehydrogenase (E.C. 1.1.1.41, locus Idh), leucine aminopeptidase (E.C. 3.4.11.1, locus Lap) and peroxidase anodic (E.C. 1.11.1.7, locus Apx). The extraction buffer (tris‐HCl pH 7·5, sucrose, PVP‐40, mercaptoethanol, ascorbic acid, diethyldithiocarbamate, bovine serum albumin, sodium metabisulphite and sodium tetraborate; Wendel and Weeden, 1989) was added to dissolve and stabilize the enzyme extracts, which were stored on filter paper wicks at –70 °C until used for analyses. The buffers (gel and electrode) used were histidine pH 5·7, and citric acid (Soltis et al., 1983). Electrophoresis was carried out at 4 °C over 6 h (constant current of 70 mA, and voltage of 200 V).
Statistical methods
The bands from each allozyme system were assigned to alleles and genotypes based on theoretical expectations and observed banding patterns. The TFPGA 1.3 package (Miller, 1997) was used to obtain the genetic estimators from the data analysis. The genotypic frequencies obtained were used to calculate observed mean heterozygosity (Ho) and allelic frequencies. Allelic frequencies at each population were used to estimate the mean number of alleles per locus (A), the average proportion of polymorphic loci (P), and expected mean heterozygosity (He), based on Hardy–Weinberg expectations (Hartl and Clark, 1997). The significance of estimators was obtained by Monte Carlo methods (Weir, 1990).
Partitioning of genetic variability was done by using F‐statistics (Wright, 1965, 1978), which were calculated according to the formula of Weir and Cockerham (1984) that estimates genetic structure by partitioning variation in the same way as a regular analysis of variance. The θ statistic (analogous to Fst) estimates populations’ divergence through allele frequencies, whereas f (similar to Fis) and F (similar to Fit) estimate heterozygote excess (<0) and deficit (>0) relative to Hardy–Weinberg expectations in local populations and the total set of populations, respectively. To determine whether f and F estimations for each locus were significantly different from zero, Chi‐square statistics [χ2 = F(2N) (k – 1)] were obtained, with k(k – 1)/2 degrees of freedom, where N is the sample size and k the number of alleles (Weir, 1990). To determine the significance of the θ statistic per locus, the chi‐square statistic was used: χ2 = (2N) θ(k – 1), with (k – 1) (n – 1) degrees of freedom, where n is the number of populations (Workman and Niswander, 1970). The confidence intervals (at 95%) of the F‐statistics were obtained by bootstrapping over loci for the multilocus estimate and jackknifing over populations for the single‐locus estimates (Weir and Cockerham, 1984; Weir, 1990). The average gene flow among populations (Nm) was estimated from θ‐values, as θ = 1/(4Nmα + 1), where α =[n/(n – 1)]2 and n is the number of populations (Crow and Aoki, 1984). Nm is interpreted as the number of migrants per generation between two given populations (Slatkin 1993, 1994).
Results
Genetic variation
The average number of alleles per locus was 1·76 ± 0·102 and 1·64 ± 0·102 for adults and seedlings of A. aromatica, respectively (Table 1). The t‐test indicated no significant differences between averages of number of alleles per locus among adults and progeny (t = 1·7, df = 44, P = 0·09). Allelic frequencies for 15 loci were scored for each individual plant (Table 1). In the adults, the percentage of polymorphic loci per population varied from 33·3 % (population 2) to 60 % (populations 1 and 3), with an average of 51·1 %. In seedling populations, the percentage of polymorphic loci varied from 46·6 % (population 3) to 60 % (populations 1 and 2), with an average of 55·5 % (Table 2).
Table 1.
Allelic frequencies of 15 enzymatic loci in individuals (adults and seedlings) of three populations of Antirhea aromatica in Veracruz, Mexico.
