Abstract
• Background and aims Kip‐related‐proteins (KRPs), negative regulators of cell division, have recently been discovered in plants but their in planta function is as yet unclear. In this study the spatial expression of all seven KRP genes in shoot apices of Arabidopsis thaliana were compared.
• Methods In situ hybridization analyses were performed on longitudinal sections of shoot apices from 2‐month‐old Arabidopsis plants.
• Key Results The study provides evidence for different expression pattern groups. KRP1 and KRP2 expression is restricted to the endoreduplicating tissues. In contrast, KRP4 and KRP5 expression is mainly restricted to mitotically dividing cells. KRP3, KRP6 and KRP7 can be found in both mitotically dividing and endoreduplicating cells.
• Conclusion The results suggest differential roles for the distinct KRPs. KRP1 and KRP2 might specifically be involved in the establishment of polyploidy. In contrast, KRP4 and KRP5 might be involved in regulating the progression through the mitotic cell cycle. KRP3, KRP6 and KRP7 might have a function in both types of cell cycle.
Key words: Arabidopsis thaliana, cell cycle, cell differentiation, endoreduplication, KRP expression
INTRODUCTION
Flowering plants are multicellular organisms where cell division plays a significant role in growth and development. The cell division cycle is regulated in yeast, animals and plants by a molecular machinery where the main drivers are cyclin‐dependent kinases (CDKs) (Norbury and Nurse, 1992; Morgan, 1997; Mironov et al., 1999). CDK activity is mediated by several mechanisms, more particularly by association with cyclins (reviewed by Pines, 1994) and phosphorylation of the Thr‐161 residue (for review, see Dunphy, 1994).
A relatively new mechanism of regulating CDK activity has been found with the identification of CDK inhibitors (CKIs; see reviews by Harper and Elledge, 1996; Pines, 1995; Sherr and Roberts, 1995, 1999). These proteins bind to cyclin/CDK complexes and inhibit CDK activity. In mammals, two CKI families have been recognized, based upon their sequence similarity and mode of action: the INK4 and the Kip/Cip families.
The CKIs of the Kip/Cip family, including p21Cip1, p27Kip1 and p57Kip2, negatively regulate cell cycle progression and enforce cell cycle arrest when expressed at high levels (Elledge and Harper, 1994; Sherr and Roberts, 1995). The Kip/Cip CKIs are involved in both G1/S and G2/M checkpoint control and the regulation of the cell cycle exit preceding cell differentiation (Zhang et al., 1999). The p21Cip1 and p27Kip1 CKIs have also been found in complexes with active cyclin‐CDKs, suggesting that CKIs may also act as positive regulators (LaBaer et al., 1997). Indeed the normal up‐regulation of CyclinD/CDK4 in mitogen‐stimulated fibroblasts depends upon p21Cip1 and p27Kip1 (Cheng et al., 1999).
Proteins of the class that act as inhibitors of CDK were unknown in plants until ICK1 was identified in Arabidopsis thaliana (Wang et al., 1997) The carboxy‐terminal domain of ICK1 shares a conserved 31 amino‐acid sequence, including part of the CDK binding domain, with the mammalian p27Kip1 kinase inhibitor. ICK1 has been shown to interact directly with both Cdc2a (CDK‐a) and CycD3 (a D‐type cyclin) by yeast two‐hybrid and in vitro binding assays (Wang et al., 1998). Actually in the A. thaliana genome seven CKI‐like genes are present, all having a region of approx. 25 amino acids that are highly conserved with the mammalian Kip/Cip proteins, hence their name, KRPs: Kip‐related proteins (De Veylder et al., 2001; Vandepoele et al., 2002). Despite their limited sequence homology with the mammalian counterparts, the KRPs have been shown to be true biochemical homologues of the Kip/Cip proteins, as recombinant KRPs are able to inhibit CDK activity in vitro (Wang et al., 1998; Lui et al., 2000), whereas their overexpression in plants results in a decrease of CDK activity in vivo (Wang et al., 2000; De Veylder et al., 2001).
