Abstract
Genetic screens have been extremely useful in identifying genes involved in hormone signal transduction. However, although these screens were originally designed to identify specific components involved in early hormone signalling, mutations in these genes often confer changes in sensitivity to more than one hormone at the whole‐plant level. Moreover, a variety of hormone response genes has been identified through screens that were originally designed to uncover regulators of sugar metabolism. Together, these facts indicate that the linear representation of the hormone signalling pathways controlling a specific aspect of plant growth and development is not sufficient, and that hormones interact with each other and with a variety of developmental and metabolic signals. Following the advent of arabidopsis molecular genetics we are beginning to understand some of the mechanisms by which a hormone is transduced into a cellular response. In this Botanical Briefing we review a subset of genes in arabidopsis that influence hormonal cross‐talk, with emphasis on the gibberellin, abscisic acid and ethylene pathways. In some cases it appears that modulation of hormone sensitivity can cause changes in the synthesis of an unrelated hormone, while in other cases a hormone response gene defines a node of interaction between two response pathways. It also appears that a variety of hormones may converge to regulate the turnover of important regulators involved in growth and development. Examples are cited of the recent use of suppressor and enhancer analysis to identify new nodes of interaction between these pathways.
Key words: Development, hormones, arabidopsis, genetic interactions
INTRODUCTION
The century‐old idea that specific substances could control plant growth and development was validated when the first hormones were identified and shown to mediated a diverse collection of plant processes (Davies, 1995). With the advent of molecular genetics and, in particular, the use of the model genetic system Arabidopsis thaliana (arabidopsis), the molecular basis of how hormone synthesis is turned into a cellular response is now being unravelled. Individual components such as receptors, signalling intermediates, e.g. kinases and phophatases, and downstream transcription factors have all been identified as playing specific roles in hormone signalling (McCourt, 1999). However, identification of individual components of hormone signal transduction pathways is of limited help in understanding how a plant uses hormones to coordinate overall growth and development. One particular conundrum is how a single hormone can affect so many unrelated responses and yet, at the same time, many different hormones can affect the same process. For example, auxin has been shown to mediate cell division, adventitious root development, apical dominance and cell expansion. However, gibberellin (GA) and brassinosteroids (BR) also appear to regulate cell expansion. Do these compounds all impinge on different aspects of cell expansion or do they all modulate the same step? Is the molecular mechanism conserved for all the processes that a single hormone affects? At first glance, genetic analysis suggests that hormones work through distinct pathways to elicit their responses and perhaps only interact distantly downstream of their primary response pathway. However, recent phenotypic analysis of hormone‐response mutants suggests that these molecules can influence each other’s synthesis and may perhaps share signalling components. The purpose of this Botanical Briefing is to outline some of the genetic approaches that have led to the belief that hormones interact or cross‐talk to form a complex web of overlapping signalling. We have limited our analysis to arabidopsis and to a few select pathways that are especially useful to the discussion of genetic interaction. We conclude with speculations of how hormones may have evolved to coordinate overall plant growth and development.
GENES THAT REGULATE HORMONE SENSITIVITY
Ethylene; I smell gas
The scrutiny of hormone signalling by the genetic eye has been most successful for understanding how the ethylene signal is perceived and transduced within arabidopsis (for a review, see Wang et al., 2002). This success mostly stems from the physiological growth assay that was used to define ethylene‐response mutants. Dark‐grown wild‐type seedlings continuously exposed to ethylene display what has been termed the ‘triple response’ phenotype: an exaggerated curvature of the apical hook, and a thick and short root and hypocotyl. Mutants that failed to generate the triple response in the presence of ethylene were classified as ethylene‐insensitive mutants, whilst plants that showed the triple response in the absence of the gas were considered either to overproduce ethylene or to be constitutive for the ethylene response.
