Abstract
Mitotic chromosomes of four Vicia species (V. sativa, V. grandiflora, V. pannonica and V. narbonensis) were subjected to in situ hybridization with probes derived from conserved plant repetitive DNA sequences (18S–25S and 5S rDNA, telomeres) and genus‐specific satellite repeats (VicTR‐A and VicTR‐B). Numbers and positions of hybridization signals provided cytogenetic landmarks suitable for unambiguous identification of all chromosomes, and establishment of the karyotypes. The VicTR‐A and ‐B sequences, in particular, produced highly informative banding patterns that alone were sufficient for discrimination of all chromosomes. However, these patterns were not conserved among species and thus could not be employed for identification of homologous chromosomes. This fact, together with observed variations in positions and numbers of rDNA loci, suggests considerable divergence between karyotypes of the species studied.
Key words: Vicia, in situ hybridization, PRINS, rRNA genes, satellite DNA, repetitive DNA, karyotype evolution
INTRODUCTION
The genus Vicia includes over 160 species with a primarily Euro‐Asiatic distribution (Kupicha, 1976; Allkin et al., 1986; Maxted, 1995), some of which, such as V. faba (field bean), V. narbonensis (narbon vetch) and V. sativa (common vetch), are economically important crops. There is a considerable variation in haploid nuclear DNA content (1·8–13·3 pg) and basic chromosome number (2n = 10, 12 or 14) between Vicia species (Raina and Narayan, 1984; Maxted, 1995; Bennett and Leitch, 1998), which makes the genus an interesting model for the study of plant genome and karyotype evolution. The karyotypes of several species have been established based on chromosome size, centromeric index and banding patterns (Cremonini, 1992; Galasso et al., 1994; Cremonini et al., 1998; Fuchs et al., 1998); however, this approach is limited by the similar morphology of chromosomes in many species. Visualization of specific DNA sequences on chromosomes using FISH (fluorescence in situ hybridization) or PRINS (primed in situ DNA labelling) can be used to overcome these limitations by providing specific labelling patterns useful for discrimination of similar chromosomes. Probes for FISH or PRINS can be derived from conserved repetitive sequences, such as 18S–25S (18S–5.8S–25S) rRNA or 5S rRNA genes, which have been used successfully for chromosome discrimination in several Vicia species (Galasso et al., 1997; Raina et al., 2001). Additional cytogenetic landmarks can be obtained using species‐ or genus‐specific satellite repeats that are often amplified to high copy numbers and form discrete bands or spots on chromosomes (Charlesworth et al., 1994; Schmidt and Heslop‐Harrison, 1998). Compared with rRNA genes, these repeats frequently occur at a higher number of genomic loci and may therefore produce signals characteristic for each chromosome within the karyotype, as was shown for V. faba (Fuchs et al., 1994, 1998) and V. sativa (Nouzová et al., 2001).
In our previous work (Macas et al., 2000) two families of satellite repeats, VicTR‐A and VicTR‐B, have been isolated. These are specific for the genus Vicia and show different levels of amplification in individual species. The copy numbers of both repeats range from undetectable to up to 106 per haploid genome; however, VicTR‐A repeats are amplified mainly in species from the taxonomic section Hypechusa and in V. narbonensis (section Narbonensis), whereas VicTR‐B sequences are most abundant in section Vicia (which lacks VicTR‐A repeats) and, interestingly, also in V. narbonensis. The aim of the present study was to use VicTR sequences in combination with conserved plant repeats (18S–25S and 5S rRNA genes) for FISH and PRINS labelling of chromosomes in four Vicia species differing in genome size and chromosome numbers (Table 1). The main objectives were: (1) to discriminate individual chromosome types and to establish karyotypes in these species; (2) to investigate whether the distribution patterns of VicTR repeats are conserved among the species; and (3) to assess the possibility of identifying homeologous chromosomes based on these hybridization patterns.
Table 1.