Although some data are available on the transcription profiles of KRPs, not much is known about their spatial pattern of expression. Preliminary expression analyses showed that the KRP1 and KRP6 genes are expressed ubiquitously in various plant organs (roots, inflorescence stems, flower buds and 3‐week‐old leaves) and in a 3‐day‐old, actively dividing suspension culture (De Veylder et al., 2001). KRP4, KRP5 and KRP7 are expressed in the same organs and culture, but mRNA clearly seems to be more abundant in tissues that display high mitotic activity (flowers and suspension cultures), with KRP4 also being abundantly present in leaves. KRP2 mRNA seems to be more abundant in flowers, and the level of KRP3 expression is high in actively dividing suspension cultures but it is not detectable, or is barely so, in intact plant organs (mainly roots and flowers) (De Veylder et al., 2001). These transcription profiles suggest that the various KRPs might play distinct roles during plant development.
Here we report the spatial expression pattern of all seven A. thaliana KRPs in the shoot apex of plants maintained in vegetative growth for 2 months in short day conditions. This material has proved to be suitable for the characterization of genes involved in the regulation of the mitotic cycle and the endoreduplication cycle (Jacqmard et al., 1999). At the cellular level the data confirm the existence of differential expression patterns for the distinct KRP genes, and on this basis a classification of the KRPs into different functional groups is suggested.
MATERIALS AND METHODS
Plant material
Arabidopsis thaliana (L.) Heynh. (ecotype Col‐o) plants were maintained in a vegetative state for 2 months by growth in short days as described in Corbesier et al. (1996). Seeds were kindly provided by C. R. Somerville (Department of Plant Biology, Carnegie Institution of Washington, Stanford, CA, USA). Apical buds were then excised for mRNA in situ hybridization analysis.
mRNA in situ hybridization
Longitudinal sections of shoot apices from 2‐month‐old Arabidopsis plants were hybridized as described by Segers et al. (1996). Non‐specific binding of the KRPs probes to the adhesive was often observed and was responsible for causing background noise in the results. We have partly succeeded in reducing this non‐specic binding of the probes by doubling the duration of the acetylation treatment. Lengthening the acetylation time did not affect the signal with the antisense probe. KRPs [35S] UTP‐labelled antisense probes were obtained from the linearized transcription vectors by in vitro transcription with T7 (KRP1, KRP2, KRP3, KRP5 and KRP7) and SP6 (KRP4 and KRP6) RNA polymerases.
RESULTS
In situ hybridizations were performed on sections of shoot apices of 2‐month‐old A. thaliana plants kept in a vegetative state when grown in short day conditions. This allowed us to characterize the KRP genes potentially involved in the regulation of the mitotic cycle and/or the endoreduplication cycle, since discrimination between dividing and endoreduplicating tissues has been established (Jacqmard et al., 1999). Without ambiguity, divisions occur in the shoot apical meristem (SAM) in just‐emerged leaf primordia up to 70 µm length, in vascular tissues and in the procambium, while endoreduplication is established in cells of the pith and the stipules. The situation is less clear regarding the leaves where, depending on their stage of maturation, both divisions and endoreduplication can be found. Indeed, DNA content in the SAM and young leaf primordia of 30–70 µm of length was mostly 2C with only a small proportion of 4C nuclei, indicating that meristematic cells are euploid (Jacqmard et al., 1999). In contrast, in maturing leaves >400 µm in length, endoreduplication is observed in all mesophyll cells while divisions still occur in vascular tissues of these leaves. An intermediate situation is observed in leaf primordia of 300–400 µm of length, where spongy mesophyll cells at the abaxial side are differentiated from the palisade layer at the adaxial side, and where cell divisions occur concomitantly with the establishment of endoreduplication in mesophyll cells.
In the in situ hybridization analysis, the KRP1 and mainly KRP2 genes were highly expressed in endoreduplicating cells of the pith and in mesophyll cells of maturing leaves (Fig. 1). Expression of both genes was also observed in cells of 300–400 µm long leaf primordia (arrows in Fig. 1, KRP1 and KRP2). KRP1 and KRP2 RNA transcripts were barely detected in the SAM, in axillary buds (not shown) and in vascular cells. The distribution of KRP1 and KRP2 transcripts in leaves varied depending on the stage of differentiation of the leaf. Transcripts of both genes were distributed in a relatively homogenous pattern in maturing leaf primordia. But in leaf primordia of 300–400 µm length, tissue‐specific patterns of expression were observed: KRP1 transcripts accumulated both in palisade cells of the mesophyll at the adaxial side and in spongy mesophyll cells at the abaxial side (arrows in Fig. 1, KRP1); KRP2 transcripts accumulated more specifically in spongy mesophyll cells (arrow in Fig. 1, KRP2). Although mitoses were still detected in leaf primordia of that stage, cell differentiation had already started.
