Abstract
• Background and Aims Cycas guizhouensis (Cycadaceae) is a rare and endangered species endemic to the southwest of China. An investigation was undertaken into the genetic variation of wild populations.
• Methods ISSR markers were used to determine the genetic variation within and between 12 extant populations of this species.
• Key Results Low genetic diversity (at population level, P = 14·21 %, HE = 0·0597; at species level, P = 35·90 %, HT = 0·1082) and a high degree of differentiation among populations (GST = 0·4321) were detected.
• Conclusions This genetic structure is considered to be due to the combined effects of slow biochemical evolution, genetic drift, inbreeding and limited gene flow between populations. Based on these findings, strategies are proposed for the genetic conservation and management of the species.
Key words: Conservation genetics, Cycas guizhouensis, genetic diversity, ISSR
INTRODUCTION
Cycads represent a very primitive group of vascular plants that have been in existence for more than 200 million years (Hendricks, 1987). Their origins can be dated to the low Permian (Zhifeng and Thomas, 1989), they were most diverse and widely dispersed in the Mesozoic era, and they were important components in the vegetation of the Triassic and Jurassic. Since then, they have been in decline and today they have only a relict distribution in tropical and subtropical regions. Field studies have shown that the majority of wild cycad populations are either threatened, critically endangered, or on the brink of extinction (Osborne, 1995). All species of cycads have been listed in the Convention on International Trade in Endangered Species of Wild Fauna and Flora. In China, all cycads have been given ‘First Grade’ conservation status.
Cycas guizhouensis is an endemic to south‐west China, mainly occurring in valleys of the Nanpan River and its tributaries in south‐west Guizhou, eastern Yunnan and north‐west Guangxi provinces at elevations of 400–1300 m a.s.l. The C. guizhouensis plants often grow on steep limestone mountains, with cylindrical trunks up to 1 m tall. In recent decades, C. guizhouensis has been threatened by rapid habitat destruction and over‐collection for food, medicine and use as an ornamental plant. Now this species has decreased dramatically in numbers, and has even become extinct in some easily accessible locations (Wang and Liang, 1996).
Understanding of the genetic variation within and between populations is essential for the establishment of effective and efficient conservation practices for rare plants. Several aspects of conservation biology, such as loss of genetic diversity in conservation programmes and restoration of threatened populations, can only be addressed by detailed population genetic studies (Hamrick and Godt, 1996). The few previous studies on the genetic variation of cycads that have been published have all been based on allozyme analysis, except for that of Pu and Chiu (1999) (Ellstrand et al., 1990; Byrne and James, 1991; Walters and Decker‐Walters 1991; Yang and Meerow, 1996, 1999; Li et al., 1999; Keppel et al., 2002). Because the number of allozyme loci that can be probed is limited, and they only correspond to coding sequences, allozyme variation may not be able to provide an accurate or complete measure of nucleotide variation in the genome (Clegg, 1990). However, new types of molecular markers allow DNA sequences other than nuclear‐coding loci to be examined, and can provide deeper insight into population genetic structures. Inter‐simple sequence repeats (ISSRs), for example, have several advantages for assessing genetic diversity (Gupta et al., 1994; Zietkiewicz et al., 1994). ISSR analyses are more specific than RAPD analyses, due to the longer SSR‐based primers, which enable higher‐stringency amplifications (Wolfe et al., 1998). The high stringency reduces problems with reproducibility, a common criticism against the low‐stringency RAPD assay (Yang et al., 1996). The shortcoming of ISSR markers, as with RAPDs, is that most bands are scored as dominant markers, giving no possibility to distinguish between homozygosis and heterozygosis directly. However, ISSR studies of natural populations have recently demonstrated the hyper‐variable nature of these markers and their potential use in population‐level studies (Ge and Sun, 1999; Culley and Wolfe, 2001).
As a first step towards investigating the genetic diversity of Cycas guizhouensis (Cycadaceae), the present study was designed to address the following questions using ISSR molecular markers. What is the level of ISSR variation in C. guizhouensis? What is the degree of between‐population differentiation in this species? What factors may have influenced its structure of genetic diversity? The answers to these questions will have important implications for the effectiveness and efficiency of any programs devised to conserve the species.