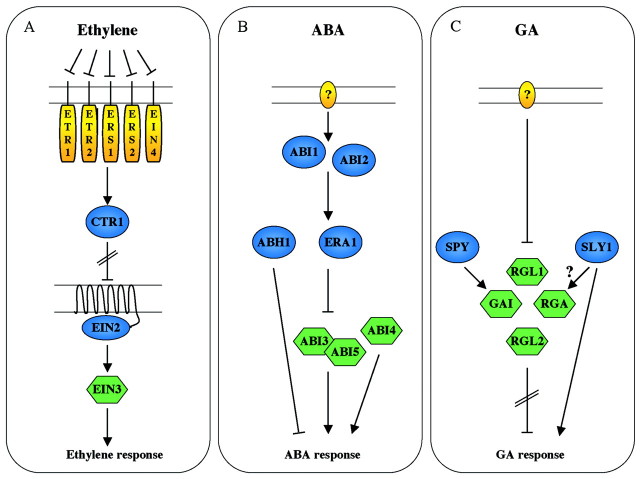
A combination of genetic, molecular and biochemical experiments has led to the following scenario being pieced together: ethylene binds to one or more members of a family of two‐component receptor kinases, ETR1, ETR2, EIN4, ERS1 and ERS2, and subsequently inactivates them (Fig. 1A). Hormone inactivation of the receptor family is transmitted via an unknown mechanism to a RAF‐like serine/threonine kinase, designated CTR1 (Kieber et al., 1993). In the presence of ethylene, inactivation of CTR1 in turn de‐represses EIN2, a positive regulator of the ethylene response pathway (Roman et al., 1995). The EIN2 gene encodes a membrane protein whose N‐terminal shows weak sequence similarity to the mammalian family of NRAMP metal transporters, with the C‐terminal region of the protein being novel (Alonso et al., 1999). The novelty of EIN2 and an inability to show any metal transport activity have made it difficult to establish the function of this protein in ethylene signalling. Finally, genetically defined downstream components of EIN2, such as EIN3, which encodes a transcription factor, and EIN5 and EIN6 which encode proteins of unknown function have also been identified (Chao et al., 1997).
Fig. 1. Three hormone signalling pathways as defined by genetic and molecular analysis. A, In the absence of ethylene the family of ethylene receptors (ETR1, ETR2, ERS1, ERS2, EIN4) activates CTR1, which in turn represses the positive membrane protein regulator EIN2. Addition of ethylene inactivates the ethylene receptors resulting in inactivation of CTR1 thereby releasing EIN2 to activate EIN3. The EIN3 transcription factor binds to regulatory sequences in the promoter of ethylene‐regulated genes inducing transcription. B, A receptor for ABA has not been defined. However, genetically downstream of ABA reception, dephosphorylation (ABI1/2), protein farnesylation (ERA1) and RNA processing (ABH1) are all required to attenuate the ABA signal. At the bottom of the pathway three transcription factors (ABI3, ABI4, ABI5) are responsible for at least seed sensitivity to ABA. C, The receptor for GA has not been defined. However, in the absence of GA a family of transcription factors (GAI, RGA, RGL1 and RGL2) inhibits various GA‐mediated responses. Through unknown mechanisms, GA antagonizes these proteins resulting in expression of GA‐regulated genes. SLY1 and SPY are also thought to regulate these transcriptional repressors. Green molecules indicate transcription factors, blue molecules indicate signalling intermediates, and yellow molecules represent receptors. Arrows represent positive regulation and bars represent negative regulation.
Abscisic acid; dwelling on the negative
Identification of genes involved in abscisic acid (ABA) signalling has usually involved screens based on inhibition of seed germination or altered gene expression to exogenously applied ABA. Broadly speaking, two classes of mutants have been identified. Mutations that confer a hypersensitive phenotype to ABA have indicated that protein farnesylation (ERA1), inositol signalling (FRY1) and RNA processing (ABH1, SAD1, HYL1) are required to attenuate the ABA response (Cutler et al., 1996; Lu and Federoff, 2000; Hugouvieux et al., 2001; Xiong et al., 2001a, b). By contrast, mutations that reduce seed ABA responsiveness suggest that dephosphorylation (ABI1, ABI2) and transcription (ABI3, ABI4, ABI5) are also important (Giraudat et al., 1992; Leung et al., 1994; Meyer et al., 1996; Leung et al., 1997; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Lopez‐Molina et al., 2000). Using a combination of suppressor and epistatic analysis it appears that ABI1/2 act at or above ERA1 and that both of these genes work at or above ABI3 and ABI5 (Fig. 1B; Parcy and Giraudat, 1997; Pei et al, 1998; S. Brady pers. comm.). Recent studies have shown that ABI3 and ABI5 interact in a yeast two‐hybrid assay for protein interaction, which is consistent with their assignment to the same genetic pathway (Nakamura et al., 2001). Results from a combination of loss‐of‐function and misexpression double mutants between ABI3, ABI4 and ABI5 suggest that these three transcription factors interact in complex ways to determine overall ABA seed sensitivity (Söderman et al., 2000). Double mutant analysis between abh1 and abi1 suggests that these genes are in separate genetic pathways, and this is supported by the observation that era1 and abh1 double mutants are additive with respect to ABA hypersensitivity (Hugouvieux et al., 2001).
