Abstract
• Backgroud and Aims The genus Bupleurum has long been recognized as a natural group, but its infrageneric classification is controversial and has not yet been studied in the light of sequence data.
• Methods Phylogenetic relationships among 32 species (35 taxa) of the genus Bupleurum were investigated by comparative sequencing of the ITS region of the 18–26S nuclear ribosomal DNA repeat. Exemplar taxa from all currently accepted sections and subsections of the genus were included, along with outgroups from four other early branching Apioideae genera (Anginon, Heteromorpha, Physospermum and Pleurospermum).
• Key Results Phylogenies generated by maximum parsimony, maximum likelihood, and neighbour‐joining methods show similar topologies, demonstrating monophyly of Bupleurum and the division of the genus into two major clades. This division is also supported by analysis of the 5.8S coding sequence alone. The first branching clade is formed by all the species of the genus with pinnate‐reticulate veined leaves and B. rigidum with a unique type of leaf venation. The other major clade includes the remaining species studied, all of which have more or less parallel‐veined leaves.
• Conclusions These phylogenetic results do not agree with any previous classifications of the genus. Molecular data also suggest that the endemic Macaronesian species B. salicifolium is a neoendemic, as the sequence divergence between the populations in Madeira and Canary Islands, and closer mainland relatives in north‐west Africa is small. All endemic north‐west African taxa are included in a single unresolved but well‐supported clade, and the low nucleotide variation of ITS suggests a recent radiation within this group. The only southern hemisphere species, B. mundii (southern Africa), is shown to be a neoendemic, apparently closely related to B. falcatum, a Eurasian species.
Key words: Anginon, Apiaceae, Apioideae, Bupleurum, Heteromorpha, ITS, molecular phylogeny, nuclear ribosomal DNA, Penninervia, subgenus nov., systematics, Umbelliferae
INTRODUCTION
Bupleurum L., with around 150 species, is one of the largest genera of the family Apiaceae (Umbelliferae). The genus includes annual and perennial species, from small herbs a few centimetres tall (e.g. B. semicompositum L., B. ranunculoides L.), to shrubs up to 3 m high (B. fruticosum L.). Woodiness in Apiaceae is rare, as woody species are found in only ten of the 400 or so genera in the family (Burtt, 1991). Bupleurum is morphologically diverse, but is easily distinguished within the family because of the almost unique feature of the leaves, which are simple and entire (Hohenackeria Fisch. & C.A.Mey. and Nirarathamnos Balf.f. also have simple and entire leaves, but are otherwise morphologically distinct from Bupleurum). All species in Bupleurum are totally glabrous; some have pinnate‐reticulate veined leaves, but most have parallel‐veined leaves (another unusual character in the family); flowers are generally yellow, sometimes purplish (only B. album Maire, from north‐west Africa, has white flowers); involucral bracts are generally present; involucel bracts (bracteoles) are always present and sometimes conspicuous; mericarps are isodiametric or slightly laterally compressed, and are generally smooth, rarely tuberculate, with five thin or narrowly winged ridges.
Bupleurum is essentially a northern hemisphere genus, occurring in Europe, North Africa, Macaronesia (Canary Islands and Madeira), Asia and North America (a single native species, B. americanum Coult. & Rose, from the Rocky Mountains). There is a notable exception to this northern distribution: the southern African endemic B. mundii Cham. & Schltdl. Despite this broad distribution, most of the species are rather rare and restricted to small areas. In terms of morphology and taxon number, the highest diversity is found in the Mediterranean, where there are many endemics. Bupleurum spp. grow in a great variety of habitats: from sea level (B. tenuissimum L.) up to 4900 m (the Himalayan B. longicaule Wall. ex DC.); through saline marshlands, calcareous, granitic or basaltic soils; in arid or mesophytic areas; from open areas to dense forests; as weeds or ruderals; and from equatorial to nearly arctic latitudes (Nasir, 1955; Tutin, 1968; Heywood, 1971; Snogerup, 1972; Cauwet‐Marc, 1976; Hara and Williams, 1979; Rechinger and Snogerup, 1987; Snogerup and Snogerup, 2001).
Bupleurum has for long been recognized as a natural group. In the 17th century, Tournefort (1694) had a concept of the genus that basically corresponds to its present delimitation. He included in Bupleurum species from all currently accepted sections (Bupleurum, Coriacea Gren. & Godr., Diaphyllum (Hoffm.) Dumort., Isophyllum (Hoffm.) Dumort. and Reticulata Gren. & Godr.), and none from different genera (Neves, 2000). Later authors, including Linnaeus (1753) and Sprengel (1820), included species from other genera in Bupleurum, or split the genus into segregate genera such as Buprestis Spreng., Diaphyllum Hoffm., Odontites Spreng., Perfoliata Fourr., Tenoria Spreng., and others (see Pimenov and Leonov, 1993). The current circumscription of Bupleurum has remained constant for more than 150 years, with the exception of Calestani (1905) who resurrected the genus Trachypleurum Rchb., a clearly artificial group that included the few Bupleurum spp. with tuberculate fruits.
Traditionally Bupleurum has been placed in subfamily Apioideae, tribe Apieae, subtribe Apiinae (Heywood, 1971). However, Downie et al. (2000b) found strong molecular evidence showing the isolated position of Bupleurum within Apiaceae and concluded that Bup leurum should be included within the monogeneric tribe Bupleureae Spreng.
Table 1 summarizes the various infrageneric classifications proposed for Bupleurum since Grenier and Godron (1848) first used sections in the genus. Wolff (1910) published the first comprehensive revision of Bupleurum, and his classification is the most widely used today. Cauwet‐Marc (1976) revised Bupleurum using phenetic and cladistic analyses of morphological characters (see also Roux et al., 1978). She proposed the subdivision of the genus into two subgenera: Bupleurum and Tenoria (Spreng.) Cauwet. However, the reliability of these results has been questioned as only a small number of characters were used and no characters were given to support the groups. In summary, there has been almost no agreement on the status or rank of the infrageneric groups within Bupleurum, with only sections Bupleurum (‘Perfoliata’) and Coriacea having some stability in terms of circumscription. The classifications of Koso‐Poljansky (1913), Cerceau‐Larrival (1962) and Cauwet‐Marc (1976) are the most discordant, with Koso‐Poljansky and Cauwet‐Marc placing great emphasis on vegetative characters.
Table 1.
Infrageneric classification of the genus Bupleurum
| Authors | Bupleurum infrageneric classification |
| Grenier and Godron(1848) | Sect. Perfoliata, sect. Reticulata, sect. Aristata, sect. Nervosa, sect. Marginata, sect. Coriacea. [The first use of section names with Bupleurum] |
| Boissier (1872) | Sect. Perfoliata, sect. Glumacea, sect. Graminea, sect. Coriacea |
| Briquet (1897) | Sect. Perfoliata (subsections: Laevia, Rugosa), sect. Reticulata, sect. Eubupleura (subsections: Aristata, Juncea, Trachycarpa, Nervosa) |
| Drude (1898) | Sect. Perfoliata, sect. Reticulata, sect. Eubupleura (subsections: Aristata, Juncea, Trachycarpa, Nervosa), sect. Rigida, sect. Coriacea |
| Calestani (1905) | Genus Bupleurum: sect. Perfoliata, sect. Reticulata, sect. Glumacea, sect. Juncea, sect. Trachycarpa, sect. Nervosa, sect. Rigida, sect. Frutescentia, sect. Spinosa, sect. Coriacea. Genus Trachypleurum |
| Wolff (1910) | Sect. Perfoliata (subsections: Laevia, Rugosa, Lophocarpa), sect. Reticulata, sect. Longifolia, sect. Eubupleura (subsections: Glumacea, Juncea, Trachycarpa, Nervosa, Marginata, Rigida), sect. Coriacea |
| Koso‐Poljanski | Subgenus Diatropa: sect. Laevia (subsections: Alophia, Lophocarpa), sect. Rugosa (subsections: Eurugosa, Granulata) |
| (1913) | Subgenus Bupleurotypus: sect. Eubupleurotypus (subsections: Plurivittata, Legitima), sect. Vittijugata, sect. Tenoria (subsections: Rigida, Coriacea) |
| Subgenus Agostana: sect. Glumacea, sect. Graminea (subsections: Lejocarpa, Trachycarpa) | |
| Cearceau‐Larrival (1962) | Section I, Graminifolia, section II [Bupleurum] |
| Tutin (1968) | Sect. Bupleurum, sect. Reticulata, sect. Diaphyllum, sect. Isophyllum (subsections: Aristata, Juncea, Trachycarpa, Nervosa, Marginata, Rigida), sect. Coriacea. [following Wolff (1910) but with nomenclatural changes] |
| Cauwet‐Mark (1976) | Subgenus Bupleurum: sect. Bupleurum (subsections: Laevia, Rugosa), sect. Bupleurotypus (subsections: Longiradiata, Nervosa), sect. Vittaejugata, sect. Agostana (subsections: Glumacea, Juncea, Trachycarpa) |
| Subgenus Tenoria: sect. Tenoria (subsections: Tenoria, Rectinervia), sect. Tyrrhenaica (subsections: Mesatlantica, Acutifolia) |
Systematic data available for Bupleurum
Many data have been gathered for Bupleurum, particularly in karyology (mainly chromosome numbers), pollen morphology, anatomy, phytochemistry and more lately molecular studies. Most of the karyological work in the genus was carried out by Cauwet‐Marc (1976, 1979a, b); she recognized five basic chromosome numbers in Bupleurum: x = 4, 6, 7, 8 and 11; the most common being x = 8, followed by x = 7 (x = 7 or 8 are uncommon in Apiaceae, where the most frequent number is x = 11). Some species exhibit various levels of ploidy, e.g. B. ranunculoides, with diploid (2n = 14), triploid (2n = 21), tetraploid (2n = 28), and hexaploid (2n = 42) populations (Küpfer, 1969; Cauwet‐Marc, 1970, 1979b). Cases of aneuploidy and dysploidy have been recorded, especially in north‐west African taxa (Cauwet‐Marc, 1979b) and in the Eurasian B. falcatum L. sensu lato (Ohta, 1991). Unfortunately, chromosome numbers alone have limited taxonomic value, especially when variation in number caused by aneuploidy and dysploidy appears common in certain species.
Pollen in Bupleurum is generally characterized as sub‐rhomboidal, and of a ‘primitive type’ (Cerceau‐Larrival, 1962, 1971; Cerceau‐Larrival and Roland‐Heydacker, 1978; Punt, 1984; Leonardis et al., 1997), and exhibits only minor variation across the many species studied.
Some anatomical work has been carried out in the genus, particularly on stems and fruits (Briquet, 1897; Panelatti, 1959; Cauwet‐Marc, 1976; Arenas and García, 1993). In some cases, additional features for species delimitation were found, but further data are needed to understand the value of these characters in the study of species relationships.
Some Bupleurum species have been included in several chemical studies, in particular concerning the distribution of secondary metabolites (see Carbonnier and Cauwet‐Marc, 1979, 1981). However, chemical information can be problematic as uncontrolled environmental factors can affect chemical production, as Berenbaum (1990) demonstrated for furacoumarins in Apiaceae.
Cauwet‐Marc et al. (1978) undertook a multidisciplinary systematic study in Bupleurum, with data from morphology, anatomy, phytochemistry, karyology, palynology and phytodermology. However, the results are problematic as data interpretation was biased by Cauwet‐Marc’s classification (Cauwet‐Marc, 1976), the study lacked a detailed account of the methods, and reasons for the proposed taxon associations were not discussed.
In recent years a few Bupleurum spp. have been included in molecular phylogenetic studies carried out on Apiaceae, using sequences of plastid genes (rbcL, matK), introns, rpoC1 and rpl16 (Kondo et al., 1996; Plunkett et al., 1996a, b, 1997; Downie et al., 1998), and chloroplast restriction site analysis (Plunkett and Downie, 1999, 2000). In all of these studies, Bupleurum consistently appears as an early branching clade within subfamily Apioideae. Choi et al. (1996) published the first ITS (internal transcribed spacers) sequences in Bupleurum for B. euphorbiodes Nakai, B. komarovianum O.A.Lincz., B. longiradiatum Turcz. and B. scorzonerifolium Willd. They also carried out an RFLP (restriction fragment length polymorphism) analysis of the same samples sequenced for ITS, and found similar taxon relationships. ITS sequences have also been obtained for B. falcatum (Lee and Rasmussen, 1998; Valiejo‐Roman et al., 1998). The three Bupleurum spp. in the analysis of Lee and Rasmussen (1998) form a separated clade, but appear in a larger group including genera such as Aciphylla J.R.Forst. & G.Forst., Daucus L., Cuminum L., Laserpitium L. and Thapsia L. This contradicts the plastid DNA tree for the rpoC1 intron (Downie et al., 1998). In contrast Valiejo‐Roman et al. (1998) showed B. falcatum in an early branching position within Apioideae, which agrees with the signal from plastid DNA. They also noted that the sequence of B. falcatum is highly divergent and that it could not be aligned unambiguously with the rest of the taxa examined. These studies have helped place Bupleurum within Apioideae; however, the number of taxa sequenced to date is small and not representative of the taxonomic, morphological, and geographical diversity in the genus. Therefore, a more extended sampling of the species is necessary to achieve a better understanding of the phylogenetic relationships in Bupleurum.
The present study sets out to provide more detailed phylogenetic resolution within the genus Bupleurum, with particular emphasis on taxa from south‐west Europe and north‐west Africa (a possible centre of origin for the genus). The increased sampling within the genus would also further test monophyly of Bupleurum. The ITS region of nuclear ribosomal DNA was chosen as it has been proved a very valuable source of phylogenetic information, particularly at the genus level and among related genera (Baldwin, 1992; Baldwin et al., 1995). The ITS region has also been successfully used in various studies in subfamily Apioideae (Downie and Katz‐Downie, 1996; Downie et al., 1998, 2000a, c; Valiejo‐Roman et al., 1998; Katz‐Downie et al., 1999).
