Abstract
The Mycobacterium tuberculosis genome encodes 11 serine/threonine protein kinases (STPKs). A similar number of two-component systems are also present, indicating that these two signal transduction mechanisms are both important in the adaptation of this bacterial pathogen to its environment. The M. tuberculosis phosphoproteome includes hundreds of Ser- and Thr-phosphorylated proteins that participate in all aspects of M. tuberculosis biology, supporting a critical role for the STPKs in regulating M. tuberculosis physiology. Nine of the STPKs are receptor type kinases, with an extracytoplasmic sensor domain and an intracellular kinase domain, indicating that these kinases transduce external signals. Two other STPKs are cytoplasmic and have regulatory domains that sense changes within the cell. Structural analysis of some of the STPKs has led to advances in our understanding of the mechanisms by which these STPKs are activated and regulated. Functional analysis has provided insights into the effects of phosphorylation on the activity of several proteins, but for most phosphoproteins the role of phosphorylation in regulating function is unknown. Major future challenges include characterizing the functional effects of phosphorylation for this large number of phosphoproteins, identifying the cognate STPKs for these phosphoproteins, and determining the signals that the STPKs sense. Ultimately, combining these STPK-regulated processes into larger, integrated regulatory networks will provide deeper insight into M. tuberculosis adaptive mechanisms that contribute to tuberculosis pathogenesis. Finally, the STPKs offer attractive targets for inhibitor development that may lead to new therapies for drug-susceptible and drug-resistant tuberculosis.
Signal transduction is an essential activity of all living cells. Broadly defined, signal transduction is the sensing of a signal or input and its conversion into an output or response that alters cell physiology. The sensor is the molecule or domain of a molecule (typically a protein) that senses the signal. The transducer is the molecule or domain that converts the signal into a response. Most commonly, signal transduction refers to the sensing of an extracellular signal that is transduced across the cytoplasmic membrane and converted into an intracellular response. Thus, signal transduction is critical for cellular adaptation to changes in the extracellular environment. In the case of bacterial pathogens, including Mycobacterium tuberculosis, these adaptive responses allow growth and/or survival in the environments encountered by the pathogen during the course of infection in the human host.
The most widely distributed and intensively studied transmembrane signaling systems in bacteria are the two-component systems (1). In these systems the sensor and transducer (referred to as the response regulator) are separate proteins, in which the sensor protein spans the cytoplasmic membrane and the response regulator is a cytoplasmic protein, usually a transcription factor that is activated in response to this phosphorylation event. Two-component systems are discussed in depth in reference 138. So-called one-component systems are cytoplasmic proteins that contain both a sensor domain and an output domain (2). The sensor domain typically senses intracellular signals via binding of small molecules, leading to effects on transcription by the output domain.
Another group of transcription regulators, the extracytoplasmic function (ECF), or group IV, sigma factors, has been referred to as the “third pillar” of bacterial signal transduction (3). The ECF sigma factors were originally described as sensing and/or regulating ECFs (4). Members of the ECF subfamily are often negatively regulated by direct interaction with an anti-sigma factor protein that serves as the sensor. Anti-sigma factors may be transmembrane proteins, e.g., M. tuberculosis RslA (the anti-sigma factor of SigL), or cytoplasmic proteins, e.g., M. tuberculosis RshA (anti-sigma factor of SigH), and thus may transduce either extra-cytoplasmic or intracellular signals (5, 6). Sigma factors and their regulatory mechanisms are discussed in depth in reference 139.
The other major mechanism of transmembrane signaling in M. tuberculosis is via the serine/threonine protein kinases (STPKs), the focus of this review. Unlike two-component systems, which are a major signaling mechanism in nearly all phyla of bacteria, STPKs are less widely distributed among different groups of bacteria. STPKs are most abundant among Acidobacteria, Actinobacteria (which includes mycobacteria), some cyanobacteria, and one order of the Deltaproteobacteria (the Myxococcales, the first bacteria in which STPKs were identified) (7, 8). In contrast to many of the widely studied bacterial pathogens and model organisms that have few or no STPKs but many two-component systems, the M. tuberculosis genome encodes 11 STPKs and a similar number of two-component systems, indicating that these two mechanisms both play important roles in signal transduction in this organism.
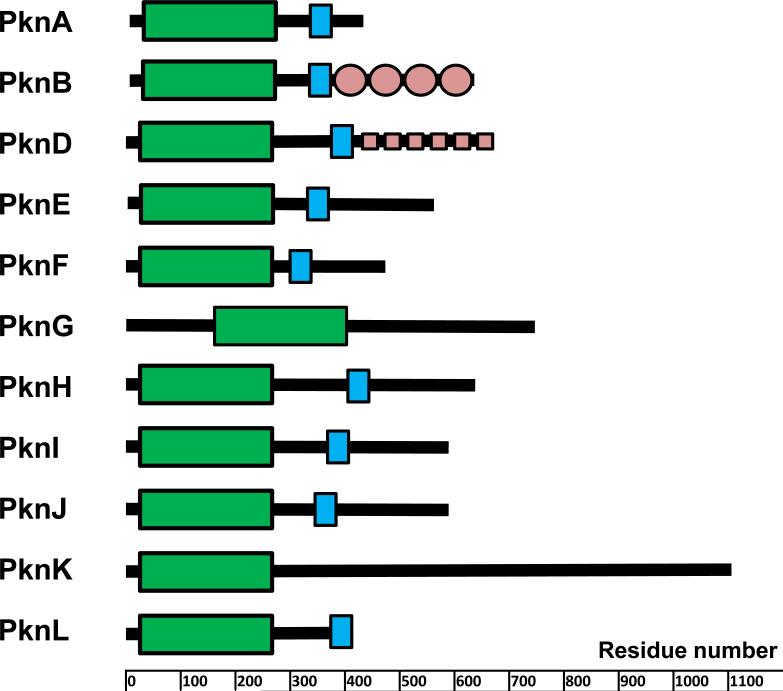
Of the 11 M. tuberculosis STPKs, all but 2 have a single transmembrane domain with an extracellular sensor domain and an intracellular kinase domain (KD) (Fig. 1). These nine transmembrane proteins can thus be classified as receptor-type kinases, in which the extracellular sensor domain senses extracytoplasmic signals and transduces this information to the intracellular KD, leading to activation of the kinase and phosphorylation of Ser or Thr residues on substrate proteins. This phosphorylation may alter protein function directly or by affecting interactions between specific pairs of proteins or within multiprotein complexes. In contrast to two-component, one-component, and ECF sigma factor signal transduction, where the usual primary output is changes in transcription, the output of Ser/Thr phosphorylation is rarely direct regulation of transcription.
