Abstract
• Background and Aims We tested whether the local differences in genome size recorded earlier in the wild barley, Hordeum spontaneum, at ‘Evolution Canyon’, Mount Carmel, Israel, can also be found in other organisms. As a model species for our test we chose the evergreen carob tree, Ceratonia siliqua.
• Methods Genome size was measured by means of DAPI flow cytometry.
• Key Results In adults, significantly more DNA was recorded in trees growing on the more illuminated, warmer, drier, microclimatically more fluctuating ‘African’ south‐facing slope than in trees on the opposite, less illuminated, cooler and more humid, ‘European’ north‐facing slope in spite of an interslope distance of only 100 m at the canyon bottom and 400 m at the top. The amount of DNA was significantly negatively correlated with leaf length and tree circumference. In seedlings, interslope differences in the amount of genome DNA were not found. In addition, the first cases of triploidy and tetraploidy were found in C. siliqua.
• Conclusions The data on C. siliqua at ‘Evolution Canyon’ showed that local variability in the C‐value exists in this species and that ecological stress might be a strong evolutionary driving force in shaping the amount of DNA.
Key words: Ceratonia siliqua, carob, DNA content, flow cytometry, genome size, ‘Evolution Canyon’
INTRODUCTION
In general, the C‐value paradox (no observed relationship between genome size diversity and organism complexity) is a largely overlooked factor in evolution and a matter of controversy (e.g. Elder and Turner, 1995). Among the different theories proposed to explain this phenomenon, two basic groups can be distinguished (Gregory, 2001). First, there are neutral mutation pressure theories (e.g. Ohno, 1972; Doolittle and Sapienza, 1980; Orgel and Crick, 1980) which state that genome size differences are due to a balance between an upward mutation pressure acting to increase DNA content in contrast to the tolerance of the ‘host’ cell to the building‐up of functionless ‘junk’ DNA (Gregory, 2001). Secondly, there are selective optimal DNA theories (e.g. Orgel et al., 1980; Cavalier‐Smith, 1982) in which DNA is considered to play both a qualitative and a quantitative role in the determination of organism fitness, with C‐values reflecting the adaptive optimized outcome of selection (Gregory, 2001).
Ecogeographically correlated intraspecific variability in genome size is documented by many authors in several plant species, including Poa annua (Grime, 1983), Milium effusum (Bennett and Bennett, 1992), Dactylis glomerata (Reeves et al., 1998) and Sesleria albicans (Lysák et al., 2000). For critical reviews of intraspecific variability of genome size see Ohri (1998) or Greilhuber (1998), and for its ecogeographical implications see Grime and Mowforth (1982) and Bennett (1987).
The ‘Evolution Canyon’ (EC) project involves evaluating the relative importance of different evolutionary forces and outcomes on local biodiversity differentiation (see reviews in Nevo, 1995, 1997, 2001). In wild barley, Hordeum spontaneum, growing in Israel, it seems that larger genome size, due to retrotransposon activity, may enable these plants to cope more effectively with aridity stress locally at EC as well as regionally (Vicient et al., 1999; Kalendar et al., 2000).
To determine whether local interslope differences in genome size are more general, as our model species we chose the carob, Ceratonia siliqua, a perennial tree species characterized by great longevity, slow growth and cross‐pollination, which contrasts to the annual, quick‐growing and mostly self‐pollinating H. spontaneum.
MATERIAL AND METHODS
Microsite
The microsite, named ‘Evolution Canyon’ I (EC I), located at Lower Nahal Oren, Mount Carmel, Israel (32°43′N, 34°58′E), is a Plio‐Pleistocene canyon (Nevo, 1995). The opposite slopes, which run in an east–west direction, are separated by 100 m at the bottom and 400 m at the top. The ‘African’ south‐facing slope (SFS) is covered by an open park forest of evergreen Ceratonia siliqua and Pistacia lentiscus with dominant savanna‐like grassland. In contrast, the ‘European’ north‐facing slope (NFS) is covered by a dense Mediterranean maquis of evergreen Quercus calliprinos and deciduous Pistacia palaestina (Nevo et al., 1999). Geology (Upper Cenomanian limestone; Karcz, 1959), regional Mediterranean climate (mean annual rainfall, approx. 600 mm; potential evapotranspiration, 1700 mm; and mean August and January temperatures 28 °C and 13 °C, respectively; Atlas of Israel, 1970), and pedology (terra rossa; Nevo et al., 1998) are the same on both slopes. Due to differences in geographic orientation in the northern hemisphere, SFSs receive, on average, more radiation per year than NFSs of the same inclination (Ayyad, 1971). Indeed, the SFS of EC I is significantly more illuminated, warmer and drier, and microclimatically and spatiotemporally more fluctuating than the opposite NFS (Pavlíček et al., 2003). These microclimate differences are reflected in strong local biodiversity differences. The SFS is, on average, richer in ‘terrestrial’ species and displays higher genetic diversity than the NFS, which is richer in ‘aquatic‐dependent’ taxa (Nevo, 1995, 1997, 2001).