MATERIALS AND METHODS
Sample collection
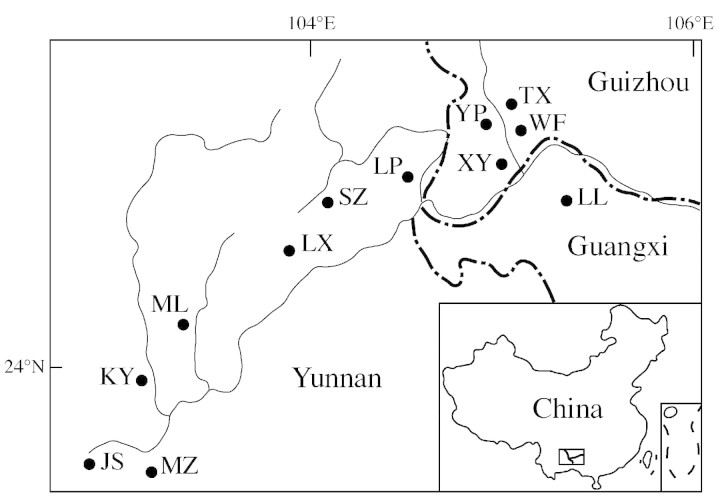
Leaf tissue was collected from 215 plants representing 12 extant populations of Cycas guizhouensis Lan & R.F. Zou (Fig. 1; Table 1). Fresh leaflets were dried with silica gel and stored at 4 °C until DNA extraction. Vouchers were collected from each population and deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Science (KUN).
Fig. 1. Map showing locations of the sampled populations of C. guizhouensis.
Table 1.
Populations of Cycas guizhouensis examined in the ISSR analysis
| Code | Population | Sample size | Latitude (N) | Longitude (E) |
| JS | Jianshui, Yunnan | 20 | 23°24′ | 102°53′ |
| KY | Kaiyuan, Yunnan | 16 | 23°50′ | 103°21′ |
| LL | Longlin, Guangxi | 20 | 24°40′ | 104°52′ |
| LP | Luoping, Yunnan | 22 | 24°43′ | 104°28′ |
| LX | Luxi, Yunnan | 16 | 24°30′ | 103°58′ |
| ML | Mile, Yunnan | 20 | 24°16′ | 103°38′ |
| MZ | Mengzi, Yunnan | 22 | 23°02′ | 103°21′ |
| SZ | Shizong, Yunnan | 6 | 24°35′ | 104°04′ |
| TX | Tingxi, Guizhou | 19 | 24°59′ | 105°18′ |
| WF | Wangfeng, Guizhou | 14 | 24°55′ | 105°03′ |
| XY | Xingyi, Guizhou | 20 | 24°52′ | 105°01′ |
| YP | Yangping, Guizhou | 20 | 24°56′ | 104°59′ |
DNA extraction and PCR amplification
Genomic DNA was extracted following the CTAB protocol (Doyle, 1991). Nuclear DNA was then PCR‐amplified using ISSR primers obtained from the University of British Columbia. Following an initial screen of 100 primers, 11 (807, 808, 810, 811, 813, 835, 836, 840, 842, 843 and 857) were selected for further analysis. Reactions were carried out in a total volume of 20 µL consisting of 20 ng of template DNA, 2·0 µL ×10 PCR buffer, 2·5 mm MgCl2, 0·1 mm dNTPs, 2 % formamide, 200 nm primer, 1·0 unit of Taq polymerase and double‐distilled water. PCR was performed with an MJ Research (Waltham, MA, USA) 96‐well thermal cycler with a hot bonnet, following Ge and Sun (1999). Amplification products were electrophoretically separated in 2·0 % agarose gels buffered with ×0·5 TBE. A 100 bp DNA ladder (New England Biolabs, Beverly, MA, USA) was used as a size marker. A negative control reaction in which the template DNA was replaced by water was performed alongside every PCR amplification in order to verify the absence of contamination. DNA fragments were identified by image analysis software for gel documentation (LabWorks Software Version 3.0; UVP, Upland, CA, USA) following staining with ethidium bromide. Only those bands that showed consistent amplification were considered. Smeared and weak bands were excluded.
Data analysis
ISSR bands were scored as presence or absence binary characters. The resulting presence/absence data matrix was analysed using POPGENE v. 1.31 (Yeh et al., 1999) to estimate two genetic diversity parameters: the percentage of polymorphic loci (P) and the expected heterozygosity (HE). At the species level, two genetic diversity measures (total gene diversity, HT, and the coefficient of gene differentiation, GST) were measured following Nei (1973), the genetic identity (I) and the genetic distance (D) between populations was also computed using the model presented in Nei (1972). Gene flow estimates (Nm) were calculated as Nm = (1– GST)/4GST (Slatkin and Barton, 1989).