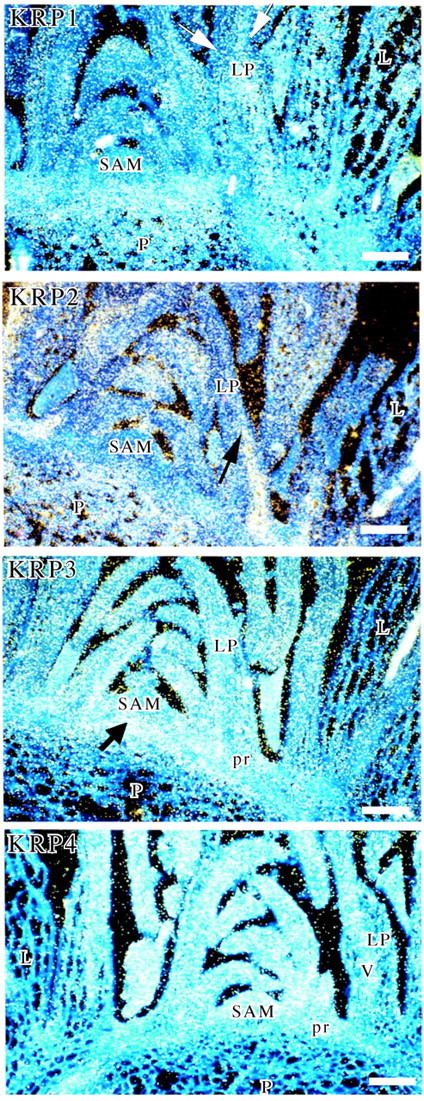
Fig. 1. mRNA in situ localization of KRPs in the shoot apex. Longitudinal sections of shoot apices of 2‐month‐old A. thaliana plants (ecotype Col‐0) hybridized with 35S‐labelled antisense riboprobes of KRP1, 2, 3 and 4. The autoradiographic signal was visualized in dark‐field illumination. L, maturing leaf of >400 µm length; LP, 300–400 µm‐long leaf primordium; P, pith; pr, procambium; SAM, shoot apical meristem; V, vascular tissue. Scale bar = 100 µm. Arrows indicate transcripts accumulation.
In contrast, KRP4 and KRP5 were expressed in all tissues where mitotic divisions occur (Fig. 1, KRP4 and Fig. 2, KRP5). The level was high in dividing cells of the SAM and in young leaf primordia of up to 400 µm. While KRP4 was also slightly expressed in the procambium (Fig. 1, KRP4), KRP5 hybridization signal was particularly strong in the just‐initiated procambial cells and in the peripheral zone of the SAM (Fig. 2, KRP5). For both genes, some expression was also detected in the vascular bundles of maturing leaves and either a very weak or no signal was observed in the pith and mesophyll cells of maturing leaves.