Model species
Ceratoniasiliqua (the carob or locust tree) is native to the eastern Mediterranean basin (Batlle and Tous, 1997). It is a large, evergreen, sclerophyllous, leguminous tree of slow growth, great longevity and an extended flowering period (Diamantoglou and Mitrakos, 1981). It is also resistant to drought and salinity (Lo Gullo and Rosso, 1986). The carob has been growing in the eastern Mediterranean since long before the start of agriculture was documented. It is known to have been growing in what is now Israel as early as 43 000 bc (Weinstein, 1982). At EC I, the carob has been present since around 6000–4000 bc (Noy, 1977). Some authors believe that the carob originated from southern Arabia and not from the eastern Mediterranean (Schweinfurth, 1894; Zohary, 1973). This is supported by the fact that C. oreothauma, the only known carob‐related species, is thought to have one of two origins, either in Oman in south‐east Arabia or around the Horn of Africa in Somalia (Hillcoat et al., 1980). Currently, the carob is cultivated widely in other regions of the world with a Mediterranean climate (Vardar et al., 1980). Relative to other taxa of the subfamily Caesalpinioideae, carob flowers are anomalous in being initially bisexual with one sex usually suppressed during late development (Tucker, 1992), and by the lack of petals. The trees are usually dioecious but can be monoecious, and occasionally hermaphroditic (Linskens and Scholten, 1980; Batlle and Tous, 1988). Male and female flowers both contain nectar, although not in equal amounts, and insects play an important role in pollination (Retana et al., 1990; Ortiz et al., 1996). The seeds’ hard testa is an effective protective structure allowing them to survive fire, and being passed through the guts of large animals which aid dispersal (Ortiz et al., 1995). It has also resulted in a persistent seed‐bank being built up in the soil. To date, the reported number of chromosomes in the carob has been 2n = 24 (de Almeida, 1948; Frahm‐Leliveld, 1957; Berger et al., 1958; Goldblatt, 1981; Yeh et al., 1986; Arista and Talavera, 1990). However, in our study we found one case each of triploidy and of tetraploidy (see Results). No intersexual differences should exist in genome size since both sexes have the same number of identical chromosomes and the sex chromosome(s) is not present.
Sample collection
The 44 seedlings analysed for genome size were collected on 15 April 2003 at EC I. They were collected directly from under the mother trees, transferred together with soil to polyethylene bags, sealed, kept at a low temperature, and delivered to the Department of Botany, Masaryk University, Brno. Measurements took place on 22 April 2003. The 81 leaf samples from the mother trees were collected on 30 and 31 May 2003, following the same procedure as for the seedlings. Measurements took place on 9 and 10 June 2003. At the same time as collecting leaves, trunk circumferences were measured. If a tree had more than one trunk, the circumference of the thicker trunk was measured. The two pods subsequently used to measure intra‐tree variability were collected on 18 September 2003. Pod 1 was sampled from a main trunk with a circumference of 230 cm, and pod 2 from a young trunk with a circumference of 10 cm from the same tree. The seeds from the pods were later extracted and germinated in a greenhouse at the Department of Botany, Masaryk University, Brno. The DNA genome size was measured in the seedlings.
Genome size measurement
A ploidy analyser (PA‐1, Partec GmbH, Münster, Germany) equipped with a HBO‐100 mercury arc lamp was used to measure genome size. Sample preparations were carried out in a two‐step procedure (Otto, 1990; Doležel and Göhde, 1995) in the Laboratory of Flow Cytometry, Department of Botany, Masaryk University, Brno. One specimen of Luzula luzuloides was used as a standard to measure the relative amount of DNA in leaves of adult trees (Barlow and Nevin (1976) reported a haploid DNA content of C = 1·15 pg for L. luzuloides) and one specimen of Medicago sativa was used to measure the relative amount of DNA in seedlings (Bennett (1972) reported a haploid DNA content of C = 1·75 pg for M. sativa). Samples from a measured individual (0·5 cm2 of leaf blade from seedlings or 0·5 cm of leaf petiole from adult trees) and from a reference standard (0·5 cm2 of leaf blade from M. sativa or 1·0 cm2 of leaf blade from L. luzuloides) were chopped with a new razor blade for about 20 s in a Petri dish containing 0·5 mL of ice‐cold Otto I buffer (4·2 g citric acid monohydrate + 1 mL 0·5 % Tween20 adjusted to 200 mL and filtered through a 0·22‐µm filter). Another 0·5 mL Otto I buffer was then added before the solution was filtered through nylon cloth (50 µm mesh size). For DNA staining, 2 mL of Otto II buffer (0·4 m disodium hydrogenphosphate dodekahydrate) including DAPI (4′,6‐diamidino‐2‐phenylindole; 4 µg mL–1 final concentration) was used. DAPI is known to form fluorescent complexes with natural double‐stranded DNA, showing fluorescence specificity preferentially for AT‐bases, and is more useable for the estimation of small infraspecific differences of DNA content than, for example, ethydium bromide. For leaves from adult trees, which contain a higher amount of secondary substances, we used only leaf petioles, not leaf blades as in seedlings. Therefore, samples from adults were incubated for 1 h (instead of 2–5 min for seedlings), and centrifuged for 15 min at 1100 g. Each sample was measured three times and the calibrated average value was included in the statistical analysis. Calibrated average relative DNA content Rsample was calculated using the following formula:
Rsample = Σi=1. . .3 (1/CVsamplei × 1/CVreferencei × F_DAPIsamplei/F_DAPIreferencei) / Σi=1. . .3 (1/CVsamplei × 1/CVreferencei)
where CV is variance coefficient and F_DAPI is relative fluorescence channel (mean of peak) for both sample and reference in the first, second and third measurements (i = 1. . . 3), i.e. each measurement of relative DNA content Rsamplei = F_DAPIsamplei/F_DAPIreferencei is included in the calculation of the average with weight equal to 1/CVsamplei × 1/CVreferencei. All measurements for both seedlings and adult trees were carried out in 2 days. Samples from the SFS and the NFS of EC I were measured alternatively. For the repeat control measurements of triploid and tetraploid samples, diploid plants of C. siliqua were used as the reference standard. For the adult plants we used a two‐step procedure with one‐step buffers, which are usually successful for plants with secondary compounds, i.e. tris‐MgCl2 buffer (Pfosser et al., 1995) and lysis buffer LB01 (Doležel et al., 1989), in combination with the following conditions: centrifuge time 5 or 15 min, g = 290 or 1100, addition of polyvinylpyrrolidone (PVP 40) in concentrations of 10, 20 or 30 g L–1 to buffer. All these methods yielded poorer results than the two‐step procedure in combination with centrifuging.
Statistical analysis
The statistical program ‘Statistica for Windows’ version 5.0 (StatSoft Inc.) was used for most of the parametric and non‐parametric tests. In order to test the effect of random events, the permutation test used was based on 500 000 random shufflings.
RESULTS
Interslope differences in genome size of adult trees
Two samples from the original data set were omitted because one was from a triploid plant and the other from a tetraploid (see below). All other sampled plants were diploid. The range of genome size variation detected among the diploid adult trees was 4·04 % relative to the smallest measured DNA content.
The mean relative genome size was greater on the SFS than on the NFS (Table 1). No deviation of the data from normality on the SFS or the NFS was detected either by visual examination of histograms or by Shapiro–Wilk’s W‐test (NFS: W = 0·9845, P < 0·915; SFS: W = 0·9893, P < 0·978). Mean variance was slightly higher on the NFS than on the SFS (see s.d. in Table 1), but not significantly different (Levene’s test, F = 1·8, P = 0·18). Therefore, the results of both the normality and the variance tests allowed us to use parametric statistics. The t‐test for independent samples showed significant interslope differences (t‐value = 6·0457, d.f. = 77, P < 0·001). The significant results of the interslope differences were confirmed by permutation tests: the H0 hypothesis of equal genome size between the slopes was rejected at P = 2.10–6.
Table 1.
Mean relative genome size of adult Ceratonia siliqua trees according to slopes and stations at ‘Evolution Canyon’ I, Mount Carmel, Israel
| Station/slope | Sample size (n) | Mean relative genome size (± s.d.) | Mean tree circumference (cm) | Mean leaf length (cm)* |
| SFS1 | 13 | 0·572 (0·003) | 49·5 | 36·4 |
| SFS2 | 15 | 0·576 (0·003) | 33·8 | 34·6 |
| SFS3 | 14 | 0·572 (0·004) | 51·8 | 37·1 |
| NFS5 | 11 | 0·565 (0·005) | 99·1 | 65·3 |
| NFS6 | 13 | 0·567 (0·003) | 41·1 | 42·0 |
| NFS7 | 13 | 0·570 (0·005) | 44·0 | 43·7 |
| Mean | ||||
| SFS ‘African’ slope | 42 | 0·573 (0·004) | 44·6 | 36·0 |
| NFS ‘European’ slope | 37 | 0·568 (0·005) | 59·4 | 50·3 |
* Data from Nevo et al., 2000.
Additional evidence of interslope differences in genome size is given by comparison of SFS with NFS stations (Table 1). The mean relative genome size was significantly higher at the three SFS stations than at the three NFS stations (Mann–Whitney U‐test, P = 0·05).
Interstation differences in adult trees
The pairwise comparisons were significant between SFS2 and NFS6 (t‐value = 7·526, d.f. = 26, P < 0·001) and between SFS3 and NFS5 (t‐value = 3·663, d.f. = 23, P = 0·001), but not between SFS1 and NFS7 (t‐value = 1·290, d.f. = 24, P = 0·209). There seems to be an upslope trend of increasing genome size from the bottom to the top of the NFS (Table 1), but not on the SFS.
Multiple regression analysis
Multiple regression analysis indicates that the most important factor is slope (β = 0·734, P = 0·0007) and the second most important is tree circumference (β = –0·186, P = 0·06), the thicker‐trunked trees having significantly smaller genomes. The stations appear not to play an important role (β = 0·251, P = 0·25). In this analysis we were not able to include leaf size since the data are only means per station.
