Abstract
A wide range of plants are grown for their edible tubers, but five species together account for almost 90 % of the total world production. These are potato (Solanum tuberosum), cassava (Manihot esculenta), sweet potato (Ipomoea batatus), yams (Dioscorea spp.) and taro (Colocasia, Cyrtosperma and Xanthosoma spp.). All of these, except cassava, contain groups of storage proteins, but these differ in the biological properties and evolutionary relationships. Thus, patatin from potato exhibits activity as an acylhydrolase and esterase, sporamin from sweet potato is an inhibitor of trypsin, and dioscorin from yam is a carbonic anhydrase. Both sporamin and dioscorin also exhibit antioxidant and radical scavenging activity. Taro differs from the other three crops in that it contains two major types of storage protein: a trypsin inhibitor related to sporamin and a mannose‐binding lectin. These characteristics indicate that tuber storage proteins have evolved independently in different species, which contrasts with the highly conserved families of storage proteins present in seeds. Furthermore, all exhibit biological activities which could contribute to resistance to pests, pathogens or abiotic stresses, indicating that they may have dual roles in the tubers.
Key words: Review, tuber, storage proteins, enzyme inhibitors, protein deposition, gene regulation
INTRODUCTION
Plant tubers as crops
Plants form the major source of dietary protein and carbohydrate for humans and livestock, being particularly important for human nutrition in developing countries where the consumption of animal products is low. Plant foods can be broadly divided into two groups. Staples provide the bulk of dietary intake and correspond to plant storage tissues (seeds and tubers) with humankind benefiting from the storage reserves laid down by the plant. In contrast, fruits and vegetables tend to be consumed in smaller amounts as part of a mixed diet. Although they may contribute essential nutrients (e.g. vitamins, minerals), they are often consumed more for their organoleptic (taste, texture) properties than nutritional quality. They vary widely in their botanical origin, and in their concentration and composition of nutrients.
The major staple crops are seeds, with cereals and legumes in particular being grown and consumed in vast quantities. In addition, five types of tuber are also considered to be staples, although only one of these (potato) is grown outside the tropics. These five species together accounted for almost 99 % of the total world production of tuber crops in 1999 (estimated as about 650 million tonnes), with potato being the most important (approx. 45 %) followed by cassava (26 %), sweet potato (20 %), yams (6 %) and taro (1 %) (FAO, 1999). In addition, numerous other types of plant tuber are consumed in small amounts in different parts of the world, particularly as vegetables. Thus, Rehm and Espig (1991) list 25 root and tuber crops, from 14 plant families, which are grown in the tropics and sub‐tropics, while readers will be aware of root vegetables consumed as part of their own diets.
Characteristics of tubers
In contrast to seeds, plant storage tubers have diverse botanical origins (Table 1). Thus, of the ‘big five’, potatoes are derived from stems, taro from corns, yams from swollen hypocotyls and cassava and sweet potato from roots. Other tubers are derived from rhizomes [e.g. turmeric (Cucuma longa, Zingiberaceae)] and swollen tap roots [e.g. yam bean (Pachyrhizus spp., Leguminosae)]. Plant tubers share one or two biological roles. The first is to store carbon and usually also nitrogen in a form that can be mobilized when required. In perennials such as cassava the storage may be long term, whereas in biennials and annuals it is from one season to the next. The second property which is shared by most, but not all, tubers is that they act as propagules, in that they are able to sprout and give rise to new plants. In this case they need to contain a sufficiently wide range of nutrients to support the requirements of the plant until it is capable of independent growth.
Table 1.
The botanical origins and protein contents of major tuber crops
| Crop | Species (family) | Origin of tubers | Approx. protein content of tubers (% d. wt) | Reference |
| Potato | Solanum tuberosum, (Solanaceae) | Stem | 3–6 | 1 |
| Sweet potato | Ipomoea batatas (Convolvulaceae) | Root | 1–10 | 2 |
| Cassava | Manihot esculenta (Euphorbiaceae) | Root | 1–2 | 3 |
| Taro | Colocasia esculenta | Corm | 2·0 | 4 |
| Cytosperma chamissonis | Corm | 0·8 | 4 | |
| Xanthosoma sagittifolium | Corm | 2·0 | 4 | |
| Alocasia macrorhiza (Araceae) | Ctem | 0·6 | 4 | |
| Yam | Dioscorea spp. (Dioscoreaceae) | Hypocotyl | 1–3 | 5 |
References: 1, calculated from Burton (1989); 2, Walter et al. (1984); 3, Clowes et al. (1995); 4, Pollock (2000); 5, Coursey (1995).
In general, plant tubers are rich in starch and indeed they are often considered solely as a source of carbohydrate for diets or for industrial use (e.g. potato, cassava). However, they do contain protein which varies in amount from about 1–2 % d. wt in cassava up to almost 10 % d. wt in yam bean (Pachyrhizus spp, Leguminosae). Nevertheless, their protein contents (Table 1) are usually substantially lower than those of seeds, whose protein contents range from about 10 % in some cereals to about 40 % in soybean. Furthermore, whereas many seeds contain stores of triacylglycerols (oil), this is not usually stored in tubers.
Do tubers contain storage proteins?
Storage proteins can be defined as proteins whose major role is to act as stores of nitrogen, sulfur and carbon. They may enable the plant to survive periods of adverse conditions or between growing seasons, and may provide nutrients to support the growth of new plants as seedlings (from seeds) or shoots (from tubers). They act as a sink for nitrogen (and probably also sulfur), accumulating in greater amounts under conditions of excess nutrient supply. They are also located in the cell in discrete deposits (protein bodies) which facilitates high‐level accumulation without any adverse effects on other cellular functions.
Seeds contain four well‐defined types of storage proteins: the prolamins, 7S globulins, 11S globulins and 2S albumins (see Shewry, 1995; also chapters in Shewry and Casey, 1999). The vast majority of proteins in these groups have no known biological activity, and are thought to function solely as storage proteins. However, they are related in their structures and evolutionary origins to groups of proteins that are biologically active, and some 2S albumins have been shown to exhibit antifungal properties (Terras et al., 1993) and to inhibit serine proteinases (trypsin, subtilisin, chymotrypsin) (Genov et al., 1997; Svendsen et al., 1989, 1994) when tested in vitro. These activities may represent a true secondary role, or roles, but they may also indicate that the storage proteins have evolved from ancestral proteins with metabolic activity.
This article is a review of work on the proteins of plant storage tubers and addresses the question of whether tubers contain true storage proteins and, if so, what are their origins and relationships. Initially, the review will focus on the ‘big five’, before considering studies of minor crops.
POTATO
Potato (Solanum tuberosum, Solanaceae), is the only major tuber crop that is grown in temperate regions. It is also the most important tuber crop in terms of production, accounting for about 45 % of the total world production of all tuber crops. The spread of the potato from its centre of origin in the high Andes of South America to other parts of the globe, and the historical consequences of this have been well documented (Messer, 2000). Potato tubers are in fact derived from swollen stems, although they are generally subterranean. Osborne and Campbell (1896) reported that the major protein in potato tubers was a globulin which they termed ‘tuberin’. More recently, Racusen and Foote (1980) reported that a glycoprotein of Mr about 45 000 accounted for about 20 % of the total soluble protein in potato and proposed the alternative name ‘patatin’, based on ‘patata’ which is the original American Indian‐derived Spanish word for potato. The name patatin has since become widely accepted.
Characteristics of patatin
Park et al. (1983) estimated the molecular mass of patatin to be about 40 000 and showed extensive heterogeneity with forms differing in electrophoretic mobility at pH 8·6 and in mobility on SDS–PAGE. Paiva et al. (1983) demonstrated that there was a linear relationship between the amount of patatin, expressed as a percentage of total soluble protein, and the logarithm of tuber weight from 0·3 to 300 g, with patatin forming about 40 % of the total soluble protein in tubers above about 200 g. Sonnewald et al. (1989b) demonstrated that patatin expressed in leaves of transgenic tobacco was glycosylated on two sites (asparagine 60 and asparagine 90), with typical small complex glycans comprising xylose, fucose, mannose and N‐acetylglucosamine in a ratio of 1 : 1 : 3 : 2, which is the same as the ratio of these sugars present in patatin isolated from potato tubers. Immunochemical studies demonstrated that patatin is located in vacuoles in tubers and in leaves induced for its expression (see below) (Sonnewald et al., 1989a), an observation which is consistent with N‐glycosylation taking place in the endoplasmic reticulum and Golgi apparatus (Kermode and Bewley, 1999).
Preliminary comparisons by N‐terminal sequencing and Ouchterlony double diffusion using polyclonal antiserum to total soluble proteins indicated that the component proteins of patatin are closely related (Park et al., 1983), and this was confirmed by the analysis of cloned cDNAs and genes (Mignery et al., 1984, 1988). This showed the existence of two classes of mRNAs and genes, with the former sharing about 98 % sequence identity. The encoded proteins showed some minor differences in sequence, particularly in the N‐terminal region, which was in agreement with the heterogeneity observed previously in directly determined N‐terminal sequences (Park et al., 1983), but also differed in the presence (class II) or absence (class I) of a 22 bp sequence within the 5′ untranslated region.
The mature class I and class II patatins comprise about 360 amino acid residues but are synthesized with N‐terminal signal sequences of 23 residues (Mignery et al., 1984). This is consistent with their transport via the endomembrane system leading to deposition in vacuoles (Sonnewald et al., 1989a).
Regulation of patatin synthesis
The patatin present in tubers is almost solely encoded by class I transcripts with class II transcripts being about 50–100 times less abundant (Mignery et al., 1988). However, Pikaard et al. (1987) showed that roots also contain small amounts of an immunologically distinct form of patatin which appears to be encoded by class II transcripts (Pikaard et al., 1987; Mignery et al., 1988).