MATERIALS AND METHODS
Sampling and molecular methods
Bupleurum taxa (the ingroup), representing all currently accepted sections and subsections of the genus, were examined for sequence variation in the ITS region of nuclear rDNA (see Table 2). Authorities for taxa are also given in Table 2. Complete ITS and 5.8S sequences for 68 accessions representing 32 species (35 taxa) of Bupleurum and two species of Anginon Raf. are reported here for the first time. Details of origin of material, GenBank accession numbers, and location of herbarium vouchers are provided in Table 2. All material used was collected and identified, either from living plants in the wild (18 accessions) or cultivated (eight), or from herbarium specimens (42). The Anginon specimens used were identified following Allison and Van Wyk (1997). All fresh material was dried in silica gel upon collection following the protocol by Chase and Hills (1991).
Table 2.
Accessions of Bupleurum and outgroup taxa examined for nuclear rDNA ITS sequence variation
| Taxon* | Section/subsection of Bupleurum† | Source and voucher | GenBank accession number |
| Anginon difforme (L.) B.L.Burtt | n/a | South Africa, Calvinia, Hantam Mountains, Farm Vanrhynshoek, 21.viii.1990, All Batten AB 1018 (E) | AF459742 |
| Anginon paniculatum (Thunb.) B.L.Burtt | n/a | South Africa, ‘Central ridge’ above Clanwilliam, dam 5 km from Clanwilliam, 18.ii.1985, H.C. Taylor 11271 (E) | AF467922 |
| Bupleurum acutifolium Boiss. [1] | Isophyllum/Rigida | Spain, Málaga, Sierra Bermeja, Peñas Blancas to Los Reales, 05.ix.1997, S.S. Neves 64 (E) | AF467925 |
| B. acutifolium [2] | Isophyllum/Rigida | Spain, Málaga, Sierra Bermeja, Estepona to Puerto de Peñas Blancas, 05.ix.1997, S.S. Neves 65 (E) | AF467926 |
| B. acutifolium [3] | Isophyllum/Rigida | Spain, Málaga, Tolóx, Sierra Parda, Majada Redonda, 06.v.1994, A. Pérez‐Latorre et al. (MGC 37708) | AF467927 |
| B. acutifolium [4] | Isophyllum/Rigida | Portugal, Baixo Alentejo, Serra do Cercal, Vila Nova de Milfontes to Cercal, 16.ix.1996, S.S. Neves 24 (E) | AF467923 |
| B. acutifolium [5] | Isophyllum/Rigida | Portugal, Baixo Alentejo, S. Luis, Serra de S. Domingos, 08.viii.1997, S.S. Neves 27 (E) | AF467924 |
| B. album Maire | (Isophyllum/Rigida) | Morocco, Anti‐Atlas, 4 km from Igherm, road to Taliouine, 10.vi.1974, Reading University/BM Expedition 532 (RNG) | AF467928 |
| B. angulosum L. | Reticulata | Pyrenees. Cultivated, Royal Botanic Garden Edinburgh, UK; RBGE Acc. No. 19861043, 20.viii.1999, S.S. Neves (E) | AF469008 |
| B. balansae Boiss. & Reut. [1] | Isophyllum/Rigida | Morocco, Oujda, Al’Youn, Machra, Homadi, Oued Senara, 09.vi.1993, J. Molero et al. JMM‐3198/5 (RNG) | AF469680 |
| B. balansae [2] | Isophyllum/Rigida | Morocco, c. 35 km S of Tetouan, along main road to Chechaouen, near Souk‐el‐Arba‐des‐Beni‐Hassan, 18.vi.1987, S.L. Jury et al. 8338 (RNG) | AF469681 |
| B. baldense Turra | Isophyllum/Aristata | Spain, Guadalajara, Checa, river Cabrillas, road from Checa to Orea, 21.vi.1995, M.A. Carrasco et al. (MA 558704) | AF469682 |
| B. barceloi Coss. ex Willk. | Isophyllum/Rigida | Balearic Islands (Spain), Mallorca, Sóller, ‘Barranco’ [cliff] 25.vii.1989, J. Orell Casasnovas (MA 474781) | AF477023 |
| B. benoistii Litard. & Maire [1] | (Isophyllum/Nervosa) | Morocco, High Atlas, S from Marrakech, N end of Jbel Oikaïmeden near azib in ski resort, 29.vii.1997, S.L. Jury et al. 18375 (E) | AF477024 |
| B. benoistii [2] | (Isophyllum/ Nervosa) | Morocco, 72 km S from Marrakech, Oukaïmeden, 03.vii.1987, S.L. Jury et al. 8858 (RNG) | AF477025 |
| B. benoistii [3] | (Isophyllum/Nervosa) | Morocco, prov. of Ksar el Souk, N High Atlas, Tizi n’Tirrecht, N of Ari n’Ayachi, near Midelt, 21.vii.1966, R.M. & A.M. Harley 766 (BM) | AF477026 |
| B. canescens Schousb. | Isophyllum/Rigida | Morocco, Immouzer Valley, N of Agadir, 28.iii.1972, D. Bramwell et al. 265 (RNG) | AF477027 |
| B. canescens var. handiense Bolle [1][syn. B. handiense (Bolle) G.Kunkel] | Isophyllum/Rigida | Canary Islands (Spain). Cultivated, Jardim Botânico de Coimbra, Portugal; seed received from the Jardín Botánico ‘Viera y Clavijo’, Canary Islands, from wild sources; 05.viii.2000, S.S. Neves Acc. No. 28 (COI) | AF477028 |
| B. canescens var. handiense [2] | Isophyllum/Rigida | Canary Islands (Spain), Lanzarote, Peñas de Chache, cliffs above Famara, 15.v.1969, D. Bramwell 1631 (E) | AF477029 |
| B. dumosum Coss. | Isophyllum/Rigida | Morocco, c. 9·5 km NNE of Asni, 5 km SSW of Tahanaoute, Gorge de Moulay Brahim, 15.iii.1994, S.L. Jury et al. 14157 (RNG) | AF477030 |
| B. falcatum L. | Isophyllum/Nervosa | Spain, Alava, Lagrán, Sierra de Cantabria, rocky crests of Recilla, 15.viii.1992, J.A. Alejandre 733/92 (MA 534085) | AF479290 |
| B. fruticescens L. subsp. fruticescens [1] | Isophyllum/Rigida | Spain, Huesca, between Baldellou and Camporrels, 30.ix.1987, G. Montserrat (MA 515853) | AF479291 |
| B. fruticescens subsp. fruticescens [2] | Isophyllum/Rigida | Spain, Murcia, Sierra de Espuña, on road to ‘Centro de Interpretación’, 24.viii.1997, S.S. Neves 52 (E) | AF479292 |
| B. fruticescens L. subsp. spinosum (Gouan) O.Bolòs & Vigo [1] | Isophyllum/Rigida | Spain, Granada, Sierra Nevada, Monachil valley, high mountain, 16.viii.1997, S.S. Neves 42 (E) | AF479293 |
| B. fruticescens subsp. spinosum [2] | Isophyllum/Rigida | Spain, Cádiz, Zahara to Grazalema, margins of road CA‐531, 22.viii.1997, S.S. Neves 47 (E) | AF479294 |
| B. fruticescens subsp.spinosum [3] | Isophyllum/Rigida | Morocco, High Atlas, S from Marrakech, ski resort of Oukaïmeden, 27.vii.1997, S.L. Jury et al. 18297 (E) | AF479295 |
| B. fruticescens subsp. spinosum [4] | Isophyllum/Rigida | Morocco, W Rif, Chefchaouene, Djebel Bouhalla to Djebel Lakraa, 23.vii.1995, M.A. Mateos et al. 6988/95 (RNG) | AF479296 |
| B. fruticosum L. [1] | Coriacea | Portugal, Estremadura, Serra da Arrábida, on road from Aldeia de Irmaõs to Casais da Serra, 10.viii.1997, S.S. Neves 33 (E) | AF479297 |
| B. fruticosum [2] | Coriacea | Spain, Málaga, Sierra Bermeja, road Ronda to Puerto de Alijar, 15.viii.1997, S.S. Neves 41 (E) | AF479298 |
| B.gerardii All. [1] | Isophyllum/Juncea | Europe. Cultivated, Jardim Botânico de Coimbra, Portugal; seeds obtained from the Jardin Botanique National de Belgique, Meise; 03.viii.1994, S.S. Neves Acc. No. 17a (E) | AF479847 |
| B. gerardii [2] | Isophyllum/Juncea | Portugal, Beira Litoral, Fátima, Valinhos, area of ‘Via Sacra’, 26.vi.1994, S.S. Neves 1 (E) | AF479848 |
| B. gerardii [3] | Isophyllum/Juncea | Spain, Madrid, ‘embalse’ [dam] of Santillana, Cerro Casal, 10.vii.1981, G. Navarro et al. (MA 310732) | AF479849 |
| B. gerardii [4] | Isophyllum/Juncea | Spain, Granada, Lobras, Barranco de los Lagartos, 07.v.1980, J. Molero Mesa (MA 214590) | AF479850 |
| B. gibraltarium Lam. [1] | Coriacea | Spain, Sevilla, near Coripe, road to Algodonales, slopes bordering river Guadalporcún, 15.viii.1997, S.S. Neves 35 (E) | AF479851 |
| B. gibraltarium [2] | Coriacea | Spain, Murcia, Sierra de Espuña, on road to ‘Centro de Interpretación’, 24.viii.1997, S.S. Neves 51 (E) | AF479852 |
| B. lancifolium Hornem. | Bupleurum/Bupleurum | Morocco, Tanger, SW of Chefchaouen, 2·3 km up road to Mokrissèt from Pont du Loukos, 21.iv.1995, S.L. Jury et al. 16552 (RNG) | AF479853 |
| B. lateriflorum Coss. ex H.Wolff [1] | Isophyllum/Rigida | Morocco, High Atlas, S from Marrakech, 4 km below Oukaïmeden, on road to Vallée de l’Ourika, 28.vii.1997, S.L. Jury et al. 18323 (E) | AF479854 |
| B. lateriflorum [2] | Isophyllum/Rigida | Morocco, High Atlas, Tizi‐n‐Test Pass, 02.x.1991, M. Ait Lafkih et al. 4939 (E) | AF479855 |
| B. longifolium L. | Diaphyllum | Germany, Baviera, Oberpfalz, Kreis Regensburg, 0·3 km WSW from Pentling, N of road from Pentling to Weichsmühl, margins of river Donau, 17.vii.1993, H. Förther 7503 (MAF 149194) | AF479856 |
| B. montanum Coss. [1] | Isophyllum/Rigida | Morocco, Chefchaouen (area 7), between Ketama and Bab‐Berret, 03.xi.1993, P. García Murillo et al. ST 251/93 (SEV) | AF479857 |
| B. montanum [2] | Isophyllum/Rigida | Morocco, High Atlas, just above El‐Ksiba, along road to Imilchil, 05.vii.1997, S.L. Jury 17456a (E) | AF479858 |
| B. montanum [3] | Isophyllum/Rigida | Morocco, Checfchaouen (area 2), Djebel Tassaot, 22.vii.1995, M.A. Mateos et al. 6914/95 (SEV) | AF479859 |
| B. mundii Cham. & Schltdl. | Isophyllum/Nervosa | South Africa, Natal. Cultivated, Royal Botanic Garden Edinburgh, UK; RBGE Acc. No. 19972669; 29.xi.1999, S.S. Neves (E) | AF479860 |
| B. odontites L. | Isophyllum/Aristata | Tunisia, Kroumirie, c. 14 km N from Jendouba (Souk el Arba) to Ain Draham, S of Tabarka, 11.v.1975, Davis & Lamond D57628 (RNG) | AF479861 |
| B. oligactis Boiss. [1] [syn. B. atlanticum Murb.] | Isophyllum/Rigida | Morocco, High Atlas, 111 km N from Errachidia (Ksar‐es‐Souk) along P21 road to Midelt, a few kms S of Tizi‐m‐Tairhemt, 12.vii.1987, S.L. Jury et al. 9240 (SEV 127166) | AF479862 |
| B. oligactis [2] | Isophyllum/Rigida | Morocco, High Atlas, about 3 km above Imilchil, on road to lake Tizlite, along El‐Ksiba to Imilchil road, 07.vii.1997, S.L. Jury 17603 (E) | AF479864 |
| B. oligactis [3] | Isophyllum/Rigida | Morocco, Middle Atlas, road from El‐Ksiba to Imilchil, c. 9 km N from Tizi‐n‐Islay, 05.vii.1997, S.L. Jury 17516 (E) | AF479863 |
| B. plantagineum Desf. | Isophyllum/Rigida | Algeria, K2, Cap Carbon, near Bejaïa (Bougie), 29.v.1991, Davis 52959 (RNG) | AF479865 |
| B. praealtum L. [1] | Isophyllum/Juncea | Spain, Lérida, Valle de Boí, 2 km from Barruera to Pont de Suert, 23.viii.1987, C. Aedo et al. 286–87 ML (MA 449973) | AF480938 |
| B. praealtum [2] | Isophyllum/Juncea | Spain, Huesca, San Juan de Plan, near ‘ermita’ [hermitage] of San Mamés, 01.viii.1981, P. Montserrat et al. (JACA 191581) | AF480939 |
| B. praealtum [3] | Isophyllum/Juncea | Spain, Teruel, Loscos, Piedrahita, river Noguera, 13.viii.1995, C. Fabregat & Lopez Udias (JACA 683295) | AF481390 |
| B. praealtum [4] | Isophyllum/Juncea | Spain, Salamanca, Montemayor del Rio, 02.viii.1983, J.L. Fernández Alonso & A. Guillen (MA 518939) | AF481391 |
| B. ranunculoides L. [1] | Isophyllum Nervosa | Europe. Cultivated, Jardim Botânico de Coimbra, Portugal; seed obtained from the Botanischer Garten Tübingen, Germany; 11.iv.1995, S.S. Neves Acc. No. 43 (E – photo) | AF481392 |
| B. ranunculoides [2] | Isophyllum/Nervosa | Europe. Cultivated, Royal Botanic Garden Edinburgh, UK; seed obtained from the Botanischer Garten der Universität Postdam, Germany; RBGE No. 19972161, 20.vii.1999, S.S. Neves (E) | AF481393 |
| B. ranunculoides [3] | Isophyllum/Nervosa | Spain, Burgos, Rebolledo de la Torre, Peña Castro, 24.vi.1990, J.A. Alejandre 1078/90 (MA 493699) | AF481394 |
| B. ranunculoides [4] [syn. B. bourgaei Boiss. & Reut.] | Isophyllum/Nervosa | Spain, Jaén, Pontones, Sierra de Banderillas, 10.viii.1982, C. Soriano (MA 462383) | AF481395 |
| B. rigidum L. subsp. rigidum [1] | Isophyllum/Marginata | Spain, Murcia, Sierra de Espuña, picnic area near ‘Centro de Interpretación’, 24.viii.1997, S.S. Neves 53 (E) | AF481396 |
| B. rigidum subsp. rigidum [2] | Isophyllum/Marginata | Spain, Málaga, Sierra Bermeja, on road from Estepona to Puerto de Peñas Blancas, 05.ix.1997, S.S. Neves 63 (E). | AF481397 |
| B. rigidum L. subsp. paniculatum (Brot.) H.Wolff [1] | Isophyllum/Marginata | Portugal, Beira Litoral, Coimbra, Quinta da Sapata, near Santa Clara, 27.vi.1994, F. Sales & S.S. Neves 3a (E) | AF481398 |
| B. rigidum subsp. paniculatum [2] | Isophyllum/Marginata | Portugal, Estremadura, Serra da Arrábida, Vale da Rasca, on road to the ‘Secil’ factory, 10.viii.1997, S.S. Neves 31 (E) | AF481399 |