Influence of sex
Sex does not seem to have influenced the interslope results of genome size. The relative genome size values of both sexes on both slopes were similar and both males and females on the SFS had larger genomes than those on the NFS (Table 2).
Table 2.
Interslope differences in genome size of adult Ceratonia siliqua trees according to sex at ‘Evolution Canyon’ I, Mount Carmel, Israel
| Slope | Sex | n | Mean (± s.d.) |
| SFS ‘African’ | Males | 28 | 0·573 (0·001) |
| SFS ‘African’ | Females | 14 | 0·574 (0·028) |
| NFS ‘European’ | Males | 18 | 0·568 (0·001) |
| NFS ‘European’ | Females | 19 | 0·567 (0·039) |
Correlation between genome size, tree circumference and leaf size
The relative genome sizes of adult trees were correlated with tree circumference. The negative correlation in all sampled trees at EC I was significant (Spearman rank order correlations, n = 79, RS = –0·2748, P = 0·0142). A significant negative correlation was also found on the SFS (n = 42, RS = – 0·2267, P = 0·0289) but not on the NFS (n = 37, RS = – 0·1402, P = 0·4079). Circumferences of tree trunks on the SFS (mean circumference, 44·6 cm) were significantly smaller than those on the NFS (mean circumference, 59·4 cm) (one way ANOVA, F = 4·01, P = 0·048). There was a significant (RS = –0·9276, P < 0·05) negative correlation between the mean relative genome contents of adult trees per station and mean leaf length per station (Table 1).
Many other significant correlations between the relative mean genome size per station and environmental factors have been reported. Examples include significantly positive correlations (RS = 0·89, P < 0·02) with mean temperature and illumination (both of which are significantly higher on the SFS: Pavlíček et al., 2003) and significant negative correlation (RS = –0·89, P < 0·02) with humidity (which is significantly higher on the NFS: Pavlíček et al., 2003).
Seedlings
The range of DNA content variation among the seedlings was 3·93 %. The mean value was higher on the NFS than on the SFS (Table 3), but differences were not significant. Mean variance was slightly higher on the NFS than on the SFS (see s.d. in Table 3), but again differences were not significant (Levene’s test, F = 0·06, P = 0·81). The permutation test did not reject the null hypothesis, as P = 0·17.
Table 3.
Mean relative genome size of Ceratonia siliqua seedlings according to slopes and stations at ‘Evolution Canyon’ I, Mount Carmel, Israel
| Station/slope | n | Mean (± s.d.) |
| SFS1 | 9 | 32·782 (0·261) |
| SFS2 | 3 | 33·132 (0·161) |
| SFS3 | 14 | 33·070 (0·215) |
| NFS5 | 13 | 33·037 (0·261) |
| NFS6 | 3 | 33·382 (0·306) |
| NFS7 | 2 | 33·332 (0·019) |
| SFS ‘African’ | 26 | 32·977 (0·263) |
| NFS ‘European’ | 18 | 33·127 (0·286) |
Intra‐tree variability
Genome sizes in seven and eight seedlings that grew from the seeds of pod 1 (sampled from a main trunk with a circumference of 230 cm) and pod 2 (sampled from a young trunk with a circumference of 10 cm), respectively, were measured. The arithmetic mean, maximum and minimum for pod 1 seedlings were 1·703 ± 0·014, 1·72, 1·68, and for pod 2 seedlings 1·702 ± 0·017, 1·72, 1·68. As the values for seedlings from both pods were almost identical, the arithmetic means were not significantly different (t‐value = 0·19, P = 0·85). No significant correlations were found either between seed weights and their DNA amounts (Spearman R = 0·014, P = 0·96) or between DNA amounts and seed positions in each pod (R = 0·36, P = 0·43 and R = 0·31, P = 0·46).
Chromosome numbers
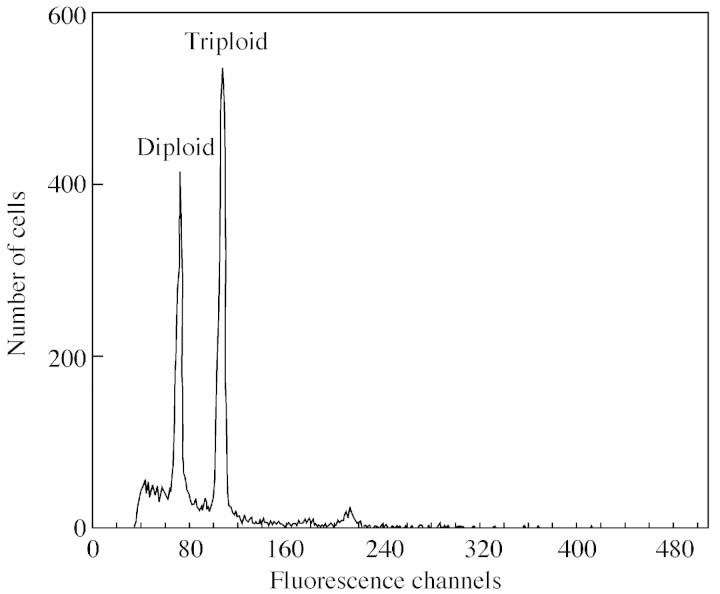
During our investigation we discovered one triploid sample (Fig. 1) and one tetraploid sample (not shown) representing trees from SFS3 and NFS7, respectively. These are the first records of triploidy and tetraploidy in C. siliqua. While the genome size among diploids varies 1·04‐fold, the amount of DNA in the triploid and tetraploid was, respectively, 1·545‐fold and 2·054‐fold more than in diploid plants with the smallest DNA content.