Shannon diversity indices were also calculated to evaluate the results. The Shannon index was calculated as HO = –∑ pilog2 pi (Lewontin, 1972), in which pi is the frequency of a given ISSR fragment. Shannon’s index of phenotypic diversity was used to measure the total diversity (Hsp) as well as the mean intra‐population diversity (Hpop). The proportion of diversity between populations was then calculated as (Hsp – Hpop)/Hsp.
In order to test for putative correlations between genetic distances (D) and geographical distances (in km) amongst the populations, a Mantel test was performed with Tools for Population Genetic Analysis (Miller, 1997) (computing 5000 permutations).
RESULTS
Genetic diversity
The 11 primers chosen for analysis produced a total of 78 reproducible ISSR bands at an average of 7·1 bands per primer. In all, there were 28 polymorphic markers (Table 2). The percentage of polymorphic loci (P) was 14·21 % on average, ranging from 8·97 % (SZ) to 20·51 % (JS) at the population level and 35·90 % at the species level (Table 3). Of the 78 bands scored, 69 % (54 bands) were found in 90 % or more of sampled individuals, 27 % (21 bands) were found in 50–89 % of the samples, 4 % (three bands) were found in less than 50 %, and no population‐specific bands were observed in the data set. ISSR divergence among natural populations of C. guizhouensis appears to have been mainly due to DNA‐fragment frequency differences rather than to the fixation of locally common or rare bands. Assuming Hardy–Weinberg equilibrium, Nei’s genetic diversity (HE) was estimated to be 0·0597, on average, at the population level and 0·1082 at the species level, while Shannon indices (HO) were 0·0864 and 0·1686, respectively. Among the 12 populations, JS exhibited the highest level of variability (HE = 0·0884; HO = 0·1270), while population SZ exhibited the lowest level (HE = 0·0366; HO = 0·0531).
Table 2.
Attributes of ISSR primers used to generate ISSR markers from 215 individuals of Cycas guizhouensis sampled from 12 populations
| Primer | Sequence, 5′ to 3′ | No. of scorable bands | No. of polymorphic bands | Poly morphism (%) |
| 807 | AGAGAGAGAGAGAGAGT | 7 | 2 | 28·6 |
| 808 | AGAGAGAGAGAGAGAGC | 8 | 3 | 37·5 |
| 810 | GAGAGAGAGAGAGAGAT | 7 | 4 | 57·1 |
| 811 | GAGAGAGAGAGAGAGAC | 8 | 1 | 12·5 |
| 813 | CTCTCTCTCTCTCTCTT | 3 | 0 | 0·0 |
| 835 | AGAGAGAGAGAGAGAGYC | 11 | 5 | 45·5 |
| 836 | AGAGAGAGAGAGAGAGYA | 7 | 1 | 14·3 |
| 840 | GAGAGAGAGAGAGAGAYT | 8 | 4 | 50·0 |
| 842 | GAGAGAGAGAGAGAGAYG | 7 | 4 | 57·1 |
| 843 | CTCTCTCTCTCTCTCTRA | 6 | 2 | 33·3 |
| 857 | ACACACACACACACACYG | 6 | 2 | 33·3 |
| Total | 78 | 28 | 35·9 |
Table 3.
Genetic variability within populations of Cycas guizhouensis, as revealed by the ISSR analysis. N, sample size; P, percentage of polymorphic loci; HE, expected heterozygosity; HO, Shannon’s diversity index
| Population | N | P | HE (s.d.) | HO (s.d.) |
| JS | 20 | 20·51 | 0·0884 (0·1796) | 0·1270 (0·2579) |
| KY | 16 | 15·38 | 0·0668 (0·1595) | 0·0961 (0·2282) |
| LL | 20 | 14·10 | 0·0551 (0·1426) | 0·0807 (0·2064) |
| LP | 22 | 15·38 | 0·0720 (0·1704) | 0·1016 (0·2402) |
| LX | 16 | 11·54 | 0·0500 (0·1405) | 0·0719 (0·2014) |
| ML | 20 | 16·67 | 0·0698 (0·1588) | 0·1013 (0·2295) |
| MZ | 22 | 15·38 | 0·0637 (0·1524) | 0·0928 (0·2206) |
| SZ | 6 | 8·97 | 0·0366 (0·1217) | 0·0531 (0·1744) |
| TX | 19 | 16·67 | 0·0687 (0·1577) | 0·1000 (0·2276) |
| WF | 14 | 10·26 | 0·0426 (0·1298) | 0·0619 (0·1865) |
| XY | 20 | 11·54 | 0·0492 (0·1396) | 0·0710 (0·1999) |
| YP | 20 | 14·10 | 0·0531 (0·1366) | 0·0789 (0·2002) |
| Mean (s.d.) | 14·21 (3·21) | 0·0597 (0·0145) | 0·0864 (0·0206) |
Genetic differentiation
There is significant differentiation among the populations of C. guizhuoensis. The coefficient of genetic differentiation between populations (GST, estimated by partitioning of the total gene diversity) was 0·4321. This finding is consistent with the type of genetic structure predicted by the Shannon’s diversity index analysis, which suggested that 48·75 % of the total variation was partitioned between populations. Genetic identities (I) between populations varied from 0·9104 to 0·9828, with a mean of 0·9473 ± 0·0176 (Table 4). The level of gene flow between populations (Nm) was estimated to be 0·3286 individuals per generation, indicating that there is a low migration rate between populations.