Chromosome numbers, DNA content and abundance of VicTR repeats in selected Vicia species
| Abundance of VicTR repeats | |||||
| Section* | Species | Chromasome number (2n) | DNA content (pg) | VicTR‐A | VicTR‐B |
| Vicia | V. sativa | 12 | 2·3 | – | 106–5 × 106 |
| V. grandiflora | 14 | 3·4 | 100–1000 | 106–5 × 106 | |
| Hypechusa | V. pannonica | 12 | 6·8 | 105–106 | ∼100 |
| Narbonensis | V. narbonensis | 14 | 7·3 | 105–106 | 105–106 |
* Assignment of species to taxonomic sections is according to Kupicha (1976) and Maxted (1995).
† Number of copies per haploid genome, according to Macas et al. (2000).
MATERIALS AND METHODS
Plant material, cell cycle synchronization and chromosome preparation
Seeds of Vicia species were obtained from the germplasm collection of the Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany (V. grandiflora Scop.), Dr I. Nianiou, Aristotelian University of Thessaloniki, Greece (V. narbonensis L. ‘IFYN574’), and from plant breeding stations at Horní Moštěnice (V. sativa L. ‘Ebena’) and Chlumec nad Cidlinou, Czech Republic (V. pannonica Crantz ‘Dětenická panonská’). Three‐day‐old seedlings (V. sativa, V. pannonica and V. narbonensis) or hairy root cultures (V. grandiflora) were used for root meristem synchronization and accumulation of cells in metaphase as described by Doležel et al. (1992) and Neumann et al. (1998), respectively, with the following modifications: hydroxyurea (ICN Biomedicals, Irvine, CA, USA) treatment was carried out for 18 h at 1 mm concentration for V. sativa, 0·75 mm for V. pannonica, 1·25 mm for V. narbonensis and 1·5 mm for V. grandiflora; the time in hydroxyurea‐free medium was 2 h (V. sativa, V. pannonica), 4·5 h (V. narbonensis) or 6 h (V. grandiflora); and metaphase accumulation was achieved using 15 µm oryzalin (Chem Service, West Chester, PA, USA) applied for 7 h (V. narbonensis) or 4 h (all other species). Synchronized root tips were used for chromosome preparations using a squashing or dropping method as described by Leitch et al. (1994) and Busch et al. (1996). Alternatively, the chromosomes of V. sativa, V. pannonica and V. narbonensis were prepared as purified chromosome suspensions (Gualberti et al., 1996) and centrifuged onto microscope slides using a Hettich Universal 32‐R centrifuge equipped with cytospin chambers.
FISH and PRINS
The different methods of chromosome preparation described above and direct or indirect probe detection systems were employed so that optimal sensitivity could be achieved and the results could be verified using alternative methods. Probes for FISH were labelled with either fluorochrome‐ (Alexa Fluor 488‐dUTP or Alexa Fluor 568‐dUTP, Molecular Probes, Eugene, OR, USA) or biotin‐conjugated dUTP (biotin‐16‐dUTP; Boehringer‐Mannheim, Mannheim, Germany). Labelling of VicTR and 5S rDNA repeats (the 5S rDNA was cloned from pea; Neumann et al., 2001) was performed in a PCR reaction using universal primers and corresponding sequences cloned into a plasmid vector as a template. Alternatively, the VicTR‐A and ‐B sequences were labelled by direct amplification from genomic DNA of V. pannonica and V. sativa as described by Macas et al. (2000). The probe for 18S–25S rDNA was prepared by random priming of the clone Ver17 (Yakura and Tanifuji, 1983). Telomeric repeats were labelled using PCR without template DNA but including the complementary primers (AGGGTTT)3 and (CCCTAAA)3.
FISH experiments were performed as described by Neumann et al. (2001). Hybridization and washing temperatures were 37 °C and 42 °C, respectively, for rDNA and telomeric probes, and 28 °C and 32 °C for VicTR sequences. Simultaneous detection of two probes with different melting temperatures was done using a two‐step hybridization protocol. The PRINS reactions were performed using pairs of sequence‐specific primers and conditions described by Macas et al. (2000). For detection of two different targets the PRINS labelling was combined with FISH (PRINS–FISH technique). In these cases, the PRINS reaction was stopped by washing slides for 1·5 min in Stop buffer (50 mm NaCl, 50 mm EDTA, pH 8·0) at room temperature. The slides were then passed through an ice‐cold ethanol series (50, 70, 96 %), air‐dried and immediately used for FISH with omission of RNase pre‐treatment, post‐fixation step and slide denaturation. When PRINS was used for detection of VicTR repeats, subsequent FISH was performed at 28 °C to avoid washing out the VicTR signals.