Fig. 1. Comparison of absorptions of diploid and triploid Ceratonia siliqua trees.
DISCUSSION
Interslope divergence in genome size
Our results on adult trees corroborate the results obtained on Hordeum spontaneum (Kalendar et al., 2000), and show that differences in genome size at EC I are distributed non‐randomly, the larger mean genome size being characteristic of the ‘African’ SFS and the smaller mean genome size of the ‘European’ NFS. Therefore, our results do not support the neutral mutation pressure theory(ies), including the neutral theory of molecular evolution (Kimura, 1983). By contrast, they do indicate that some environmental factors might play an important role in generating and, perhaps, maintaining interslope divergence of genome size at EC I. It might also be possible to reject the nucleoskeletal theory, which suggests that nuclear size is selected to meet the needs of the cell for balanced growth (e.g. Cavalier‐Smith, 1985, 1991), since leaf cells on SFS trees contain more DNA but are probably smaller than those on NFS trees.
Selection for higher genome size in carob?
Unfortunately, we do not know whether similarities between H. spontaneum and C. siliqua go beyond the similar interslope patterns in genome size at EC I. The most intriguing problem is that we did not find support for the interslope differences in seedling genome size. Conversely, our results might point to selection of seedling genome size in C. siliqua as suggested (but not proved) by the significant negative correlations between genome size and both tree circumference and leaf length. However, we cannot exclude the possibility that interslope differences may be caused by ontogenetic changes (e.g. by a differential transposition of somatically active transposable elements, which is known to occur in invertebrates and vertebrates: Emmons and Yesner, 1984; Woodruff et al., 1999) or by associations of genome size changes with other ontogenetic processes. Also, it should be remembered that processes not sufficiently understood have led to environmentally induced changes of genome size during cell culturing (e.g. Blundy et al., 1987; Quemada et al., 1987; Zheng et al., 1987) and that heritable changes within a single generation, involving changes both at the phenotypic level and in DNA, have been observed in annual flax (Cullis, 1990). However, there is no indication that parallel processes should necessarily occur in perennial trees.
Correlates of genome size
Many authors have studied, mainly at the cellular and physiological levels, the effect of DNA quantity on morphological features. Similar results to our findings of significant negative correlations between genome size and leaf size or tree circumference in C. siliqua have been found, for example, in leaf length of the grass Dasypyrum villosum (Caceres et al., 1998). However, among legumes, Chung et al. (1998) detected a positive correlation between leaf size and nuclear DNA content in soybeans while Minelli et al. (1996) found a negative correlation between height of the main stem and nuclear DNA content in Vicia faba, a similar pattern to that detected between leaf size and tree circumference in C. siliqua. We can probably exclude from consideration the intra‐plant differences in DNA contents, as described between seeds of Helianthus annuus (Cavallini et al., 1989), as we did not find any significant intra‐tree correlations between DNA content and the age of trunks, seed weight or seed position in the pod.
C. siliqua as a model to understand interslope biodiversity differences at EC I
The observed pattern of interslope differences in genome size probably fits the more general pattern of higher biodiversity on the ‘African’ SFS than on the ‘European’ NFS (Nevo, 1995, 1997, 2001). At the drier, more climatically stressful SFS we documented higher genetic variability (summarized in Nevo, 2001), a three‐fold higher mutation rate in the fungus Sordaria fimicola (Saleem et al., 2001), and a four‐fold higher male recombination rate in Drosophila melanogaster (Derzhavets et al., 1997). The common denominator of all genetic interslope differences, including genome size, could be ecological stress which, according to Nevo (2001), is the most probable driving force generating global‐scale adaptive genome and phenome strategies at both microscales and macroscales, reinforcing homeostasis and fitness and suggesting continuity between microevolution and macroevolution.
Polyploidization in C. siliqua
Randomly naturally occurring triploids and tetraploids have been detected among Leguminosae trees in the genus Acacia (Blakesley et al., 2002). With the carob tree, it may be interesting to investigate large‐scale ploidy levels and possible ecological correlates.
Conclusions
Our data from C. siliqua at EC I, together with circumstantial evidence, indicate that ecology might be a strong evolutionary driving force in shaping quantities of DNA. In addition, we discovered that C. siliqua is not only diploid, but can also be triploid and tetraploid.