Table 4.
Nei’s (1972) original measures of genetic identity (above diagonal) and genetic distance (below diagonal)
| JS | KY | LL | LP | LX | ML | MZ | SZ | TX | WF | XY | YP | |
| JS | – | 0·9319 | 0·9315 | 0·9403 | 0·9214 | 0·9345 | 0·9339 | 0·9393 | 0·9104 | 0·9394 | 0·9237 | 0·9269 |
| KY | 0·0705 | – | 0·9277 | 0·9420 | 0·9477 | 0·9444 | 0·9314 | 0·9462 | 0·9406 | 0·9637 | 0·9607 | 0·9660 |
| LL | 0·0710 | 0·0751 | – | 0·9264 | 0·9577 | 0·9527 | 0·9258 | 0·9359 | 0·9572 | 0·9572 | 0·9448 | 0·9583 |
| LP | 0·0616 | 0·0598 | 0·0765 | – | 0·9390 | 0·9718 | 0·9214 | 0·9340 | 0·9436 | 0·9567 | 0·9501 | 0·9402 |
| LX | 0·0819 | 0·0537 | 0·0432 | 0·0629 | – | 0·9549 | 0·9288 | 0·9671 | 0·9521 | 0·9781 | 0·9744 | 0·9685 |
| ML | 0·0678 | 0·0572 | 0·0484 | 0·0286 | 0·0462 | – | 0·9307 | 0·9493 | 0·9469 | 0·9700 | 0·9705 | 0·9557 |
| MZ | 0·0684 | 0·0710 | 0·0771 | 0·0819 | 0·0739 | 0·0718 | – | 0·9414 | 0·9163 | 0·9449 | 0·9285 | 0·9245 |
| SZ | 0·0626 | 0·0553 | 0·0663 | 0·0682 | 0·0334 | 0·0520 | 0·0604 | – | 0·9282 | 0·9785 | 0·9619 | 0·9546 |
| TX | 0·0939 | 0·0613 | 0·0438 | 0·0581 | 0·0490 | 0·0545 | 0·0874 | 0·0745 | – | 0·9561 | 0·9600 | 0·9669 |
| WF | 0·0625 | 0·0369 | 0·0437 | 0·0443 | 0·0222 | 0·0304 | 0·0567 | 0·0217 | 0·0449 | – | 0·9828 | 0·9748 |
| XY | 0·0794 | 0·0401 | 0·0568 | 0·0512 | 0·0259 | 0·0300 | 0·0742 | 0·0389 | 0·0408 | 0·0174 | – | 0·9789 |
| YP | 0·0759 | 0·0346 | 0·0426 | 0·0617 | 0·0320 | 0·0453 | 0·0785 | 0·0465 | 0·0336 | 0·0255 | 0·0213 | – |
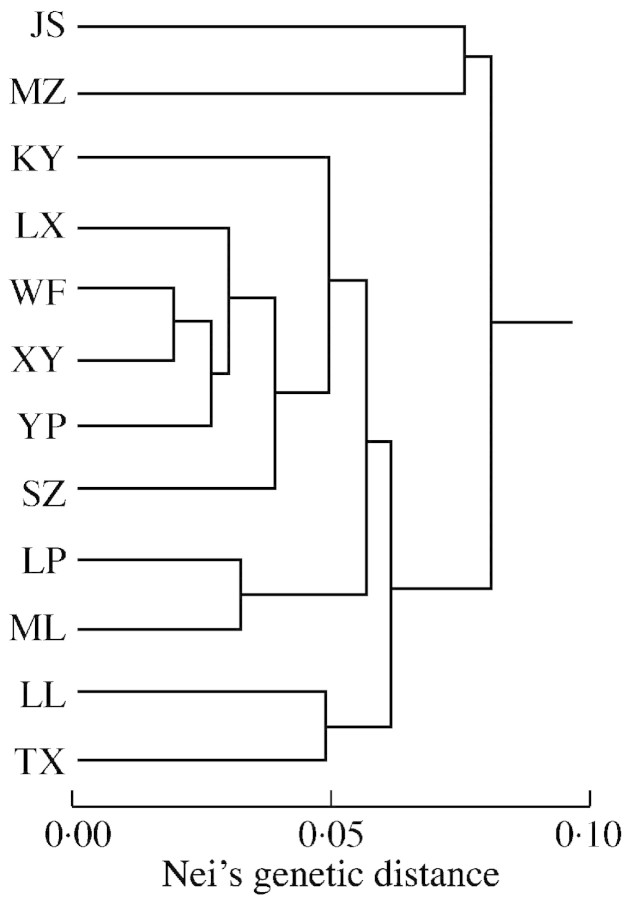
There was no significant correlation between genetic differentiation and geographical distance (r = 0·5132, P = 0·008) except for the three populations WF, XY and YP (r = 0·9727, P = 0·008) (Fig. 2), for which the separating distances were only about 7 km.