Detection of biotin‐labelled probes was done using streptavidin‐fluorescein (Leitch et al., 1994) or using the Tyramide Signal Amplification Indirect system (NEN Life Sciences Products, Boston, MA, USA) with streptavidin‐fluorescein at the final step. Probes labelled with Alexa‐dUTP were visualized directly. Chromosomes were counterstained with DAPI (4′,6‐diamidino‐2‐phenylindole) and observed using a Nikon Eclipse‐600 epifluorescence microscope equipped with a CCD camera. Signals were collected separately using UV‐2A, B‐2A and G‐2A filter sets, and processed using LUCIA software (Laboratory Imaging, Prague, Czech Republic).
Establishment of the karyotypes
Chromosome measurements were made using slides prepared by the squashing or dropping methods and subjected to FISH or PRINS with appropriate probe(s). At least eight complete metaphase figures with clearly distinguishable chromosomes were measured using LUCIA software, and average values of relative sizes of the chromosomes and their arms were used to construct idiograms. The rules for numbering chromosomes within idiograms generally followed those used by Hollings and Stace (1974).
RESULTS AND DISCUSSION
Vicia sativa
Vicia sativa has the smallest genome among the species investigated in this study (2·3 pg/1C). It possesses six pairs of chromosomes (one metacentric, four subacrocentrics and one acrocentric), two of which bear 18S–25S rRNA genes. The genes for 5S rRNA were detected at two pericentromeric loci on chromosome 3 (Fig. 1A). In V. sativa, only one of the VicTR repeats, VicTR‐B, is amplified, and is present in large clusters with intercalary and (sub)terminal localization (Fig. 1B). VicTR‐B signals corresponded to most of the DAPI‐positive bands visible on mitotic chromosomes and produced patterns specific for each chromosome.
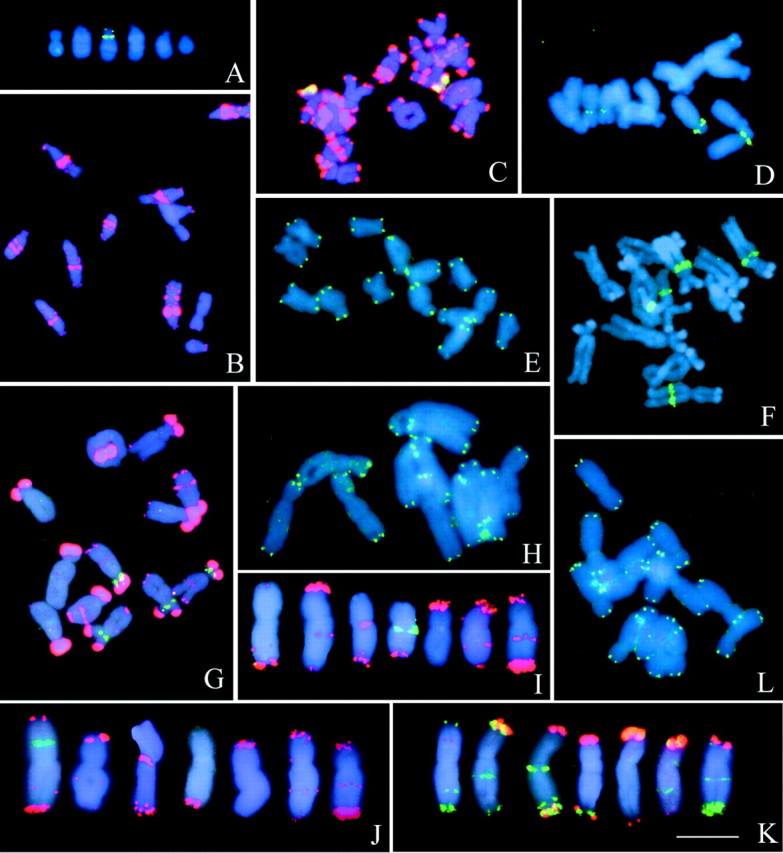
Fig. 1. Localization of repetitive sequences on metaphase chromosomes of Vicia sativa (A, B), V. grandiflora (C–E), V. pannonica (F–H) and V. narbonensis (I–L). 5S rDNA (A) and VicTR‐B (B) sequences on V. sativa chromosomes. VicTR‐B (red) and 18S–25S rDNA (green) (C), 5S rDNA (D), and telomeric probes (E) on V. grandiflora. 18S–25S rDNA (F), VicTR‐A (red) and 5S rDNA (green) (G), and telomeric sequences (H) on V. pannonica. VicTR‐B (red) and 18S–25S rDNA (green) (I), VicTR‐B (red) and 5S rDNA (green) (J), VicTR‐A (red) and VicTR‐B (green) (K), and telomeric repeats (L) on V. narbonensis chromosomes. Sequences were localized using FISH except for VicTR repeats in B, C and G, which were visualized using PRINS. Chromosomes were counterstained with DAPI (blue). Bar = 10 µm.