Unlike the triple response assay, which appears to be a specific output response to ethylene application in arabidopsis, germination can be influenced by a myriad of external and internal cues. This is perhaps not surprising given that germination is a terminal response and it might therefore be advantageous for a plant to have multiple inputs lined up before committing to this irreversible process. For example, several hormones such as GA, BR and ethylene can promote germination of arabidopsis, and mutations that affect each of these pathways reduce the germination capacity in the presence of exogenous ABA (Steber et al., 1998; Beaudoin et al., 2000; Ghassemian et al., 2000; Steber and McCourt, 2001). Hence, the complexity of the output can confound the specificity of the genetic screen.
The lack of specificity when using germination to identify ABA signalling components can be partially overcome by using more specific outputs such as hormone‐specific gene expression. However, an alternative approach is to use more sophisticated ABA chemistry. A number of chemical isomers of ABA exist and, by taking advantage of a differential germination response between two different ABA stereoisomers, mutations in arabidopsis were identified that conferred an increased insensitivity to one isomer vs. the other (Nambara et al., 2002). As expected, the screen identified old ABA‐insensitive genes such as ABI3, ABI4 and ABI5 but also uncovered loss‐of‐function mutations in two new genes designated CHO1 and CHO2. The fact that these new genes were not identified in saturating ABA‐insensitivity screen bodes well for the use of more sophisticated chemistry as a new approach to finding mutations in pathways that may only confer subtle phenotypes due to genetic redundancy or may play only a minor role in the process under study.
Gibberellins; bigger is better
GA affects a variety of processes ranging from seed germination, leaf expansion, stem elongation, flower and trichome initiation, and flower and fruit development. Using genetic approaches two classes of GA‐response mutants were identified based on their vegetative phenotype and response to GA (Harberd et al., 1998). The first group comprises GA‐insensitive dwarf mutants which resemble mutants that are deficient in GA biosynthesis. These mutations result in plants that are stunted, have dark green leaves and show defects in flower development and timing of flowering, but unlike GA auxotrophs these mutants are not rescued by GA application. In arabidopsis, the first clearly characterized GA‐response mutants were semi‐dominant mutations in a gene designated GAI (Peng et al., 1997). Recessive mutations in GAI conferred, at best, only subtle phenotypes to the plant, suggesting that this gene was genetically redundant. This was verified by the identification of recessive mutations in the RGA gene that partially rescued the GA phenotype of a GA‐biosynthetic mutant (Silverstone et al., 1997). The RGA gene, which encodes a transcription factor, turned out to be a homologue of GAI, and three more GAI/RGA homologues have recently been identified in the arabidopsis genome (Silverstone et al., 1998; Lee et al., 2002; Wen and Chang, 2002). Interestingly, loss‐of‐function mutations in two of these genes, RGL1 and RGL2, have been shown to be negative regulators of germination and may have other roles in GA‐dependent processes. Based on the genetics of these studies it appears that members of the GAI/RGA/RGL1/RGL2 gene family of transcription factors act as negative regulators of various aspects of GA‐dependent processes, and that the function of GA is to inhibit these inhibitors (Fig. 1C; Harberd et al., 1998). Consistent with this idea, application of GA appears to increase the turnover of RGA (Dill et al., 2001; Silverstone et al., 2001). However, similar experiments with RGL1 did not show a GA‐dependent turnover indicating that although these transcription factors appear to have overlapping functions with respect to GA signalling they may be regulated differently (Wen and Chang, 2002).
The second group of GA‐response mutations appears to confer a GA‐independent phenotype to the plant and, of these, mutations in the SPY gene are the best characterized (Jacobsen and Olszewski, 1993). Loss‐of‐function mutations in SPY mimic GA‐treated wild‐type plants in that they show slender, elongated stems and are early‐flowering. Since loss‐of‐function mutations partially suppress the gai dwarf phenotype, formally this gene acts genetically at or downstream of GAI (Peng et al., 1997). However, since SPY is an O‐linked N‐acetyl‐glucosamine transferase, it could influence GA signalling by glycosylating GAI‐like proteins directly (Jacobsen et al., 1996).