| B. rotundifolium L. | Bupleurum/ Bupleurum | Europe. Cultivated, Jardim Botânico de Coimbra, Portugal; seed obtained from the Botanischer Garten St. Gallen, Switzerland, 03.viii.1994, S.S. Neves Acc. No. 4 (E) | AF481400 |
| B. salicifolium R.Br. ex Buch [1] | Isophyllum/Rigida | Madeira (Portugal), between Pico do Arieiro and Pico Ruivo, 29.xi.1989, L. Chilton & N.J. Turland 135 (BM) | AF481926 |
| B. salicifolium [2] | Isophyllum/Rigida | Canary Islands (Spain). Cultivated, Jardim Botânico de Coimbra, Portugal; seed received from the Jardín Botánico ‘Viera y Clavijo’, Canary Islands, from wild sources; 05.viii.2000, S.S. Neves Acc. No. 29 (COI) | AF481927 |
| B. salicifolium [3] | Isophyllum/Rigida | Canary Islands (Spain), Gran Canaria, Cruz de Tejeda, between Tejeda and Roque Rublo, 19.vi.1995, M.F. Gardner & S.G. Knees 5750 (E) | AF481928 |
| B. semicompositum L. | Isophyllum/Trachycarpa | Spain, Ciudad Real, Daimiel, Tablas de Daimiel, Isla del Morenillo, 12.v.1992, S. Cirujano (MA 552469) | AF481929 |
| B. stellatum L. | Reticulata | Switzerland, Canton du Valais, Col du Simplon, ‘sous la statue de l’Aigle’, 10.viii.1988, B. de Retz 88690 (MAF 145370) | AF481930 |
| B. subspinosum Maire & Weiller | (Isophyllum/Rigida) | Morocco, High Atlas, S slope of Jbel Angour, 21.vii.1976, C.J. & A.R. Humphries 99 (BM) | AF481931 |
| B. tenuissimum L. | Isophyllum/Trachycarpa | Portugal, Beira Litoral, Figueira da Foz, Vila Verde, 16.ix.1994, S.S. Neves 22 (E) | AF481932 |
| Heteromorpha arborescens (Spreng.) Cham. & Schltdl. | n/a | S/C Africa. Cultivated, University of Illinois at Urbana‐Champaign, USA, S. Downie 42 (ILL) – Sequences from Downie and Katz‐Downie (1996) | U27578 (ITS1) U30314 (ITS2) |
| Physospermum cornubiense (L.) DC. | n/a | Ukraine, Crimea, Alikat‐Bogaz Pass. Cultivated, Moscow State University Botanical Garden, Russia; Pimenov & Tomkovich s.n. (MW) – Sequences from Downie et al. (1998) | U78382 (ITS1) U78442 (ITS2) |
| Pleurospermum foetens Franch. | n/a | China, Yunnan. Cultivated, Royal Botanic Garden Edinburgh, UK; RBGE Acc. No. 19910914, Chungtien, Lijiang & Dali Expedition (1990) 1181 (E) – Sequences from Katz‐Downie et al. (1999) | AF008604 (ITS1) AF009083 (ITS2) |
Sequences of Heteromorpha, Physospermum and Pleurospermum were obtained from the literature. Herbarium acronyms provided according to Holmgren et al. (1990). Some herbaria have their own accession numbers of specimens; in such cases this number is given after herbarium abbreviation. Author name abbreviation follows Brummit and Powell (1992).
* Numbers in square brackets are used in Figs 1–3 and correspond to the different samples of a single species or subspecies.
† Classification essentially follows Wolff (1910), with minor corrections (nomenclature of sections) introduced by Tutin (1968). For species described after Wolff’s publication, section and subsection names (placed in brackets) are those suggested by the species authors.
Relationships of Bupleurum to other early branching genera in Apioideae are uncertain and a sister group is currently unknown. The ITS sequences of Bupleurum are highly divergent and difficult to align with other members of the family (Downie et al., 1998; Valiejo‐Roman et al., 1998; Katz‐Downie et al., 1999). As choice of outgroups used for tree rooting is critical in the generation of a tree topology (Watrous and Wheeler, 1981; Maddison et al., 1984), multiple taxa were selected as outgroups, including four other early branching Apioideae genera that have appeared more closely related to Bupleurum in previous molecular phylogenetic studies (Plunkett et al., 1996a, b; Downie et al., 1998). The outgroup taxa included: Anginon difforme, A. paniculatum, Heteromorpha arborescens (ITS1 and ITS2 sequences from Downie and Katz‐Downie, 1996), Physospermum cornubiense (Downie et al., 1998) and Pleurospermum foetens (Katz‐Downie et al., 1999). For analysis of the 5.8S coding region alone, the two Anginon spp. were used as the outgroup (5.8S has not been sequenced for the other outgroup taxa). In spite of possible close relationships, all the outgroup genera used here are clearly distinct from Bupleurum, in both morphological and molecular characters, and so there seems to be no risk that any one of them is nested within Bupleurum.
Fresh or herbarium material (generally leaves, sometimes stems, flowers or fruits) were used for total DNA extraction using a modified CTAB (cetyltrimethyl‐ammonium bromide) method from Doyle and Doyle (1987) with no additional cleaning. In a few cases where DNA extraction or PCR (polymerase chain reaction) amplification was problematic, the DNeasy Plant Mini Kit (QIAGEN Ltd, Crawley, West Sussex, UK) was used.
Double‐stranded DNA of the entire ITS region (covering ITS1, 5.8S and ITS2) was amplified with primers ‘ITS 5P’ (Möller and Cronk, 1997; the same as ‘modified ITS 5’ in Downie and Katz‐Downie, 1996) and ‘ITS 4’ (White et al., 1990) in an equimolar ratio and 1·5 mM MgCl2. The PCR profile followed Möller and Cronk (1997). In a few cases when PCR product was in low concentration, PCR amplification from original DNA extract was repeated with 40 cycles. Successful PCR amplification produced a single DNA band of approx. 700 bp long on a 1·5 % agarose gel. PCR products were subsequently purified using the QIAquick PCR Purification Kit (QIAGEN). DNA concentration of purified PCR products was estimated on gel by comparison to a ladder of known concentration (we used the 123 bp DNA ladder from Sigma, St Louis, MO, USA).
Purified PCR products were sequenced using the primer strategy of Möller and Cronk (1997) and the dRhodamine Terminator Cycle Sequencing Kit (ABI Prism™; Perkin Elmer Applied Biosystems, Warrington, Cheshire, UK), with Ampli Taq® DNA polymerase FS. Sequence fragments were analysed on an ABI Prism 377 DNA automated sequencer, following the manufacturer’s instructions. For each sampled specimen, forward and reverse sequencing reactions were performed for sequence confirmation.
Sequence fragments obtained for each sample were inspected and edited using Factura™, version 1.2.Or6 (Applied Biosystems, Foster City, CA, USA), and later assembled using the Clustal option of multiple alignment in Sequence Navigator™, version 1.0.1 (Applied Biosystems). Ambiguous positions were verified against the original electropherograms. The complete sequence of the ITS region for each sampled specimen was then stored as a separated text file, and later deposited with GenBank (see accession numbers in Table 2). Sequence boundaries of ITS1, 5.8S and ITS2 were determined following Yokota et al. (1989).
Clustal X, version 1.8 (Thompson et al., 1997), was used for multiple alignment of complete sequences. SeqPup (version 0.6f, 1995, D. G. Gilbert, University of Indiana, USA) was used for final manual editing on the alignment and for storage of multiple sequence files; aligned sequences can be obtained from the authors.
Pairwise distances (sequence divergence) between taxa and base frequencies (G + C content) were determined using PAUP* (Swofford, 2000). Transition : transversion ratios (Ti : Tv) over a subset of the maximally parsimonious trees were calculated using MacClade version 3.05 (Maddison and Maddison, 1992).
Phylogenetic analysis
Phylogenetic analyses were performed using PAUP*, versions 4.0b4 and 4.0b8. Characters were unordered and equally weighted (‘Fitch parsimony’; Fitch, 1971). Gaps were treated as missing data and multistate data interpreted as uncertainty. Maximally parsimonious (MP) trees were sought using the heuristic search option with starting tree obtained by stepwise addition, 1000 random‐addition replicates, TBR (tree bisection–reconnection) branch‐ swapping, ‘MultTrees’ (= MultPars) and steepest descent options in effect, with ACCTRAN optimization. Inde pendent parsimony analysis of the 5.8S coding region was also performed. To evaluate clade support, bootstrap (Felsenstein, 1985) and jackknife (Lanyon, 1985) analyses were performed in PAUP* using a full heuristic search with 1000 replicates, simple addition sequence, and TBR branch‐swapping. To reduce computational time, no more than 10 000 or 1000 trees were saved per replicate for bootstrap or jackknife, respectively. As a measure of clade support based on the original data without perturbation, decay indices (Bremer, 1988, 1994; Donoghue et al., 1992) were obtained using AutoDecay (Eriksson, 1999), version 4.0.2, and the reverse constrain option in PAUP*, performing a heuristic search for each node of the MP strict consensus tree, with random‐addition of 100 replicates, TBR branch‐swapping, and a limit of 10 000 trees per replicate. Measures of character fit to the phylogenetic trees, such as the consistency index (CI) (Kluge and Farris, 1969), the retention index (RI) and the rescaled consistency index (RC) (Farris, 1989), were also calculated using PAUP*.
Maximum likelihood (ML) analyses were performed using PAUP*, with settings corresponding to the Hasegawa–Kishino–Yano model, HKY85 (Hasegawa et al., 1985), and using gamma distribution (Yang, 1993, 1996). The Rogers–Swofford approximation method (Rogers and Swofford, 1998) was used to calculate initial branch lengths. Starting likelihood parameters (Ti : Tv ratio, nucleotide frequencies, proportion of invariable sites, and shape parameter of gamma distribution) were estimated from an initial set of trees from the MP analysis; empirical values of nucleotide frequencies were also alternatively used. Likelihood scores were used to perform ML heuristic searches with stepwise addition, addition sequence ‘as is’ or random (ten replicates), TBR branch‐swapping, and ‘MultTrees’ option in effect. Likelihood scores were estimated for each of the ML trees found and these new parameters used for further searches.
Distance trees were obtained from neighbour‐joining (NJ) analyses (Saitou and Nei, 1987) using three of the distance measures available in PAUP*: Jukes and Cantor (1969), Kimura’s two parameter (K2P) (Kimura, 1980), and HKY85 models. For K2P and HKY85, substitution rates were also assumed to follow gamma distribution (shape parameter set at various values). Neighbour‐joining bootstrap values were calculated from 10 000 replicates.
RESULTS
Sequence analysis
Of the 71 accessions examined (68 new sequences), only 61 were included in the final analyses, because some of the sequences obtained from the same taxon/population were identical, and thus only one sequence was used for each of these. This is represented in Figs 1–3 as more than one number appearing after the species names in the trees. Identical sequences were obtained from different samples in B. benoistii (two identical out of three), B. fruticosum, B. gibraltarium, B. oligactis (two out of three), and in the two distinct populations of B. acutifolium from Spain (three samples with identical sequences) and Portugal (two identical sequences). However, the Spanish and Portuguese populations of B. acutifolium produced considerably different sequences (15 mutations), the latter population including a 35 bp deletion in ITS1 (37 bp indel when aligned with all sequences). Some other samples for a single species showed little variation (1–3 bp), but all of these sequences were used in the analyses (e.g. the three samples of B. montanum). Sequences obtained for B. fruticescens subsp. fruticescens (endemic to Spain) and subsp. spinosum (Spanish samples) were all identical (north‐west African samples of ‘B. spinosum’ showed several differences). Identical sequences were also obtained for taxa that are morphologically distinct species (e.g. B. benoistii and B. lateriflorum), and little variation was found between several of the north‐west African endemic taxa (e.g. B. benoistii, B. lateriflorum, B. montanum and B. plantagineum).