Vicia grandiflora
Although V. grandiflora belongs to the same taxonomic section as V. sativa (Kupicha, 1976; Maxted, 1995), it is divergent in chromosome number as well as in location of rDNA and VicTR‐B sequences. Its genome consists of seven chromosome pairs (one metacentric, three submetacentrics and three subacrocentrics). Only one chromosome pair (chromosome 2) possesses a secondary constriction and gave a signal with the 18S–25S rDNA probe (Fig. 1C). 5S rRNA genes were detected at one locus on each of chromosomes 1 and 5 (Fig. 1D). As in V. sativa, the majority of DAPI bands showed hybridization with VicTR‐B probe; however, these bands were located more at chromosome termini and were less intense within the chromosome arms (Fig. 1C).
Vicia pannonica
The six chromosome pairs of this species include four submetacentrics and two subacrocentrics, similar in size and DAPI banding. Hybridization sites of 18S–25S rDNA probe corresponded to positions of NOR regions on chromosomes 1 and 2 (Fig. 1F), and 5S rDNA was observed on chromosomes 1 and 4 (Fig. 1G). Strong hybridization signals of VicTR‐A sequences were found at the terminal regions of all chromosomes, and additional minor intercalary bands were visible on all chromosomes except chromosome 6 (Fig. 1G). Similarly to VicTR repeats in other species, the signals of VicTR‐A were located at heterochromatic regions detectable after DAPI staining.
Vicia narbonensis
Vicia narbonensis represents a species with a relatively large genome (7·3 pg/1C) and is unique in its abundance of both families of VicTR repeats. Its complement consists of three metacentric and four submetacentric chromosomes, bearing one NOR on chromosome 4 (Fig. 1I) and one locus of 5S rDNA on chromosome 1 (Fig. 1J). VicTR‐A repeats were located in the distal regions of all chromosomes except chromosome 1 (Fig. 1K); only a minor signal was observed on this chromosome and, in contrast to all other VicTR‐A signals, it was located in an intercalary position within its short arm. A large number of VicTR‐B sequences showed a similar distribution to those of VicTR‐A, and the two repeats co‐localized at the (sub‐) telomeric regions of several chromosomes (Fig. 1K). However, the termini of some chromosomes hybridized to only one of these probes, and additional VicTR‐B signals were observed as bands with intercalary and submetacentric locations. The preferential distribution of VicTR repeats at chromosome termini corresponds with the positions of heterochromatic regions detected by Cremonini et al. (1998) using C‐banding and Hoechst 33258 staining.