GA‐response mutants have also been identified by taking advantage of the antagonistic interactions that occur between this hormone and ABA at the level of arabidopsis germination. Screens to identify mutations that suppress ABA‐dependent inhibition of germination uncover loss‐of‐function mutations in GA biosynthesis and GA perception, and the inability of GA auxotrophs to germinate is suppressed by mutations that reduce ABA biosynthesis or responsiveness (Koornneef et al., 1982; Steber et al., 1998). Furthermore, spy mutations also show reduced ABA sensitivity at the level of germination (Steber et al., 1998). Simplistically, these observations suggest that regulation of the germination response in arabidopsis is a balance between ABA and GA action. However, this antagonism does not extend to other aspects of GA‐ or ABA‐regulated functions. For example, loss of ABA biosynthesis does not counteract GA auxotrophic defects such as reduced cell expansion, just as reduction of GA levels does not rescue the wilty phenotype of an ABA auxotroph. Once again it is apparent that hormone interactions are often developmentally constrained in time and space.
GENETIC INTERACTIONS
Who’s on first?
The hormone sensitizing screens mentioned above were developed to find genes that specify a hormone signalling pathway and to this end they have proved successful. For example, mutations that were originally identified by an altered ethylene triple response in arabidopsis uncovered ETR1, which was subsequently shown to encode an ethylene receptor (Schaller et al., 1995). However, the specificity of the ethylene‐response mutants seems somewhat at odds with the plethora of physiological studies that show changes in a single hormone can manifest changes in the synthesis or response of other hormones. Some of these differences can be explained by the artificiality of hormone application inherent to physiological experiments. However, some of these discrepancies are also explained by a lack of detailed phenotypic analysis of the original hormone mutants when they were first identified. For example, mutations in ETR1 or other ethylene‐response genes also reduced the sensitivity of roots to exogenous ABA (Beaudoin et al., 2000; Ghassemian et al., 2000). More perplexingly, these same ethylene‐response mutations increase the sensitivity of the seed to ABA. Therefore, genetic analysis of ethylene and ABA action suggests that these hormones antagonize each other at the level of germination, act additively with respect to root growth, but do not appear to interact in processes such as the triple response or stomatal closure (Hugouvieux et al., 2001).
Loss‐of‐function mutations in the EIN2 gene have also been recovered in screens using auxin transport inhibitors or resistance to cytokinin application. In this latter case, the cytokinin insensitivity of ein2 mutants arises because the rate‐limiting step in ethylene synthesis (ACC synthase) is positively regulated by cytokinin. As a consequence, many of the growth defects attributed to cytokinin application are the result of ethylene overproduction; hence, mutants insensitive to ethylene appear insensitive to exogenous cytokinin (Vogel et al., 1998). Although, this example demonstrates clearly how addition of one hormone can influence the biosynthesis of another, mutational analysis of ethylene action also suggests that specific components of the pathway may be shared with other signalling pathways. For example, molecular analysis of EIN2 has shown that this protein has independent ethylene and jasmonic acid signalling domains (Alonso et al., 1999). Because of their biochemical nature it is not difficult to envisage how promiscuous signalling components could have multiple targets in different signalling pathways so that certain components could act as nodes for informational transfer between various pathways. However, unlike hormone synthesis, which can influence other pathways non‐cell autonomously, signalling component nodes can only interact in a cell‐autonomous manner.
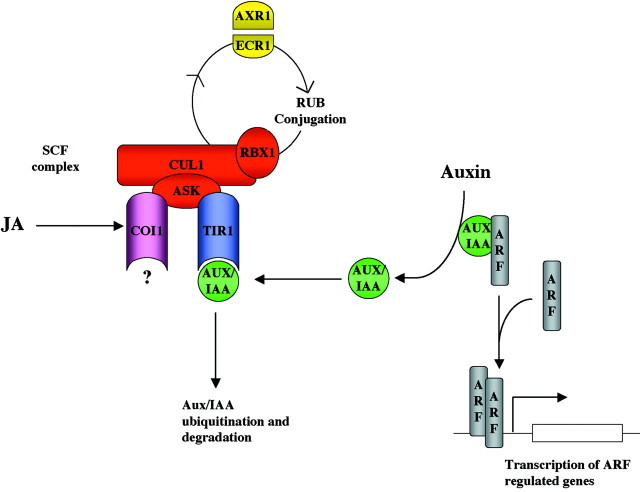
From the above examples it appears that hormones can interact at a number of levels, from influencing each other’s synthesis to sharing signalling components to create nodes of interaction. Furthermore, these inter‐relationships have a developmental context which means that the interactions have both temporal and spatial patterns. Given this information, one intriguing question that arises is how all this cross‐talk evolved. Were there separate hormone signalling pathways that somehow attained shared components? Is it possible for one hormone‐dependent pathway to be influenced by a second hormone if the first pathway picks up a protein motif that is influenced by the second hormone? For example, recent molecular genetic analysis of the auxin response in arabidopsis suggests that this hormone functions by modulating specific IAA‐regulated genes through the