| Populations | ||||
| Locus | Allele | 1 | 2 | 3 |
| Mdh1 | ||||
| Adults | a | 0·1714 | 0·1471 | 0·7778 |
| b | 0·8286 | 0·0588 | 0·0370 | |
| Seedlings | c | 0·0000 | 0·7941 | 0·1852 |
| a | 0·6282 | 0·1719 | 0·9375 | |
| b | 0·3718 | 0·1094 | 0·0625 | |
| c | 0·0000 | 0·7188 | 0·0000 | |
| Mdh2 | ||||
| Adults | a | 0·8649 | 0·1029 | 0·2115 |
| b | 0·1351 | 0·0588 | 0·0769 | |
| c | 0·0000 | 0·8382 | 0·7115 | |
| Seedlings | a | 0·6875 | 0·1000 | 1·0000 |
| b | 0·3000 | 0·2333 | 0·0000 | |
| c | 0·0125 | 0·6667 | 0·0000 | |
| Est1 | ||||
| Adults | a | 0·3194 | 0·9559 | 0·8889 |
| b | 0·0833 | 0·0441 | 0·1111 | |
| c | 0·5972 | 0·0000 | 0·0000 | |
| Seedlings | a | 0·7143 | 0·3750 | 0·0667 |
| b | 0·2857 | 0·6250 | 0·9333 | |
| Est2 | ||||
| Adults | a | 0·5294 | 0·9722 | 0·6800 |
| b | 0·4706 | 0·0278 | 0·3200 | |
| Seedlings | a | 0·7639 | 0·1452 | 0·8667 |
| b | 0·2361 | 0·8548 | 0·1333 | |
| Est3 | ||||
| Adults | a | 0·1111 | 0·0714 | 0·5000 |
| b | 0·8889 | 0·9286 | 0·5000 | |
| Seedlings | a | 0·6447 | 0·3833 | 0·8235 |
| b | 0·3553 | 0·6167 | 0·1765 | |
| Pgi1 | ||||
| Adults | a | 0·1351 | 0·9857 | 0·3000 |
| b | 0·8649 | 0·0143 | 0·1200 | |
| c | 0·0000 | 0·0000 | 0·5800 | |
| Seedlings | a | 0·5811 | 0·1452 | 0·9375 |
| b | 0·4189 | 0·8548 | 0·0625 | |
| Pgi2 | ||||
| Adults | a | 0·8243 | 0·9286 | 0·2500 |
| b | 0·1757 | 0·0714 | 0·7500 | |
| Seedlings | a | 0·6389 | 0·3833 | 0·0938 |
| b | 0·3611 | 0·6167 | 0·9063 | |
| Got | ||||
| Adults | a | 0·7027 | 0·6250 | 0·3077 |
| b | 0·2973 | 0·3750 | 0·6923 | |
| Seedlings | a | 0·6410 | 0·6563 | 0·8000 |
| b | 0·3590 | 0·3438 | 0·2000 | |
| Pgm | ||||
| Adults | a | 0·0735 | 0·9714 | 0·2222 |
| b | 0·9265 | 0·0286 | 0·2593 | |
| c | 0·0000 | 0·0000 | 0·5185 | |
| Seedlings | a | 0·5921 | 0·2813 | 0·9706 |
| b | 0·4079 | 0·7187 | 0·0294 | |
| 6Pgd | ||||
| Adults | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Seedlings | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Dia 1 | ||||
| Adults | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Seedlings | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Dia 2 | ||||
| Adults | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Seedlings | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Idh | ||||
| Adults | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Seedlings | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Lap | ||||
| Adults | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Seedlings | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Apx | ||||
| Adults | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Seedlings | a | 1·0000 | 1·0000 | 1·0000 |
| b | 0·0000 | 0·0000 | 0·0000 | |
| Mean no. of alleles/locus (± SD) | ||||
| Adults | 1·67 (0·617) | 1·73 (0·704) | 1·87 (0·834) | |
| Seedlings | 1·67 (0·617) | 1·73 (0·704) | 1·53 (0·516) | |
Table 2.
Levels of genetic variation for adult and seedling populations of A. aromatica. A, mean number of alleles per locus; P, percentage of polymorphic loci; Ni, average sample size; Ho and He are the observed and expected mean heterozygosity, respectively.
| Population | A | P | N i | H o | H e |
| 1 | |||||
| Adults | 1·67 | 60·00 | 36·33 | 0·1549 | 0·1884 |
| Seedlings | 1·67 | 60·00 | 38·53 | 0·2044 | 0·2682 |
| 2 | |||||
| Adults | 1·73 | 33·33 | 35·73 | 0·0847 | 0·1056 |
| Seedlings | 1·73 | 60·00 | 31·46 | 0·2309 | 0·2466 |
| 3 | |||||
| Adults | 1·87 | 60·00 | 26·46 | 0·1696 | 0·2607 |
| Seedlings | 1·53 | 46·66 | 16·40 | 0·0775 | 0·0952 |
| Mean (± SD) | |||||
| Adults | .1·76 (0·102) | .51·11 (15·40) | .32·84 (5·53) | .0·1364 (0·045) | .0·1849 (0·078) |
| Seedlings | .1·64 (0·102) | .55·55 (7·70) | .28·80 (11·30) | .0·1709 (0·082) | .0·2033 (0·094) |
Observed mean heterozygosity was 0·14 ± 0·04 (range 0·08–0·17) and 0·17 ± 0·08 (range 0·07–0·23) for the adult and seedling populations, respectively. Expected mean heterozygosity was 0·18 ± 0·08 vs 0·20 ± 0·09, for the adult and seedling populations, respectively (Table 2).