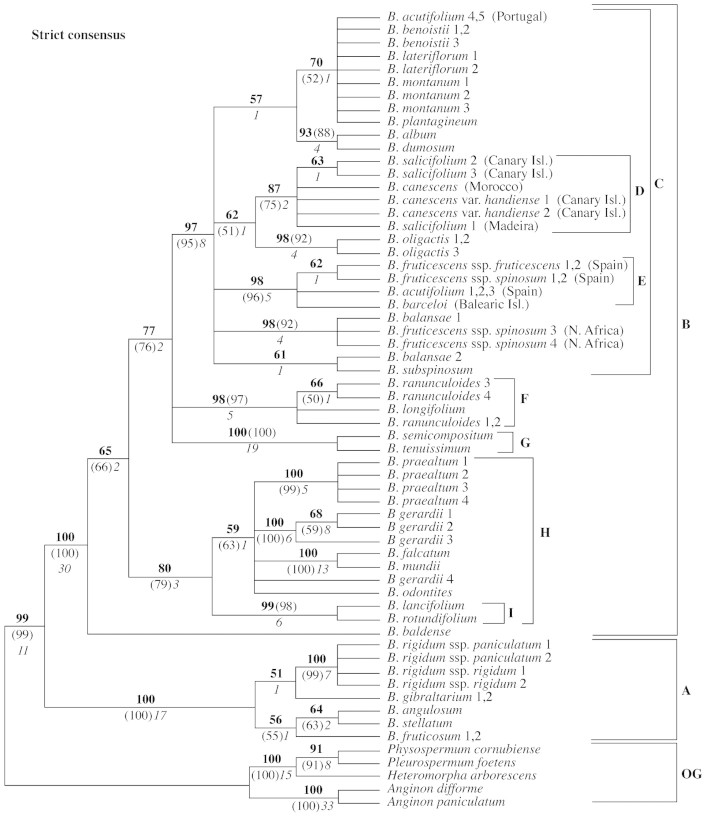
Fig. 1. Strict consensus tree of 128 most‐parsimonious trees of 772 steps derived from equally weighted maximum parsimony analysis of sequences of the ITS region of Bupleurum and outgroup (OG) taxa (CI = 0·58; RI = 0·85). Bootstrap percentages appear above the branches (boldface); jackknife percentages are given in brackets; values <50 % are not indicated. Decay indices are below the branches in italics. Clade A = subgenus Penninervia; clade B = subgenus Bupleurum; clade C = the ‘NW African’ group; clade D = ‘Macaronesian’ group; clade E = the ‘Iberian and Balearic’ group; clade F = the ‘Ranunculoides’ group; clade G = the ‘Trachycarpa’ group; clade I = the ‘Perfoliata’ group.
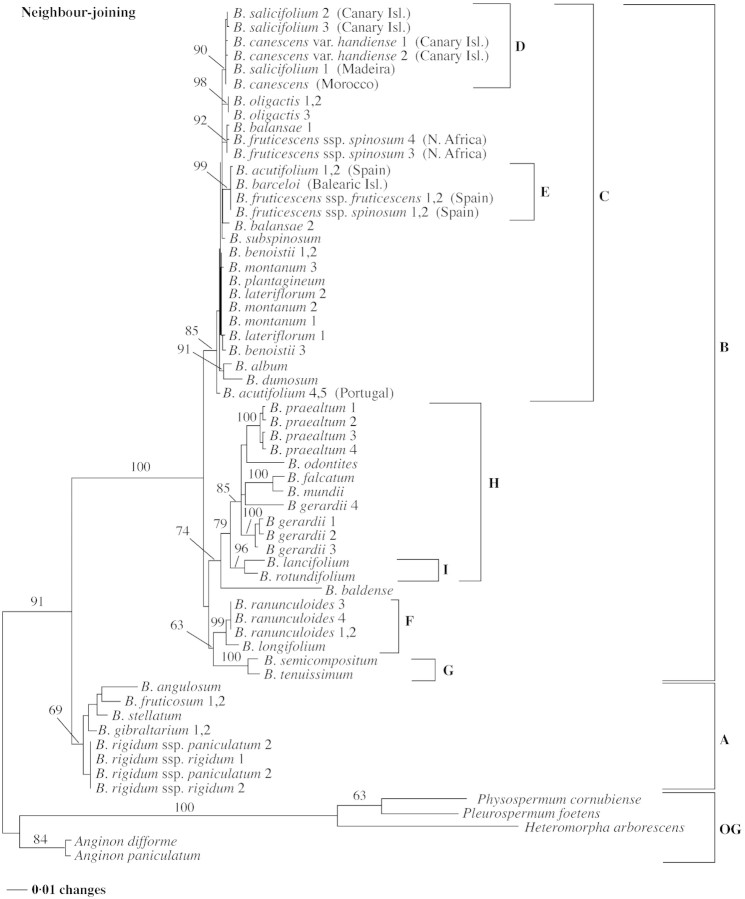
Fig. 3. Single tree obtained from neighbour‐joining (NJ) analysis of substitution rates on sequences of the ITS region of Bupleurum and outgroup (OG) taxa. Branch lengths are proportional to distances (see scale bar) estimated from the two‐parameter method of Kimura (1980), with substitution rates assumed to follow gamma distribution (∝ = 0·5). Numbers above branches are NJ bootstrap estimates; values <50 % are not indicated. The letters A–I represent the same Bupleurum groups referred to in Fig. 1.
The primers used were perfect matches to the sequences in Bupleurum, with the exception of primer ‘ITS 2G’, which sequence should have been: 5′‐GTG ACA CCC AGG CAG ACG T‐3′. Alignment of all sequences resulted in a matrix of 646 positions, including ITS1, 5.8S and ITS2. Characteristics of these sequences, including length, G + C content, number of constant, autapomorphic and parsimony‐informative characters are summarized in Table 3. ITS1 ranged from 172 bp (Anginon difforme and A. paniculatum) to 217 bp (Bupleurum rigidum and Physospermum cornubiense), and ITS2 from 215 bp (Pleurospermum foetens) to 234 bp (Anginon paniculatum). The total length of ITS1 + ITS2 ranges from 404 bp (Bupleurum acutifolium, Portuguese population, and Anginon difforme) to 447 bp (Bupleurum angulosum). The 5.8S is 164 bp long in all taxa. ITS1 is slightly shorter and more variable in length than ITS2. Major gaps (indels (insertions/deletions): a 37 bp long indel in B. acutifolium, from Portugal, and a 44 bp indel in Anginon) occur in a GC‐rich region that is located between two broadly conserved sequence motifs (‘c1’ and ‘c2’) of ITS1 (Hershkovitz et al., 1999; Hershkovitz and Zimmer, 2000). A region of 46 bp at the start of ITS2 was excluded from further analysis, because an unambiguous alignment could not be obtained. As expected, the 5.8S gene showed little variation, with a maximum of 6·1 % divergence across all taxa (4·9 % in Bupleurum). The overall of ITS divergence was approx. 29 % within Bupleurum.
Table 3.
Sequence characteristics of the internal transcribed spacers, ITS1 and ITS2, and of the 5.8S subunit of nuclear rDNA of Bupleurum and outgroup genera (Anginon, Heteromorpha, Physospermum and Pleurospermum)
| Sequence characteristics | ITS1 | ITS2 | 5.8S* |
| Length range in all taxa (bp) | 172–217 | 215–234 | 164 |
| Length range in Bupleurum (bp) | 180–217 | 222–231 | 164 |
| Mean length in Bupleurum (bp) | 213·9 | 225·5 | 164 |
| Length range in outgroup (bp) | 172–217 | 215–234 | 164 |
| Mean length in outgroup (bp) | 198 | 224·6 | 164 |
| Aligned length (bp) | 229 | 253 | 164 |
| G + C content range (mean) in Bupleurum (%) | 53·9–65·4 (60·7) | 53·9–69·4 (61·4) | 53·1–54·9 (54·1) |
| G + C content range (mean) in all taxa (%) | 50·2–69·2 (60·5) | 53·9–73·9 (61·5) | 53·1–56·7 (54·2) |
| Sequence divergence in Bupleurum (%)† | 0–29·7 | 0–29·3 | 0–4·9 |
| Sequence divergence in all taxa (%)† | 0–37·7 | 0–37·9 | 0–6·1 |
| Size of indels in Bupleurum (bp)† | 1–37 | 1–5 | 0 |
| Size of indels in all taxa (bp)† | 1–44 | 1–5 | 0 |
| Number of excluded sites | 0 | 46 | 0 |
| Number of sites after exclusion | 229 | 207 | 164 |
| Number (and %) of constant sites† | 73 (31·9) | 83 (40·1) | 148 (90·2) |
| Number (and %) of variable sites† | 156 (68·1) | 124 (59·9) | 16 (9·8) |
| Number (and %) of parsimony informative sites† | 128 (55·9) | 99 (47·8) | 11 (6·7) |
| Number (and %) of autapomorphic sites† | 28 (12·2) | 25 (12·1) | 5 (3·1) |
| Transitions (minimum)† | 217 | 166 | 17 |
| Transversions (minimum)† | 176 | 150 | 6 |
| Transitions : transversions† (Ti : Tv) | 1·23 | 1·11 | 2·83 |
* 5.8S sequence data refers only to Bupleurum and Anginon.
† Based on alignment excluding ambiguous sites.
Phylogenetic analysis
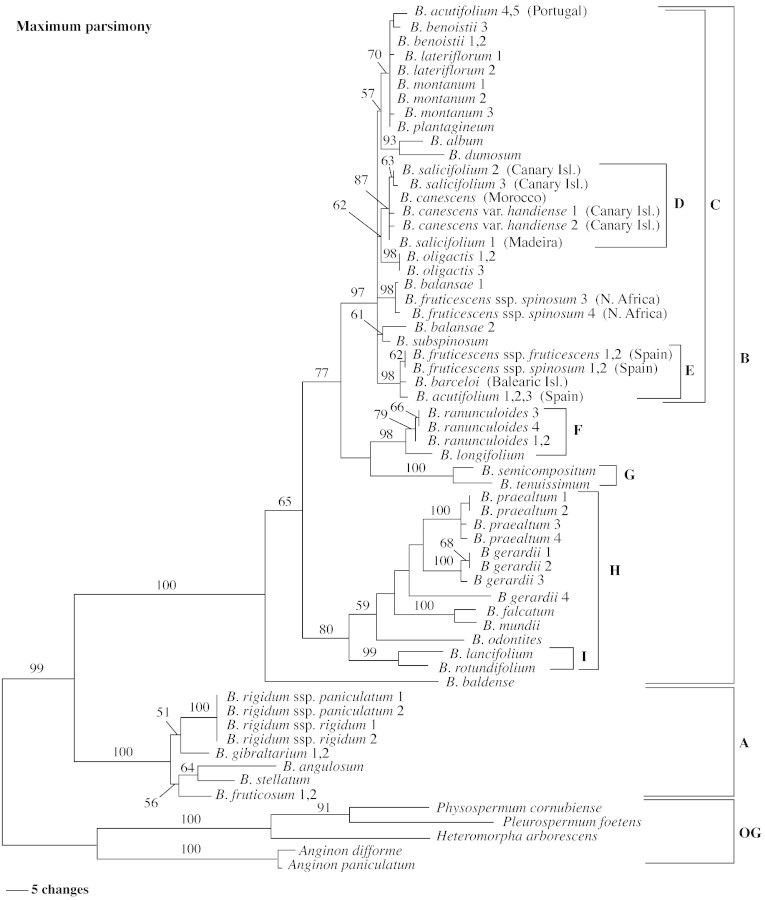
Parsimony analysis of 600 characters, including ITS1 and ITS2 (46 characters excluded in the latter) and the 5.8S region, resulted in 128 most‐parsimonious (MP) trees, each 772 steps long, consistency index (CI) 0·58 (excluding uninformative characters) and retention index (RI) of 0·85. The strict consensus of these trees, with accompanying bootstrap, jackknife and decay values, is shown in Fig. 1. One of the most‐parsimonious trees is presented in Fig. 2. In all these trees, Bupleurum forms a strongly supported clade (A plus B), branch length 31 steps, with 99 % bootstrap and jackknife support, and a decay value of 11. The Bupleurum clade is then subdivided into two major clades (A and B), both with 100 % bootstrap and jackknife support and high decay values (17 for clade A, and 30 for clade B); there are 27 and 48 bp substitutions supporting, respectively, clades A and B. The relationships within the two major Bupleurum clades are not fully resolved, but some of the clades are strongly supported and, in two cases, correspond to taxonomically delimited groups in previous classifications: clade I corresponds to section Bupleurum, and clade G to subsection Trachycarpa of section Isophyllum.
Fig. 2. One of 128 most‐parsimonious trees of 772 steps derived from equally weighted maximum parsimony analysis of sequences of the ITS region of Bupleurum and outgroup (OG) taxa (CI = 0·58; RI = 0·85). Branch lengths are proportional to the number of substitutions (see scale bar). Numbers represent bootstrap values; values <50 % are not indicated. The letters A–I represent the same Bupleurum groups referred to in Fig. 1.
Although final parsimony analyses used five taxa as the outgroups, analyses were also performed using different outgroup combinations, which included several other Apioideae genera (sequences from GenBank). The exclusion of all outgroups except Heteromorpha resulted in 32 MP trees, whose strict consensus was virtually identical to the tree given in Fig. 1. Tree statistics were also similar, although slightly higher (CI = 0·67, RI = 0·87), due to the reduction of the number of taxa and so of possibly conflicting characters. Similar results were also obtained when using Physospermum and Pleurospermum as the outgroup (96 MP trees; CI = 0·66, RI = 0·87). The use of the two species of Anginon as the outgroup resulted in 16 MP (CI = 0·67, RI = 0·88), whose strict consensus was a tree similar in topology to that presented in Fig. 3 (NJ tree), with the exception of the position of B. baldense, which appeared as the first‐branching taxon in clade B, as happened in the MP trees (Figs 1 and 2). In summary, the use of alternative outgroups did not affect the main clades of the Bupleurum ITS phylogeny. Only two clades changed position, and both of these had an unresolved relationship in the MP strict consensus tree (clades F and G).