Establishment of karyotypes and their comparative analysis
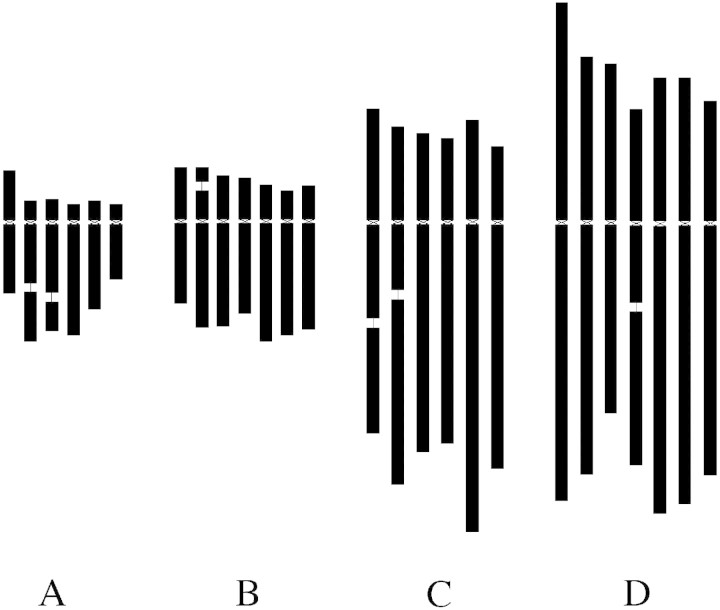
The probes for VicTR repeats and rDNA sequences produced signals which allowed for the construction of karyotypes in all four species (Fig. 2). As all chromosomes were clearly discriminated, their relative sizes could be determined based on measurements of chromosome lengths of mitotic chromosomes subjected to FISH or PRINS with suitable probes (Table 2; Fig. 3). Establishment of the karyotypes also allowed us to address the question of identification of homologous chromosomes among the species. The hybridization patterns obtained after FISH with repetitive probes have been successfully used for this task in several plant genera (Lubaretz et al., 1996; Cuadrado and Jouve, 1997; Lim et al., 2000; Hizume et al., 2002). However, contrary to these reports, no homologous chromosomes could be unambiguously identified between the four Vicia species investigated here. Even the species from the same taxonomic section (V. sativa and V. grandiflora) displayed significant differences in chromosome morphology as well as in numbers and positions of hybridization signals (Fig. 2). This may be a consequence of a higher rate of karyotype evolution or a greater evolutionary distance among the species under study. The results of a comparative analysis of repetitive sequences (Nouzová et al., 2001) and morphological and isozyme diversity (Leht and Jaaska, 2002) indicate that the latter explanation is more likely.

Fig. 2. Idiograms of Vicia sativa (A), V. grandiflora (B), V. pannonica (C) and V. narbonensis (D). Presence and distribution of 18S–25S rRNA genes (orange), 5S rRNA genes (yellow), VicTR‐A (red), VicTR‐B (green) and telomeres (white) is indicated.
Table 2.
Relative lengths and centromeric indexes of chromosomes
| Vicia sativa | Vicia grandiflora | Vicia pannonica | Vicia narbonensis | |||||
| Arm* | Relative length ± s.d.† | Centromeric index ± s.d.‡ | Relative length ± s.d.† | Centromeric index ± s.d.‡ | Relative length ± s.d.† | Centromeric index ± s.d.‡ | Relative length ± s.d.† | Centromeric index ± s.d.‡ |
| 1p | 7·49 ± 0·63 | 5·17 ± 0·33 | 4·66 ± 0·27 | 7·62 ± 0·59 | ||||
| q | 10·31 ± 0·43 | 1·39 ± 0·15 | 8·10 ± 0·40 | 1·58 ± 0·16 | 12·54 ± 0·62 | 2·70 ± 0·17 | 9·93 ± 0·77 | 1·28 ± 0·11 |
| 2p | 2·87 ± 0·48 | 4·23 ± 0·52 | 5·59 ± 0·55 | 5·79 ± 0·54 | ||||
| q | 16·40 ± 1·01 | 5·90 ± 1·19 | 10·43 ± 0·63 | 2·50 ± 0·36 | 9·99 ± 0·57 | 1·80 ± 0·19 | 8·87 ± 0·73 | 1·53 ± 0·20 |
| 3p | 3·08 ± 0·39 | 4·42 ± 0·33 | 4·34 ± 0·52 | 5·59 ± 0·45 | ||||
| q | 14·46 ± 0·88 | 4·78 ± 0·82 | 10·47 ± 0·80 | 2·39 ± 0·35 | 11·42 ± 0·90 | 2·65 ± 0·31 | 6·60 ± 0·55 | 1·18 ± 0·13 |
| 4p | 2·41 ± 0·47 | 4·17 ± 0·45 | 4·06 ± 0·53 | 3·95 ± 0·71 | ||||
| q | 16·69 ± 1·14 | 7·17 ± 1·51 | 9·07 ± 0·55 | 2·20 ± 0·32 | 10·95 ± 0·80 | 2·72 ± 0·30 | 8·25 ± 0·84 | 2·09 ± 0·23 |
| 5p | 2·91 ± 0·45 | 3·52 ± 0·48 | 4·99 ± 0·52 | 5·12 ± 0·50 | ||||
| q | 12·78 ± 0·62 | 4·51 ± 0·83 | 12·02 ± 0·49 | 3·47 ± 0·50 | 15·42 ± 0·94 | 3·11 ± 0·31 | 9·94 ± 0·62 | 1·94 ± 0·17 |