Genetic structure
The Wright’s F‐statistics, F (similar to Fit) and f (similar to Fis), were positive and significantly different from zero for all polymorphic loci (P < 0·05) in both adult and seedling populations, indicating inbreeding (Table 3). Similarly, all polymorphic loci showed values of θ (similar to Fst) significantly different from zero (P < 0·05). The mean F was higher for the adult than for the seedling population (0·64 ± 0·049 vs.0·46 ± 0·04; F(1,28) = 1330, P < 0·00001). Similarly, the f‐statistic was higher for adult than for seedling populations (F(1,28) = 101, P < 0·00001) (Table 3).
Table 3.
Wright’s F‐statistics for adult and seedling populations of Antirhea aromatica in Veracruz, Mexico.
| Loci | F | q | f |
| Mdh1 | |||
| Adults | 0·6565** | 0·4867** | 0·3308** |
| Seedlings | 0·4563* | 0·3261* | 0·1932** |
| Mdh2 | |||
| Adults | 0·6386** | 0·4961** | 0·2827* |
| Seedlings | 0·4503* | 0·3229* | 0·1881* |
| Est1 | |||
| Adults | 0·6640** | 0·5089** | 0·3158** |
| Seedlings | 0·4686** | 0·3489 | 0·1838* |
| Est2 | |||
| Adults | 0·6397** | 0·5267* | 0·2387* |
| Seedlings | 0·4477** | 0·3238* | 0·1832* |
| Est3 | |||
| Adults | 0·6420** | 0·5216** | 0·2517** |
| Seedlings | 0·4620** | 0·3666* | 0·1506* |
| Pgi1 | |||
| Adults | 0·6213** | 0·4735* | 0·2807** |
| Seedlings | 0·4531* | 0·3331* | 0·1800** |
| Pgi2 | |||
| Adults | 0·6277** | 0·5086** | 0·2423** |
| Seedlings | 0·4764* | 0·3584* | 0·1840** |
| Got | |||
| Adults | 0·6379* | 0·5439** | 0·2060** |
| Seedlings | 0·4980* | 0·3760* | 0·1955* |
| Pgm | |||
| Adults | 0·6037** | 0·4729* | 0·2481** |
| Seedlings | 0·4744* | 0·3467* | 0·1955* |
| 6Pgd | |||
| Adults | 0·6370 | 0·5050 | 0·2666 |
| Seedlings | 0·4654 | 0·3450 | 0·1838 |
| Dia 1 | |||
| Adults | 0·6370 | 0·5050 | 0·2666 |
| Seedlings | 0·4654 | 0·3450 | 0·1838 |
| Dia 2 | |||
| Adults | 0·6370 | 0·5050 | 0·2666 |
| Seedlings | 0·4654 | 0·3450 | 0·1838 |
| Idh | |||
| Adults | 0·6370 | 0·5050 | 0·2666 |
| Seedlings | 0·4654 | 0·3450 | 0·1838 |
| Lap | |||
| Adults | 0·6370 | 0·5050 | 0·2666 |
| Seedlings | 0·4654 | 0·3450 | 0·1838 |
| Apx | |||
| Adults | 0·6370 | 0·5050 | 0·2666 |
| Seedlings | 0·4654 | 0·3450 | 0·1838 |
| Mean | |||
| Adults | 0·6384 | 0·5107 | 0·2690 |
| Seedlings | 0·4673 | 0·3477 | 0·1841 |
| Standard deviation | |||
| Adults | 0·0491 | 0·0664 | 0·1085 |
| Seedlings | 0·0439 | 0·0534 | 0·0374 |
| Confidence interval to 95% | |||
| Adults | 0·5410–0·7258 | 0·3496–0·6126 | 0·0740–0·4492 |
| Seedlings | 0·3582–0·5368 | 0·2319–0·4336 | 0·1275–0·2553 |
* P < 0·05; ** P < 0·01.
The average genetic differentiation among adult populations (θ = 0·51) was higher than for seedling populations (θ = 0·35). Thus 51 and 35 % of the genetic variation for adults and seedlings, respectively, is due to differences among populations of A. aromatica (Table 3). Also, the exact tests for population differentiation of Raymond and Rousset (1995), indicated significant differences among adult (χ2 = 178·3; df = 18; P < 0·00001), and seedling (χ2 = 161·2; df = 18; P < 0·00001) populations. The θ‐values were different among loci in both populations (adults, range 0·47–0·54; and seedlings, range 0·32–0·38), and suggest that genetic drift and inbreeding have been the dominant differentiating processes. Both estimations are significantly different from zero, and there were differences between them (F(1,28) = 688, P = 0·00001).