While this paper has been in press, an ITS sequence for Hohenackeria exscapa (Steven) Koso‐Pol. has become available (GenBank Accession Nos AF337178 and AF337186) from the work of Valiejo‐Roman et al. (2002). Preliminary results of an analysis of this sequence in our dataset shows that this species is included within clade B (subgenus Bupleurum). This shows that Hohenackeria would not be an appropriate outgroup for Bupleurum, as it appears to belong to the ingroup, and that our choice of outgroup remains acceptable. Preliminary results also suggest that the inclusion of Hohenackeria in the dataset does not affect the main clades of Bupleurum, although it does reduce the resolution within clade B.
Parsimony analysis of the 5.8S sequences (11 informative characters) produced 16 MP trees, each 21 steps long, with three substitutions separating Bupleurum from the outgroup (Anginon). The dichotomy of Bupleurum was also shown by analysis of the 5.8S, with four substitutions supporting the separation of clade A from clade B (one substitution supporting clade A and three for clade B). The tree statistics of the 5.8S MP trees were high (CI = 0·91, RI = 0·98) indicating that despite the small number of variable characters (16 out of 164), homoplasy was low.
The likelihood scores of the MP trees were similar regarding estimated base frequencies and Ti : Tv ratios (1·48–1·49), but showed larger ranges of values for the proportion of invariable sites (0·07–0·21), and shape parameter of gamma distribution (∝ = 0·59–0·94). Values used were: proportion of invariable sites = 0·08, 0·12 and 0·2, and ∝ = 0·5, 0·6 and 0·8. The ML tree (length 774 steps, CI = 0·62, RI = 0·85) found in all analyses was highly congruent with the MP trees and so is not shown.
One NJ tree is presented in Fig. 3. The NJ tree topology was not affected by different models of sequence evolution (Jukes‐Cantor, K2P or HKY85); however, slight differences were produced with higher gamma distribution or if substitution rates were set as equal. The NJ tree produced when assuming equal rates of substitution is similar to the MP trees (e.g. Fig. 2), and this tree is also obtained for values of gamma distribution larger than two.
In all the MP, ML and NJ analyses, Bupleurum appears as a strongly supported monophyletic group. The division of the genus into two major clades (A and B in Figs 1–3) is consistent among all trees. The separation of these two clades is also supported by three indels: a 3 bp insertion in ITS1, a 4 bp insertion in ITS2, and a 1 bp deletion at the start of the 26S coding gene. These are all synapomorphies of clade A. Indels were not included in the various analyses and therefore provide further support to the results obtained by analysis of nucleotide substitutions alone.
Other clades that have high statistical support are also present in the trees of all analyses: clades C (mostly north‐west African endemic species), D (‘Macaronesian’ group), E (‘Iberian and Balearic’ group), F (B. ranunculoides and B. longifolium), G (Trachycarpa group) and I (Perfoliata group). The main difference between the MP, ML and NJ trees is the position of B. baldense. This species appears as the first‐branching Bupleurum species in the MP trees (Figs 1 and 2), but as sister of clade H in some of the NJ trees (e.g. Fig. 3) and in the ML tree. As B. baldense appeared in conflicting positions in the various analyses, searches were also performed excluding this species, but overall the resulting trees were not affected.
DISCUSSION
ITS characteristics
The length and levels of variability of ITS1 and ITS2 were in line with those reported in other studies of Apiaceae (Downie et al., 1998; Katz‐Downie et al., 1999). The region of ITS2 that was excluded due to difficulty in alignment has been found in the sequences of ITS2 in several plant families (e.g. Apiaceae, see Downie and Katz‐Downie, 1996; Gesneriaceae, see Möller and Cronk, 1997), and it corresponds to the variable region ‘v1’ as referred in Hershkovitz and Zimmer (1996), and also discussed by Mai and Coleman (1997) and Denduangboripant and Cronk (2001). ITS divergence (approx. 29 %) in Bupleurum was higher than that found in many other genera (usually under 16 %, although divergence up to 30 % has been recorded in a few other genera; Hershkovitz et al., 1999). This high level of divergence is more often found in taxa of higher rank (e.g. Lee and Downie, 1999; Downie et al. 2000a).
Monophyly of Bupleurum
Regardless of method (parsimony, likelihood or neighbour‐joining) or outgroup, all phylogenetic analyses of ITS confirmed that Bupleurum is monophyletic, and considerably divergent from the other early branching genera of Apioideae. However, the recently published ITS sequence of Hohenackeria exscapa raises the possibility that this genus should be included within Bupleurum. Hohenackeria is a genus of two species distributed in the western Mediterranean, the Caucasus and south‐west Asia. It is traditionally classified alongside Bupleurum, differentiated by its white flowers, sessile umbels, and conspicuous calyx teeth, and the lack of bracteoles and of a carpophore (mericarps do not separate). It is expected that further work on this genus of simple‐leaved plants will show that these species should be included within Bupleurum.
Bupleurum has long been recognized as a natural group, mainly due to its characteristic simple and entire leaves, a synapomorphy of the genus. Another synapomorphy of Bupleurum seems to be its characteristic sub‐rhomboidal pollen type (Cerceau‐Larrival, 1962, 1971; Punt, 1984), found in all the species studied, with the exception of two Chinese species (B. sibiricum Vest ex Roem. & Schult. and B. chinense DC.), which have suboval and sub‐rectangular pollen types (Merrgen, 1994). The typical seedlings of Bupleurum, with linear, single‐veined and glabrous cotyledons, appear to be unique in the family (Cerceau‐Larrival, 1962), and thus are probably also synapomorphic to the genus. Cerceau‐Larrival recorded hairy cotyledons in seedlings of some species in Bupleurum, but this has not been confirmed in our work with the same species, where all seedlings were found to be glabrous (Neves, 2000).
Relationships of Bupleurum with other genera
As mentioned above, some difficulties were experienced unambiguously aligning the Bupleurum ITS sequences to those from other Apioideae taxa, including other early branching genera in the subfamily (Downie et al., 1998; Valiejo‐Roman et al., 1998; Katz‐Downie et al., 1999). However, sequences from clade B members (e.g. B. fruticosum) are not so disparate from those of other early branching Apioideae genera, such as Heteromorpha, and unambiguous alignment is possible for most of ITS. The only sequence region excluded in the present analysis (46 characters in ITS2) was difficult to align not only between the different genera, but also within Bupleurum.
ITS sequences of Anginon are distinct from Bupleurum (24–35 % sequence divergence) and from Heteromorpha (approx. 30 % sequence divergence between Anginon and Heteromorpha). Previous molecular studies have suggested that Anginon and Heteromorpha may be closely related (Plunkett et al., 1996a, b, 1997; Downie et al., 1998, 2000b, c; Downie and Katz‐Downie, 1999; Plunkett and Downie, 1999, 2000), but the ITS results indicate that they are considerably divergent.
Published molecular studies, mainly using the plastid genome, have demonstrated that Bupleurum is an early‐branching genus in subfamily Apioideae (Plunkett et al., 1996a, b, 1997; Downie et al., 1998, 2000b, c; Downie and Katz‐Downie, 1999; Plunkett and Downie, 1999, 2000). This confirms the long‐held belief that Bupleurum is among the ‘primitive’ or ‘relict’ groups in the family (Drude, 1898; Wolff, 1910; Cerceau‐Larrival, 1962, 1971; Cauwet‐Marc, 1976). Bupleurum has been considered an ancestral group in Apiaceae mainly because it includes woody species and some of the earliest pollen fossil records known for the family (Gruas‐Cavagnetto and Cerceau‐Larrival, 1978). Woody habit is sometimes regarded as ancestral in the family, as inferred by the close relationship to the mostly woody Araliaceae (Plunkett et al., 1996a, b). Secondary woodiness is known to occur in Apiaceae (Oskolski, 2001), but the wood in Bupleurum exhibits some ancestral characteristics (Rodriguez, 1957; A. Oskolski, Botanical Museum, St Petersburg, pers. comm.). It also appears that the annual habit has evolved at least twice within Bupleurum: in clade H, which includes mostly annuals, the only perennial herbs being B. falcatum and B. mundii (B. baldense, an annual, may also belong to this clade); and in clade G (Trachycarpa), which includes only annuals.
Bupleurum diverged early in the history of Apioideae, but its relationships to other genera remain uncertain. In the present study, none of the genera used as the outgroup (Anginon, Heteromorpha, Physospermum and Pleurospermum) seemed to be closely related, and all are clearly divergent from Bupleurum. There are no clear morphological or anatomical characters associating the genus to these or other early‐branching genera in Apioideae. The present ITS data are in agreement with previous molecular studies that have found Bupleurum to be an isolated group in the family treated as a monogeneric tribe Bupleureae (Downie et al., 2000b). Inclusion of clade A members of Bupleurum in future molecular studies may help to clarify the relationships of the genus to other early branching genera in Apioideae.
Infrageneric classification
The phylogenetic results of the current study do not agree with any published infrageneric classifications of Bupleurum (see Table 1). No previous classifications have suggested the subdivision of the genus into the two main groups shown by analysis of ITS. Of the five sections in Wolff’s classification (Wolff, 1910), only two are likely to be monophyletic: section Bupleurum (the species with perfoliate leaves: clade I; Figs 1–3) and section Reticulata (B. angulosum and B. stellatum). The most species‐rich group, section Isophyllum, is paraphyletic, with sections Bupleurum (clade I) and Diaphyllum (B. longifolium) nested within this large group (clade B). Section Coriacea (B. fruticosum, B. gibraltarium) may also be paraphyletic, but its status is uncertain in the present study, and B. foliosum (the only species not sequenced from section Coriacea), should be included in future studies. The status of section Diaphyllum could not be assessed, because only one of its two species was included in the analysis (B. longiradiatum was not sampled).
Cauwet‐Marc’s classification (Cauwet‐Marc, 1976) also does not agree with the present phylogenetic analysis, with her subgenera Bupleurum and Tenoria being paraphyletic. Her subgenus Tenoria includes the species of section Coriacea (in clade A) and many species from section Isophyllum (clade B), and her subgenus Bupleurum includes species in section Bupleurum (clade B), section Reticulata (clade A), section Diaphyllum (clade B) and the remaining species of section Isophyllum (the majority from clade B, and B. rigidum from clade A).
A major finding of the current study is that Bupleurum shows an early division into two strongly supported groups (clades A and B, Figs 1–3). Maximum likelihood and distance analyses (NJ) also confirm this main dichotomy within the genus, as did analysis of the 5.8S sequences alone.
Although the number of species sampled in the genus is small (32 species out of approx. 150), the taxa studied represent a good sample of the morphological variation seen within Bupleurum. Representatives of all the sections and subsections of the currently accepted classification are included in the study (Table 2), and coverage is also good when considering other alternative classifications, such as that of Cauwet‐Marc (1976). Although western Mediterranean and Macaronesian species are particularly well represented (only one species missing), many of the species sequenced have distributions that extend into eastern Europe and Asia, and three of the species are from outside the main area of study: B. longifolium (from central Europe extending to eastern Russia), B. mundii (southern Africa) and B. stellatum (European Alps and Corsica). We are, therefore, confident that the implications on the classification of the genus can be extrapolated beyond the taxa studied.
There has been considerable discussion on the need to use several molecular markers in phylogenetic investigations, criticizing the interpretation of single‐gene trees as species trees (Doyle, 1992, 1997; Maddison, 1997). Our study is limited in this sense, but with the exception of the study of Choi et al. (1996), who sampled four species, the present study is the first systematic investigation within the genus using molecular data. Previous researchers have included a small number of DNA sequences from Bupleurum, but their aim was to investigate generic and suprageneric relationships in Apiaceae (e.g. Plunkett et al., 1996a, b, 1997; Downie et al., 1998; Valiejo‐Roman et al., 1998). Although, there is no doubt that more evidence from other genes is needed, the degree of confidence in the two major clades obtained from the current study is strong. Therefore, we formally propose the subdivision of Bupleurum into two subgenera, as follows.
Subgenus Penninervia S.S.Neves & M.F.Watson subgenus nov.
Subgenus specierum foliis penninervis aut ± parallelinervis diende nervis marginalibus et costa valde crassa, frutices vel herbae perennes, distributionis mediterraneae.
Type species: Bupleurum fruticosum L., Sp. Pl.: 238 (1753).
Other included species: B. angulosum L., B. gibraltarium Lam., B. rigidum L. and B. stellatum L.
This early branching clade (Clade A) is sister to a larger clade that includes the vast majority of the species in Bupleurum. Penninervia is formed, as the name suggests, by all the species in the genus with pinnate‐reticulate veined leaves, plus B. rigidum (which itself has a unique type of leaf venation). This subgenus includes shrubs and perennial herbs, all native to the Mediterranean region. Subgenus Penninervia includes the species previously placed in sections Coriacea (B. foliosum, B. fruticosum and B. gibraltarium), and Reticulata (B. angulosum and B. stellatum), and one species from section Isophyllum subsect. Marginata (B. rigidum).
No single morphological character has yet been found that is common to all the species of subgenus Penninervia, but the molecular signal from ITS is strong, not only by analysis of the nucleotide substitution pattern, but also by the presence of three indels (described above) that are synapomorphic to this clade.