ACKNOWLEDGEMENTS
This study was supported by the Israel Discount Bank Chair of Evolutionary Biology, the Ancell‐Teicher Research Foundation for Genetics and Molecular Evolution, and the Ministry of Education of the Czech Republic (research project MSM 143100010 Spatial and Temporal Biodiversity Dynamics in Ecosystems of Central Europe). Our thanks go to Professor A. B. Korol for help with the permutation test and to Mr Rosovsky and Robin Permut for editing.
Received: 7 November 2003; Returned for revision: 26 December 2003; Accepted: 22 January 2004 Published electronically: 16 March 2004
References
- de AlmeidaJLF.1948. Sobre a cariologia de Ceratonia siliqua L. Agronomia Lusitana 10: 263–277. [Google Scholar]
- AristaM, Talavera S.1990. Números cromosómicos para la flora Española 620–629. Lagascalia 16: 323–328. [Google Scholar]
- Atlas of Israel..1970. Amsterdam: Ministry of Labour (various paginations). [Google Scholar]
- AyyadM.1971. A study of solar radiation on sloping surfaces at Alexandria. United Arab Republic Journal of Botany 14: 65–72. [Google Scholar]
- BarlowPW, Nevin D.1976. Quantitative karyology of some species of Luzula Plant Systematics and Evolution 125: 77–86. [Google Scholar]
- BatlleI, Tous J.1988. Lineas de investigación sobre el algarrobo (Ceratonia siliqua L.) en el IRTA, Cataluña (España). In: Brito de Carvalho JH, ed. I Encorto Linhas de Investigaçao de Alfarroba AIDA, Oeiras: AIDA, 92–104. [Google Scholar]
- BatlleI, Tous J.1997.Carob tree. Ceratonia siliqua L. Promoting the conservation and use of underutilized and neglected crops. 17 Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome, 1–79. [Google Scholar]
- BennettMD.1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London, Series B—Biological Sciences, 181: 109–135. [DOI] [PubMed] [Google Scholar]
- BennettMD.1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106(Supplement 1): 177–200. [Google Scholar]
- BennettST, Bennett MD.1992. Variation in nuclear DNA amount between wild and cultivated populations of Milium effusum (2n = 28). Genome 35: 1050–1053. [Google Scholar]
- BergerCA, Witkus ER, McMahon RM.1958. Cytotaxonomic studies in Leguminosae. The Bulletin of the Torrey Botanical Club 85: 405–414. [Google Scholar]
- BlakesleyD, Allen A, Pellny TK, Roberts AV.2002. Natural and induced polyploidy in Acacia dealbata Link. and Acacia mangium Willd. Annals of Botany 90: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BlundyKS, Cullis CA, Hepburn AG.1987. Ribosomal DNA methylation in a flax genotroph and a crown gall tumor. Plant Molecular Biology 8: 217–225. [DOI] [PubMed] [Google Scholar]
- CaceresME, De Pace C, Scarascia Mugnozza GT, Kotsonis P, Ceccarelli M, Cionini PG.1998. Genome size variation within Dasypyrum villosum: correlations with chromosomal traits, environmental factors and plant phenotypic characteristic and behaviour in reproduction. Theoretical and Applied Genetics 96: 559–567. [Google Scholar]
- Cavalier‐SmithT.1982. Skeletal DNA and the evolution of genome size. Annual Review of Biophysics and Bioengineering 11: 273–302. [DOI] [PubMed] [Google Scholar]
- Cavalier‐SmithT.1985. Cell volume and the evolution of genome of eukaryote genome size. In: Cavalier‐Smith T, ed. The evolution of genome size Chichester: John Wiley & Sons, 104–184. [Google Scholar]
- Cavalier‐SmithT.1991. Coevolution of vertebrate genome, cell and nuclear sizes. In: Ghiara G, Angelini F, Ettore O, eds. Selected symposia and monographs UZI Vol. 4. Modena: Mucchi editore, 51–86. [Google Scholar]
- CavalliniA, Zolfino C, Cionini G, Cremonini R, Natali L, Sassoli O, Cionini PG.1986. Nuclear DNA changes within Helianthus annus L: Cytophotometric, karyological and biochemical analyses. Theoretical and Applied Genetics 73: 20–26. [DOI] [PubMed] [Google Scholar]
- CavalliniA, Zolfino C, Natali L, Cionini G, Cionini PG.1989. Nuclear DNA changes within Helianthus annus L: Origin and control mechanism. Theoretical and Applied Genetics 77: 12–16. [DOI] [PubMed] [Google Scholar]
- ChungJ, Lee J‐H, Arumuganathan K, Graef GL, Specht JE.1998. Relationships between nuclear DNA content and seed and leaf size in soybean. Theoretical and Applied Genetics 96: 1064–1068. [Google Scholar]
- CullisCA.1990. DNA rearrangements in response to environmental stress. Advances in Genetics 28: 73–97. [DOI] [PubMed] [Google Scholar]