Gene flow
Indirect estimates of gene flow (Nm) for A. aromatica indicate that an average of 0·26 ± 0·08 migrant individuals per generation between populations pairs. These are relatively low values of gene flow with respect to other plant species with similar reproductive systems. The lowest Nm value was obtained between populations 1 and 2 (Nm = 0·16) separated by 15·55 km, and the highest one between populations 2 and 3 (Nm = 0·31) separated by 2·74 km.
Discussion
Tropical endemic trees are vulnerable to forest fragmentation because of their low densities, complex demographic dynamics, high genetic differentiation (θ ≈ 1), and self‐incompatibility systems (Bawa et al., 1985; Barrett and Kohn, 1991; Alvarez‐Buylla and Garay, 1994; Schemske et al., 1994; Alvarez‐Buylla et al., 1996b). Also, tropical forest fragmentation is likely to decrease gene flow, increase inbreeding and therefore produce a high differentiation among remnant populations (Alvarez‐Buylla et al., 1996b; González‐Astorga and Núñez‐Farfán, 2001). This would be particularly the case for A. aromatica where one would expect a low genetic diversity. Contrary to this expectation, we found high levels of genetic diversity for A. aromatica, for both adults and seedlings. In fact, the values found in this species are among the highest ones recorded for tropical forest trees (Eguiarte et al., 1992; Alvarez‐Buylla and Garay, 1994; Cardoso et al., 1998; Loveless et al., 1998; Lee et al., 2002; Ledig et al., 2002), and especially with respect to those found in fragmented environments (Hall et al., 1996; Cascante et al., 2002).
The mean percentage of polymorphic loci in A. aromatica was higher (51·1 and 55·5 %, adults and seedlings, respectively) than that reported for other long‐lived perennial and endemic plant species (48·1 %; Hamrick and Godt, 1996a). The mean expected heterozygosity within populations for A. aromatica was also higher (0·185 and 0·203, adults and seedlings, respectively), than that reported for other regionally distributed (sensu Hamrick and Godt, 1989), tropical long‐lived trees, and even higher than those for temperate long‐lived trees (0·125 and 0·145, respectively; Hamrick et al., 1994). It is also higher than those of other long‐lived perennial and endemic species (0·105; Hamrick and Godt, 1996a). This shows that A. aromatica has exceptionally high genetic diversity and variability, despite its low population density (cf. Young et al., 1996; González‐Astorga and Núñez‐Farfán, 2001). This phenomenon is likely to be associated with the reproductive system. It has been reported that other Rubiaceae have a pre‐zygotic self‐incompatibility crossing system (Anderson, 1973; Richards, 1997; Faivre and McDade, 2001) that reduces inbreeding, and loss of genetic variation. The heterostyly observed in A. aromatica (J. González‐Astorga, personal observation) denotes the existence of a self‐incompatibility reproductive system (Sobrevila et al., 1983; Richards and Kortur, 1993; Riveros et al., 1995), even though outcrossing rate [t = (1 – f)/(1 + f); sensu Allard et al., 1968] in adult populations of A. aromatica indicates that 57 % of the offspring are a product of exogamy. This result contrasts with an average outcrossing rate of 88 ± 12 % for of 30 species of tree and shrubs reported by Eguiarte (1990), and 90 ± 5 % reported by Boshier (2000) for seven tropical trees. However, because the t‐value is an indirect estimate, caution has to be taken in its interpretation (Ledig et al., 1997).
Pollinator efficiency allows the establishment of a stable genetic neighbourhood supporting an adequate genetic variability within populations, and is reflected in the high genetic diversity found in A. aromatica populations (P = 51·1 and He = 0·185 in adults, and P = 55·5 and He = 0·203 in seedlings).
The inbreeding values found in A. aromatica were high when compared to other tropical species (Eguiarte, 1990; Boshier, 2000). These values were significantly greater in adults that in seedlings. The relatively low inbreeding observed in seedlings suggests that the individuals sampled came from an exceptional fruit‐setting year for many adult trees, which could be due to a random sampling effect from the gene pools among cohorts (Husband and Schemske, 1996), or a past cornucopia effect (cf. Sazima et al., 2001; Leite and da Encarnacao, 2002).
On the other hand, the high genetic differentiation found among populations (θ = 0·51, adults; and θ = 0·35, seedlings) may be due to two processes: in the immediate‐term to reduced pollinator‐efficiency as a result of flowering asynchrony between populations (cf. Murren, 2002), and in the longer‐term to reduced spatial distribution and increased population isolation due to fragmentation (Young et al., 1996), which in turn could restrain gene flow and disrupt the demographic structure of formerly stable populations (Loveless and Hamrick, 1984). Alternatively, the reduced gene flow detected in A. aromatica adult populations may be due to low seed dispersal efficiency. Frugivore bats deposit massive amounts of seeds of the same mother tree under very few resting trees, such as Brosinium alicastrum, Bursera simaruba, Hyperbaena mexicana, Manilkara zapota and Protium copal (Castillo‐Campos and Lorence, 1985; Castillo‐Campos, 1995). Although we did not evaluate gene flow in seedlings, we would expect a similar pattern to the one observed in the adults if we assume that most of the seedlings would eventually reach maturity.