Fig. 2. UPGMA dendrogram based on Nei’s (1972) genetic distance.
Discussion
The genetic structure of natural populations is strongly affected by both intrinsic factors, such as migratory capabilities and mating systems, and extrinsic factors including the ecological characteristics of their habitats and historical events. As long‐lived, dioecious gymnosperms, the levels of genetic variation in cycads should theoretically be relatively high within populations and relatively low between populations (Hamrick et al, 1992). However, in contrast to expectations, the genetic structure of C. guizhouensis found in this study was characterized by low genetic variation within populations and high genetic differentiation between populations. Similar results have been found in most previous studies on cycads, such as Macrozamia communis (HT = 0·08, GST = 0·270, Ellstrand et al., 1990), Zamia pumila (HT = 0·047, Walters and Decker‐Walters, 1991), Cycas pectinata (HT = 0·126, GST = 0·387, Yang and Meerow, 1996), Cycas panzhihuaensis (HT = 0·049, GST = 0·345, Yang and Meerow, 1999) and Cycas seemannii (HT = 0·138, FST = 0·594, Keppel et al., 2002).
Low genetic diversity
In this study, low genetic diversity was found both at the population level and at the species level in C. guizhouensis (0·1686 for Shannon indices and 0·1082 for Nei’s expected heterozygosity at the species level, with mean values of 0·0864 and 0·0597, respectively, at the population level). In comparison with ISSR diversity, lower genetic variation had been detected in three sampled populations of C. guizhouensis at the allozyme level (HE = 0·100; Yang and Meerow, 1999). Our data support the hypothesis that low genetic variation within populations is biologically typical for cycads, unlike other gymnosperms (Walters and Decker‐Walters, 1991).
There are several possible explanations for the low genetic diversity revealed in C. guizhouensis. First, biochemical evolution within cycads may be slow compared to that of other seed plants (Walters and Decker‐Walters, 1991), which is supported by many previous studies on cycads (Yang and Meerow, 1996; 1999). Second, C. guizhouensis has a limited geographical range, with rather small and isolated populations. This distributional pattern may be an effect of Pleistocene glaciations. At the glacial maxima, it must have been restricted to refuges with an endurable temperature, and such areas are expected to have been small. Two major genetic consequences of small population size for long periods of time are high levels of genetic drift and inbreeding (Barrett and Kohn, 1991; Ellstrand and Elam, 1993). Both of these factors could be, at least partially, responsible for the low genetic diversity of this species. Genetic drift in particular could result in the loss of low‐frequency alleles in populations. In C. guizhouensis, we observed only three electrophoretic bands (4 %) with a frequency lower than 50 %, indicative of such stochastic processes of genetic drift. The Mantel test and the UPGMA dendrogram (Fig. 2) provided further support for the conclusions that there is no significant correlation between genetic distance and geographical distance, and the genetic differentiation does not fit with spatial division. These results provide further evidence for the action of genetic drift (Dodd and Helenurm, 2002). Turning to the likelihood of inbreeding, most of the extant species of Cycas are restricted to local flora with relict distributions and small population sizes due to habitat fragmentation (Lin et al., 2000; Huang et al., 2001). Completely isolated population fragments are prone to elevated rates of inbreeding. Furthermore, the large, gravity‐dispersed seeds (2·1–2·6 × 1·7–2·2 cm, Wang 2000) of C. guizhouensis may promote mating between individuals in close proximity within populations (Keppel et al., 2002). Thus, inbreeding presumably occurs frequently through consanguineous mating in C. guizhouensis.