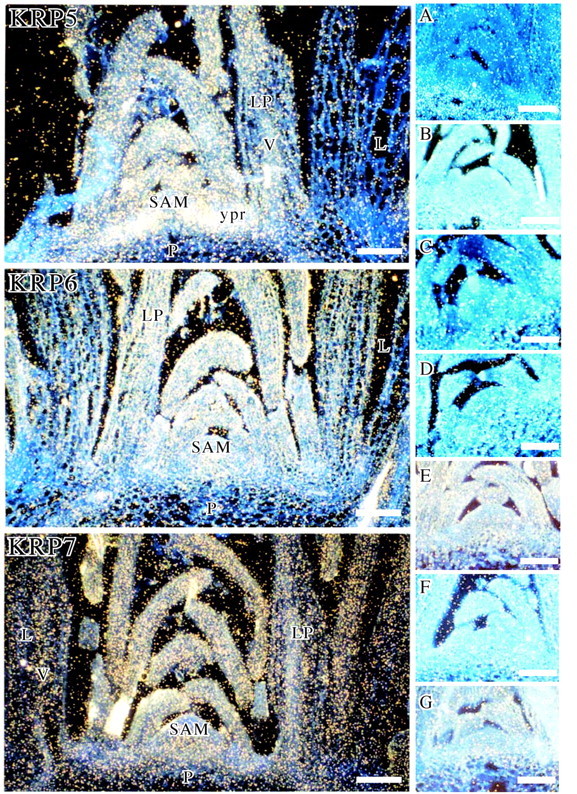
Fig. 2. mRNA in situ localization of KRPs in the shoot apex. Longitudinal sections of shoot apices of 2‐month‐old A. thaliana plants (ecotype Col‐0) hybridized with 35S‐labelled antisense riboprobes of KRP5, 6 and 7, or sense riboprobes of (A) KRP1, (B) KRP2, (C) KRP3, (E) KRP5, (F) KRP6 and (G) KRP7. (D) Section pretreated with RNase and hybridized with 35S‐labelled antisense riboprobes of KRP3. The autoradiographic signal was visualized in dark‐field illumination. L, maturing leaf of >400 µm length; LP, 300–400 µm‐long leaf primordium; P, pith; SAM, shoot apical meristem; V, vascular tissue; ypr, young procambium. Scale bar = 100 µm.
The KRP3, KRP6 and KRP7 genes were expressed in both dividing cells of the emerged leaf primordia and endoreduplicating cells of the pith and maturing leaves within the shoot apex (Fig. 1, KRP3 and Fig. 2, KRP6 and 7). KRP3 transcripts accumulated particularly in the upper cells of the pith just produced by the rib meristem (arrow in Fig. 1, KRP3). The KRP3 and KRP7 signals were absent from the SAM while that of KRP6 was weakly detected. The KRP3, KRP6 and KRP7 hybridization signals were low in dividing procambial cells of the stem (Fig. 1, KRP3 and Fig. 2, KRP6 and 7). A signal was not observed in control hybridizations with sense probes for six KRPs (shown for KRP2, KRP3, KRP5, KRP6 and KRP7, but not shown for KRP4) (Fig. 2A, B, C, E, F and G, respectively). A very low signal was detected in the pith with the sense KRP1 probe but it is much lower than with the antisense probe (compare Fig. 1, KRP1 and Fig. 2A). Also, a signal was not observed in control hybridizations with antisense probes pretreated with RNase (shown for KRP3 in Fig. 2D).
DISCUSSION
Analysis of the expression patterns of the seven KRPs of A. thaliana in the shoot apex by mRNA in situ hybridization suggests differential functions for the distinct KRPs in the control of the mitotic division cycle and/or endoreduplication cycle. The expression data presented (summarized in Table 1) provide evidence for different expression pattern groups. The first group comprises KRP1 and KRP2, which are highly expressed in endoreduplicating tissues such as the pith cells and mesophyll cells of maturing leaves, but are not present in the mitotic dividing cells of the SAM or the vascular cells of the shoot apex. Therefore, KRP1 and KRP2 might specifically be involved in the shift of the mitotic cycle to the endoreduplication cycle in the shoot apex, or even perhaps in the control of the endocycle itself. A role for the KRPs in controlling the ploïdy level is evident from transgenic plants overexpressing the KRP1 and KRP2 genes. In comparison with control plants, these transgenic plants display a decrease in their ploïdy level (De Veylder et al., 2001; Zhou et al., 2002). Curiously, whereas KRP1 mRNA was distributed equally over the whole of the leaf primordia, KRP2 transcripts accumulated preferentially in the differentiating spongy mesophyll cells. This expression profile is complementary to that reported for CYCD3;1, a D‐type cyclin shown to be rate‐limiting for cell division in dividing calli starved of cytokinin (Riou‐Khamlichi et al., 1999). Its specific accumulation pattern in the shoot suggests a role for KRP2 in the onset of cell differentiation.
Table 1.