ubiquitination of a collection of short‐lived repressors termed AUX/IAA proteins (Fig. 2; for reviews, see Hellmann and Estelle, 2002 and Kepinski and Leyser, 2002). The ubiquitination of AUX/IAA proteins is dependent on an F‐box protein that associates with the SCF complex (for SKP1, Cullin/CDC53, F‐box protein). The specificity of the system is determined by the F‐box proteins and these proteins have now been implicated in flower morphogenesis, photocontrol of circadian clocks and leaf senescence, implying a large spectrum of functions for the SCF pathway in plant development (Ingram et al, 1997; Yang et al., 1999; Somers et al., 2000; Dieterle et al., 2001; Woo et al., 2001). More importantly, the COI1 and SLY1 genes both encode F‐box proteins suggesting that SCF‐dependent protein turnover is essential for correct signalling in jasmonate and GA pathways (Xie et al., 1998; C. M. Steber, pers. comm.). Although it appears that F‐box proteins determine specificity, the recent report that mutations in the AXR1 gene, which was originally identified as an auxin response gene, produce plants that are also insensitive to jasmonate demonstrates how this ubiquitination complex can act as a node of interaction between two hormone pathways (Fig. 2; Tiryaki and Staswick, 2002). From this example it is not difficult to envisage how an already existing developmental pathway might acquire, through evolution, a protein component that is sensitive to ubiquitination. By coming under the control of the SCF complex it is possible that the developmental pathway could become linked to auxin, jasmonate and GA signalling indirectly. On a related note, a number of mutations that increase ABA sensitivity are defective in RNA processing. This suggests that ABA may also regulate the level of specific proteins by regulating RNA stability (Hugouvieux et al., 2001). Hence, if hormones control the lifetime of specific signalling proteins they could, in principle, coordinate disparate signalling pathways to coordinate overall plant development.
Fig. 2. Hormone signalling can be regulated by the turnover of signalling components. The SCF‐complex is composed of four subunits (CUL1, ASK1, RBX1 and an F‐box protein). The cullin (CUL1) requires RUB modification mediated by AXR1‐ECR1 for normal activity of the complex. By interacting with specific substrates the F‐box proteins confer specificity to the degradation machinery. Loss‐of‐function mutations in the genes encoding the F‐box proteins TIR1, COI1 and SLY1 confer impaired sensitivity only to a single hormone, in this case auxin, JA and GA, respectively. In contrast, mutations in the ARX1 gene, which encodes the activating enzyme of the RUB complex, affect a variety of hormone responses such as auxin, JA and ABA. In the example shown, the AUX/IAA proteins associate with the TIR1 F‐box protein allowing them to be ubiquitinated by the SCF complex. This targets the AUX/IAA proteins for degradation. The removal of these proteins allows dimerization of ARF transcription factors allowing transcription of auxin response genes. JA signalling follows a similar mechanism, except that the F‐box protein is COI1.
What’s on second?
If hormones can co‐opt an already existing pathway, the expectation would be that genetic screens originally developed to identify genes in hormone‐independent processes might also identify genes in hormone signalling. Nowhere has this been more apparent than in genetic screens designed to identify mutations that affect sugar signalling. The premise of these screens was to identify mutants that were able to germinate and grow on media containing sugar concentrations that normally inhibit growth and development of wild‐type seedling. Surprisingly, most of the mutations identified were new alleles of known ABA‐biosynthetic genes, mutations in a subset of the ABA‐response loci, and mutations in genes involved in the ethylene response (for a review, see Gibson, 2000). Interestingly, not all sugar‐response mutants are in one hormone pathway and not all genes identified in a single hormone pathway are sugar‐response mutants. For example, only mutations in ABI4 and ABI5 confer an altered sugar response in arabidopsis. If the level of sugar sensitivity is determined through an ABA signalling pathway, why doesn’t the screen also identify mutations in ABI1, ABI2 and ABI3? Consistent with this, reconstruction experiments with known mutations in these genes did not confer an altered sugar response. However, these reconstruction experiments were limited to the use of single mutant alleles for each of these genes. More recently, using a larger collection of abi3 mutants, it has been shown that certain alleles do confer an altered glucose sensitivity (Nambara et al., 2002). Thus, the role of ABI3 in sugar–ABA interaction seems to be allele‐specific, and caution should be exercised in placing a gene outside of a particular function based solely on single alleles.