High population differentiation
Plant species differ markedly in the way that genetic diversity is partitioned between populations. In the present study, both the Nei’s genetic diversity and Shannon’s diversity index analyses of the ISSR data gave similar indications about the nature of the genetic structure of the C. guizhouensis populations. The coefficient of gene differentiation (GST) derived for the species was 0·4321, and the proportion of diversity between populations based on the Shannon indices was similarly high: 48·75 %. By contrast, three C. guizhouensis populations examined using allozyme analysis by Yang and Meerow (1999) appeared to have much lower levels of genetic differentiation (GST = 0·080). In other studies, estimates of genetic differentiation between populations based on molecular markers (such as RAPD and RFLP) are generally higher than estimates based on allozyme analysis (e.g. Zhang et al., 1993; Buso et al., 1998; Ayres and Ryan, 1999; Francisco‐Ortega et al., 2000). Thus, the level of polymorphism we found at ISSR loci of 12 populations is likely to give a more accurate estimate of GST than the previous estimate derived from analysis of a single allozyme polymorphism in just three populations.
The genetic differentiation of plant populations reflects interactions amongst a range of different processes, including the long‐term evolutionary history of the species (e.g. shifts in distribution, habitat fragmentation and population isolation), mutation, genetic drift, mating systems, gene flow and selection (Schaal et al., 1998). The strong differentiation in C. guizhouensis may be mainly due to genetic drift and limited gene flow. The small populations of C. guizhouensis, which have been isolated from one another since after the Pleistocene, are likely to have been subjected to strong genetic drift, as mentioned above. Genetic drift changes the distribution of genetic variation in two ways, by reducing variation within populations and by increasing differentiation between populations (Ellstrand and Elam, 1993). The limited seed and pollen dispersal contributes to the low level of gene flow and the high level of inter‐population differentiation (Wallace, 2002). Beetles and rodents serve as dispersal agents for the pollens and seeds of C. guizhouensis, respectively (L.‐Q. Xiao, pers. obs., also see Wang, 1995; Schneider et al., 2002). The level of gene flow via seed and pollen is constrained by the low migratory capabilities of the pollinators and the seed‐carriers. The gene flow distance between local populations in Cycas was estimated to be only 2–7 km in a study by Yang and Meerow (1996). However, the distances between extant populations of C. guizhouensis are mostly >30 km. So, gene exchange between populations is hindered by the distance between them in this species. A migration rate of 0·5 is considered sufficient to overcome the diversifying effects of random drift (Ellstrand and Elam, 1993). The estimate of Nm (0·3286) derived in the present study is too low to effectively prevent stochastic differentiation by genetic drift.
It is worth mentioning that although the overall correlation between genetic differentiation and geographical distance was weak, there was a strong correlation for three populations (WF, XY and YP) that are separated by a distance of only about 7 km (Mantel test, r = 0·9727, P = 0·008). This indicates that C. guizhouensis has the capacity to exchange genes between populations within a certain geographical distance. Our results suggest that this limit is close to approx. 7 km, which is in agreement with Yang and Meerow’s (1996) estimate that the gene flow distance between members of local populations is 2–7 km in cycads.
Considerations for conservation
The primary objective in nature conservation is to preserve as much as possible of the evolutionary potential of species through maintaining as much genetic diversity as possible. Knowledge of the genetic variation between and within populations of rare and endangered species plays a significant role in the formulation of appropriate management strategies directed towards their conservation (Milligan et al., 1994). Our results further confirm the hypothesis that low intra‐population genetic variation with relatively high spatial differentiation is a biological and evolutionary characteristic of cycads (Walters and Decker‐Walters, 1991). These population genetic‐structure characteristics have significant implications for conservation strategies. Low genetic diversity may reduce the potential of species or populations to survive in a changing environment (Ellstrand and Elam, 1993). There is an urgent need to take effective measures to protect this species against further loss of genetic diversity. Considering the high genetic differentiation of C. guizhouensis, preservation of any one population would be insufficient to conserve all the variation in the species. Thus, the priority must be to protect all the existing populations in situ and prevent anthropogenic destruction, allowing them to propagate and increase in size through natural regeneration. If ex situ conservation is required, samples should be collected from as many populations as possible, especially from those harbouring relatively high genetic diversity, such as population JS in this study. Because of its long life‐cycle, the seed and germplasm collections in botanical gardens or other institutions should be of practical value for the conservation of genetic diversity in C. guizhouensis.
ACKNOWLEDGEMENTS
We thank Dr Bo Tian for his great help in collecting the samples. This work was supported by the Natural Science Foundation of China (No. 30070081).