Expression levels of seven KRPs of A. thaliana in the shoot apex
| Shoot apical | Young leaf primordia | Mature leaf | |||||
| Type of KRPs | mersitem | Vascular cells | Pith | Cortex | Palisade mesophyll | Spongy mesophyll | primordia |
| KRP1 | O | O | XX | X | XX | XX | XX |
| KRP2 | O | O | XXX | X | XX | XXX | XX |
| KRP3 | X | X | XX | X | XX | XX | X |
| KRP4 | XX | X | O | O | XX | XX | O |
| KRP5 | XXX | XX | X | X | XX | XX | X |
| KRP6 | XX | X | X | X | XX | XX | XX |
| KRP7 | X | XX | XX | XX | XX | XX | XX |
Abbreviations: O = not expressed; X = slightly expressed; XX = expressed; XXX = highly expressed.
The second class of KRPs comprises KRP4 and KRP5, which are highly expressed in the dividing cells of the SAM, young leaf primordia, procambium and vascular tissue of the shoot apex. Since the mitotic cyclin CYCB1;1 is also expressed in these cells (Segers et al., 1996), we hypothesize that KRP4 and KRP5 might be involved in regulating the progression through the mitotic cell cycle. There is an apparent discrepancy between our data for KRP4 and those previously published by De Veylder et al. (2001), who have observed from transcription profiles a KRP4 expression in 3‐day‐old leaves where most cells are elongating and are probably endoreduplicating. Since divisions occur in the vascular cells of these leaves, we cannot exclude the possibility that KRP4 might be part of regulating the progression through the mitotic cell cycle. Curiously, the KRP5 hybridization signal is particular high in just‐initiated procambial cells and in the peripheral zone of the SAM, suggesting a more specific role for KRP5 in the divisions that occur in the zone where leaf primordia initiate and in the zone where the periphery of the stem is constructed.
The third group of KRPs comprises KRP3, KRP6 and KRP7, for which transcripts can be detected in both mitotically dividing and endoreduplicating cells. The KRP3 transcripts accumulate particularly in the upper cells of the pith, just below the L3 layer. It would be of interest to determine if KRP3 interacts with either gene involved in the maintenance of the SAM state (Sharma and Fletcher, 2002).
At this time it is still unclear why A. thaliana has so many different KRPs that are expressed in the same tissues. It is probable that KRPs might participate in regulating CDK activity in response to different developmental or environmental signals. This hypothesis is supported by the fact that KRP1 expression is induced by abscisic acid (Wang et al., 1998). Moreover, the KRP4 expression pattern in the shoot apex is highly similar to that of CYCD3;1 (Riou‐Khamlichi et al., 1999). Since CYCD3 is specifically induced by cytokinin, KRP4 might also be under the control of this mitogen. In order to further understand the functions of the KRP genes it would be interesting to identify the distinct mitogenic and environmental factors regulating their expression.
ACKNOWLEDGEMENTS
The authors thank Gilbert Engler for critical reading of the text, Marleen Brunain and Nathalie Detry for technical assistance, and Martine De Cock for help in preparing the manuscript. This work was supported by grants from the Interuniversity Poles of Attraction Programme (Belgian State, Prime Minister’s Office‐Federal Office for Scientific, Technical and Cultural Affairs; P4/15, P5/13) and the ‘Fonds de la Recherche Fondamentale et Collective’ (no. 2.4524.96). S.O. is indebted to the ‘Fonds National pour la Recherche Scientifique’ for a grant of ‘Collaborateur Scientifique’. L.D.V. is a postdoctoral fellow of the Fund for Scientific Research (Flanders).