The close relationship between the species of section Coriacea and section Reticulata was suspected, as they share some morphological characters (pinnate‐reticulate leaves, and large fruits with narrowly winged ridges). However, the inclusion of B. rigidum in this group was unexpected because this species is morphologically distinct from the other members of the subgenus: B. rigidum has more or less parallel‐veined leaves, with thick and prominent veins, delicate and diffusely branching inflorescences, small and narrow bracteoles, and fruits with filiform ridges, while the other members of the subgenus have pinnate leaves with a thick midrib (but other veins are delicate), thicker flowering branches, larger bracts and bracteoles, and narrowly winged fruits. Nevertheless, the traditional association based on inflorescence structure of B. rigidum to species of section Isophyllum (all of them included in clade B) has been controversial, and some classifications treat it in a subsection of its own (subsection Marginata). The leaves of B. rigidum are by far the longest in the genus (up to 45 cm), and its prominent leaf veins are unique in Bupleurum with no obvious affinities with other species. The new placement of this enigmatic taxon is well supported by molecular data for each of the four different accessions, representing the two subspecies of B. rigidum, collected from different natural populations. These produced virtually identical ITS sequences (note branch length in Fig. 2). It is possible that the similar morphologies between B. rigidum and some species of clade B may be due to ancient hybridization between members of the two subgenera (clades A and B), but due to the mechanisms of concerted evolution (Dover, 1982; Elder and Turner, 1995), only one of the parental sequences was kept in the evolutionary lineage of B. rigidum. However, this is a complex explanation with no other evidence to support it. It is simpler to accept that there is only apparent morphological similarity (convergence or parallelism) between the inflorescence of B. rigidum and those of some other perennial herbs of subgenus Bupleurum, such as B. acutifolium or B. oligactis.
The basic chromosome number for subgenus Penninervia appears to be x = 7, with all species being 2n = 14, with the exception of B. rigidum, which also has records of 2n = 16. Cauwet‐Marc (1979b) assumed that the basic number for B. rigidum was x = 8, but when counts of 2n = 14 were found for the species, she interpreted this as dysploidy, with change from eight to seven. Considering the present molecular evidence, it appears that x = 7 was ancestral.
The clades within subgenus Penninervia have low support and further research is needed to clarify the relationships between these taxa. If the main subdivision of subgenus Penninervia shown in the strict consensus tree is confirmed (Fig. 1), a group defined by the presence of pinnate‐reticulate leaves would be paraphyletic: B. gibraltarium appears to be more closely related to B. rigidum than to B. fruticosum, B. angulosum and B. stellatum. Pinnate venation is probably plesiomorphic, and a classification based on this character would be artificial.
Despite the low statistical support (64 % bootstrap, 63 % jackknife; Fig. 1) of the clade formed by B. angu losum and B. stellatum (sect. Reticulata), the group is probably monophyletic. Bupleurum angulosum (endemic to the Pyrenees) and B. stellatum (endemic to the Alps and Corsica) are both perennial herbaceous species that are morphologically similar. The only fundamental difference between them is the degree of fusion of the bracteoles in the involucel (free in B. angulosum, and fused at least in two‐thirds of the total length in B. stellatum). The morphological congruence of these species is such that it seems unlikely that it would have arisen independently.
We were unable to sequence B. foliosum (the only species missing from sect. Coriacea). The DNA extracts were degraded and amplification failed in all attempts. However, B. foliosum is expected to be a member of subgenus Penninervia as this species is morphologically close to B. gibraltarium. Future sequencing of ITS is needed to confirm the inclusion of the species in this subgenus.
Subgenus Bupleurum.
This large group (Clade B) is formed by most of the species of the genus (including the type species, B. rotundifolium), which typically have ± parallel‐veined leaves. The plants of this subgenus are shrubs, subshrubs, perennial or annual herbs, and have a worldwide distribution.
The conservation of B. rotundifolium as the type species of Bupleurum [Taxon 41: 572 (1992); Taxon 44: 611 (1995)] is still awaiting confirmation [Taxon 48: 373–374, 398 (1999)]. We follow the current usage of B. rotundifolium as the generitype, because enforcing the use of the earlier ‘mechanical’ type designation (B. rigidum) would be disruptive both to the traditional and to our new infrageneric classification.
Most of the species in this subgenus appear to have x = 8 as the basic chromosome number, but x = 7 occurs in some species (B. ranunculoides: 2n = 14, 28, 42, 56), as do other numbers, such as x = 6 (B. barceloi: 2n = 24). Dysploidy and aneuploidy are also common in some of the species, such as B. falcatum and B. oligactis (syn. B. atlanticum), which further complicates the understanding of chromosome number evolution in this group.
The phylogeny of subgenus Bupleurum is not fully resolved (Fig. 1). In the MP trees, B. baldense is sister to all remaining species of the subgenus, and terminates a long branch (43 or 44 steps), suggesting that this clade may be an artefact of sampling. Statistical support for the dichotomy is also low (65 % bootstrap and 66 % jackknife). Moreover, the position of B. baldense changes when the ITS data is analysed by ML or NJ (Fig. 3) methods. We have found no conspicuous morphological evidence to suggest that B. baldense is evolutionarily isolated within the genus. This problem may be resolved by adding more taxa, as there are some eastern Mediterranean annual species that may be more closely related to B. baldense.
Although incompletely resolved, subgenus Bupleurum shows some clades that have strong statistical support and are likely to be monophyletic. We withhold attributing formal taxonomic rank to these groups because the relationships of the various clades need further clarification.
The ‘NW African’ group.
Clade C is strongly supported (97 % bootstrap, 95 % jackknife, decay index 8) and includes all the endemic species in north‐west Africa and also two Iberian species (B. acutifolium and B. fruticescens), a species endemic to the Balearic Islands (B. barceloi) and the endemic Macaronesian taxa (B. salicifolium and B. canescens var. handiense). This is referred to as the ‘NW African’ group. The taxa from outside north‐west Africa are morphologically close to some of the endemics in the area: B. acutifolium and B. barceloi are clear allies of B. oligactis; B. fruticescens subsp. spinosum also occurs in north‐west Africa, and B. salicifolium is undoubtedly close to B. canescens. Bupleurum dianthifolium Guss. (a subshrub endemic to the island of Marettimo, near Sicily) was not included in the current study, but has morphological affinities to some of the taxa in the group (such as B. acutifolium and B. barceloi), and is therefore included in this group.
The variation in ITS nucleotide sequence within the ‘NW African’ group is generally low (in Fig. 2 branch lengths are proportional to the number of substitutions), with some morphologically distinct species having identical sequences (e.g. B. benoistii and B. lateriflorum). This could indicate that the group has radiated recently. Bateman (1999) has shown that in recently evolved groups, sequences may show little variation and could be unsuitable for delimiting species. This low rate of variation could also be explained by biased gene conversion (Hillis et al., 1991) against new variants of a sequence, but generally the ITS region tends to diverge between isolated species, as has been shown in many studies, including those in Apioideae (e.g. Downie et al., 2000a).
The ‘Macaronesian’ group.
The Macaronesian endemics, B. salicifolium (Madeira and Canary Islands) and B. canescens var. handiense (syn. B. handiense: eastern Canary Islands) appear together in Clade D with B. canescens sensu stricto (endemic to south Morocco). These taxa are also morphologically similar and are herein referred to as the ‘Macaronesian’ group. Bupleurum canescens sensu stricto occurs in an area of Morocco that is recognized by some authors as ‘the Macaronesian enclave’ in north‐west Africa (Sunding, 1979). ITS sequence data show a ‘Macaronesian clade’ with good branch support (bootstrap = 87%), but a branch that is only two steps long.
The ‘Iberian and Balearic’ group.
Clade E includes B. acutifolium (Spanish population), B. fruticescens and ‘B. spinosum’ (Spanish populations), and B. barceloi (endemic to the Balearic Islands). It is strongly supported (98 % bootstrap; 96 % jackknife), and has a fairly reliable branch length (6 steps; 5 decay index). We refer to this clade as the ‘Iberian and Balearic’ group. This clade is one of five unresolved clades within clade C (Fig. 1), which is predominantly formed by north‐west African species, and may have a north‐west African origin.
The ‘Trachycarpa’ group.
Clade G is strongly supported (100 % bootstrap and jackknife support; 19 decay index), and it is traditionally classified as subsection Trachycarpa of section Isophyllum. It includes annual herbs, with linear to linear‐lanceolate leaves, and papillose fruits. The affinities of the ‘Trachycarpa’ are not clear from the current study. In the various analyses, it appears either as a sister to the ‘NW African’ group or as sister to the ‘Ranunculoides’ group.
The ‘Ranunculoides’ group.
The group formed by B. ranunculoides and B. longifolium (clade F) is strongly supported in the analyses (98 % bootstrap; 97 % jackknife; 5 decay index). There are undoubtedly morphological affinities between these species, but more taxa need to be included to confirm the monophyly of the group, in particular perennial herbs from Asia, such as B. longicaule Wall. ex DC., B. candollei Wall. ex DC., and taxa of the complex ‘B. falcatum’ group.
The ‘Perfoliata’ group.
Clade I corresponds to section Bupleurum of previous classifications (Tutin, 1968). It is characterized by the absence of involucral bracts and the presence of perfoliate upper leaves; both synapomorphies of the group. Phylogenetic analysis confirms that this is a monophyletic group (99 % bootstrap, 98 % jackknife, 6 decay index), as has long been regarded by taxonomists. There is also a 5 bp deletion in ITS1 synapomorphic to these taxa, which provides further support for the group. The other four species with perfoliate leaves (B. croceum Fenzl, B. heldreichii Boiss. & Balansa, B. lophocarpum Boiss. & Balansa, and B. schistosum Woronow: all endemic to Turkey; Snogerup, 1972) should be sequenced in future studies to verify if they also fall within this group.
Species delimitation in Bupleurum
Bolòs and Vigo (1974) treated B. fruticescens and B. spinosum as a single species, by recognizing B. spinosum as a subspecies of B. fruticescens. At the time they did not explain the basis for this taxonomic decision, but having studied morphological variation in the herbarium and in the field (Spain), intermediate plants are easily found and we agree with their treatment. The ITS analyses in the current study further endorses their treatment as a single species, with all sequences from Iberian material being invariant. However, the sequences obtained from the North African ‘B. spinosum’ showed several differences (12 substitutions and three indels of 1 bp each) in relation to those of the Iberian material of B. fruticescens. Morphologically, the Spanish and African populations of ‘B. spinosum’ are almost identical and should be considered part of the same species. The divergence in ITS sequences may have arisen because of the long geographical separation of the populations. However, the African populations of ‘B. spinosum’ have an ITS sequence that is much closer to that of another North African species, B. balansae, than to its Iberian relatives. This could be an indication of gene introgression (i.e. that hybridization may have occurred between B. balansae and the North African ‘B. spinosum’). Both species are shrubs and are found in the same geographical area. If this was the case, we may have only detected the ‘balansae’ ITS type, which could have replaced, or be more common in the genome, than the ‘spinosum’ ITS. In our research, only one ITS type of sequence was found in each species, but only extensive sequencing of ITS repeats by cloning could demonstrate that there is a single type of sequence in the genome. Distinct ITS copies (paralogues) in a single genome have been found in several plant groups, in some cases clearly as a result of interspecific hybridization (e.g. Sang et al., 1995).
The low bootstrap value (62 %) shown for the clade of the Spanish B. fruticescens (part of clade E in Fig. 1), which have identical sequences, reflects the fact that there is little difference between the sequences of these taxa and those of B. acutifolium (Spain) and B. barceloi.
Bupleurum bourgaei has traditionally been considered a distinct species, endemic to the mountains of south‐east Spain. From a morphological study of herbarium material (Neves, 2000) we considered that this taxon was not sufficiently distinct from B. ranunculoides and should be included as a synonym within it. The sample of B. ranunculoides No. 4 (province of Jaén) corresponds to material previously attributable to B. bourgaei, but the sequence of this sample differs in only one ambiguous nucleotide to that of B. ranunculoides No. 3, from the province of Burgos in north‐east Spain. ITS sequence data thus support the decision based on morphology alone.
The two populations of B. acutifolium (Portuguese and Spanish) show minor morphological differences, but these cannot be relied upon, as they are considerably plastic. However, the ITS sequences obtained for the two populations are considerably different. The sequence of the Portuguese material shows a fairly large deletion in ITS1 (35 bp), which is likely to result from a single mutation, but there are also 14 other single nucleotide substitutions. In addition, the two B. acutifolium types of sequences arise in different parts of the trees. The geographic separation of the two populations is also quite large, and interbreeding has probably not occurred for some considerable time. ITS data indicates that the two populations of B. acutifolium have diverged significantly and could now be considered different species, even though the morphological differences are not so clear cut (this will form the subject of future work). There is also the possibility that hybridization may have occurred between the Spanish population of B. acutifolium and B. fruticescens (a Spanish ‘neighbour’, which is morphologically a distinct species). However, the two species are not sympatric, have distinct ecological preferences (the Spanish B. acutifolium grows on serpentine soil, whereas B. fruticescens is found on calcareous soil), and there is no evidence of morphological introgression between them. Hybridization is rare in the family and these two species merit further study.
Bupleurum balansae is another taxon with considerable variation in ITS sequence data between populations, but in this case the morphological characters used to delimit the taxon may be inadequate. The main characters used to delimit this species are the sessile or subsessile flowers and fruits, and the slightly prominent veins in both surfaces of the leaves. However, there is a great deal of variation in shape and length of leaves, and even in prominence of veins. Future work may reveal that it is possible to distinguish more than one species in these populations of ‘shrubs with sessile flowers’. Again, the possibility of reticulate evolution in some of the North African taxa needs to be investigated.
One of the sequences obtained for B. gerardii (No. 4) does not group with the others, indicating a problem in species delimitation. Morphological delimitation of B. gerardii and B. praealtum is also problematic, and difficulties are often experienced in distinguishing them. In Fig. 2, B. gerardii No. 4 appears as sister to the clade including B. gerardii and B. praealtum. One of the possibilities is that hybridization has occurred between B. gerardii and B. praealtum, which would explain the problems of morphological characterization of the taxa. A second possibility is that we are including two different species under the name B. gerardii, which is the opinion of other authors (Snogerup and Snogerup, 2001). A third possibility is that B. gerardii and B. praealtum are a single species. Therefore, a more detailed study of B. gerardii and B. praealtum is necessary, including material from the rest of the area of distribution (Europe and west Asia), and also other allied species, such as B. commutatum Boiss. & Balansa, and B. trichopodum Boiss. & Spruner.