In conclusion, our results show that the three extant populations of A. aromatica present a relatively high genetic diversity when compared with other plants with similar attributes. The conservation implications for the species are evident, since the species has only been found in three forest patches of a very scarce vegetation type in Mexico. In this region the human population has continuously increased since the 16th century (de la Madrid et al., 1988). This has resulted in the extensive cultivation of agricultural crops, initially such as sugar cane and later mango and coffee plantations, with the subsequent fragmentation of the original distribution range of A. aromatica (Castillo‐Campos, 1995). The isolation and reduction of the species’ populations have reduced intrapopulation gene flow and have generated a systematic process of genetic isolation. However, our results suggest that A. aromatica has genetically viable populations, and at present the main threats are primarily associated with changes in the environment due to human activities.
We suggest that this tree species should be considered an indicator species (sensu Noss, 1990) with unexplored economic potential (Given, 1994). The preservation of the extant populations of A. aromatica through the creation of a nature reserve would be ideal. In addition, we would recommend the implementation of management practices by local people in order to reinforce conservation of the species, such as is the case for the conservation of the cycad Dioon edule by means of sustainable utilization in other regions close to our study area (Vovides and Iglesias, 1994; Vovides et al., 2002).
Acknowledgments
This study was supported partially by four grants: CONACyT 34077‐N 902‐10, CONACyT‐SEMARNAT 2002‐C01‐0183, FMCN, A.C. B‐2·00/013 to J.G.‐A., and 902‐14 to G. C.‐C., of the Instituto de Ecología, AC. We thank Jesús Vargas García for technical assistance in the laboratory. Our sincere gratitude is expressed to Mikael Hedrén, Luis Eguiarte, Daniel Piñero, Andrew P. Vovides, Andrea Cruz‐Angón and two anonymous reviewers for their comments to improve the manuscript. Alejandro Flores‐Palacios, Miriam Ferrer, Maria Luisa Martínez, Víctor Luna, José García‐Franco and Israel Acosta provided tireless assistance in the field.
Received: 25 April 2003; Returned for revision: 16 October 2003; Accepted: 23 December 2003 Published electronically: 31 March 2004
References
- AllardRW, Jain SK, Workman PL.1968. The genetics of inbreeding populations. Advances of Genetics 14: 55–131. [Google Scholar]
- Alvarez‐BuyllaER, Garay AA.1994. Population genetic structure of Cecropia obtusifolia, a tropical pioneer species. Evolution 48: 437–453. [DOI] [PubMed] [Google Scholar]
- Alvarez‐BuyllaER, Chaos A, Piñero D, Garay AA.1996a. Demographic genetics of a pioneer tropical tree species: patch dynamics, seed dispersal, and seed banks. Evolution 50: 1155–1166. [DOI] [PubMed] [Google Scholar]
- Alvarez‐BuyllaER, García‐Barrios R, Lara‐Moreno C, Martínez‐Ramos M.1996b. Demographic and genetic models in conservation biology: Applications and perspectives for tropical rain forest tree species. Annual Review of Ecology and Systematics 27: 387–421. [Google Scholar]
- AndersonWR.1973. A morphological hypothesis for the origin of heterostyly in the Rubiaceae. Taxon 22: 537–542. [Google Scholar]
- AntonovicsJ, Via S.1987. Genetic influences on the distribution and abundance of plants. In: Devy AJ, Hutchings MJ, Watkinson AR, eds. Perspectives on plant population biology Sunderland, MA: Sinauer Associates, 185–203. [Google Scholar]
- BarrettSCH, Kohn JR.1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE, eds. Genetic and conservation of rare plants New York: Oxford University Press, 3–30. [Google Scholar]
- BatistaF, Sosa PA.2002. Allozyme diversity in natural populations of Viola palmensis Webb & Berth. (Violaceae) from La Palma (Canary Islands): implications for conservation genetics. Annals of Botany 90: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BawaKS, Perry DR, Beach JH.1985. Reproductive biology of tropical lowland rainforest trees. 1. Sexual systems and incompatibility mechanisms. American Journal of Botany 72: 331–345. [Google Scholar]