Tubers are usually formed from underground stolons but can also form above ground from auxiliary buds as a result of injury, disease or removal of stolons and tubers. These tubers accumulate patatin to similar levels to those in tubers, i.e. approx. 40–45 % of total soluble proteins (Paiva et al., 1983). Removal of tubers and auxiliary buds can result in the accumulation of patatin, other tuber proteins and starch in stems and petioles, without any swelling or tuber formation (Paiva et al., 1983). The expression of class I patatin genes and accumulation of patatin are also induced in leaves incubated with high concentrations of sucrose (Paiva et al., 1983; Rosahl et al., 1986; Rocha‐Sosa et al., 1989; Jefferson et al., 1990), but patatin gene expression in tubers is inhibited by wounding (Logemann et al., 1988) and in whole plants and induced stem cuttings by treatment with gibberellic acid (Hannapel et al., 1985). Detailed studies of the 5′ upstream sequences of a patatin gene have been reported by Holdsworth et al. (1992) and Grierson et al. (1994), aimed at identifying specific sequences and trans‐acting factors that determine the developmental regulation and sucrose‐inducibility. This has led to the identification of a new type of DNA binding protein, called Storekeeper (STK) which is thought to regulate patatin gene expression (Zourelidou et al., 2002).
Functional properties of patatin
Potatoes are a major source of starch for food and industrial uses, with the tuber proteins forming a by‐product. The proteins are usually recovered in an aggregated denatured state which limits their use to low value feed for livestock. However, undenatured potato proteins have promising functional properties (e.g. formation and stabilization of emulsions and foams) (Holm and Eriksen, 1980; Wojnowska et al., 1981; Jackman and Yada, 1988; Ralet and Gueguen, 2000) as well as good nutritional quality (Kapoor et al., 1975; Liedl et al., 1987). Consequently, a number of studies have been carried out on the structure and properties of patatin, particularly on its stability and thermal aggregation in relation to the production of functional proteins on an industrial scale.
Pots et al. (1999b) showed that patatin comprised ten peaks by reversed‐phase high pressure liquid chromato graphy that could be separated into four pools by ion‐exchange chromatography. These pools represented 62 % (A), 26 % (B), 5 % (C) and 7 % (D) of the total fraction, and each comprised isoforms with masses of about 40 400 and 41 600, which were considered to be due to differences in glycosylation. No differences in the properties or conformational stability of the pools were observed. In other studies, the same group has investigated the effects of pH and temperature on the stability and aggregation of whole patatin fractions (Pots et al., 1998a, b, 1999a, c).
Biological activity of patatin
The first indication that patatin exhibits enzymic activity came from studies of Galliard (1971), who purified an enzyme from potato tubers that catalysed the deacylation of a range of lipid substrates (mono‐ and diacylphospholipids, galactosyl diglycerides, mono‐ and diglycerides). Sub sequent studies demonstrated that this acyl hydrolase activity was due to patatin (Racusen, 1984), and that it also acts as an esterase against PNP laurate, PNC acetate, α‐naphthyl laurate, β‐naphthyl acetate, α‐naphthyl acetate and phenyl acetate substrates (Racusen, 1986).
The specificity of the acyl hydrolase has since been studied in more detail (Andrews et al., 1988; Anderson et al., 2002), particularly its activity as a phospholipase on phospholipid and lysophospholipid substrates (Senda et al., 1996; Hirschberg et al., 2001). The esterase activity has also been confirmed by expression in transgenic tobacco plants (Rosahl et al., 1987). This showed only minor differences in the activity of the products of class I and class II genes, the former being identical to those of the form present in potato tubers (Höfgen and Willmitzer, 1990).
A further type of hydrolytic activity has also been described recently for patatin, as an acidic β‐1,3‐glucanase (Tonón et al., 2001). β‐1,3‐Glucanases are thought to contribute to plant defence to fungal pathogens, by digesting β‐1,3‐glycans in hyphal cell walls, and often form part of the pathogenesis‐related (PR) protein response (Shewry and Lucas, 1997; van Loon and van Strien, 1999). This may imply that patatin plays a role in the defence of potato tubers.
A role of patatin in defence against pests and pathogens is also indicated by two other observations. First, the inclusion of patatin in artificial diets resulted in inhibition of growth of larvae of corn rootworm, Diabrotica spp. (Strickland et al., 1995). Treatment of patatin with di‐isopropylfluorophos phate inhibited its phospholipase, galactolipase and acyl hydrolase activities, and also eliminated its negative effect on larval growth. Comparison of the enzymatic and inhibitory properties of patatin fractions from different cultivars showed that galactolipase activity was correlated with growth inhibition, but not phospholipase or acyl hydrolase activity. Inhibitory activity was also reduced by provision of cholesterol in the diet. It was concluded that patatin may provide defence against the insect pests by effects on lipid metabolism. A patatin‐like protein with galactolipase activity is also induced by drought stress in leaves of cowpea (Vigna unguiculata) (Matos et al., 2000, 2001), indicating that patatins may play a wider role in stress response.
The second indication that patatins may play a role in plant defence comes from studies of tobacco leaves infected with tobacco mosaic virus (Dhondt et al., 2000). Three genes encoding patatin‐like proteins were rapidly induced on infection, with one of the proteins exhibiting phospholipase A2 (PLA2) activity. The increase in PLA2 occurred before the accumulation of fatty acid‐derived defence signals (12‐oxophytodienoic and jasmonic acids), and it is suggested that PLA2 initiates the synthesis of these by releasing fatty acid substrates from membrane lipids.
An intriguing report, whose significance is still not understood, is that the gene responsible for the STURDY mutant of arabidopsis encodes a patatin‐like protein (Huang et al., 2001). This activation‐tagged mutant is characterized by a stiff inflorescence stem, thick leaves, short siliques, large seeds, round flowers and delayed growth.
Allergenicity of patatin
Potato may elicit allergenic responses in humans and children, either when consumed as food or by skin contact with raw potatoes. Seppälä et al. (1999) showed that patatin bound to IgE (a class of immoglobulins specific for allergenic responses) from children with a positive skin‐prick test to raw potato, and also showed that purified patatin gave positive skin‐prick tests in allergenic children. This was subsequently confirmed by more detailed studies including skin exposure tests and oral challenge (Majamaa et al., 2001), and patatin has been given the allergen designation Sol t 1. Heat treatment of potato results in decreased allergenicity, which appears to result from aggregation with other potato proteins rather than denaturation of patatin itself (Koppelman et al., 2002).
Earlier work had shown that a major allergen of latex, called Hev b 7, was an Mr 43 000 protein with sequence homology to patatin (Kostyal et al., 1998; Sowka et al., 1998; Breiteneder et al., 1999) and it came as no surprise that patients with allergy to natural rubber latex also show in vitro reactivity of IgE to patatin (Seppälä et al., 2000). Similarly, a related allergenic protein is also present in tomato (Reche et al., 2001).
SWEET POTATO
Sweet potato (Ipomoea batatus, Convolvulaceae) is a dicotyledonous species with tubers derived from swollen roots. Apparently, it has its origin in South America but may have spread into Polynesia in pre‐Columbian times followed by post‐Columbian spread into Asia and Africa. ‘Crude’ protein content has been reported to vary between 1–3 % and 10 %, but this includes 10–15 % non‐protein nitrogenous components (Walter et al., 1984). The major storage protein is reported to account for over 80 % of the total protein (Maeshima et al., 1985). It was initially called ipomoein (Jones and Gersdorff, 1931) but is now known as sporamin.
Characteristics of sporamin
Maeshima et al. (1985) reported the purification of two major sporamins, termed A and B, which had similar masses by SDS–PAGE (approx. 25 000) and similar amino acid compositions, immunological properties and peptide maps. The two ‘proteins’ also differed when separated without reduction of disulfide bonds with dithiothreitol, with sporamin A migrating to a position consistent with a mass of 31 000 and sporamin B to a mass of 22 000. Comparison of the staining intensities of these bands indicated a ratio of about 2 : 1 of sporamin A to sporamin B in the mature tubers. N‐terminal amino acid sequencing of sporamin A showed the presence of at least two variants.
The existence of two major sub‐families of sporamins was confirmed by detailed molecular studies reported by Nakamura and colleagues at Nagoya (Murakami et al., 1986; Hattori et al., 1985, 1989). They sequenced 49 cDNA clones and showed that 22 corresponded to sporamin A and 27 to sporamin B (Hattori et al., 1989), with at least five different sequences within each subfamily. Similarities in coding sequence ranged from 94 to 98 % within members of a subfamily and from 82 to 84 % between subfamilies (Fig. 1). The mature proteins comprised about 180 residues (Murakami et al., 1986). All of the sequences contained four cysteine residues in conserved positions. Comparison with the Kunitz‐type trypsin inhibitors, which are homologues of sporamin (see below), would suggest that these form two intra‐chain disulfide bonds. It is not, therefore, possible to explain the differences in behaviour of sporamins A and B when separated under non‐reducing conditions.

Fig. 1. Alignment of the amino acid sequences of sporamins from sweet potato, G2 globulins from taro (Colocasia esculenta), miraculin from miracle berry (Richardella dulcifera) and Kunitz trypsin inhibitor from soybean. The sporamin sequences include the N‐terminal propeptides (residues 1–15); other sequences are mature proteins. NCBI accession numbers are: sporamin A, P10917; sporamin B, P10965; taro G2b, BAA03723; taro G2a, BAA03724; miraculin, A33872; Kunitz inhibitor, P01070.
Biological activity of sporamin
The sequence homology with Kunitz‐type trypsin inhibitors (Fig. 1) led Yeh et al. (1997a) to test sporamin for inhibitory activity, using recombinant protein expressed in Escherichia coli. The recombinant protein inhibited trypsin in a gel‐based assay, with no difference in activity between the mature form, the proform (i.e. not processed, see below) and the preproform (with the signal peptide). The authors concluded that processing of the protein was not required for inhibitory activity. More recently, the same group have developed a structural model for sporamin, based on the structures of other plant Kunitz trypsin inhibitors (Fig. 2), and used this to design mutants affecting the putative active site loop and to replace one of the cysteine residues involved in an intra‐chain disulfide bond (Yao et al., 2001). Expression of the recombinant mutant proteins in E. coli demonstrated that three mutations (Asp70Val, Glu72Arg and Ser73Ile) had greatly reduced inhibitory activity, to only 2–4 % of the wild‐type protein activity, confirming the position of the inhibitory site. However, a fourth loop mutation, Ala69Ser, did not have much effect nor did a mutation designed to destabilize the loop (Arg51Pro). Elimination of a single cysteine residue (Cys45Leu) did result in decreased activity, to about 12 % of the wild‐type protein, confirming the importance of inter‐chain disulfide bonds for stabilization. The activity of sporamin as a trypsin inhibitor may account for its ability to confer resistance to the lepidopteran pest, Spodoptera litura, when expressed in transgenic tobacco (Yeh et al., 1997b).