Supplementary Material
Received: 1 September 2003; Returned for revision: 1 December 2003; Accepted: 17 December 2003 Published electronically: 22 March 2004
References
- ChengM, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ.1999. The p21Cip1 and p27Kip1 CDK ‘inhibitors’ are essential activators of cyclin D‐dependent kinases in murine fibroblasts. EMBO Journal 18: 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CorbesierL, Gadisseur I, Silvestre G, Jacqmard A, Bernier G.1996. Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. The Plant Journal 9: 947–952. [DOI] [PubMed] [Google Scholar]
- De VeylderL, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D.2001. Functional analysis of cyclin‐dependent kinase inhibitors of Arabidopsis The Plant Cell 13: 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DunphyWG.1994. The decision to enter mitosis. Trends in Cell Biology 4, 202–207. [DOI] [PubMed] [Google Scholar]
- ElledgeSL, Harper JW.1994. CDK inhibitors: on the threshold of check points and development. Current Opinion in Cell Biology 6: 847–852. [DOI] [PubMed] [Google Scholar]
- HarperJW, Elledge SJ.1996. Cdk inhibitors in development and cancer. Current Opinion in Genetics & Development 6: 56–64. [DOI] [PubMed] [Google Scholar]
- JacqmardA, De Veylder L, Segers G, de Almeida Engler J, Bernier G, Van Montagu M, Inzé D.1999. CKS1At expression in Arabidopsis thaliana suggests a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207: 496–504. [DOI] [PubMed] [Google Scholar]
- LaBaerJ, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E.1997. New functional activities for the p21 family of CDK inhibitors. Genes and Development 11: 847–862. [DOI] [PubMed] [Google Scholar]
- LuiH, Wang H, DeLong C, Fowke LC, Crosby WL, Fobert PR.2000. The Arabidopsis Cdc2a‐interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin‐dependent kinase activity in vitro The Plant Journal 21: 379–385. [DOI] [PubMed] [Google Scholar]
- MironovV, De Veylder L, Van Montagu M, Inzé D.1999. Cyclin‐dependent kinases and cell division in plants‐ the nexus. The Plant Cell 11: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MorganDO.1997. Cyclin‐dependent kinases: engines, clocks, and micro‐processors. Annual Review of Cell and Developmental Biology 13: 261–291. [DOI] [PubMed] [Google Scholar]
- NorburyC, Nurse P.1992. Animal cell cycles and their control. Annual Review of Biochemistry 61: 440–470. [DOI] [PubMed] [Google Scholar]
- PinesJ.1994. Protein kinases and cell cycle control. Seminars in Cell Biology 5: 399–408. [DOI] [PubMed] [Google Scholar]
- PinesJ.1995. Cyclins and cyclin‐dependent kinases: a biochemical view. Biochemical Journal 308: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou‐KhamlichiC, Huntley R, Jacqmard A, Murray JAH.1999. Cytokinin activation of Arabidopsis cell division through a D‐type cyclin. Science 283: 1541–1544. [DOI] [PubMed] [Google Scholar]
- SegersG, Gadisseur I, Bergounioux C, de Almeida Engler J, Jacqmard A, Van Montagu M, Inzé D.1996. The Arabidopsis cyclin‐dependent kinase gene cdc2bt is preferentially expressed during S and G2 phases of the cell cycle. The Plant Journal 10: 601–612. [DOI] [PubMed] [Google Scholar]
- SharmaVK, Fletcher JC.2002. Maintenance of shoot and floral meristem cell proliferation and fate. Plant Physiology 129: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SherrCJ, Roberts JM.1995. Inhibitors of mammalian G1 cyclin‐dependent kinases. Genes and Development 9: 1149–1163. [DOI] [PubMed] [Google Scholar]
- SherrCJ, Roberts JM.1999. CDK inhibitors: Positive and negative regulators of G1‐phase progression. Genes and Development 13: 1501–1512. [DOI] [PubMed] [Google Scholar]
- Van de poeleK, Reas J, De Veylder L, Rouzé P, Rombauts S, Inzé D.2002. Genome‐wide analysis of core cell cycle genes in Arabidopsis. The Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WangH, Fowke LC, Crosby WL.1997. A plant cyclin‐dependent kinase inhibitor gene. Nature 386: 451–452. [DOI] [PubMed] [Google Scholar]
- WangH, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC.1998. ICK1, a cyclin‐dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. The Plant Journal 15: 501–510. [DOI] [PubMed] [Google Scholar]
- WangH, Zhou Y, Gilmer S, Whitwill S, Fowke LC.2000. Expression of the plant cyclin‐dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. The Plant Journal 24: 613–623. [DOI] [PubMed] [Google Scholar]
- ZhangP, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ.1999. p21CIP1 and p57KIP2 control muscle differentiation at the myogenin step. Genes and Development 13: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhouY, Fowke LC, Wang H.2002. Plant CDK inhibitors: studies of interactions with cell cycle regulators in the yeast two‐hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Reports 20: 967–975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