Received: 7 August 2003; Returned for revision: 14 January 2004; Accepted: 15 March 2004. Published electronically: 14 May 2004
References
- AyresDR, Ryan FJ.1999. Genetic diversity and structure of the narrow endemic Wyethia reticulata and its congener W bolanderi (Asteraceae) using RAPD and allozyme techniques. American Journal of Botany 87: 344–353. [PubMed] [Google Scholar]
- BarrettSCH, Kohn JK.1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: DA Falk, KE Holsinger, eds. Genetics and conservation of rare plants New York: Oxford University Press, 3–30. [Google Scholar]
- BusoGSC, Rangel PH, Ferreira ME.1998. Analysis of genetic variability of South American wild rice populations (Oryza glumaepatula) with isozymes and RAPD markers. Molecular Ecology 7: 107–117. [Google Scholar]
- ByrneM, James SH.1991. Genetic diversity in the cycad Macrozamia riedlei Heredity 67: 35–39 [Google Scholar]
- CleggMT.1990. Molecular diversity in plant populations. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding and genetic resources Massachusetts: Sinauer Associates, Inc., 98–115. [Google Scholar]
- CulleyTM, Wolfe AD.2001. Population genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR molecular markers. Heredity 86: 545–556. [DOI] [PubMed] [Google Scholar]
- DoddSC, Helenurm K.2002. Genetic diversity in Delphinium variegatum (Ranunculaceae): a comparison of two insular endemic subspecies and their widespread mainland relative. American Journal of Botany 89: 613–622. [DOI] [PubMed] [Google Scholar]
- DoyleJ.1991. DNA protocols for plants – CTAB total DNA isolation. In: Hewitt GM, Johnston A, eds. Molecular techniques in taxonomy Berlin: Springer, 283–293. [Google Scholar]
- EllstrandNC, Elam DR.1993. Population genetic consequences of small population size: implication for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- EllstrandNC, Ornduff R, Clegg JM.1990. Genetic structure of the Australian cycad, Macrozamia communis (Zamiaceae). American Journal of Botany 77: 677–681. [Google Scholar]
- Francisco‐OrtegaJ, Santos‐Guerra A, Kim SC, Crawford DJ.2000. Plant genetic diversity in the Canary Islands: a conservation perspective. American Journal of Botany 87: 909–919. [PubMed] [Google Scholar]
- GeXJ, Sun M.1999. Reproductive biology and genetic diversity of a cryptoviviparous mangrove Aegiceras corniculatum (Myrsinaceae) using allozyme and intersimple sequence repeat (ISSR) analysis. Molecular Ecology 8: 2061–2069. [DOI] [PubMed] [Google Scholar]
- GuptaM, Chyi YS, Romero‐Severson J, Owen JL.1994. Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple‐sequence repeats. Theoretical and Applied Genetics 89: 998–1006. [DOI] [PubMed] [Google Scholar]
- HamrickJL, Godt MJW.1996. Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL, eds. Conservation genetics: case histories from nature. New York: Chapman and Hall, 281–304. [Google Scholar]
- HamrickJL, Godt MJW,Sherman‐Broyles SL.1992. Factors influencing levels of genetic diversity in woody plant species. New Forest 6: 95–124. [Google Scholar]
- HendricksJG.1987. The Gondwanan Cycas Encephalartos 10: 24–25. [Google Scholar]
- HuangS, Chiang YC, Schaal BA, Chou CH, Chiang TY.2001. Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Molecular Ecology 10: 2669–2681. [DOI] [PubMed] [Google Scholar]
- JonesDL.1993.Cycads of the World. Washington D.C.: Smithsonian Institution Press. [Google Scholar]
- KeppelG, Lee SW, Hodgskiss PD.2002. Evidence for long isolation among populations of a Pacific cycad: genetic diversity and differentiation in Cycas seemannii A. Br. (Cycadaceae). Journal of Heredity 93: 133–139. [DOI] [PubMed] [Google Scholar]
- LewontinRC.1972. The apportionment of human diversity. Evolutionary Biology 6: 381–398. [Google Scholar]
- LiCL, Wang Q, Jiang SY, Ge S, Wang KQ.1999. Genetic diversity of allozymes in populations of Cycas panzhihuaensis L. Zhou & S.Y. Yang. In: Chen CJ, eds. Biology and conservation of cycads. Proceedings of the Fourth International Conference on Cycad Biology Beijing: International Academic Publishers, 323–327. [Google Scholar]
- LinTP, Sun YC, Lo HC, Cheng YP.2000. Low genetic diversity of Cycas taitungensis (Cycadaceae), an endemic species in Taiwan, revealed by allozyme analysis. Taiwan Journal of Forestry Science 14: 35–42. [Google Scholar]