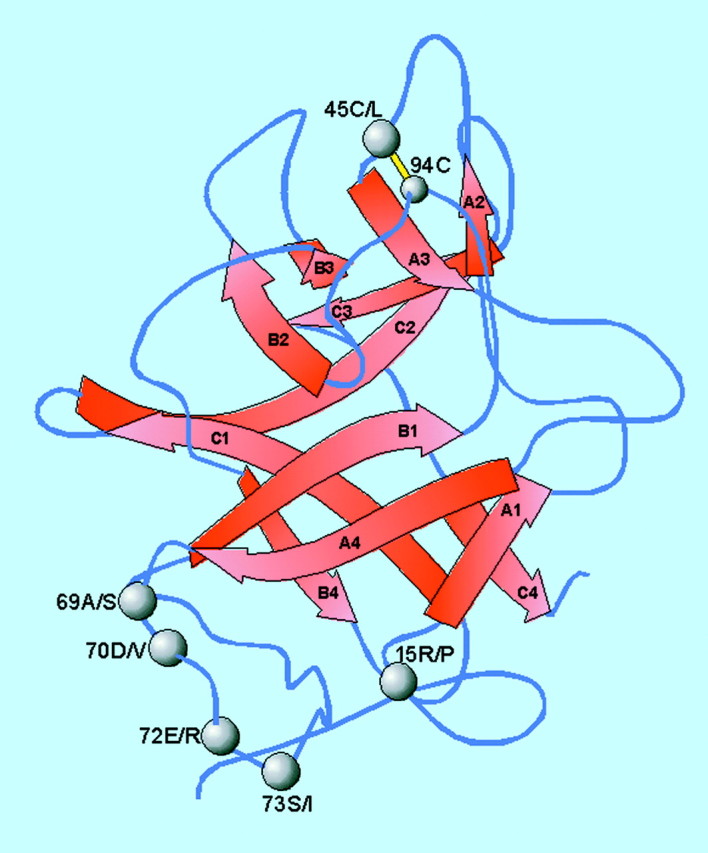
Fig. 2. A structural model for the sweet potato protein sporamin, based on homology with Kunitz trypsin inhibitors. Modification of six residues (shown by spheres and labelled with the wild‐type and mutant amino acids separated by a slash) resulted in different effects on the inhibitory activity, as discussed in the text. The first disulfide bond Cys45–Cys94 is indicated in yellow. Red arrows indicate beta‐sheet structure. Standard single letter abbreviations for amino acids are used: A, alanine; C, cysteine; D, aspartic acid; E, glutamic acid; I, isoleucine; L, leucine; P, proline; R, arginine; S, serine; V, valine. Redrawn from Yao et al. (2001) with permission.
Hou and Lin (1997) also reported that sporamin had antioxidant activity, acting as a dehydroascorbate reductase and monodehydroascorbate reductase, and that this was associated with intermolecular thiol/disulfide exchange. Furthermore, it is also able to scavenge against both 1,1‐diphenyl‐2‐picrylhydrazl radicals and hydroxyl radicals (Hou et al., 2001a). The biological significance of these observations is not clear. An in vivo role in regulating protease activity is also indicated by the demonstration that sporamin inhibits an endogenous serine proteinase from sweet potato tubers (Hou and Lin, 2002).
Targeting and processing of sporamin
Sporamins are deposited in vacuoles of tuber cells (Hattori et al., 1988), and are initially synthesized on the endoplasmic reticulum (ER) as preproproteins. An N‐terminal signal sequence of 21 residues is cleaved co‐translationally as the nascent protein is transported into the lumen of the ER, while an N‐terminal prosequence of 16 residues is then cleaved after transport into the vacuole (Matsuoka et al., 1990; Matsuoka and Nakamura, 1991). Detailed studies in transgenic tobacco have shown that a short sequence present within this propeptide, Asn–Pro–Ile–Arg–Leu (NPIRL), is sufficient to ensure targeting of spormain to the vacuole, even when it is attached to the C‐terminal rather than the N‐terminal end of the protein (Koide et al., 1997). Replacement of Asn, Pro, Ile and Leu with other residues demonstrated that the large alkyl side chains of isoleucine and leucine were particularly important determinants of the vacuolar sorting signal (Matsuoka and Nakamura, 1999). However, the sporamin expressed in the transgenic tobacco cells differed from that present in sweet potato, in that some glycosylation of serine residues (O‐glycosylation) occurred in the Golgi apparatus during the trafficking of the protein from the ER lumen to the vacuole (Matsuoka et al., 1995).
Regulation of sporamin synthesis
Sporamin is not detectable, or present only at very low levels, in any organs except the tuberous roots of plants when grown under normal field conditions (Maeshima et al., 1985; Hattori et al., 1990). However, it is synthesized in stems of plants grown in vitro (Hattori et al., 1990) and in excised leaves and petioles (Hattori et al., 1991), provided high levels of sucrose (approx. 3 %) are provided. Similar sucrose‐inducible expression also occurred when the 5′ upstream region of a sporamin A gene was fused to the chloramphenicol acetyl transferase (CAT) gene and transferred to transgenic tobacco (Hattori et al., 1990).
Ohto et al. (1992) reported that wounding of sweet potato leaves was capable of inducing sporamin expression in the wounded leaves, the petioles and even in remote leaves, but that this did not occur reproducibly. However, treatment with polygalacturonic acid and chitosan did result in reproducible induction of gene expression. These compounds are known to induce the expression of other defence‐related proteins (e.g. proteinase inhibitors, chitinases, β‐glucanases) so the results are consistent with sporamin playing a defensive role in the vegetative tissues of the plant. More recently, Yeh et al. (1997b) have shown that both local and systemic induction of sporamin gene expression occurs when leaves are wounded, and a 1·2 kb 5′ upstream region also confers wound‐induced expression in leaves and stems (but not roots) of transgenic tobacco plants when fused to the β‐glucuronidase (GUS) reporter gene (Wang et al., 2002).
Thus, sporamin may play several roles—storage, defence and regulation of endogenous proteinases—in tuberous roots and a single defence role in leaves and stems.
YAMS
Yam (Dioscorea spp., Dioscoreaceae) is classified as monocotyledonous but is considered to be closely related to dicotyledonous plants as a second cotyledon remains undeveloped in the embryo (Lawton and Lawton, 1967). The storage organ is probably a swollen hypocotyl (Lawton and Lawton, 1969), but is often described as a swollen root. A number of species are grown widely in the humid tropics with D. rotundata and D. cayenensis being of most importance, followed by D. alata and D. esculenta (Akoroda, 1993). These are all of African or East Asian origin, with only the minor species D. trifida being of American origin (Brücher, 1989). The tubers contain about 1–3 % protein on a dry weight basis (Coursey, 1995).
Characteristics of dioscorin
Harvey and Boulter (1983) reported that a major group of proteins accounted for about 85 % of the total protein content of the tuber of D. rotundata, and concluded that they corresponded to the major tuber storage proteins. They also demonstrated the presence of protein deposits within vacuoles and as ‘cytoplasmic protein aggregates’ but did not establish the identity of these. Harvey and Boulter (1983) also showed that the major protein comprised a number of isoforms with molecular masses of about 31 000 and was not glycosylated.
Little further work was carried out on yam tuber proteins until Conlan et al. (1995) reported the cloning of cDNAs for the major tuber proteins from D. cayenensis. Two classes of cDNA were identified which encoded proteins of about 70 % sequence similarity. The mature proteins encoded by the clones were calculated to have masses of about 28 000–29 000, and their correspondence to the major storage protein (called dioscorin) was established by comparison with partial amino acid sequences. Harvey and Boulter (1983) reported that dioscorins usually contain a single disulfide bond, but Conlan et al. (1998) subsequently showed that the presence or absence of an intra‐chain disulfide bond could be used to discriminate between the major groups of components. Comparison of the sequences of class A and class B dioscorins showed that both contained three cysteine residues but that only two of these were in conserved positions in the two proteins. The positions of the third cysteine residues could therefore account for differences in the ability of class A and class B dioscorins to form an intra‐chain disulfide bond. Conlan et al. (1998) also showed that antibodies raised against dioscorin reacted with protein deposited in vacuoles of the tuber cells, confirming the location of the protein.
Biological activity of dioscorin
Hewett‐Emmett and Tashian (1996) noted that the dioscorin sequences reported by Conlan et al. (1995) are related to those of α‐carbonic anhydrase enzymes (α‐CAs) from various sources. This was reiterated by Conlan et al. (1998), who aligned the sequences of human and mouse α‐CAs with dioscorin A and B and noted that identical amino acids were present in at least one dioscorin and one α‐CA at over 12 % of the total positions (Fig. 3). Also included in Fig. 3 are the amino acid sequences of two putative carbonic anhydrase‐related proteins encoded by the arabidopsis genome.

Fig. 3. Alignment of the amino acid sequences of mature forms of dioscorins and carbonic anhydrases and related precursor proteins predicted from the arabidopsis genome sequence. The sequences and NCBI accession numbers are as follows: human carbonic anhydrase I (P00915); mouse carbonic anhydrase I (P13634); D. cayensensis (Dc) dioscorin A (S57766); D. cayenensis (Dc) dioscorin (CAA53781); D. alata (Da) dioscorin A (AAF63334); D. alata (Da) dioscorin B (AAF44711); Arabidopsis thaliana protein (At unknown) (AAF79837); and Arabidopsis thaliana (At) carbonic anhydrase‐like protein (CAB79100).