- BoshierDH.2000. Mating systems. In: Young A, Boshier D, Boyle T, eds. Forest conservation genetics Australia: CABI Publishing, 63–79. [Google Scholar]
- CardosoMA, Provan J, Powell W, Ferreira PCG, de Oliveira DE.1998. High genetic differentiation among remnant populations of the endangered Caesalpinia echinata Lam. (Leguminoseae‐Caesalpinioideae). Molecular Ecology 7: 601–608. [Google Scholar]
- CascanteA, Quesada M, Lobo JJ, Fuchs EA.2002. Effects of dry tropical forest fragmentation on the reproductive success and genetic structure of the tree Samanea saman Conservation Biology 16: 137–147. [DOI] [PubMed] [Google Scholar]
- Castillo‐CamposG.1995.Ecología del paisaje del municipio de Jalcomulco, Veracruz. MSc thesis. UNAM, México. [Google Scholar]
- Castillo‐CamposG, Lorence DH.1985.Antirhea aromatica (Rubiaceae, Guettaradeae), a new species for Veracruz, México. Annals of Missouri Botanical Garden 72: 268–271. [Google Scholar]
- CrowJF, Aoki K.1984. Group selection for a polygenic behavioral trait: estimating the degree of population subdivision. Proceedings of the National Academy of Sciences of the United States of America 81: 6073–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la MadridHM, Bartlett DM, Gutierrez BF, Olmedo RC, González AS.1988.Los Municipos de Veracruz. México: Colección, Encyclopedia de los Municipios de México. [Google Scholar]
- DelgadoP, Piñero D, Chaos A, Pérez‐Nasser N, Alvarez‐Buylla ER.1999. High population differentiation and genetic variation in the endangered Mexican pine Pinus rzedowskii (Pinaceae). American Journal of Botany 86: 669–676. [PubMed] [Google Scholar]
- DolanRW, Yahr R, Menges E S, Halfhill MD.1999. Conservation implications of genetic variation in three rare species endemic to Florida rosemary scrub. American Journal of Botany 86: 1556–1562. [PubMed] [Google Scholar]
- EguiarteLE.1990.Genética de poblaciones de Astrocaryum mexicanum Liebm. en los Tuxtlas, Veracruz Ph.D. thesis. UNAM, México. [Google Scholar]
- EguiarteLE, Perez‐Nasser N, Piñero D.1992. Genetic structure, outcrossing rate and heterosis in Astrocaryum mexicanum (tropical palm): implications for evolution and conservation. Heredity 69: 217–228. [Google Scholar]
- EvansMEK, Dolan RW, Menges ES, Gordon DR.2000. Genetic diversity and reproductive biology in Warea carteri (Brassicaceae), a narrowly endemic Florida scrub annual. American Journal of Botany 87: 372–381. [PubMed] [Google Scholar]
- FaivreAE, McDade LA.2001. Population‐level variation in the expression of heterostyly in three species of Rubiaceae: does reciprocal placement of anthers and stigmas characterize heterostyly. American Journal of Botany 88: 841–853. [PubMed] [Google Scholar]
- FrankhamR.1995. Conservation genetics. Annual Review of Genetics 29: 305–327. [DOI] [PubMed] [Google Scholar]
- GarcíaE.1988.Modificaciones del sistema de clasificación climática de Köeppen México, DF: Instituto de Geografía, UNAM. [Google Scholar]
- GitzendannerMA, Soltis PS.2000. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany 87: 783–792. [PubMed] [Google Scholar]
- GivenDR.1994.Principles and practice of plant conservation. Portland Oregon, Timber Press. [Google Scholar]
- González‐AstorgaJ, Núñez‐Farfán J.2001. Effect of habitat fragmentation on the genetic structure of the narrow endemic Brongniartia vazquezii Evolutionary Ecology Research 3: 861–872. [Google Scholar]
- González‐AstorgaJ, Vovides AP, Ferrer MM, Iglesias C.2003. Population genetics of Dioon edule Lindl. (Zamiaceae, Cycadales): biogeographical and evolutionary implications. Biological Journal of the Linnean Society 80: 457–467. [Google Scholar]
- HallP, Walker S, Bawa KS.1996. Effect of forest fragmentation on genetic diversity and mating system in a tropical tree, Pithecellobium elegans Conservation Biology 10: 757–768. [Google Scholar]
- HartlDL, Clark AG.1997.Principles of population genetics. 3rd edn. Sunderland, MA: Sinauer Associates. [Google Scholar]
- HamrickJL, Godt MJW.1989. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding and genetic resources Sunderland, MA: Sinauer, 43–63. [Google Scholar]