- MillerMP.1997.Tools for population genetic analysis (TFPGA), Version 1.3. Arizona, USA: Department of Biological Science, Northern Arizona University. [Google Scholar]
- MilliganBG, Leebens‐Mack J, Strand AE.1994. Conservation genetics: beyond the maintenance of marker diversity. Molecular Ecology 12: 844–855. [Google Scholar]
- NeiM.1972. Genetic distance between populations. American Naturalist 106: 283–292. [Google Scholar]
- NeiM.1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PuH, Chiu WL.1999. Genetic variation in Cycas Section Stangerioides using anchored microsatellite primers. In: Chen CJ, ed. Biology and conservation of cycads. Proceedings of the Fourth International Conference on Cycad Biology Beijing: International Academic Publishers, 334–344. [Google Scholar]
- OsborneR.1995. The world cycad census and a proposed revision of the threatened species status for cycad taxa. Biological Conservation 71: 1–12. [Google Scholar]
- SchaalBA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA.1998. Phylogeographic studies in plants: problems and prospects. Molecular Ecology 7: 465–474. [Google Scholar]
- SchneiderD, Wink M, Sporer F.2002. Cycads: their evolution, toxins, herbivores and insect pollinators. Naturwissenschaften 89: 281–294. [DOI] [PubMed] [Google Scholar]
- SlatkinM, Barton NH.1989. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368. [DOI] [PubMed] [Google Scholar]
- WallaceLE.2002. Examining the effects of fragmentation on genetic variation in Platanthera leucophaea (Orchidaceae): inferences from allozyme and random amplified polymorphic DNA markers. Plant Species Biology 17: 37–49. [Google Scholar]
- WaltersTW, Decker‐Walters DS.1991. Patterns of allozyme diversity in the West Indies cycad Zamia pumila (Zamiaceae). American Journal of Botany 78: 436–445. [Google Scholar]
- WangDY.1995. A preliminary study of the Cycas micholitzii complex. Encephalartos 44: 31–38. [Google Scholar]
- WangDY.2000.Studies on morphology, anatomy, taxonomy and evolution of Cycadaceae. PhD Dissertation. Nanjing: Nanjing Forestry University. [Google Scholar]
- WangFX, Liang HB.1996.Cycads in China. Guangzhou: Guangdong Science and Technology Press. [Google Scholar]
- WatkinsonAR, Powell JC.1997. The life history and population structure of Cycas armstrongii in monsoonal northern Australia. Oecologia 111: 341–349. [DOI] [PubMed] [Google Scholar]
- WolfeAD, Xiang QY, Kephart SR.1998. Diploid hybrid speciation in Penstemon (Scrophulariaceae). Proceedings of the National Academy of Sciences of the USA 95: 5112–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YangSL, Meerow AW.1996. The Cycas pectinata (Cycadaceae) complex structure and gene flow. International Journal of Plant Sciences 157: 468–483. [Google Scholar]
- YangSL, Meerow AW.1999. Genetic variation in Chinese cycad population. In: Chen CJ, ed. Biology and conservation of cycads. Proceedings of the Fourth International Conference on Cycad Biology Beijing: International Academic Publishers, 175–186. [Google Scholar]
- YangWA, De‐Oliveira AC, Godwin I, Schertz K, Bennetzen JL.1996. Comparison of DNA marker technologies in characterizing plant genome diversity: variability in Chinese sorghums. Crop Science 36: 1669–1676. [Google Scholar]
- YehFC, Yang RC, Boyle T.1999.POPGENE Microsoft Windows‐based freeware for population genetic analysis. Release 1.31. Edmonton: University of Alberta. [Google Scholar]
- ZhangQ, Saghai Maroof MA, Kleinhofs A.1993. Comparative diversity analysis of RFLPs and isozymes within and among populations of Hordeum vulgare ssp. spontaneum Genetics 134: 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhifengG, Thomas BA.1989. A review of fossil cycad megasporophylls, with new evidence of Crossozamia pommel and its associated leaves from the lower Permian of Taiyuan, China. Review of Paleobotany and Palynology 60: 205–223. [Google Scholar]
- ZietkiewiczE, Rafalski A, Labuda D.1994. Genome fingerprinting by simple sequence repeat (SSR)‐anchored polymerase chain reaction amplification. Genomics 20: 176–183. [DOI] [PubMed] [Google Scholar]