Hou et al. (1999b, 2000) subsequently showed that dioscorins purified from D. batatas, D. alata and D. pseudojaponica all exhibited carbonic anhydrase activity. However, they did not apparently require zinc, which is usually required by α‐CAs. This is consistent with the absence of one of the three putative Zn‐liganding sites (a histidine residue) from the sequence reported by Conlan et al. (1995). Hou et al. (1999b) also reported that dioscorin from D. batatas reacted with antibody raised against trypsin inhibitor (presumably sporamin) from sweet potato tubers, although the two proteins have no known sequence homology, and showed that dioscorins from three species (D. batatas, D. alata, D. pseudojaponica) showed low activity as trypsin inhibitors. They also reported that the three dioscorins were glycosylated, based on ConA‐peroxidase activity staining, which contrasts with the results of Harvey and Boulter (1983) on D. rotundata.
Hou et al. (1999a, 2001b) also showed that dioscorin from D. batatas has antioxidant properties, with activity as dehydroascorbate reductase and mono‐dehydroascorbate reductase, and an ability to scavenge against both 1,1‐diphenyl‐2‐picrylhydrazl (DPPH) radicals and hydroxyl radicals. The reduction of dehydroascorbate was also associated with intermolecular thiol‐disulfide exchanges of the dioscorin. The authors demonstrated similar activity for the trypsin inhibitor of sweet potato (see above). It was therefore suggested that the consumption of yam proteins or tubers could have health benefits.
The activity of dioscorin as carbonic anhydrase is readily explained on the basis of its sequence relationship to α‐CAs, while its reductase and antioxidant activities may result from disulfide/thiol exchanges. However, its activity as a trypsin inhibitor was low and cannot at present be explained on the basis of homology with other inhibitors. Similarly, the reactive (inhibitory) site of dioscorin has not been identified. Hsu et al. (2002) also demonstrated that dioscorin and its peptic hydrolysates were capable of inhibiting angiotensin converting enzyme (ACE), which is a target for pharmacological agents used in treatment of hypertension.
TARO
Taro is the generic name for four related species of the family Araceae (aroids). These are Colocasia esculenta (taro), Cyrtosperma chamissonis (giant swamp taro) and Xanthosoma sagittifolium in which the corms are eaten, and Alocasia macrorhiza (giant taro) in which the edible part is the thickened underground stem (Pollock, 2000). It has been a staple in the Pacific for 3000–4000 years but is now also widely grown in other parts of the wet tropics (Pollock, 2000). Taro (i.e. Colocasia) varieties generally contain from about 1–4·5 % protein (on a dry weight basis) with 11·7 % being reported for one cultivar (Splittstoesser, 1977). Pollock (2000) has quoted values of 2 % protein for Colocasia and Xanthosen, 0·8 % for Cyrtosperma and 0·6 % for Alocasia. Sumathi and Pattabiraman (1977) purified a trypsin/chymotrypsin inhibitor from corms of Alocasia, which was subsequently sequenced and cloned (Argall et al., 1994; Mathews et al., 1996). The mature protein comprises 188 amino acids with a mass of about 25 000. Hammer et al. (1989) also showed that inhibitors with similar properties were present in corms of Cyrtosperma and Colocasia, and the latter species has since been studied in some detail.
Characteristics of G2 globulin
de Castro et al. (1992) and Monte‐Neshich et al. (1995) showed that two major proteins of mass approx. 24 000 and 22 000, with pIs close to 7·5, accumulated in developing corms of Colocasia, and called these G2a and G2b (G meaning globulins). G2a was slightly more abundant in the variety used, and gel filtration studies suggested that the proteins were present as dimers. Partial amino acid sequencing confirmed their identity as related to trypsin inhibitors. Densitometric analysis of SDS–PAGE separations showed that G2 globulins accounted for about 40 % of the total soluble tuber protein, and immunocytochemical analysis showed that they were present in protein deposits within vacuoles of the parenchyma cells (Monte‐Neshich et al., 1995). Thus, they appear to be true storage proteins.
Hirai et al. (1993) reported the amino acid sequences of two highly homologous G2 globulins, deduced from the nucleotide sequences of cDNA clones, but did not determine whether these corresponded to G2a or G2b.
The amino acid sequences of the Alocasia and Colocasia inhibitors are clearly related, with about 81 % sequence similarity (Mathews et al., 1996). Wider comparisons show that they belong to the Kunitz family of proteinase inhibitors, which includes the sweet potato storage protein sporamin (see above) (Fig. 1). de Castro et al. (1992) also reported sequence similarity to the taste‐modifying protein, miraculin, from fruits of the miracle berry plant (Richardella dulcifera) (Theeraslip et al., 1989).
Characteristics of tarin (G1 globulin)
Corms of Colocasia also contain a second major storage protein fraction called tarin or G1 globulin, which also accounts for about 40 % of the total soluble proteins (Monte‐Neshich et al., 1995). Tarin consists of about ten isoforms, including five which are most abundant, with masses of about 12 500 by SDS–PAGE and pIs ranging from about 5·5 to 9·5. Immunocytochemistry demonstrated that the protein is present in the same vacuolar deposits within parenchyma cells as the G2 globulin (trypsin inhibitor).
N‐terminal amino acid sequencing of four tarin isoforms showed two types (G1a and G1c, G1b and G1d) which shared about 25 % sequence identity. These were subsequently shown to be synthesized as single proproteins, with G1b/d present at the N‐terminus and G1a/c at the C‐terminus (Hirai et al., 1993; Bezerra et al., 1995). It is possible that proteolytic cleavage of the proprotein to release the two mature tarins occurs in the vacuole, by analogy with the processing of albumin and globulin storage proteins in seeds (see Kermode and Bewley, 1999). However, in seeds this proteolysis is catalysed by a specific family of cysteine proteinase, called legumains, which cleave at the C‐terminal side of asparagine residues (Müntz and Shutov, 2002). Asparagine residues are not present adjacent to the N‐termini of G1a/c in the proproteins reported by Hirai et al. (1993) and Bezerra et al. (1995), with arginine residues being present instead. Hence it is more likely that a trypsin‐like serine proteinase is responsible. The proprotein is also initially synthesized with an N‐terminal signal sequence, which is consistent with its entering the ER lumen and subsequent transport to the vacuole.
The two tarin proteins encoded by the proprotein each have two cysteine residues, but the pattern of disulfide bonds formed (i.e. inter‐chain or intra‐chain) has not been determined. However, gel filtration chromatography under non‐denaturing conditions gave a mass of about 28 000 (Monte‐Neshich et al., 1995), leading to the suggestion that the two proteins present in the proprotein form a dimer after proteolytic processing.
Sequence comparisons showed that tarins from Colocasia had sequence homology with mannose‐binding lectins (Fig. 4), including 40 % identity with the snowdrop (Galanthus nivalis) lectin (Bezerra et al., 1995). Furthermore, Van Damme et al. (1995) demonstrated that the major protein present in tubers of Arum maculatum (wild arum) was a lectin comprising two different Mr 12 000 subunits synthesized as a single preprotein. Gel filtration and SDS–PAGE under reducing and non‐reducing conditions indicated that the protein was a tetramer (Mr 50 000), which was not stabilized by inter‐chain disulfide bonds. It is of interest that Carneiro et al. (1990) also initially showed that the Colocasia protein had a native Mr of about 50 000, which suggests that it too may be tetrameric rather than dimeric as subsequently reported. Van Damme et al. (1995) confirmed that related two‐chain lectins were present in tubers of Colocasia and Xanthosoma sagittifolia and demonstrated that the proteins from Arum, Colocasia and Xanthosoma all agglutinated erythrocytes of rabbits but not humans, and that this activity was inhibited by mannose but not by other monosaccharides, although this inhibition occurred only at relatively high mannose concentrations (100 mm) and was transient. It has also been reported that tubers of Alocasia contain a major lectin (Singh et al., 1993). It can be concluded, therefore, that major storage proteins from taro, including tarin/G1 globulin from Colocasia and proteins from Xanthosoma and Alocasia, are mannose‐binding lectins and may therefore have defensive as well as storage roles.

Fig. 4. Alignment of the amino acid sequences of tarin (G1 globulin) from taro (Colocasia esculenta), leaf lectin from arum lily (Arum maculatum) and curculin from Curculago latifolia. The N‐termini of the two tarin subunits are Leu1 and Asn117 and of the lectin subunits Val1 and Asn117. NCBI accession numbers are: tarin, S56688; lectin (LECAMA1), AAC48997; curculin, P19667.
Bezera et al. (1995) reported that tarin/G1 globulin has about 45 % identity with curculin, the taste‐modifying protein from fruits of Curculago latifolia (Hypoxidaceae) (Yamashita et al., 1990) (Fig. 4). The demonstration that both major storage proteins of Colocasia are related in sequence to taste‐modifying proteins (miraculin and curculin) is intriguing but it should be noted that neither of the taro proteins has so far been tested for taste‐modifying properties.
Regulation of synthesis of Colocasia storage proteins
The two Colocasia storage proteins show similar patterns of accumulation during corm development and are broken down in older corms, as the formation of new cormels occurs (Monte‐Neshich et al., 1995). Whether their expression is entirely restricted to corms is still not resolved as a low level of mRNA for tarin has been reported in roots but not leaves of Colocasia (de Castro et al., 1992).
The promoter region of a tarin gene shows homology with the promoters of patatin genes of potato, with the region from –558 to –684 bp downstream of the transcription start site of Tar1 being 65 % identical to regions in the pgT12 and pgT16 patatin genes (Guimarães et al., 2001). Trans formation of potato with a 5·7 kb genomic fragment containing the entire Tar1 gene (including 2·7 kb of 5′ flanking region) resulted in expression of tarin in potato tubers (Guimarães et al., 2001). Furthermore, expression was restricted to the tuber in glasshouse‐grown plants, but also occurred in stems, but not in roots or leaves, when plants were grown in vitro. Expression in stems was also eliminated when the sucrose concentration in the medium was increased from 1 % to 3 % or 6 %. The authors concluded that the expression of Tar1 in potatoes followed the pattern for patatin synthesis.