| 6p | 2·41 ± 0·38 | 2·92 ± 0·26 | 3·77 ± 0·19 | 5·01 ± 0·49 | ||||
| q | 8·17 ± 0·56 | 3·48 ± 0·69 | 11·33 ± 0·52 | 3·91 ± 0·41 | 12·27 ± 0·76 | 3·26 ± 0·29 | 9·70 ± 0·66 | 1·94 ± 0·22 |
| 7p | 3·43 ± 0·35 | 4·17 ± 0·48 | ||||||
| q | 10·73 ± 0·41 | 3·16 ± 0·38 | 8·96 ± 0·87 | 2·15 ± 0·28 | ||||
* Short (p) and long (q) chromosome arms.
† Average (± s.d.) relative lengths in % of haploid genome with standard deviations.
‡ Centromeric indexes calculated as chromosome arm ratios (q/p) and supplied with standard deviations.
Fig. 3. Schematic comparison of chromosome sizes among Vicia sativa (A), V. grandiflora (B), V. pannonica (C) and V. narbonensis (D). Relative chromosome lengths were derived from the data in Table 2 and scaled according to the nuclear DNA content of the species (Table 1).
Differential amplification of repetitive sequences may cause progressive changes of karyotypes in related species (Uozu et al., 1997; De la Herrán et al., 2001). Whereas amplification of retroelement sequences, which are generally dispersed in the genome (Pearce et al., 1996; Neumann et al., 2001), is likely to increase the size of all chromosomes within the karyotype in an approximately equal manner (Uozu et al., 1997), amplification of satellite DNA localized in large clusters has greater potential for increasing the asymmetry of the karyotypes (Uozu et al., 1997; De la Herrán et al., 2001). Moreover, satellite repeats are known to undergo rapid changes in copy numbers, resulting in their accumulation in a single species or a genus (Macas et al., 2002). As the copy numbers of VicTR‐A and ‐B sequences vary by over six orders of magnitude (Macas et al., 2000), it can be supposed that this variability may have had an impact on differentiation of karyotypes in Vicia. A comparison of chromosome location of VicTR repeats in individual species revealed that the VicTR‐B family is more variable compared with the VicTR‐A family, which occurs almost exclusively at terminal regions of chromosome arms. It is interesting to note that on a level of nucleotide sequences of the repeat monomers, VicTR‐B appears to be more conserved than VicTR‐A (Macas et al., 2000). However, more sequence data from different species are needed to confirm these findings and to study any possible correlation between sequence variability and genomic localization of these repeats.
As the majority of chromosomes displayed hybridization signals of VicTR probes at the termini of at least one of their arms, we investigated the presence of the Arabidopsis‐type telomeric repeats (Richards and Ausubel, 1988) at these regions. We also searched for additional hybridization signals in intercalary or centromeric regions that could point to chromosome rearrangements. In contrast to Vicia faba, in which such interstitial signals were located on several chromosomes (Schubert, 1992), the telomeric sequences in all Vicia species tested in this work were detected only at terminal regions of their chromosomes (Fig. 1E, H, L).