The reason that ABA‐biosynthetic mutants and some ABA‐insensitive mutants have an altered sugar response is at this time unclear, and more perplexing is the observation that the inhibitory effect of high sugar levels is only confined to approx. 2 d after germination in arabidopsis. Again this demonstrates the importance of a developmental context in determining sensitivities of tissues. This case suggests that following germination a sugar‐sensitive developmental window appears that requires ABA (Gibson, 2000). After the seedling becomes photosynthetically competent, it is possible that sensitivity to ABA decreases resulting in a plantlet insensitive to sugar‐induced ABA synthesis. Interestingly, ABI5 expression studies suggest that this transcription factor functions in a short post‐germination developmental window (Lopez‐Molina et al., 2001). This time frame may establish the stage at which arabidopsis seedlings are sensitive to high sugar concentrations.
Early seedling sugar sensitivity can also be influenced by ethylene since mutants that overproduce ethylene and constitutive ethylene‐signalling mutants are insensitive to high glucose concentrations, whereas ethylene‐insensitive mutants are glucose‐hypersensitive (Zhou et al., 1998; Gibson et al., 2001). These results suggest that increased ethylene action works to decrease the sensitivity of young seedlings to glucose. Moreover, evidence that the effects of ethylene on sugar responses require ABA synthesis indicates that ABA functions at or downstream of the ethylene signalling pathway during early seedling development (Zhou et al., 1998). Mutations that reduce ethylene signalling could increase ABA levels in the germinating seed, thereby inhibiting subsequent seedling growth. In this scenario, high sugar levels may signal through the ethylene pathway to regulate ABA biosynthesis. Alternatively, sugar‐induced ABA synthesis or sensitivity may be antagonized by ethylene action. In either case, these interactions between hormone signalling and primary metabolism suggest that it may be difficult in the future to place a hierarchy of control on signalling pathways in plants.
FUTURE PROSPECTS
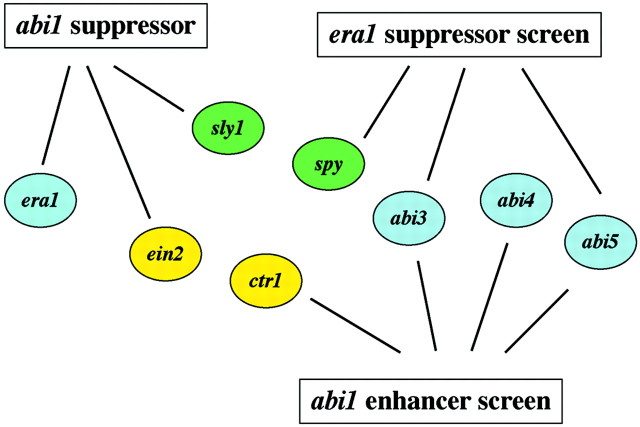
The approach of taking physiological assays that are influenced by a particular hormone and isolating mutants that do not respond correctly has led to the identification of genes required for hormone action. However, close phenotypic inspection of these mutants has also uncovered the complexity of hormone action and the difficulty of making simple linear pathways of inputs and outputs. Perhaps this inability to map hormone pathways into the two‐dimensional space of a diagram is a reflection of the complexity of the four‐dimensional space and time which a cell inhabits. Thus, the drawing of pathways with arrows and hierarchies shown in this review may be conceptually limiting. An alternative and less mechanistic representation of hormone interactions would be a diagram of the pathway based only on genetic interactions in the absence of a hierarchy. An example, using suppressor and enhancer analysis of era1 and abi1 mutants as a starting point, shows new patterns between hormone pathways (Fig. 3). Suppressors of era1 identify expected downstream ABA‐response genes, such as ABI3 and ABI5, but also the GA‐signal transduction component SPY1. Enhancers of abi1 identify downstream ABI transcription factors (ABI3, ABI4 and ABI5) and mutations in CTR1, which is involved in ethylene action. Furthermore, suppressors of abi1 identify genetic interactions with ABA‐response genes (ERA1), ethylene‐response genes (EIN2) and genes involved in GA signalling (SLY1). This genetic interaction map represents hormone signalling as more akin to a spider’s web of nodes and lines. Interestingly, the overall oscillation of a spider’s web is more important to the spider than the function of any individual node, as all nodes work in unison to give the correct oscillation. It is possible that the overall oscillation caused by genes being active or inactive due to hormonal cues may determine whether an arabidopsis seed will germinate or not, and that this information travels throughout the web by many different routes. In the future, comparing genetic interaction maps with patterns based on transcript profiling and other genomics technologies may allow a clearer representation of hormone interactions within the cell.