CASSAVA
Casava (Manihot esculenta, Euphorbiaceae), also called manioc, tapioca or yuca, is one of the most important food crops in the humid tropics, being particularly suited to conditions of low nutrient availability and able to survive drought. It was initially domesticated in Central and South America but was carried to West Africa in the late 15th century, probably by Portuguese traders. It subsequently spread to south‐east Asia and the Pacific Islands, and all these regions remain major producers.
Although cassava leaves are sometimes consumed, the major harvested organ is the tuber, which is actually a swollen root and is not able to act as a propagule, the plant being propagated from stem cuttings or seeds. Cassava tubers contain only about 1–2 % of protein on a dry weight basis with a low content of sulfur‐containing amino acids (Yeoh and Chew, 1977). The tubers are therefore consumed and fed to livestock as a source of starch and also used to produce tapioca starch for industrial and food use.
Souza et al. (1998) reported the isolation of a major protein of Mr about 22 000 which was restricted to the parenchyma rather than the peel of the tuber. However, no detailed characterization of this protein was carried out and its role as a storage protein was not established. Light microscopy also failed to identify any major protein deposits within the tuber cells (Shewry et al., 1993). The existence of true storage proteins in cassava tuberous roots therefore remains to be established.
OTHER TUBEROUS CROPS
A wide range of minor crops are cultivated for their tubers, particularly in the tropics (O’Hair, 1990; Rehm and Espig, 1991). Very few of these have been studied in detail and the following discussion is restricted further to those which have been analysed for protein content and composition.
A number of tuberous legumes are cultivated, including winged bean (Psophocarpus tetragonolobus), African yam bean (Sphenostylis stenocarpa) and Pachyrhizus. Of these only the latter has been studied in detail.
Pachyrhizus is native to South and Central America with several species being cultivated. P. tuberosus is cultivated in Bolivia, Peru, Equador and Brazil, P. erosus (Mexican yam bean) in Central America and the Caribbean and P. ahipa in the Andean valleys of Bolivia and northern Argentina (Sørensen et al., 1997; Barnes and Gomes, 1998). Pachyrhizus tubers have been reported to have protein contents between 5 and 15 % d. wt, but a recent study of six accessions of P. ahipa and one of P. erosus grown under glasshouse conditions gave lower values of 4·8–8·4 % d. wt for the former and 2·7 % d. wt for the latter (Forsyth and Shewry, 2002). Gomes et al. (1997) reported that two proteins of Mr about 26 000 and 28 000 accounted for over 70 % of the total soluble protein in tubers of P. erosus and suggested that these corresponded to storage proteins. N‐terminal sequencing showed homology to cysteine proteinases of the papain family and both proteins also showed limited proteolysis of an azocasein substrate. Binding to a concanavalin A affinity column and oxidation of gel separations with periodate followed by binding with biotin‐hydrazide indicated that both were glycoproteins.
In contrast, Forsyth and Shewry (2002) failed to identify any major storage proteins in tubers of P. ahipa, either as deposits in vacuoles observed by light microscopy or as major components of the soluble protein fraction (which accounted for about 93 % of the total tuber nitrogen). A protein of Mr about 30 000 was shown to have N‐terminal sequence homology to the major cysteine proteinases identified in P. erorus by Gomes et al. (1997), but this only accounted for about 5–6 % of the total fraction. The existence of genuine storage proteins in tubers of Pachyrhizus species therefore remains to be established conclusively.
Recently, a novel storage protein has been reported from oca (Oxalis tuberosa, Oxalidaceae), an Andean tuberous crop dating from pre‐Columbian times (Hodge, 1957). This Mr 18 000 protein, termed ocatin, accounts for 40–60 % of the total soluble protein (Flores et al., 2002). Partial amino acid sequencing revealed homology to proteins of the Bet v 1 (birch pollen allergen)/PR (pathogenesis‐related) 10/MLP (major latex protein) protein family and in vitro tests showed inhibition of growth of several pathogenic bacteria (Pseudomanas aureofaciens, Serratia marcescens, Agrobacterium spp.) and fungi, including ascomycetes (Nectria hematococcus, Fusarium oxysporum), oomycetes (Phytophthora cinnamami) and basidiomycetes (Rhizoctonia solani). Western blot analysis showed that the protein was restricted to the tuber, but only to the pith, sub‐epidermal and epidermal regions, not being present in the cortex. The subcellular location was not determined. Thus, although ocatin clearly has at least some of the characteristics of a storage protein, this role needs to be established by further studies.
Finally, Jerusalem artichoke is a species of sunflower (Helianthus tuberosus, Compositae) that is cultivated for its tubers which are rich in inulin, a fructosan polysaccharide. Tubers have been reported to accumulate three major polysaccharides of Mr about 16 000, 16 500 and 18 000, which have been described as storage proteins (Mussigmann and Ledoigt, 1989). However, these have not been characterized in detail and their biological role remains to be established.
CONCLUSIONS: COMMON PROPERTIES OF TUBER STORAGE PROTEINS
Tuber storage proteins provide a fascinating example of how a diverse range of proteins can fulfil the same biological function. Whereas seed storage proteins fall into four major groups (2S albumins, 7S globulins, 11S globulins and prolamins), tuber storage proteins have widely different origins. In fact, of the types described here, only two have any relationships to each other: sporamin and G2 globulin from taro, both of which belong to the Kunitz family of trypsin inhibitors. This diversity may in part reflect the diverse botanical origins of the tubers themselves, being derived from swollen roots, stems or hypocotyls. There is no evidence for the expression of typical seed storage proteins in tubers, or indeed in any other non‐seed tissues, but both trypsin inhibitors and lectins do also act as storage proteins in some seeds (e.g. lectins in Phaseolus spp.).
Despite their diverse origins it is possible to recognize some common features of tuber storage proteins. First, they are all polymorphic mixtures of components, which is also a common feature of seed storage proteins. In the case of patatin, sporamin and dioscorin, it is also possible to recognize two distinct groups of proteins and/or genes which, in the case of patatin, are differentially regulated. Secondly, the major storage proteins of potato, sweet potato, yams and taro all exhibit biological activities that are consistent with a role in protecting the tubers against pests, pathogens and perhaps also abiotic stresses, i.e. as hydrolases, enzyme inhibitors, lectins and antioxidants. It is therefore probable that all play a dual role in storage and protection. It is also possible that all initially had roles in defence, with a storage role being acquired as a secondary function.
It is also of interest that some tubers, notably cassava, appear to lack true storage proteins. This may be related to whether the tubers act as propagules or solely for storage. Tubers of yam, sweet potato, potato and taro can all act as propagules and so may require storage proteins to be broken down to support sprouting. In contrast, cassava roots are unable to act a propagules, but remain on the plant as a flexible store of carbon to facilitate survival through periods of environmental stress.
The major tuber crops discussed here represent only a small proportion of the vast number of tubers that are grown or harvested from the wild and consumed around the world. Emerging studies on a wider range of species should add new information and insights into the distributions and properties of tuber storage proteins.
ACKNOWLEDGEMENTS
I am grateful to Dr Nigel Halford for preparing the sequence alignments in Figs 1, 3 and 4, and to Mrs Lynda Castle for Fig. 2. Long Ashton Research Station receives grant‐aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Table 2.