Phylogeny and biogeography
Several clades in the ITS analysis in Bupleurum are congruent with the geographical distribution of the taxa. The clearest case is the large group designated here as ‘NW African’, a clade in which all the endemics to north‐west Africa are included. Some of these endemics are morphologically quite distinct, and they have not previously been classified into a single group. As discussed above, two clades that are included within the NW African group, the ‘Macaronesian’ and the ‘Iberian and Balearic’ clades also reflect geographical areas.
Two clades of subgenus Bupleurum include species that have essentially a Eurasian distribution: (1) the clade including ‘Perfoliata’ and the groups of B. falcatum and B. gerardii; and (2) the clade of B. ranunculoides and B. longifolium. Bupleurum lancifolium and B. odontites (included in clade B) occur in North Africa, but their original (native) area of distribution is not clear as both species have been introduced and are ruderals or weeds in cultivated land. The only exception to this Eurasian distribution is B. mundii, a species endemic to southern Africa, whose closest relative appears to be B. falcatum (Figs 1–3). There are also morphological affinities between these two species.
Evidence from the current study suggests that the genus Bupleurum originated somewhere in the western Mediterranean. First, the first branching clade in Bup leurum is formed by species that fundamentally only occur in the western Mediterranean. Bupleurum fruticosum is the only species with a distribution that extends across the Mediterranean, but this can be explained as subsequent dispersal. Also, the shrubby plants of the genus are restricted to the western Mediterranean (the only exception, again, is B. fruticosum). This is particularly relevant if woodiness is regarded as an ancestral state (Plunkett et al., 1996b), which appears to be supported by wood anatomy in Bupleurum. Also, the western Mediterranean is undoubtedly richer in the ‘basic’ morphological patterns found in Bupleurum (Neves, 2000).
The eastern Mediterranean has high species diversity in annuals (e.g. Snogerup, 1972), but it is likely that these have evolved more recently: ITS analysis indicates that the annual habit is a derived state (possibly having evolved more than once in the genus). A shorter life cycle increases the chances of evolutionary change, so a shorter period of time is required to explain increased diversity (molecular and morphological) in annual plants. This could explain why in the ITS phylogeny (Figs 2 and 3) the groups including only herbs and mainly annuals (such as clades G, H and I) appear to have longer branches than those comprised by perennials only (e.g. clade C).
There is little difference between the ITS sequences of B. salicifolium (endemic to Madeira and Canary Islands) and those of its closer continental relatives, in particular B. canescens. Therefore, B. salicifolium is clearly a neoendemic species, with a recent arrival in the islands.
Another neoendemic is the disjunct species B. mundii (endemic to southern Africa). Our data refute the notion that this is a remnant species from a past larger distribution of Bupleurum in Africa. This has been questioned before, in particular considering the relationships of ‘primitive’ woody genera of Apiaceae, most of which only occur in central and southern Africa (Cerceau‐Larrival, 1971; Burtt, 1991; Downie and Katz‐Downie, 1999).
CONCLUSIONS
The results of the present study have confirmed the monophyly of Bupleurum and provided resolution of major groups within the genus. Light has also been shed on the origins of this widespread genus, and on the biogeography of some of the enigmatic species and species groups. Inevitably the analyses have given rise to new ideas and hypotheses that will require further work. The phylogenetic signal from the taxa studied will be strengthened by the use of other genes (nuclear and plastid), and the inclusion of new taxa from the eastern Mediterranean and Asia will give a more comprehensive sampling. Bupleurum foliosum and B. dianthifolium need to be sequenced for verification of their putative placement in subgenus Penninervia and the ‘NW African’ group, respectively. The use of genes with slower rates of mutation than ITS may help to resolve some of the clades in Bupleurum (e.g. the plastid genes trnL‐trnF and matK have also been useful at generic and infrageneric levels; Soltis and Soltis, 1998).
The ‘NW African’ group (clade C) showed very low nucleotide variation in the ITS region. At present, the ITS spacers are two of the fastest evolving regions sequenced for phylogenetic analysis. Other loci with rapid rates of evolution are known, but sequence homology and alignment have been problematic. Therefore, to resolve the relationships of the taxa in this group, we will need methods of analysis that can detect molecular variation at lower levels, e.g. isozymes, AFLPs and microsatellites that have been particularly useful in detecting molecular polymorphism at the species level and below.
ACKNOWLEDGEMENTS
This work was funded by a doctoral scholarship to Susana Neves (BD/4057/94) from the program Praxis XXI, Fundação para a Ciência e a Tecnologia, Lisbon (Portugal). We thank the Royal Botanic Garden Edinburgh (RBGE) and the Institute of Cell and Molecular Biology (ICMB), University of Edinburgh, UK, for ample use of their research facilities and technical assistance. We are especially grateful to Jill Preston and Caroline Guihal (RBGE) and Nicola Preston (ICMB) for help with sequencing, Michael Möller and James Richardson (RBGE) for advice on phylogenetic analysis, and Fátima Sales (Departamento de Botânica, Universidade de Coimbra, Portugal) and Ian Hedge (RBGE) for many valuable discussions during the development of this project. Robert Mill is thanked for checking the Latin diagnosis, and Charlie Jarvis for his advice on the typification of Bupleurum. We thank an anonymous reviewer, Mike Fay and particularly Gregory Plunkett, for helpful comments on the manuscript. The Davis Expedition Fund (University of Edinburgh) provided financial support for a field collection trip in Spain. We also thank all the institutions that provided the herbarium and/or living material used in this study: Botanischer Garten Tübingen (Germany); Botanischer Garten der Universität Potsdam (Germany); Botanischer Garten St Gallen (Switzerland); Departamento de Biología Vegetal, Universidad de Málaga (Spain); Departamento de Botânica, Universidade de Coimbra; Departamento de Botánica, Universidad de Sevilla (Spain); Department of Botany, The Natural History Museum, London (UK); Department of Botany, University of Reading (UK); Facultad de Farmacia, Universidad Complutense, Madrid (Spain); Instituto Pirenaico de Ecología, Jaca (Spain); Jardín Botánico ‘Viera y Clavijo’, Canary Islands; Jardin Botanique National de Belgique, Meise (Belgium); Real Jardín Botánico de Madrid; and Royal Botanic Garden Edinburgh.
Received: 25 June 2002; Returned for revision: 20 February 2003; Accepted: 8 January 2004; Published electronically: 23 February 2004
References
- AllisonI, Van Wyk BE.1997. A revision of the genus Anginon (Apiaceae). Nordic Journal of Botany 17: 561–577. [Google Scholar]
- ArenasJA, García F.1993. Atlas carpológico y corológico de la subfamilia Apioidea Drude (Umbelliferae) en España peninsular y Baleares. Ruizia 12. [Google Scholar]
- BaldwinBG.1992. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Molecular Phylogenetics and Evolution 1: 3–16. [DOI] [PubMed] [Google Scholar]
- BaldwinBG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ.1995. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden 82: 247–277. [Google Scholar]
- BatemanRM.1999. Integrating molecular and morphological evidence of evolutionary radiations. In: Hollingsworth PM, Bateman RM, Gornall RJ, eds. Molecular systematics and plant evolution The Systematics Association Special Vol. 57 London and New York: Taylor & Francis, 432–471. [Google Scholar]
- BerenbaumMR.1990. Plant consumers and plant secondary chemistry: past, present and future. 4. A case study: furanocoumarins in Apiaceae Oxford Surveys in Evolutionary Biology 7: 285–307. [Google Scholar]
- BoissierE.1872.Flora Orientalis Vol. 2. Geneva and Basel. [Google Scholar]
- BolòsO, Vigo J.1974. Notes on plant taxonomy and nomenclature I. Butlletí de la Institució Catalana d’Història Natural 38 [Secció de Botànica, 1]: 83. [Google Scholar]
- BremerK.1988. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42: 795–803. [DOI] [PubMed] [Google Scholar]
- BremerK.1994. Branch support and tree stability. Cladistics 10: 295–304. [Google Scholar]
- BriquetJ.1897. Monographie des buplèvres des Alpes Maritimes. In: Burnat E, ed. Matériaux pour servir à l’histoire de la flore des Alpes Maritimes. Geneva and Basel: Georg & Co, Libraires‐Éditeurs. [Google Scholar]
- BrummitRK, Powell CE.1992.Authors of plant names. Richmond: Royal Botanic Gardens, Kew. [Google Scholar]
- BurttBL.1991. Umbelliferae of Southern Africa: an introduction and annotated check‐list. Edinburgh Journal of Botany 48: 133–282. [Google Scholar]
- CalestaniV.1905. Contributo alla sistematica delle Ombellifere d’Europa. Webbia 1: 89–170. [Google Scholar]
- CarbonnierJ, Cauwet‐Marc AM.1979. Constituants du genre Bupleurum (Umbelliferae): mise au point des connaissances actuelles. Bulletin du Muséum National d’Histoire Naturelle (Paris). Série 4, section B, 1: 213–263. [Google Scholar]
- CarbonnierJ, Cauwet‐Marc AM.1981. A comparative phytochemical investigation of the genus Bupleurum L. (Umbelliferae). Taxon 30: 617–627. [Google Scholar]
- Cauwet‐MarcAM.1970. Races chromosomiques, écologie et biologie du Bupleurum ranunculoides L. dans la partie orientale des Pyrénées. Bulletin de la Société Botanique de France 117: 373–384. [Google Scholar]
- Cauwet‐MarcAM.1976.Biosystématique des Espèces Vivaces de Bupleurum L. (Umbelliferae) du Bassin Méditerranéen Occidental. PhD Thesis (3 vols). Université des Sciences et Techniques du Languedoc, Perpignan, France. [Google Scholar]
- Cauwet‐MarcAM.1979a. Connaissances caryologiques actuelles sur le genre Bupleurum L. (Umbelliferae): nombres chromosomiques et nombres de base. Bulletin du Muséum National d’Histoire Naturelle (Paris). Série 4, section B, 1: 191–211. [Google Scholar]
- Cauwet‐MarcAM.1979b. Contribution de la caryologie à la connaissance de la systématique et de la phylogénie du genre Bupleurum L. Candollea 34: 49–86. [Google Scholar]
- Cauwet‐MarcAM, Carbonnier J, Cerceau‐Larrival MT, Dodin R, Guyot M.1978. Contribution à l’étude multidisciplinaire du genre Bupleurum L. In: Cauwet‐Marc AM, Carbonnier J, eds. 1982. Les Ombellifères: contributions pluridisciplinaires à la systématique: actes du 2ème Symposium International sur les Ombellifères, Centre Universitaire de Perpignan, 18–21 Mai 1977. St Louis, MO: Missouri Botanical Garden, 623–651. [Google Scholar]
- Cerceau‐LarrivalMT.1962. Plantules et pollens d’Ombellifères. Mémoires du Muséum d’Histoire Naturelle (Paris). Série B, Botanique, 14: 1–166. [Google Scholar]
- Cerceau‐LarrivalMT.1971. Morphologie pollinique et correlations phylogénétiques chez les Ombellifères. In: Heywood VH, ed. The biology and chemistry of the Umbelliferae London: Academic Press, 112–113. [Google Scholar]
- Cerceau‐LarrivalMT, Roland‐Heydacker F.1978. Apport de la palynologie à la connaissance des Ombellifères actuelles et fossiles. In: Cauwet‐Marc AM, Carbonnier J, eds. 1982. Les Ombellifères: contributions pluridisciplinaires à la systématique: actes du 2ème Symposium International sur les Ombellifères, Centre Universitaire de Perpignan, 18–21 Mai 1977. St Louis, MO: Missouri Botanical Garden, 213–229. [Google Scholar]
- ChaseMW, Hills HG.1991. Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40: 215–220. [Google Scholar]
- ChoiHK, Kim HJ, Shin H, Kim Y.1996. Phylogeny and ribosomal DNA variation of Bupleurum (Umbelliferae). Korean Journal of Plant Taxonomy 26: 219–233. [Google Scholar]
- DenduangboripantJ, Cronk QCB.2001.Evolution and alignment of the hypervariable arm 1 of Aeschynanthus (Gesneriaceae) ITS2 nuclear ribosomal DNA. Molecular Phylogenetics and Evolution 20: 163–172. [DOI] [PubMed] [Google Scholar]
- DonoghueMJ, Olmstead RG, Smith JF, Palmer JD.1992. Phylogenetic relationships of Dipsacales based on rbcL sequences. Annals of the Missouri Botanical Garden 79: 333–345. [Google Scholar]
- DoverG.1982. Molecular drive: a cohesive mode of species evolution. Nature 299: 111–117. [DOI] [PubMed] [Google Scholar]
- DownieSR, Katz‐Downie DS.1996. A molecular phylogeny of Apiaceae subfamily Apioideae: evidence from nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany 83: 234–251. [PubMed] [Google Scholar]
- DownieSR, Katz‐Downie DS.1999. Phylogenetic analysis of chloroplast rps16 intron sequences reveals relationships among woody southern African Apiaceae subfamily Apioideae. Canadian Journal of Botany 77: 1120–1135. [Google Scholar]
- DownieSR, Katz‐Downie DS, Spalik K.2000a. A phylogeny of Apiaceae tribe Scandiceae: evidence from nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany 87: 76–95. [PubMed] [Google Scholar]