- DerzhavetsE, Korol A, Pavlíček T, Nevo E.1997. Adaptation to stressful environment and mutation rate: a case study in Drosophila melanogaster Drosophila Information Service 80: 73. [Google Scholar]
- DiamantoglouS, Mitrakos K.1981. Leaf longevity in Mediterranean evergreen sclerophylls. In: Margaris NS, Mooney HA, eds. Components of productivity of Mediterranean climate regions. Basic and applied aspects The Hague: Junk Publishers, 17–19. [Google Scholar]
- DoleželJ, Binarová P, Lucretti S.1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 31: 113–120. [Google Scholar]
- DoleželJ, Göhde W.1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high‐resolution flow cytometry. Cytometry 19: 103–106. [DOI] [PubMed] [Google Scholar]
- DoolittleWF, Sapienza C.1980. Selfish genes, the phenotype peradigm and genome evolution. Nature 284: 601–603. [DOI] [PubMed] [Google Scholar]
- ElderJF, Turner BJ.1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Quarterly Review of Biology 70: 297–320. [DOI] [PubMed] [Google Scholar]
- EmmonsSW, Yesner L.1984. High‐frequency excision of transposable element Tc1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell 36: 599–605. [DOI] [PubMed] [Google Scholar]
- Frahm‐LeliveldJA.1957. Observations cytologiques sur quelques Légumineuses tropicales et subtropicales. Revue de Cytologie et de Biologie Végétales‐le Botaniste 18: 273–287. [Google Scholar]
- GaliliE, Schick T.1990. Basketry and a wooden bowl from the pottery of a Neolithic submerged site of Kefar Samir. Journal of the Israel Prehistoric Society 23: 142–151. [Google Scholar]
- GoldblattP.1981. Cytology and phylogeny of the Leguminosae. In: Polhill RM, Raven PH, eds. Advances in legume systematics Vol. 2. Kew: Royal Botanic Gardens, 427–464. [Google Scholar]
- GregoryTR.2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C‐value enigma. Biological Review 76: 65–101. [DOI] [PubMed] [Google Scholar]
- GreilhuberJ.1998. Intraspecific variation in genome size: a critical reassessment. Annals of Botany 82(Supplement A): 27–35. [Google Scholar]
- GrimeJP.1983. Prediction of weed and crop responce to climate based on measurement of DNA content. Aspects of Applied Biology 4: 87–98. [Google Scholar]
- GrimeJP, Mowforth MA.1982. Variation in genome size – an ecological interpretation. Nature 299: 151–153. [Google Scholar]
- HillcoatD, Lewis G, Verdcourt B.1980. A new species of Ceratonia (Leguminosae‐Caesalpinoideae) from Arabia and the Somali Republic. Kew Bulletin 35: 261–271. [Google Scholar]
- KalendarR, Tanskanen J, Immonen S, Nevo E, Schulman AH.2000.Genome evolution of wild barley (Hordeum spontaneum) by BARE‐1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KarczY.1959. The structure of the northern Carmel. Bulletin of the Research Council of Israel 8G: 119–130. [Google Scholar]
- KimuraM.1983.The neutral theory of molecular evolution. Cambridge: University Press. [Google Scholar]
- LinskensHS, Scholten W.1980. The flower of carob. Portugaliae Acta Biologica 16: 95–102. [Google Scholar]
- Lo GulloMA, Rosso R.1986. Drought avoidance strategy in Ceratonia siliqua L. a mesomorphic‐leaved tree in the xeric Mediterranean area. Annals of Botany 58: 745–756. [Google Scholar]
- LysákMA, Rostková A, Dixon JM, Rossi G, Doležel J.2000. Limited genome size variation in Sesleria albicans Annals of Botany 86: 399–403. [Google Scholar]
- MinelliS, Moscariello P, Ceccarelli M, Cionini PG.1996. Nucleotype and phenotype in Vicia faba Heredity 76: 524–530. [Google Scholar]
- NevoE.1995. Asian, African and European biota meet at ‘Evolution Canyon’ Israel: Local test of global biodiversity and genetic diversity patterns. Proceedings of the Royal Society of London (Series B) 262: 149–155. [Google Scholar]
- NevoE.1997. Evolution in action across phylogeny caused by microclimatic stresses at ‘Evolution Canyon’. Theoretical and Population Biology 52: 231–243. [DOI] [PubMed] [Google Scholar]
- NevoE.2001.Evolution of genome‐phenome diversity under environmental stress. Proceedings of the National Academy of Sciences USA 98: 6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NevoE, Travleev A, Belova NA, Tsatskin A, Pavlíček T, Kulik AF, Tsvetkova NN, Yemshanov DC.1998. Edaphic interslope and valley bottom divergence at ‘Evolution Canyon’, Lower Nahal Oren, Mount Carmel, Israel. Catena 340: 241–254. [Google Scholar]