Properties of major tuber storage proteins
| Potato | Sweet potato | Taro (Colocasia) | Yam | ||
| Protein | Patatin | Sporamin | Tarin (G1 globulin) | Trypsin inhibitor (G2 globulin) | Dioscorin |
| % soluble tuber protein mass | 40 | 80 | 40 | 40 | 85 |
| Approximate mass | 40 000 | 25 000 | 12 500 | 22 000–24 000 | 28 000–29 000 |
| Post‐translational modifications | N‐glycosylation | Removal of prosequence in vacuole | Two subunits released from precursor proteins | Possibly glycosylated | |
| Gene families | Two subfamilies (class I/II) | Two subfamilies (A/B) | Two major types of each subunit (G1a/c, G1b/d) | Two major types (G2a/G2b) | Two subfamilies (A/B) |
| Tissue location | Tubers, also induced in stemsleaves, petioles | Tubers, also induced in stems, leaves, petioles | Only corms | Corms, possibly also roots | Tubers |
| Cell location | Vacuoles | Vacuoles | Vacuoles | Vacuoles | Vacuoles |
| Related proteins | Virus‐induced proteins in potato/drought stress protein in cowpea/ allergens in latex and tomato/arabidopsis STURDY mutant protein | Kunitz trypsin inhibitors | Mannose‐binding lectins/curculin | Kunitz trypsin inhibitors/miraculin/sporamin | α‐Carbonic anhydrase |
| Biological activity | Esterase/lipid acylhydrolase/β‐1,3‐glucanase | Trypsin inhibition/antioxidant/radical scavenger | Agglutination of erythrocytes | Trypsin inhibition? | Carbonic anhydrase/antioxidant/trypsin inhibition/radical scavenger |
| Other biological roles | Defence | Defence | Defence | Defence | Defence |
| Other properties | Allergenic | – | – | – | – |
Supplementary Material
Received: 6 December 2002; Returned for revision: 20 January 2003; Accepted: 29 January 2003 Published electronically: 9 April 2003
References
- AkorodaMO.1993. Genetic improvement of vegetable crops: yam (Dioscorea spp.). In: Kalloo G and Bergh BO (eds). Genetic improvement of vegetable crops Oxford: Pergamon Press, 717–733. [Google Scholar]
- AndersonC, Pinsirodom P, Parkin KL.2002. Hydrolytic selectivity of patatin (lipid acyl hydrolase) from potato (Solanium tuberosum L.) tubers towards various lipids. Journal of Food Biochemistry 26: 63–74. [Google Scholar]
- AndrewsDL, Beames B, Summers MD, Park WD.1988. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochemical Journal 252: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ArgallME, Bradbury JH, Shaw DC.1994. Amino‐acid sequence of a trypsin/chymotrypsin inhibitor from giant taro (Alocasia macrorrhiza). Biochimica et Biophysica Acta 1204: 189–194. [DOI] [PubMed] [Google Scholar]
- BarnesJA, Gomes AV.1998. The yam bean: a plant of promise. The Biologist (London) 45: 221–224. [Google Scholar]
- BezerraIC, de Castro LAB, Neshich G, de Almeida ERP, Grossi de Sá MF, Mello LV, Monte‐Neshich DC.1995. A corm‐specific gene encodes tarin, a major globulin of taro (Colocasia esculenta L. Schott). Plant Molecular Biology 28: 137–144. [DOI] [PubMed] [Google Scholar]
- BreitenederH, Sowka S, Wagner S, Krebitz M, Hafner C, Kinaciyan T, Yeang HY, Scheiner O.1999. Cloning of the patatin‐like latex allergen Hev b 7, its expression in the yeast Pichia pastoris and its immunological characterization. International Archives of Allergy and Immunology 118: 309–310. [DOI] [PubMed] [Google Scholar]
- BrücherH.1989. Tubers/corms/rhizomes. In: Brücher H, ed. Useful plants of neotropical origin and their wild relatives Berlin: Springer‐Verlag, 4–53. [Google Scholar]
- BurtonWG.1989.The potato. Harlow: Longman. [Google Scholar]
- CarneiroM, Rodriques CA, de Castro LAB, de Silva MC, Coutinho MV.1990. Isolation and characterization of the major albumin from Colocasia esculenta corms. Plant Science 67: 39–46. [Google Scholar]
- ClowesAEE, Tatham AS, Beeching JR, Shewry PR.1995. Characterisation of cassava root proteins. In: Proeedings of the Second International Scientific Meeting of the Cassava Biotechnology Network, Bogor, Indonesia, 22–26 August, CIAT, Cali, Colombia. Vol. II, 716–728. [Google Scholar]
- ConlanS, Griffiths L‐A, Turner M, Fido R, Tatham A, Ainsworth C, Shewry PR.1998. Characterisation of the yam tuber storage protein dioscorin. Journal of Plant Physiology 153: 25–31. [Google Scholar]
- ConlanRS, Griffiths L‐A, Napier JA, Shewry PR, Mantell S, Ainsworth C.1995. Isolation and characterisation of cDNA clones representing the genes encoding the major tuber storage protein (dioscorin) of yam (Dioscorea cayenensis Lam.). Plant Molecular Biology 28: 369–380. [DOI] [PubMed] [Google Scholar]
- CourseyDG.1995.Yams: an account of the nature, origins, cultivation and utilisation of the useful members of the dioscoreaceae. Harlow: Longman. [Google Scholar]
- de CastroLAB, Carneiro M, Monte‐Neshich DC, de Paiva GR.1992. Spatial and temporal gene expression patterns occur during corm development. Plant Cell 4: 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DhondtS, Geoffroy P, Stelmach BA, Legrand M, Heitz T.2000. Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus‐infected tobacco leaves and is contributed by patatin‐like enzymes. The Plant Journal 23: 431–440. [DOI] [PubMed] [Google Scholar]
- FAO.1999.Production yearbook, Vol. 53. Rome: FAO. [Google Scholar]
- FloresT, Alape‐Girón A, Flores‐Díaz M, Flores HE.2002. Ocatin. A novel tuber storage protein from the Andean tuber crop oca with antibacterial and antifungal activities. Plant Physiology 128: 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ForsythJL, Shewry PR.2002. Characterization of the major proteins of tubers of yam bean (Pachyrhizus ahipa). Journal of Agricultural and Food Chemistry 50: 1939–1944. [DOI] [PubMed] [Google Scholar]
- GalliardT.1971. The enzymic deacylation of phospholipids and galactolipids in plants: purification and properties of a lipolytic acyl‐hydrolase from potato tubers. Biochemical Journal 121: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenovN, Goshev I, Nikolova D, Georgieva DN, Filippi B, Svendsen I.1997. A novel thermostable inhibitor of trypsin and subtilisin from the seeds of Brassica nigra: amino acid sequence, inhibitory and spectroscopic properties and thermostability. Biochimica et Biophysica Acta 1341: 157–164. [DOI] [PubMed] [Google Scholar]
- GomesAV, Grace S, Barnes JA.1997. Major proteins of yam bean tubers. Phytochemistry 46: 185–193. [DOI] [PubMed] [Google Scholar]
- GriersonC, Du J‐S, de Torres Zabala M, Beggs K, Smith C, Holdsworth M, Bevan M.1994. Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. The Plant Journal 5: 815–826. [DOI] [PubMed] [Google Scholar]
- GuimarãesRL, Marcellino LH, Grossi de Sá MF, de Castro Monte D.2001. A storage protein gene from taro shows tuber‐specific expression in transgenic potato. Physiologia Plantarum 111: 182–187. [Google Scholar]
- HammerBC, Shaw DC Bradbury JH.1989. Trypsin inhibitors from Colocasia esculenta, Alocasia macrorhiza and Cyrtosperma chamissonis Phytochemistry 28: 3019–3026. [PubMed] [Google Scholar]
- HannapelDJ, Miller JC, Park WD.1985. Regulation of potato tuber protein accumulation by gibberellic acid. Plant Physiology 78: 700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HarveyPJ, Boulter D.1983. Isolation and characterization of the storage protein of yam tubers (Dioscorea rotundata). Phytochemistry 22: 1687–1693. [Google Scholar]
- HattoriT, Nakagawa T, Maeshima M, Nakamura K, Asahi T.1985. Molecular cloning and nucleotide sequence of cDNA for sporamin, the major soluble protein of sweet potato tuberous roots. Plant Molecular Biology 5: 313–320. [DOI] [PubMed] [Google Scholar]
- HattoriT, Matsuoka K, Nakamura K.1988. Subcellular localization of the sweet potato tuberous root storage protein. Agricultural Biological Chemistry 52: 1057–1059. [Google Scholar]
- HattoriT, Yoshida N, Nakamura K.1989. Structural relationship among the members of a multigene family coding for the sweet potato tuberous root storage protein. Plant Molecular Biology 13: 563–572. [DOI] [PubMed] [Google Scholar]
- HattoriT, Nakagawa S, Nakamura K.1990. High level expression of tuberous root storage protein genes of sweet potato in stems of plantlets grown in vitro on sucrose medium. Plant Molecular Biology 14: 595–604. [DOI] [PubMed] [Google Scholar]
- HattoriT, Fukumoto H, Nakagawa S, Nakamura K.1991. Sucrose‐induced expression of genes coding for the tuberous root storage protein, sporamin, of sweet potato in leaves and petioles. Plant and Cell Physiology 32: 79–86. [Google Scholar]
- Hewett‐EmmettD, Tashian RE.1996. Functional diversity, conervation, and convergence in the evolution of the α‐, β‐, and γ‐carbonic anhydrase gene families. Molecular Phylogenetics and Evolution 5: 50–77. [DOI] [PubMed] [Google Scholar]
- HiraiM, Nakamura K, Imai T, Sato T.1993. cDNAs encoding for storage proteins in the tubers of taro (Colocasia esculenta Schott). Japanese Journal of Genetics 68: 229–236. [DOI] [PubMed] [Google Scholar]
- HirschbergHJHB, Simons J‐WFA, Dekker N, Egmond MA.2001. Cloning, expression, purification and characterization of patatin, a novel phospholipase A. European Journal of Biochemistry 268: 5037–5044. [DOI] [PubMed] [Google Scholar]
- HodgeWH.1957. Three native tubers of the high Andes. Economic Botany 5: 185–201. [Google Scholar]
- HöfgenR, Willmitzer L.1990. Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato (Solanum tuberosum). Plant Science 66: 221–230. [Google Scholar]
- HoldsworthMJ, Grierson C, Schuch W, Bevan M.1992. DNA‐binding properties of cloned TATA‐binding protein from potato tubers. Plant Molecular Biology 19: 455–464. [DOI] [PubMed] [Google Scholar]
- HolmF, Eriksen S.1980. Emulsify properties of undenatured potato protein concentrate. Journal of Food Technology 15: 71–83. [Google Scholar]