- HamrickJL, Godt MJW.1996a. Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL, eds. Conservation genetics. Case histories from nature New York: Chapman & Hall, 281–304. [Google Scholar]
- HamrickJL, Godt MJW.1996b. Effects of the history traits on genetic diversity in plants. Philosophical Transactions of the Royal Society of London Biological Sciences 351: 1291–1298. [Google Scholar]
- HamrickJL, Schnabel A, Wells PV.1994. Distribution of genetic diversity within and among populations of Great Basin conifers. In: Harper KT, St Clair LL, Thorne KH, Hess WW, eds. Natural history of the Colorado Plateau and Great Basin Niwot, CO, USA: University of Colorado Press, 147–161. [Google Scholar]
- HannanGL, Orick MW.2000. Isozyme diversity in Iris cristata and the threatened glacial endemic I. lacustris (Iridaceae). American Journal of Botany 87: 293–301. [PubMed] [Google Scholar]
- HedrickPW.2001. Conservation genetics: where are we now? Trends in Ecology and Evolution 16: 629–636. [Google Scholar]
- HolsingerKE, Mason‐Gamer RJ, Whitton J.1999. Genes, demes, and plant conservation. In: Landweber LA, Dobson AP, eds. Genetic and extinction of species Princeton, NJ: Princeton University Press, 23–46. [Google Scholar]
- HusbandBC, Schemske DW.1996. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution 50: 54–70. [DOI] [PubMed] [Google Scholar]
- KöeppenW.1948.Climatología. México: Fondo de Cultura Económica. [Google Scholar]
- LandeR.1999. Extinction risks from anthropogenic, ecological, and genetic factors. In: Landweber LA, Dobson AP, eds. Genetic and extinction of species. Princeton, NJ: Princeton University Press, 1–22. [Google Scholar]
- LeeD, Park EJ, Cuendent M, Axelrod F, Chavez PI, Fong HHS, Pezzuto JM, Kinghorn, AD.2001. Cyclooxygenase‐ inhibitor and antioxidant constitutes of aerial parts of Antirhea acutata Bioorganic & Medicinal Chemistry Letters 11: 1565–1568. [DOI] [PubMed] [Google Scholar]
- LedigFT. Jacob‐Cervantes V, Hodgskiss PD, Eguialuz‐Pineda T.1997. Recent evolution and divergence among populations of rare Mexican endemic, Chihuahua spruce, following Holocene climatic warming. Evolution 51: 91–99. [DOI] [PubMed] [Google Scholar]
- LedigFT, Conkle MT, Bermejo‐Vázquez B, Eguialuz‐Pineda T, Hodgskiss PD, Johnson DR, Dvorak WS.1999. Evidence for an extreme bottleneck in a rare Mexican pinyon: genetic diversity, disequilibrium, and the mating system in Pinus maximartinezii Evolution 53: 91–99. [DOI] [PubMed] [Google Scholar]
- LedigFT, Hodgskiss PH, Jacob‐Cervantes V.2002. Genetic diversity, mating system, and conservation of a Mexican subalpine relict, Picea mexicana Martínez. Conservation Genetics 3: 113–122. [Google Scholar]
- LeeSL, Ng KKS, Saw LG, Norwati A, Salwana MHS, Lee CT, Norwati M.2002. Population genetics of Intsia palembanica (Leguminoseae) and genetic conservation of Virgin Jungle in Peninsular Malaysia. American Journal of Botany 89: 447–459. [DOI] [PubMed] [Google Scholar]
- LeiteIRD, da Encarnacao CRF.2002. Phenology of coconut on the Coastal Zone of Pernambuco, Brazil. Pesquisa Agropecuaria Brasileira 37: 745–752. [Google Scholar]
- LovelessMD, Hamrick JL.1984. Ecological determinants of genetic structure in plant populations. Annual Review of the Ecology and Systematics 15: 65–95. [Google Scholar]
- LovelessMD, Hamrick JL, Foster RB.1998. Population structure and mating system in Tachigali versicolor, a monocarpic neotropical tree. Heredity 81: 134‐ 143. [Google Scholar]
- MakiM, Asada Y.1998. High genetic variability revealed by allozymic loci in the narrow endemic fern Polystichum otomasui (Dryopteridaceae). Heredity 80: 604–610. [Google Scholar]
- Martínez‐PalaciosA, Eguiarte LE, Furnier GR.1999. Genetic diversity of the endangered endemic Agave victoriae‐reginae (Agavaceae) in the Chihuahuan desert. American Journal of Botany 86: 1093–1098. [PubMed] [Google Scholar]
- Mateu‐AndrésI, Segura‐Moragues G.2000. Population subdivision and genetic diversity in two narrow endemics of Antirrhinum L. Molecular Ecology 9: 2081–2087. [DOI] [PubMed] [Google Scholar]
- MillerMP.1997. Tools for population genetic analyses (TFPGA) 1.3: a Windows program for the genetic data. Computer software distributed by author. [Google Scholar]