ACKNOWLEDGEMENTS
We thank Dr I. Nianiou (Aristotelian University of Thessaloniki, Greece) and Dr U. Pich (IPK Gatersleben, Germany) for seeds of Vicia species, Ms H. Štěpančíková and Ms O. Šonková for technical assistance, and Ms S. Rafelski for assistance in preparation of the manuscript. This work was supported by grant no. AVOZ 5051902 from the Academy of Sciences of the Czech Republic.
Supplementary Material
Received: 9 December 2002; Returned for revision: 17 February 2003; Accepted: 5 March 2003
References
- AllkinR, Goyder DJ, Bisby FA, White RJ.1986.Names and synonyms of species and subspecies in the Vicieae: Issue 3. Vicieae Database Project, Publication No 7 (ISSN 0263‐8517), University of Southampton, UK. [Google Scholar]
- BennettMD, Leitch IJ.1998. Angiosperm DNA C‐values database. http://www.rgbkew.org.uk/cval/homepage.html [Google Scholar]
- BuschW, Herrmann RG, Houben A, Martin R.1996. Efficient preparation of plant metaphase spreads. Plant Molecular Biology Reporter 14: 149–155. [Google Scholar]
- CharlesworthB, Sniegowski P, Stephan W.1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- CremoniniR.1992. The chromosomes of Vicia faba: banding patterns and in situ hybridizations. Biologisches Zentralblatt 111: 188–203. [Google Scholar]
- CremoniniR, Miotto D, Ngu MA, Tota D, Pignone D, Blangiforti S, Venora G.1998. Cytology of Vicia species. 5. Nuclear chromatin structure, karyomorphological analysis and DNA content in newly discovered relatives of Vicia faba L.: Vicia kalakhensis Khattab, Maxted et Bisby and Vicia eristalioides Maxted. Cytologia 63: 371–379. [Google Scholar]
- CuadradoA, Jouve M.1997. Distribution of highly repeated DNA sequences in species of the genus Secale Genome 40: 309–317. [DOI] [PubMed] [Google Scholar]
- De la HerránR, Robles F, Cunado N, Santos JL, Rejón MR, Garrido‐Ramos MA, Rejón CR.2001. A heterochromatic satellite DNA is highly amplified in a single chromosome of Muscari (Hya cinthaceae). Chromosoma 110: 197–202. [DOI] [PubMed] [Google Scholar]
- DoleželJ, Číhalíková J, Lucretti S.1992. A high‐yield procedure for isolation of metaphase chromosomes from root‐tips of Vicia faba L. Planta 188: 93–98. [DOI] [PubMed] [Google Scholar]
- FuchsJ, Pich U, Meister A, Schubert I.1994. Differentiation of field bean heterochromatin by in situ hybridization with a repeated FokI sequence. Chromosome Research 2: 25–28. [DOI] [PubMed] [Google Scholar]
- FuchsJ, Strehl S, Brandes A, Schweizer D, Schubert I.1998. Molecular‐cytogenetic characterization of the Vicia faba genome – heterochromatin differentiation, replication patterns and sequence localization. Chromosome Research 6: 219–230. [DOI] [PubMed] [Google Scholar]
- GalassoI, Sonnante G, Tota DG, Pignone D.1997. Comparison of molecular cytogenetic and genetic analyses in accessions of the two biotypes of Vicia benghalensis L. Annals of Botany 79: 311–317. [Google Scholar]
- GalassoI, Piergiovanni AR, Cremonini R, Perrino P, Pignone D.1994. Cytology of Vicia species. 3. Characterization of the chromosomal chromatin of some species of the section Cracca. Cytobios 77: 175–182. [Google Scholar]
- GualbertiG, Doležel J, Macas J, Lucretti S.1996. Preparation of pea (Pisum sativum L.) chromosome and nucleus suspensions from single root tips. Theoretical and Applied Genetics 92: 744–751. [DOI] [PubMed] [Google Scholar]
- HizumeM, Shibata F, Matsusaki Y, Garajova Z.2002. Chromosome identification and comparative karyotypic analyses of four Pinus species. Theoretical and Applied Genetics 105: 491–497. [DOI] [PubMed] [Google Scholar]
- HollingsE, Stace CA.1974. Karyotype variation and evolution in the Vicia sativa aggregate. New Phytologist 73: 195–208. [Google Scholar]