- HouW‐C, Lin Y‐H.1997. Dehydroascorbate reductase and mono dehydroascorbate reductase activities of trypsin inhibitors, the major sweet potato (Ipomoea batatas [L.] Lam) root storage protein. Plant Science 128: 151–158. [Google Scholar]
- HouW‐C, Chen H‐J, Lin Y‐H.1999a. Dioscorins, the major tuber storage proteins of yam (Dioscorea batatas Decne), with dehydroascorbate reductase and monodehydroascorbate reductase activities. Plant Science 149: 151–156. [Google Scholar]
- HouW‐C, Liu JS, Chen H‐J, Chen TE, Chang CF, Lin Y‐H.1999b. Dioscorin, the major tuber storage protein of yam (Dioscorea batatas Decne) with carbonic anhydrase and trypsin inhibitor activities. Journal of Agricultural and Food Chemistry 47: 2168–2172. [DOI] [PubMed] [Google Scholar]
- HouW‐C, Chen H‐J, Lin Y‐H.2000. Dioscorins from different Dioscorea species all exhibit both carbonic anhydrase and trypsin inhibitor activities. Botanical Bulletin of Academia Sinica 41: 191–196. [Google Scholar]
- HouW‐C, Chen Y‐C, Chen H‐J, Liu Y‐H, Yang L‐L, Lee M‐H.2001a. Antioxidant activities of a 33KDa root storage protein of sweet potato (Ipomoea batatas (L.) Lam cv. Tainong 57). Journal of Agricultural and Food Chemistry 49: 2978–2981. [DOI] [PubMed] [Google Scholar]
- HouW‐C, Lee M‐H, Chen H‐J, Liang W‐L, Han C‐H, Liu Y‐W, Lin Y‐H.2001b. Antioxidant activities of dioscorin, the storage protein of yam (Dioscorea batatas Decne) tuber. Journal of Agricultural and Food Chemistry 49: 4956–4960. [DOI] [PubMed] [Google Scholar]
- HouW‐C, Lin Y‐H.2002. Sweet potato (Ipomoea batatas (L.) Lam) trypsin inhibitors, the major root storage proteins, inhibit one endogenous serine protease activity. Plant Science 163: 733–739. [Google Scholar]
- HsuFL, Lin YH, Lee MH, Lin CL, Hou W‐C.2002. Both dioscorin, the tuber storage protein of yam (Dioscorea alata cv. Tainong No. 1), and its peptic hydrolysates exhibited angiotensin converting enzyme inhibitory activities. Journal of Agricultural and Food Chemistry 50: 6109–6113. [DOI] [PubMed] [Google Scholar]
- HuangS, Cerny RE, Bhat DS, Brown SM.2001. Cloning of an Arabidopsis patatin‐like gene, STURDY, by activation T‐DNA tagging. Plant Physiology 125: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JackmanRL, Yada RY.1988. Functional properties of whey‐potato protein composite blends in model system. Journal of Food Science 53: 1427–1432. [Google Scholar]
- JeffersonRJ, Goldsbrough A, Bevan M.1990. Transcriptional regulation of a patatin‐1 gene in potato. Plant Molecular Biology 14: 995–1006. [DOI] [PubMed] [Google Scholar]
- JonesDB, Gersdorff CEF.1931. Ipomoein, a globulin from sweet potatoes, Ipomoea batatas Isolation of a secondary protein derived from ipomoein by enzymic action. Journal of Biological Chemistry 93: 119–126. [Google Scholar]
- KapoorA, Desborough SL, Li PH.1975. Potato tuber proteins and their nutritional quality. Potato Research 18: 469–478. [Google Scholar]
- KermodeAR, Bewley JD.1999. Synthesis, processing and deposition of seed proteins: the pathway of protein synthesis and deposition in the cell. In: Shewry PR, Casey R, eds. Seed proteins Dordrecht: Kluwer Academic Publishers, 807–841. [Google Scholar]
- KoideY, Hirano H, Matsuoka K, Nakamura K.1997. The N‐terminal propeptide of the precursor to sporamin acts as a vacuole‐targeting signal even at the C terminus of the mature part in tobacco cells. Plant Physiology 114: 863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KoppelmanSJ, van Koningsveld GA, Knulst AC, Gruppen H, Pigmans IGAJ, de Jongh HHJ.2002. Effect of heat‐induced aggregation on the IgE binding of patatin (Sol t 1) is dominated by other potato proteins. Journal of Agricultural and Food Chemistry 50: 1562–1568. [DOI] [PubMed] [Google Scholar]
- KostyalDA, Hickey VL, Noti JD, Sussman GL, Beezhold DH.1998. Cloning and characterization of a latex allergen (Hev b 7): hoology to patatin, a plant PLA(2). Clinical and Experimental Immunology 112: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LawtonJR, Lawton JRS.1967. The morphology of the dormant embryo and young seedling of five species of Dioscorea from Nigeria. Proceedings of the Linnean Society of London 178: 153–159. [Google Scholar]
- LawtonJR, Lawton JRS.1969. The development of the tuber in seedling of five species of Dioscorea from Nigeria. Botanical Journal of the Linnean Society 62: 223–232. [Google Scholar]
- LiedlBE, Kosier T, Desborough SL.1987. HPLC isolation and nutritional value of a major potato tuber protein. American Potato Journal 64:545–557. [Google Scholar]
- LogemannJ, Mayer JE, Schell J, Willmitzer L.1988. Differential expression of genes in potato tubers after wounding. Proceedings of the National Academy of Sciences USA 84: 1136–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MaeshimaM, Sasaki T, Asahi T.1985. Characterization of major proteins in sweet potato tuberous roots. Phytochemistry 24: 1899–1902. [Google Scholar]
- MajamaaH, Seppälä U, Palosuo T, Turjanmaa K, Kalkkinen N, Reunala T.2001. Positive skin and oral challenge responses to potato and occurrence of immunoglobulin e antibodies to patatin (Sol t 1) in infants with atopic dermatitis. Paediatric Allergy and Immunology 12: 283–288. [DOI] [PubMed] [Google Scholar]
- MathewsA, Llewellyn DJ, Wu Y, Dennis ES.1996. Isolation and characterisation of full‐length cDNA clones of the giant taro (Alocasia macrorrhiza) trypsin/chymotrypsin inhibitor. Plant Molecular Biology 30: 1035–1039. [DOI] [PubMed] [Google Scholar]
- MatosAR, d’Arcy‐Lameta A, França M, Zuily‐Fodil Y, Pham‐Thi AT.2000. A patatin‐like protein with galactolipase activity is induced by drought stress in Vigna unguiculata leaves. Biochemical Society Transactions 28: 779–781. [PubMed] [Google Scholar]
- MatosAR, d’Arcy‐Lameta A, França M, Pêtres S, Edelman L, Kader J‐C, Zuily‐Fodil Y, Pham‐Thi AT.2001. A novel patatin‐like gene stimulated by drought stress encodes a galactolipid acyl hydrolase. FEBS Letters 491: 188–192. [DOI] [PubMed] [Google Scholar]
- MatsuokaK, Nakamura K.1991. Propeptide of a precursor to a plant vacuolar protein required for vacuolar targeting. Proceedings of the National Academy of Sciences USA 88: 834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MatsuokaK, Nakamura K.1999. Large alkyl side‐chains of isoleucine and leucine in the NPIRL region constitute the core of the vacuolar sorting determinant of sporamin precursor. Plant Molecular Biology 41: 825–835. [DOI] [PubMed] [Google Scholar]
- MatsuokaK, Matsumoto S, Hattori T, Machida S, Nakamura K.1990. Vacuolar targeting and post‐translational processing of the precursors to the sweet potato tuberous root storage protein in heterologous plant cells. Journal of Biological Chemistry 265: 19750–19757. [PubMed] [Google Scholar]
- MatsuokaK, Watanabe N, Nakamura K.1995.O‐glycosylation of a precursor to a sweet potato vacuolar protein, sporamin, expressed in tobacco cells. The Plant Journal 8: 877–889. [DOI] [PubMed] [Google Scholar]
- MesserE.2000. Potatoes (white). In: Tipple KF, Ornelas KC, eds. The Cambridge world history of food Cambridge: Cambridge University Press, 187–201. [Google Scholar]
- MigneryGA, Pikaard CS, Hannapel DJ, Park WD.1984. Isolation and sequence analysis of cDNAs for the major potato tuber protein, patatin. Nucleic Acids Research 12: 7987–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MigneryGA, Pikaard CS, Park WD.1988. Molecular characterization of patatin multigene family of potato. Gene 62: 27–44. [DOI] [PubMed] [Google Scholar]
- Monte‐NeshichDC, Rocha TL, Guimarãres RL, Santana EF, Loureiro ME, Valle M, Grossi de Sá MF.1995. Characterization and spatial localization of the major globulin families of taro (Colocasia esculenta L. Schott) tubers. Plant Science 112: 149–159. [Google Scholar]
- MüntzK, Shutov AD.2002. Legumains and their functions in plants. Trends in Plant Science 7: 340–344. [DOI] [PubMed] [Google Scholar]
- MurakamiS, Hattori T, Nakamura K.1986. Structural differences in full‐length cDNAs for two classes of sporamin, the major soluble protein of sweet potato tuberous roots. Plant Molecular Biology 7: 343–355. [DOI] [PubMed] [Google Scholar]
- MussigmannC, Ledoigt G.1989. Major storage proteins in Jerusalem artichoke tubers. Plant Physiology and Biochemistry 27: 81–86. [Google Scholar]
- O’HairSK.1990. Tropical root and tuber crops. In: Janick J, ed. Horticultural reviews, Vol. 12 Portland, OR: Timber Press, 157–196. [Google Scholar]
- OhtoM, Nakamura‐Kito K, Nakamura K.1992. Induction of expression of genes coding for sporamin and β‐amylase by polygalacturonic acid in leaf‐petiole cuttings of sweet potato. Plant Physiology 99: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OsborneTB, Campbell GF.1896. The proteides of the potato. Journal of the American Chemical Society 18: 575–582. [Google Scholar]
- PaivaE, Lister RM, Park WD.1983. Induction and accumulation of major tuber proteins of potato stems and petioles. Plant Physiology 71: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ParkWD, Blackwood C, Mignery GA, Hermodson MA, Lister RM.1983. Analysis of the heterogeneity of the 40 000 molecular weight tuber glycoprotein of potatoes by immunological methods and by NH2‐terminal sequence analysis. Plant Physiology 71: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PikaardCS, Brusca JS, Hannapel DJ, Park WD.1987. The two classes of genes for the major tuber protein, patatin, are differentially expressed in tubers and roots. Nucleic Acids Research 15: 1979–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PollockNJ.2000. Taro. In: Tipple KF, Ornelas KC, eds. The Cambridge world history of food Cambridge: Cambridge University Press, 218–230. [Google Scholar]