- DownieSR, Katz‐Downie DS, Watson MF.2000b. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: towards a suprageneric classification of subfamily Apioideae. American Journal of Botany 87: 273–292. [PubMed] [Google Scholar]
- DownieSR, Ramanath S, Katz‐Downie DS, Llanas E.1998. Molecular systematics of Apiaceae subfamily Apioideae: phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacer and plastid rpoC1 intron sequences. American Journal of Botany 85: 563–591. [PubMed] [Google Scholar]
- DownieSR, Watson MF, Spalik K, Katz‐Downie DS.2000c. Molecular systematics of Old World Apioideae (Apiaceae): relationships among some members of tribe Peucedaneae sensu lato, the placement of several island‐endemic species, and resolution within the apioid superclade. Canadian Journal of Botany 78: 506–528. [Google Scholar]
- DoyleJJ.1992. Gene trees and species trees: molecular systematics as one‐character taxonomy. Systematic Botany 17: 144–163. [Google Scholar]
- DoyleJJ.1997. Trees within trees: genes and species, molecules and morphology. Systematic Biology 46: 537–553. [DOI] [PubMed] [Google Scholar]
- DoyleJJ, Doyle JL.1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- DrudeO.1898. Umbelliferae. In: Engler A, Prantl K, eds. Die natürlichen Pflanzenfamilien Vol. 3, Part 8 Leipzig: Wilhelm Engelman, 63–250. [Google Scholar]
- ElderJF, Turner BJ.1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Quarterly Review of Biology 70: 297–320. [DOI] [PubMed] [Google Scholar]
- ErikssonT.1999.AutoDecay 4·0 [program distributed by the author]. Bergius Foundation, Royal Swedish Academy of Sciences, Stockholm. [Google Scholar]
- FarrisJS.1989. The retention index and the rescaled consistency index. Cladistics 5: 417–419. [DOI] [PubMed] [Google Scholar]
- FelsensteinJ.1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- FitchWM.1971. Toward defining the course of evolution: minimal change for a specific tree topology. Systematic Zoology 20: 406–416. [Google Scholar]
- GrenierM, Godron M.1848.Flore de France. Vol. 1. Paris. [Google Scholar]
- Gruas‐CavagnettoC, Cerceau‐Larrival MT.1978. Presence de pollens d’Ombellifères fossiles dans le Paleogene du Bassin Anglo‐Parisien: premiers resultats. In: Cauwet‐Marc AM, Carbonnier J, eds. 1982. Les Ombellifères: contributions pluridisciplinaires à la systématique: actes du 2ème Symposium International sur les Ombellifères, Centre Universitaire de Perpignan, 18–21 Mai 1977. St Louis, MO: Missouri Botanical Garden, 255–267. [Google Scholar]
- HaraH, Williams HJ.1979.An Enumeration of the Flowering Plants of Nepal, Vol. 2 London: Trustees of the British Museum (Natural History), 184–185. [Google Scholar]
- HasegawaM, Kishino H, Yano T.1985. Dating of the human‐ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 22: 160–174. [DOI] [PubMed] [Google Scholar]
- HershkovitzMA, Zimmer EA.1996. Conservation patterns in angiosperm rDNA ITS2 sequences. Nucleic Acids Research 24: 2857–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HershkovitzMA, Zimmer EA.2000. Ribosomal DNA evidence and disjunctions of Western American Portulacaceae. Molecular Phylogenetics and Evolution 15: 419–439. [DOI] [PubMed] [Google Scholar]
- HershkovitzMA, Zimmer EA, Hahn WJ.1999. Ribosomal DNA sequences and angiosperm systematics. In: Hollingsworth PM, Bateman RM, Gornall RJ, eds. Molecular Systematics and Plant Evolution The Systematics Association Special Vol. 57 London and New York: Taylor & Francis, 268–326. [Google Scholar]
- HeywoodVH.1971. Systematic survey of Old World Umbelliferae. In: Heywood VH, ed. The biology and chemistry of the Umbelliferae London: Academic Press, 31–41. [Google Scholar]
- HillisDM, Moritz C, Porter CA, Baker RJ.1991. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251: 308–310. [DOI] [PubMed] [Google Scholar]
- HolmgrenPK, Holmgren NH, Barnett LC.1990.Index herbariorum Part I. The herbaria of the world, 8th edn. New York: New York Botanical Garden. [Google Scholar]
- JukesTH, Cantor CR.1969.Evolution of protein molecules. In: Munro HN, ed. Mammalian protein metabolism Vol. 3. New York: Academic Press, 21–132. [Google Scholar]
- Katz‐DownieDS, Valiejo‐Roman CM, Terentieva EI, Troitsky AV, Pimenov MG, Lee B, Downie SR.1999. Towards a molecular phylogeny of Apiaceae subfamily Apioideae: additional information from nuclear ribosomal DNA ITS sequences. Plant Systematics and Evolution 216: 167–195. [Google Scholar]
- KimuraM.1980. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- KlugeAG, Farris JS.1969. Quantitative phyletics and the evolution of anurans. Systematic Zoology 18: 1–32. [Google Scholar]
- KondoK, Terabayashi S, Okada M, Yuan C, He S.1996. Phylogenetic relationship of medicinally important Cnidium officinale and Japanese Apiaceae based on rbcL sequences. Journal of Plant Research 109: 21–27. [Google Scholar]
- Koso‐PoljanskyBM.1913. Epitome Bupleurorum Rossiae. Acta Horti Petropolitani 30: 135–333. [Google Scholar]
- KüpferP.1969. Recherches cytotaxinomiques sur la flore des montagnes de la Péninsule Ibérique. Bulletin de la Société Neuchateloise des Sciences Naturelles 92: 31–48. [Google Scholar]
- LanyonSM.1985. Detecting internal inconsistencies in distance data. Systematic Zoology 34: 397–403. [Google Scholar]
- LeeBY, Downie SR.1999. A molecular phylogeny of Apiaceae tribe Caucalideae and related taxa: inferences based on ITS sequence data. Systematic Botany 24: 461–479. [Google Scholar]
- LeeSB, Rasmussen SK.1998.Phylogenetic analysis of rDNA internal transcribed spacer of Angelica, Bupleurum and Peucedanum species Risø‐R‐1091(EN) [internal report]. Roskilde, Denmark: Riso National Laboratory, 5–13. [Google Scholar]
- LeonardisW, Fichera G, Longhitano N, Marletta G, Fiumara PMR, Zizza A.1997. Indagine palinologica su alcune specie di Bupleurum L. e Smyrnium L. (Umbelliferae) in relazione alla posizione sistematica. Allionia 35: 219–223. [Google Scholar]
- LinnaeusC.1753.SpeciesPlantarum. Vol. 1 Stockholm [Bupleurum in pp. 236–239]. [Google Scholar]
- MaddisonWP.1997. Gene trees in species trees. Systematic Biology 46: 523–536. [Google Scholar]
- MaddisonWP, Maddison, DR.1992.MacClade: analysis of phylogeny and character evolution, Version 3. Sunderland, MA: Sinauer Associates. [Google Scholar]
- MaddisonWP, Donoghue MJ, Maddison DR.1984. Outgroup analysis and parsimony. Systematic Zoology 33: 83–103. [Google Scholar]
- MaiJC, Coleman AW.1997. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. Journal of Molecular Evolution 44: 258–271. [DOI] [PubMed] [Google Scholar]
- MerrgenWQ.1994. On the pollen morphology of Bupleurum L. from Nei Menggu and its classification. Acta Scientiarium Naturalium Universitatis Neimenggu 25: 554–560. [Google Scholar]
- MöllerM, Cronk QCB.1997. Origin and relationships of Saintpaulia (Gesneriaceae) based on ribosomal DNA internal transcribed spacer (ITS) sequences. American Journal of Botany 84: 956–965. [PubMed] [Google Scholar]
- NasirE.1955. The bupleura (Umbelliferae) of North‐West Himalaya. University of California Publications in Botany 27: 417–444. [Google Scholar]
- NevesSS.2000.Taxonomy and evolution in Bupleurum L. (Umbelliferae) in the W Mediterranean and Macaronesia. PhD Thesis, University of Edinburgh, UK. [Google Scholar]
- OhtaS.1991. Cytogenetical study on the speciation of Bupleurum falcatum L. (Umbelliferae). Journal of Science of Hiroshima University , Ser. B, Div. 2 (Botany) 23: 273–348. [Google Scholar]
- OskolskiAA.2001. Phylogenetic relationships within Apiales: evidence from wood anatomy. Edinburgh Journal of Botany 58: 201–206. [Google Scholar]
- PanelattiJ.1959. Contribution a l’étude anatomique du genre Bupleurum L. au Maroc. Travaux de l’Institut Scientifique Chérifien , Série Botanique 15. [Google Scholar]
- PimenovMG, Leonov MV.1993.The genera of the Umbelliferae: a nomenclator. Richmond and Moscow: Royal Botanic Gardens Kew and Botanical Garden of Moscow State University. [Google Scholar]
- PlunkettGM, Downie SR.1999. Major lineages within Apiaceae subfamily Apioideae: a comparison of chloroplast restriction site and DNA sequence data. American Journal of Botany 86: 1014–1026. [PubMed] [Google Scholar]
- PlunkettGM, Downie SR.2000. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Systematic Botany 25: 648–667. [Google Scholar]
- PlunkettGM, Soltis DE, Soltis PS.1996a. Higher level relationships of Apiales (Apiaceae and Araliaceae) based on phylogenetic analysis of rbcL sequences. American Journal of Botany 83: 499–515. [Google Scholar]
- PlunkettGM, Soltis DE, Soltis PS.1996b.Evolutionary patterns in Apiaceae: inferences based on matK sequence data. Systematic Botany 21: 477–495. [Google Scholar]
- PlunkettGM, Soltis DE, Soltis PS.1997. Clarification of the relationship between Apiaceae and Araliaceae based on matK and rbcL sequence data. American Journal of Botany 84: 565–580. [PubMed] [Google Scholar]
- PuntW.1984. Umbelliferae. In: Punt W, Clarke GC, eds. The Northwest European Pollen Flora, Vol. 4 (37) Amsterdam: Elsevier Science, 155–363. [Google Scholar]
- RechingerKH, Snogerup S.1987.Bupleurum In: Hedge IC, Lamond JM, Rechinger KH et al. eds. Umbelliferae. In: Rechinger KH, ed. Flora Iranica, Vol. 162 Graz, Austria: Akademische Druck‐ u. Verlagsanstalt, 269–297. [Google Scholar]
- RodriguezRL.1957. Systematic anatomical studies on Myrrhidendron and other woody Umbellales. University of California Publications in Botany 29: 145–318. [Google Scholar]
- RogersJS, Swofford DL.1998. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Systematic Biology 47: 77–89. [DOI] [PubMed] [Google Scholar]
- RouxM, Carbonnier J, Cauwet‐Marc AM.1978. Un exemple d’analyse cladistique: Le genre Bupleurum L. (Umbelliferae). In: Cauwet‐Marc AM, Carbonnier J, eds. 1982. Les Ombellifères: contributions pluridisciplinaires à la systématique: actes du 2ème Symposium International sur les Ombellifères, Centre Universitaire de Perpignan, 18–21 Mai 1977. St Louis, MO: Missouri Botanical Garden, 575–592. [Google Scholar]
- SangT, Crawford DJ, Stuessy TF.1995. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences USA 92: 6813–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SaitouN, Nei M.1987. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- SnogerupS.1972.Bupleurum In: Davis PH, ed. Flora of Turkey, Vol. 4 Edinburgh: Edinburgh University Press, 393–416. [Google Scholar]
- SnogerupS, Snogerup B.2001.Bupleurum L. (Umbelliferae) in Europe. 1. The annuals, B. sect. Bupleurum and sect. Aristata Willdenowia 31: 205–308. [Google Scholar]
- SoltisDE, Soltis PS.1998. Choosing an approach and an appropriate gene for phylogenetic analysis. In: Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants II: DNA sequencing Boston: Kluwer Academic Publishers, 1–42. [Google Scholar]
- SprengelK.1820. Umbelliferae. In: Roemer JJ, Schultes JA, eds. Systema vegetabilium, Vol. 6. Sttutgart, Germany. [Google Scholar]
- SundingP.1979. Origins of the Macaronesian flora. In: Bramwell, D. ed. Plants and islands London: Academic Press, 13–40. [Google Scholar]
- SwoffordDL.2000.PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- ThompsonJD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG.1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TournefortJP.1694.Élémens de botanique, Vol 1. Paris: L’Imprimerie Royale. [Google Scholar]
- TutinTG.1968.Bupleurum In: Tutin TG, Heywood VG, Burges NA, Moore DM, Valentine DH, Walter SM, Webb DA, eds. Flora Europaea, Vol. 2 Cambridge: Cambridge University Press, 345–350. [Google Scholar]
- Valiejo‐RomanCM, Pimenov MG, Terentieva EI, Downie SR, Katz‐Downie DS, Troitsky AV.1998. Molecular systematics of the Umbelliferae: using nuclear ribosomal DNA internal transcribed spacer sequences to resolve issues of evolutionary relationships. Botanicheskii Zhurnal 83: 1–22. [Google Scholar]
- Valiejo‐RomanCM, Terentieva EI, Samigullin TH, Pimenov MG.2002. Relationships among genera in Saniculoideae and selected Apioideae (Umbelliferae) inferred from nrITS sequences. Taxon 51: 91–101. [Google Scholar]
- WatrousLE, Wheeler QD.1981. The out‐group comparison method of character analysis. Systematic Zoology 30: 1–11. [Google Scholar]
- WhiteTJ, Bruns T, Lee S, Taylor J.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols, a guide to methods and applications San Diego, London: Academic Press, 315–322. [Google Scholar]
- WolffH.1910.Bupleurum. In: Engler A, ed. Das Pflanzenreich Regnis Vegetabilis Conspectus, IV.228 Leipzig, Germany: Verlag von Wilhelm Engelmann, 36–173. [Google Scholar]
- YangZ.1993. Maximum‐likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Molecular Biology and Evolution 10: 1396–1401. [DOI] [PubMed] [Google Scholar]
- YangZ.1996. Among‐site rate variation and its impact on phylogenetic analyses. Trends in Ecology and Evolution 11: 367–372. [DOI] [PubMed] [Google Scholar]
- YokotaY, Kawata T, Iida Y, Kato A, Tanifuji S.1989. Nucleotide sequences of the 5·8S rRNA gene and internal transcribed spacer regions in carrot and broad bean ribosomal DNA. Journal of Molecular Evolution 29: 294–301. [DOI] [PubMed] [Google Scholar]