Fig. 3. Hormone signalling as described by genetic interaction. Using the sensitized genetic backgrounds of era1 (increased sensitivity to ABA) and abi1 (decreased sensitivity to ABA) as a starting point, second site suppressor and enhancer mutations were identified. Lines connect second site mutations to the original mutation that was being suppressed or enhanced. Screens used seed germination as the assay for ABA sensitivity. Blue molecules represent known ABA‐response genes, green molecules represent known GA‐response genes and yellow molecules represent known ethylene response genes.
ACKNOWLEDGEMENT
We wish to acknowledge Shelley Lumba for helpful commentary and editing of the manuscript.
Supplementary Material
Received: 8 November 2002; Returned for revision: 11 December 2002; Accepted: 20 January 2003 Published electronically: 6 March 2003
References
- AlonsoJM, Hirayama T, Roman G, Nourizadeh S, Ecker JR.1999. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- BeaudoinN, Serizet C, Gosti F, Graudat J.2000. Interaction between abscisic acid and ethylene signalling cascades. The Plant Cell 12: 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ChaoQ, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR.1997. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE‐INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- CutlerS, Ghassemian M, Bonetta D, Cooney S, McCourt P.1996. A protein farnesyl transferase involved abscisic acid signal trasduction in Arabidopsis. Science 273: 1239–1241. [DOI] [PubMed] [Google Scholar]
- DaviesPJ.1995. Plant hormones; physiology, biochemistry and molecular biology. Dordrecht: Kluwer. [Google Scholar]
- DieterleM, Zhou Y‐C, Schäfer E, Funk M, Kretsch T.2001. EID1, an F‐box protein involved in phytochrome A‐specific light signalling. Genes and Development 15: 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DillA, Jung HS, Sun TP.2001. The DELLA motif is essential for gibberellin‐induced degradation of RGA. Proceedings of the National Academy of Sciences of the USA 98: 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FinkelsteinRR, Lynch TJ.2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FinkelsteinRR, Wang ML, Lynch, TJ, Rao S, Goodman HM.1998. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. The Plant Cell 10: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GhassemianM, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P.2000. Regulation of abscisic acid signalling by the ethylene response pathway in Arabidopsis. The Plant Cell 12: 1117–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GibsonSI.2000. Plant sugar response pathways. Part of a complex regulatory web. Plant Physiology 124: 1532–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GibsonSI, Laby RJ, Kim D.2001. The sugar‐insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1 Biochemical and Biophysical Research Communications 280: 196–203. [DOI] [PubMed] [Google Scholar]
- GiraudatJ, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM.1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell 4: 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HarberdNP, King KE, Carol RJ, Richards DE.1998. Gibberellin: inhibitor of an inhibitor of…? Bioessays 20: 1001–1008. [DOI] [PubMed] [Google Scholar]
- HellmannH, Estelle M.2002. Plant development: regulation by protein degradation. Science 297: 793–797. [DOI] [PubMed] [Google Scholar]
- HugouvieuxV, Kwak JM, Schroeder JI.2001. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487. [DOI] [PubMed] [Google Scholar]
- IngramGC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES.1997. Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum EMBO Journal 16: 6521–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JacobsenSE, Olszewski NE.1993. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. The Plant Cell 5: 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JacobsenSE, Binkowski KA, Olszewski NE.1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 93: 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KepinskiS, Leyser O.2002. Ubiquitination and auxin signaling: a degrading story. The Plant Cell S81‐S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KieberJJ, Rothenberg M, Roman G, Feldmann KA, Ecker J.1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the RAF family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- KoornneefM, Jorna ML, Brinkhorst‐van der Swan DLC, Karssen CM.1982. The isolation and analysis of abscisic acid (ABA)‐deficient mutants by selection of induced revertants in non‐germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theoretical and Applied Genetics 61: 385–393. [DOI] [PubMed] [Google Scholar]
- LeeS, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J.2002. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA‐like gene whose expression is up‐regulated following imbibition. Genes and Development 16: 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeungJ, Merlot S, Giraudat J.1997. The Arabidopsis ABSCISIC ACID‐INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. The Plant Cell 9: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeungJ, Bouvier‐Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J.1994. Arabidopsis ABA response gene ABI1: features of a calcium‐modulated protein phosphatase. Science 264: 1448–1452. [DOI] [PubMed] [Google Scholar]