- NevoE, Fragman O, Dafni A, Beiles A.1999. Biodiversity and interslope divergence of vascular plants caused by microclimatic differences at ‘Evolution Canyon’, Lower Nahal Oren, Mount Carmel, Israel. Israel Journal of Plant Sciences 47: 49–59. [Google Scholar]
- NoyT.1977. Nahal Oren. In: Avi‐Yona M, Stern E, eds. Encyclopedia of archeological excavations in the Holy Land Vol. 3. Givatayim: The Israeli Exploratory Society and Masada Publishers, 903. [Google Scholar]
- OhnoS.1972. So much ‘‘junk’’ DNA in our genome. In: Smith HH, ed. Evolution of genetic systems New York: Gordon & Breach, 366–370. [Google Scholar]
- OhriD.1998.Genome size variation and plant systematics. Annals of Botany 82(Supplement A): 75–83. [Google Scholar]
- OrgelLE, Crick FHC.1980. Selfish DNA: the ultimite parasite. Nature 284: 604–607. [DOI] [PubMed] [Google Scholar]
- OrgelLE, Crick FH, Sapienza C.1980. Selfish DNA. Nature 288: 645–646. [DOI] [PubMed] [Google Scholar]
- OrtizPL, Arista M, Talavera S.1995. Germination ecology of Ceratonia siliqua L. (Caesalpinaceae), a Mediterranean tree. Flora 190: 89–95. [Google Scholar]
- OrtizPL, Arista M, Talavera S.1996. Producción de nectar y frecuencia de polinizadores en Ceratonia siliqua L. (Caesalpinaceae). Anales de Jardín Botánico Madrid 54: 540–646. [Google Scholar]
- OttoF.1990. DAPI staining of fixed cells for high‐resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z, eds. Methods in cell biology Vol. 33. New York: Academic Press, 105–110. [DOI] [PubMed] [Google Scholar]
- PavlíčekT, Sharon D, Kravchenko V, Saaroni H, Nevo E.2003. Microclimatic interslope differences underlying biodiversity contrasts in ‘Evolution Canyon’, Mt. Carmel, Israel. Journal of Earth Sciences 52: 1–9. [Google Scholar]
- PfosserA, Amon A, Lelley T, Heberle‐Bors E.1995. Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat‐rye addition lines. Cytometry 21: 387–393. [DOI] [PubMed] [Google Scholar]
- QuemadaH, Roth EJ, Lark KG.1987. Changes in methylation of tissue cultured soybean cells detected by digestion with the restriction enzymes HpaII and MspI. Plant Cell Reports 6: 63–66. [DOI] [PubMed] [Google Scholar]
- ReevesG, Francis D, Davies MS, Rogers HJ, Hodkinson TR.1998.Genome size is negatively correlated with altitude in natural populations of Dactylis glomerata Annals of Botany 82 (Supplement A): 99–105. [Google Scholar]
- RetanaJ, Ramoneda J, García del Pino F.1990. Importancia de los insectos en la polinización del algarrobo. Boletin de Sanidad Vegetal Plagas 16: 143–153. [Google Scholar]
- SaleemM, Lamb BC, Nevo E.2001. Inherited differences in crossing‐over and gene conversion frequencies between wild strains of Sordaria fimicola from ‘Evolution Canyon’. Genetics 159: 1573–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SchweinfurthG.1894. Sammlungarabish‐aethiopischer Pflanzen, Ergebnisse von Reisen in den Jahren 1881, 1888–89, 1891,–92. Bulletin de l’herbier Boissier 2: 1–114. [Google Scholar]
- TuckerSC.1992. The developmental basis for sexual expression in Ceratonia siliqua (Leguminosae: Caesalpinioideae: Cassieae). American Journal of Botany 79: 318–327. [Google Scholar]
- VardarY, Seçmen Ö, Öztürk M.1980. Some distributional problems and biological characteristics of Ceratonia in Turkey. Portugaliae Acta Biologica 16: 75–86. [Google Scholar]
- VicientCM, Suoniemi A, Anamthawat‐Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH.1999. Retrotransposon BARE‐1 and its role in genome evolution in the genus Hordeum The Plant Cell 11: 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WeinsteinM.1982.The paleoecology of the early Würm in the Hulah Basin. PhD Thesis. Tel Aviv University, Israel (in Hebrew). [Google Scholar]
- WoodruffRC, Thompson JN, Barker JSF, Huai H.1999. Transposable DNA elements and life history traits: II. Transposition of P DNA element in somatic cells reduce fitness, mating activity, and locomotion of Drosophila melanogaster Genetica 107: 261–269. [PubMed] [Google Scholar]
- YehMS, Yuasa H, Maekawa F.1986. Chromosome numbers in the Leguminosae. Science Report of the Research Institute of Evolutionary Biology 3: 57–71. [Google Scholar]
- ZhengKL, Castiglione S, Biasini MG, Biroli A, Morandi C, Sala F.1987. Nuclear DNA amplification in cultured cells of Oryza sativa Theoretical and Applied Genetics 74: 65–70. [DOI] [PubMed] [Google Scholar]
- ZoharyM.1973.Geobotanical foundations of the Middle East. 2 Volumes. Stuttgart: G. Fisher. [Google Scholar]