- KupichaFK.1976. The infrageneric structure of Vicia Notes of the Royal Botanic Garden, Edinburgh 34: 287–326. [Google Scholar]
- LehtM, Jaaska V.2002. Cladistic and phenetic analysis of relationships in Vicia subgenus Vicia (Fabaceae) by morphology and isozymes. Plant Systematics and Evolution 232: 237–260. [Google Scholar]
- LeitchAR, Schwarzacher T, Jackson D, Leitch IJ.1994. In situ hybridization: a practical guide. RMS microscopy handbooks, No. 27. Oxford: Bios Scientific Publishers Ltd. [Google Scholar]
- LimKY, Matyášek R, Lichtenstein CP, Leitch AR.2000. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma 109: 245–258. [DOI] [PubMed] [Google Scholar]
- LubaretzO, Fuchs J, Ahne R, Meister A, Schubert I.1996. Karyotyping of three Pinaceae species via fluorescent in situ hybridization and computer‐aided chromosome analysis. Theoretical and Applied Genetics 92: 411–416. [DOI] [PubMed] [Google Scholar]
- MacasJ, Mészáros T, Nouzová M.2002. PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18: 28–35. [DOI] [PubMed] [Google Scholar]
- MacasJ, Požárková D, Navrátilová A, Nouzová M, Neumann P.2000. Two new families of tandem repeats isolated from genus Vicia using genomic self‐priming PCR. Molecular and General Genetics 263: 741–751. [DOI] [PubMed] [Google Scholar]
- MaxtedN.1995. An ecogeographical study of Vicia subgenus Vicia Systematic and ecogeographic studies on crop genepools. 8. Rome: International Plant Genetic Resources Institute. [Google Scholar]
- NeumannP, Nouzová M, Macas J.2001. Molecular and cytogenetic analysis of repetitive DNA in pea (Pisum sativum L.). Genome 44: 716–728. [PubMed] [Google Scholar]
- NeumannP, Lysák M, Doležel J, Macas J.1998. Isolation of chromosomes from Pisum sativum L. hairy root cultures and their analysis by flow cytometry. Plant Science 137: 205–215. [Google Scholar]
- NouzováM, Neumann P, Navrátilová A, Galbraith DW, Macas J.2001. Microarray‐based survey of repetitive genomic sequences in Vicia spp. Plant Molecular Biology 45: 229–244. [DOI] [PubMed] [Google Scholar]
- PearceSR, Harrison G, Li D, Heslop‐Harrison JS, Kumar A, Flavell AJ.1996. The Ty1‐copia group retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal localisation. Molecular and General Genetics 250: 305–315. [DOI] [PubMed] [Google Scholar]
- RainaSN, Narayan RKJ.1984. Changes in DNA‐composition in the evolution of Vicia species. Theoretical and Applied Genetics 68: 187–192. [DOI] [PubMed] [Google Scholar]
- RainaSN, Mukai Y, Kawaguchi K, Goel S, Jain A.2001. Physical mapping of 18S–5·8S–26S and 5S ribosomal RNA gene families in three important vetches (Vicia species) and their allied taxa constituting three species complexes. Theoretical and Applied Genetics 103: 839–845. [Google Scholar]
- RichardsEJ, Ausubel FM.1988. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana Cell 53: 127–136. [DOI] [PubMed] [Google Scholar]
- SchmidtT, Heslop‐Harrison JS.1998. Genomes, genes and junk: the large‐scale organization of plant chromosomes. Trends in Plant Science 3: 195–199. [Google Scholar]
- SchubertI.1992. Telomeric polymorphism in Vicia faba Biologisches Zentralblatt 111: 164–168. [Google Scholar]
- UozuS, Ikehashi H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K.1997. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza Plant Molecular Biology 35: 791–799. [DOI] [PubMed] [Google Scholar]
- YakuraK, Tanifuji S.1983. Molecular cloning and restriction analysis of EcoRI‐fragments of Vicia faba rDNA. Plant and Cell Physiology 24: 1327–1330. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.