- MirandaF, Hernández X.1963. Los tipos de vegetación de México y su clasificación. Boletín de la Sociedad Botánica de México 28: 29–179. [Google Scholar]
- MurrenCJ.2002. Effects of habitat fragmentation on pollination: pollinators, pollinia viability and reproductive success. Journal of Ecology 90: 100–107. [Google Scholar]
- NossRF.1990. Indicators of monitoring biodiversity: a hierarchical approach. Conservation Biology 4: 355–364. [Google Scholar]
- RankerTA.1994. Evolution of high genetic variability in the rare Hawaiian fern Adenophorus periens and implications for conservation management. Biological Conservation 70: 19–24. [Google Scholar]
- RaymondM, Rousset F.1995. An exact test for population differentiation. Evolution 49: 1280–1283. [DOI] [PubMed] [Google Scholar]
- RichardsAJ.1997.Plant breeding systems. 2nd edn. London, UK: Chapman & Hall. [Google Scholar]
- RichardsJH, Kortup S.1993. Floral variation and distyly in Guettarda scabra (Rubiaceae). American Journal of Botany 80: 31–40. [Google Scholar]
- RiverosGM, Barría OR, Humaña PAM.1995. Self‐compatibility in distylous Hedyotis salzmannii Plant Systematics and Evolution 194: 1–8. [Google Scholar]
- SavolainenO, Kuittien H.2000. Small populations processes. In: Young A, Boshier D, Boyle T, eds. Forest conservation genetics. Collingwood: CABI Publishing, 91–100. [Google Scholar]
- SazimaM, Vogel S, do Prado AL, de Oliveira DM, Franz G, Sazima I.2001. The sweet jelly of Combretum lanceolatum flowers (Combretaceae): a cornucopia resource for bird pollinators in the Pantanal, western Brazil. Plant Systematics and Evolution 227: 195–208. [Google Scholar]
- SchemskeDW,Husband BC, Ruckelshaus MH, Goodwillie C, Parker IM, Bishop JG.1994. Evaluation approaches to the conservation of rare and endangered plants. Ecology 75: 584–606. [Google Scholar]
- SlatkinM.1993. Isolation by distance in equilibrium and non‐equilibrium populations. Evolution 47: 264–279. [DOI] [PubMed] [Google Scholar]
- SlatkinM.1994. Gene flow and population structure. In: Real L, ed. Ecological genetics New Jersey: Princeton University Press, 3–17. [Google Scholar]
- SobrevilaC, Ramirez N, de Enrech NX.1983. Reproductive biology of Palicourea fendleri and P. petiolaris (Rubiaceae), heterostylous of a tropical cloud forest in Venezuela. Biotropica 15: 161–169. [Google Scholar]
- SoltisDE, Haufler CH, Darrow DC, Gastony GJ.1983. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. American Fern Journal 73: 9–27. [Google Scholar]
- VovidesAP, Iglesias CG.1994. An integrated conservation strategy for the cycad Dioon edule Lindl. Biodiversity and Conservation 3: 137–141. [Google Scholar]
- VovidesAP, Iglesias CG, Pérez‐Farrera M, Vazquez‐Torres M, Schippmann U.2002. Peasant Nurseries: a concept for an integrated conservation strategy for cycads in Mexico. In: Maunder M, Clubbe C, Hankamer C, Groves M, eds. Plant conservation in the tropics Kew, UK: Royal Botanic Gardens, 421–444. [Google Scholar]
- WallerDM, O’Malley DM, Gawler SC.1987. Genetic variation in the extreme endemic Pedicularis furbishiae (Scrophulariaceae). Conservation Biology 1: 335–340. [Google Scholar]
- WeirBS.1990.Genetic data analysis. Sunderland, MA: Sinauer Associates. [Google Scholar]
- WeirBS, Cockerham CC.1984. Estimating F‐statistics for the analysis of populations structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- WendelJF, Weeden NF.1989. Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS, eds. Isozymes in plant biology Portland, Oregon, USA: Discorides, 5–45. [Google Scholar]
- WorkmanPL, Niswander JD.1970. Population studies on southwestern Indian tribes II. Local differentiation in the Papago. American Journal of Human Genetics 22: 24–29. [PMC free article] [PubMed] [Google Scholar]
- WrightS.1951. The genetic structure of populations. Annals of Eugenetics 16: 97–159. [Google Scholar]
- WrightS.1965. The interpretation of population structure by F‐statistics with spatial regard to system of mating. Evolution 19: 355–420. [Google Scholar]
- WrightS.1978.Evolution and the genetics of populations. Vol. 4. Chicago and London: University of Chicago Press. [Google Scholar]
- YoungA, Boyle T, Brown T.1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution 11: 413–418. [DOI] [PubMed] [Google Scholar]