- PotsAM, de Jongh HHJ, Gruppen H, Hamer RJ, Voragen AGJ.1998a. Heat‐induced conformational changes of patatin, the major potato tuber protein. European Journal of Biochemistry 252: 66–72. [DOI] [PubMed] [Google Scholar]
- PotsAM, de Jongh HHJ, Gruppen H, Hessing M, Voragen AGJ.1998b. The pH dependence of the structural stability of patatin. Journal of Agricultural Food Chemistry 46: 2546–2553. [Google Scholar]
- PotsAM, ten Grotenhuis E, Gruppen H, Voragen AGJ, de Kruif KG.1999a. Thermal aggregation of patatin studied in situ. Journal of Agricultural Food Chemistry 47: 4600–4605. [DOI] [PubMed] [Google Scholar]
- PotsAM, Gruppen H, Hessing M, van Boekel MAJS, Voragen AGJ.1999b. Isolation and characterization of patatin isoforms. Journal of Agricultural Food Chemistry 47: 4587–4592. [DOI] [PubMed] [Google Scholar]
- PotsAM, Gruppen H, de Jongh HHJ, van Boekel MAJS, Walstra P, Voragen AGJ.1999c. Kinetic modeling of the thermal aggregation of patatin. Journal of Agricultural Food Chemistry 47: 4593–4599. [DOI] [PubMed] [Google Scholar]
- RacusenD.1984. Lipid acyl hydrolase of patatin. Canadian Journal of Botany 62: 1640–1644. [Google Scholar]
- RacusenD.1986. Esterase specificity of patatin from two potato cultivars. Canadian Journal of Botany 64: 2104–2106. [Google Scholar]
- RacusenD, Foote M.1980. A major soluble glycoprotein of potato tubers. Journal of Food Biochemistry 4: 43–52. [Google Scholar]
- RaletM‐C, Guéguen J.2000. Fractionation of potato proteins: solubility, thermal coagulation and emulsifying properties. Lebensmittel‐Wissenschaft und‐Technologie 33: 380–387 [Google Scholar]
- RecheM, Pascual CY, Vicente J, Caballero T, Martin‐Munoz F, Sanchez S, Martin‐Esteban M.2001. Tomato allergy in children and young adults: cross‐reactivity with latex and potato. Allergy 56: 1197–1201. [DOI] [PubMed] [Google Scholar]
- RehmS, Espig G.1991.The cultivated plants of the tropics and subtropics: cultivation, economic value, utilization. Weckersheim Verlag Josef Margraf (Scientific Books). [Google Scholar]
- Rocha‐SosaM, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer I.1989. Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO Journal 8: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RosahlS, Eckes P, Schell J, Willmitzer I.1986. Organ‐specific gene expression in potato: isolation and characterization of tuber‐specific cDNA sequences. Molecular and General Genetics 202: 368–373. [Google Scholar]
- RosahlS, Schell J, Willmitzer L.1987. Expression of a tuber‐specific storage protein in transgenic tobacco plants: demonstration of an esterase activity. EMBO Journal 6: 1155–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SendaK, Yoshioka H, Doke N, Kawakita K.1996. A cytosolic phospholipase A2 from potato tissues appears to be patatin. Plant Cell Physiology 37: 347–353. [DOI] [PubMed] [Google Scholar]
- SeppäläU, Alenius H, Turjanmaa K, Reunala T, Palòsuo T, Kalkkinen N.1999. Identification of patatin as a novel allergen for children with positive skin prick test responses to raw potato. Journal of Allergy and Clinical Immunology 103: 165–171. [DOI] [PubMed] [Google Scholar]
- SeppäläU, Palòsuo T, Seppälä U, Kalkkinen N, Ylitalo L, Reunala T, Turjanmaa K, Reunala T.2000. IgE reactivity to patatin‐like latex allergen, Hev b 7, and to patatin of potato tuber, Sol t 1, in adults and children allergic to natural rubber latex. Allergy 55: 266–273. [DOI] [PubMed] [Google Scholar]
- ShewryPR.1995. Plant storage proteins. Biological Review 70: 375–426. [DOI] [PubMed] [Google Scholar]
- ShewryPR, Lucas JA.1997. Plant proteins that confer resistance to pests and pathogens. In: J. Callow ed. Advances in botanical research, Vol. 26 London: Academic Press, 135–192. [Google Scholar]
- ShewryPR, Casey R.1999.Seed proteins. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- ShewryPR, Clowes A, Tatham AS, Beeching J.1993. Opportunities for manipulating the amount and composition of the proteins of cassava tuberous roots. Proceedings of 1st International Scientific Meeting of the Cassava Biotechnology Network, Cartagena de Indias, Colombia, September 1992 CIAT, Cali, Colombia, 251–254. [Google Scholar]
- SinghJ, Kamboj SS, Sandhu RS, Shangary S, Kamboj KK.1993. Purification and characterization of a tuber lectin from Alocasia indica Phytochemistry 33: 979–983. [Google Scholar]
- SonnewaldU, Studer D, Rocha‐Sosa M, Willmitzer L.1989a. Immunocytochemical localization of patatin, the major glyco protein in potato (Solanum tuberosum L.) tubers. Planta 178: 176–183. [DOI] [PubMed] [Google Scholar]
- SonnewaldU, Sturm A, Chrispeels MJ, Willmitzer L.1989b. Targeting and glycosylation of patatin, the major tuber protein in leaves of transgenic tobacco. Planta 179: 171–180. [DOI] [PubMed] [Google Scholar]
- SørensenM, Døygaards S, Estella JE, Kvist LP, Nielsen PE.1997. Status of the South American tuberous legume Pachyrhizus tuberosus (Lam.) Spreng. Biodiversity and Conservation 6: 1581–1625. [Google Scholar]
- SouzaPAS, Gomes E, Compos FAP.1998. Tissue distribution and deposition pattern of a cellulosic parenchyma‐specific protein from cassava roots. Brazilian Archives of Biology and Technology 41: 1–9. [Google Scholar]
- SowkaS, Wagner S, Krebitz M, Arija‐Mad‐Arif S, Yusof F, Kinaciyan T, Brehler R, Scheiner O, Breiteneder H.1998. cDNA cloning of the 43‐kDa latex allergen Hev b 7 with sequence similarity to patatins and its expression in the yeast Pichia pastoris. European Journal of Biochemistry 255: 213–219. [DOI] [PubMed] [Google Scholar]
- SplittstoesserWE.1977. Protein quality and quantity of tropical roots and tubers. Hortscience 12: 294–298. [Google Scholar]
- StricklandJA, Orr GL, Walsh TA.1995. Inhibition of Diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers. Plant Physiology 109: 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SumathiS, Pattabiraman TN.1977. Natural plant enzyme inhibitors. V. A trypsin/chymotrypsin inhibitor from Alocasia macrorhiza tuber. Biochimica et Biophysica Acta 485: 167–178. [DOI] [PubMed] [Google Scholar]
- SvendsenI, Nicolova D, Goshev I, Genov N.1989. Isolation and characterisation of a trypsiin inhibitor from the seeds of khorhabi (Brassica napus var. Rapifera) belonging to the napin family of storage proteins. Carlsberg Research Communication 54: 231–239. [DOI] [PubMed] [Google Scholar]
- SvendsenI, Nicolova D, Goshev I, Genov N.1994. Primary structure, spectroscopic and inhibitory properties of a two‐chain trypsin inhibitor from the seeds of charlock (Sinapis arvensis L.), a member of the napin protein family. International Journal of Peptide and Protein Research 43: 425. [DOI] [PubMed] [Google Scholar]
- TerrasFRG, Schoofs HME, Thevissen K, Osborn RW, Vanderleyden J, Cammue BPA, Broekaert WF.1993. Synergistic enhancement of the antifungal activity of wheat and barley thionins by radish and oilseed rape 2S albumins and by barley trypsin inhibitors. Plant Physiology 103: 1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TheeraslipS, Hitotsuya H, Nakajo S, Nakaya K, Nakamura Y, Kurlhara Y.1989. Complete amino acid sequence and structural characterization of the tast‐modifying protein, miraculin. Journal of Biological Chemistry 264: 6655–6659. [PubMed] [Google Scholar]
- TonónC, Daleo G, Oliva C.2001. An acidic β‐1,3 glucanase from potato tubers appears to be patatin. Plant Physiology and Biochemistry 39: 849–854. [Google Scholar]
- Van DammeEJ, Goossens K, Smeets K, Torrekens S, Van Leuven F, Verhaert P, Peumans WJ.1995. The major tuber storage protein of Araceae species is a lectin. Characterization and molecular cloning of the lectin from Arum maculatum L. Plant Physiology 107: 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van LoonLC, van Strien EA.1999. The families of pathogenesis‐related proteins, their activities and comparative analysis of PR‐1 type proteins. Physiological and Molecular Plant Pathology 55: 85–97. [Google Scholar]
- WalterAMJr, Collins WW, Purcell AE.1984. Sweet potato protein: a review. Journal of Agriculture and Food Chemistry 32: 695–699. [Google Scholar]
- WangS‐J, Lan Y‐C, Chen S‐F, Chen Y‐M, Yeh K‐W.2002. Wound‐response regulation of the sweet potato sporamin gene promoter region. Plant Molecular Biology 48: 223–231. [DOI] [PubMed] [Google Scholar]
- WojnowskaI, Poznanski S, Bednarski W.1981. Processing of potato protein concentrates and their properties. Journal of Food Science 47: 167–172. [Google Scholar]
- YamashitaH, Theeraslip S, Aluchy T, Nakaya K, Nakamura N, Kurlhara Y.1990. Purification and complete amino acid sequence of a new type of sweet protein with tast‐modifying activity, curculin. Journal of Biological Chemistry 265: 15770–15775. [PubMed] [Google Scholar]
- YaoP‐L, Hwang M‐J, Chen Y‐M, Yeh K‐W.2001. Site‐directed mutagenesis evidence for a negatively charged trypsin inhibitory loop in sweet potato sporamin. FEBS Letters 496: 134–138. [DOI] [PubMed] [Google Scholar]
- YehK‐W, Chen JC, Lin MI, Chen Y‐M, Lin CY.1997a. Functional activity of sporamin from sweet potato (Ipomoea batatas Lam): a tuber storage protein with trypsin inhibitory activity. Plant Molecular Biology 33: 565–570. [DOI] [PubMed] [Google Scholar]
- YehK‐W, Lin ML, Tuan SJ, Chen Y‐M, Lin CY, Kao SS.1997b. Sweet potato (Ipomoea batatas) trypsin inhibitors expressed in transgenic tobacco plants confer resistance against Spodoptera litura Plant Cell Reports 16: 696–699. [DOI] [PubMed] [Google Scholar]
- YeohHH, Chew MY.1977. Protein content and acid composition of cassava seed and tuber. Malaysian Agricultural Journal 51: 1–6. [Google Scholar]
- ZourelidouM, de Torres‐Zabala M, Smith C, Bevan MW.2002. Storekeeper defines a new class of plant‐specific DNA‐binding proteins and is a putative regulator of patatin expression. The Plant Journal 30: 489–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