- Lopez‐MolinaL, Chua NH.2000. A null mutation in a bZIP factor confers ABA‐insensitivity in Arabidopsis thaliana. Plant and Cell Physiology 41: 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez‐MolinaL, Mongrand S, Chua NH.2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LuC, Fedoroff N.2000. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. The Plant Cell 12: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourtP.1999. Genetic analysis of hormone signalling. Annual Review of Plant Physiology and Plant Molecular Biology 50: 219–243. [DOI] [PubMed] [Google Scholar]
- MeyerK, Leube MP, Grill E.1994. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana Science 264: 1452–1455. [DOI] [PubMed] [Google Scholar]
- NakamuraS, Lynch TJ, Finkelstein RR.2001. Physical interactions between ABA response loci of Arabidopsis. Plant Journal 26: 627–635. [DOI] [PubMed] [Google Scholar]
- NambaraE, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P.2002. A screen for genes that function in abscisic acid signalling in Arabidopsis thaliana Genetics 161:1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ParcyF, Giraudat J.1997. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant Journal 11: 693–702. [DOI] [PubMed] [Google Scholar]
- PengJ, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP.1997. The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes and Development 11: 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PeiZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI.1998. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282:287–290. [DOI] [PubMed] [Google Scholar]
- RomanG, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR.1995. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SchallerGE, Bleecker AB.1995. Ethylene‐binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811. [DOI] [PubMed] [Google Scholar]
- SilverstoneAL, Ciampaglio CN, Sun T.1998. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. The Plant Cell 10: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SilverstoneAL, Mak PY, Martinez EC, Sun TP.1997. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana Genetics 146: 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SilverstoneAL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP.2001. Repressing a repressor: gibberellin‐induced rapid reduction of the RGA protein in Arabidopsis. The Plant Cell 13: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SödermanEM, Brocard, IM, Lynch TJ, Finkelstein RR 2000. Regulation and function of the Arabidopsis ABA‐insensitive4 gene in seed and abscisic acid response signalling networks. Plant Physiology 124: 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SomersDE, Schultz TF, Milnamow M, Kay SA.2000. ZEITLUPE encodes a novel clock‐associated PAS protein from Arabidopsis Cell 101: 319–329. [DOI] [PubMed] [Google Scholar]
- SteberCM, McCourt P.2001. A role for brassinosteroids in germination in arabidopsis. Plant Physiology 125: 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SteberCM, Cooney SE, McCourt P.1998. Isolation of the GA‐response mutant sly1 as a suppressor of ABI1‐1 in Arabidopsis thaliana Genetics 149: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TiryakiI, Staswick PE.2002. An arabidopsis mutant defective in jasmonate response is allelic to the auxin‐signalling mutant axr1 Plant Physiology 130: 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VogelJP, Woeste KE, Theologis A, Kieber JJ.1998. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proceedings of the National Academy of Sciences of the USA 95: 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WangKL, Li H, Ecker JR.2002. Ethylene biosynthesis and signalling networks. The Plant Cell 14: S131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WenCK, Chang C.2002. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. The Plant Cell 14: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WooHR, Chung KM, Park J‐H, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG.2001. ORE9, an F‐box protein that regulates leaf senescence in Arabidopsis The Plant Cell 13: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XieDX, Feys BF, James S, Nieto‐Rostro M, Turner JG.1998. COI1: an Arabidopsis gene required for jasmonate‐regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- XiongL, Lee Bh, Ishitani M, Lee H, Zhang C, Zhu JK.2001a FIERY1 encoding an inositol polyphosphate 1‐phosphatase is a negative regulator of abscisic acid and stress signalling in Arabidopsis. Genes and Development 15: 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XiongL, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK.2001b Modulation of abscisic acid signal transduction and biosynthesis by an Sm‐like protein in Arabidopsis. Developmental Cell 1: 771–781. [DOI] [PubMed] [Google Scholar]
- YangM, Hu Y, Lodhi M, McCombie R, Ma H.1999. The Arabidopsis SKP1‐LIKE1 gene is essential for male meiosis and may control homologue separation. Proceedings of the National Academy of Sciences of the USA 96: 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhouL, Jang JC, Jones TL, Sheen J.1998. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose‐insensitive mutant. Proceedings of the National Academy of Sciences of the USA 95: 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.