FIGURE 1.
Domain organization of STPKs from M. tuberculosis. Domains were predicted using the SMART algorithm (136, 137). Kinase domains are shown as green boxes, trans-membrane portions are in blue, and some known extracellular domains are in light red. doi:10.1128/microbiolspec.MGM2-0006-2013.f1
As will be discussed below, for several of the M. tuberculosis STPKs, at least some of the signals sensed and some of the proteins targeted are known. There remains a great deal to be learned, however, about the exact mechanisms and functions of the STPKs in regulating M. tuberculosis physiology. In this article we highlight some of the major findings and current state of knowledge regarding the role of Ser/Thr and Tyr phosphorylation-mediated signal transduction in M. tuberculosis. The rapid expansion of the literature in this field, however, makes it impossible to note every observation regarding protein phosphorylation and its functional effects in mycobacteria.
SEQUENCE CHARACTERISTICS AND COMPARATIVE GENOMICS OF MYCOBACTERIAL STPKs
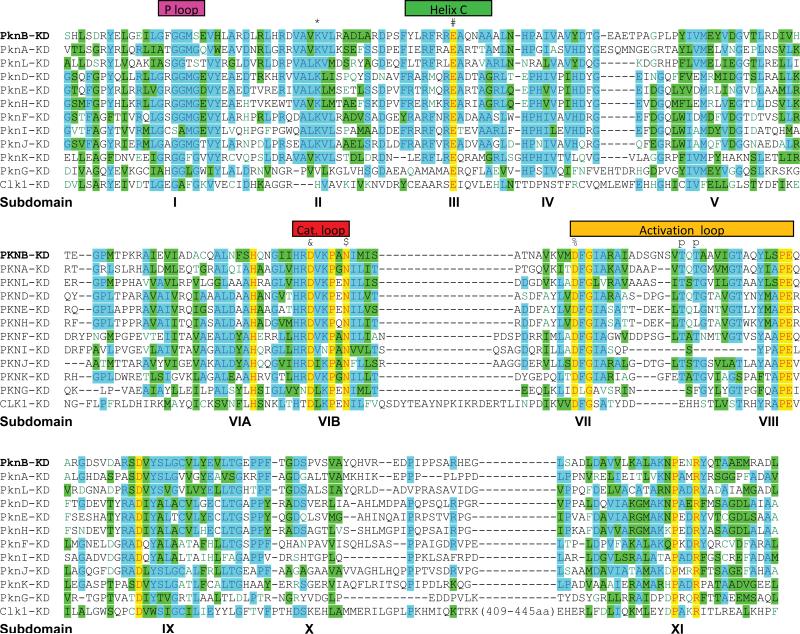
The M. tuberculosis STPKs were first described as “eukaryotic-like” protein kinases based on their sequence similarity to eukaryotic STPKs (9). The protein sequence similarity among the KDs of eukaryotic kinases led Hanks and Hunter in 1995 to identify a “superfamily” of protein kinases containing 11 subdomains (10). These subdomains contain conserved residues and motifs present in members of the superfamily, with specific functions attributable to each subdomain. With the massive expansion in the number of eukaryotic protein kinase sequences in the genomic era, this subdomain organization has remained valid, and sequence alignments have indicated the presence of many subfamilies of functionally and/or structurally related kinases. Subdomains 1 through 4 and part of 5 comprise the N-terminal lobe of the KD (see below), responsible for ATP binding and alignment, while subdomains 5 through 11 are responsible for substrate binding and phosphate transfer. Comparison of the M. tuberculosis STPKs to eukaryotic protein kinases demonstrates that the M. tuberculosis proteins incorporate each of the 11 Hanks subdomains, despite relatively limited sequence identity (Fig. 2).
FIGURE 2.
Sequence alignment of STPKs from M. tuberculosis. Kinase domains predicted by the SMART algorithm (see Fig. 1) were aligned and grouped using AlignX software (Life Technologies). Human Clk1 kinase is also included for comparison. Major features are noted. Selected conserved residues are labeled with the following symbols (residue numbers from PknB): *, Lys40; #, Glu59; &, Asp138; $, Asn143; %, Asp156; p, major phosphorylation sites in the activation loop. doi:10.1128/microbiolspec.MGM2-0006-2013.f2
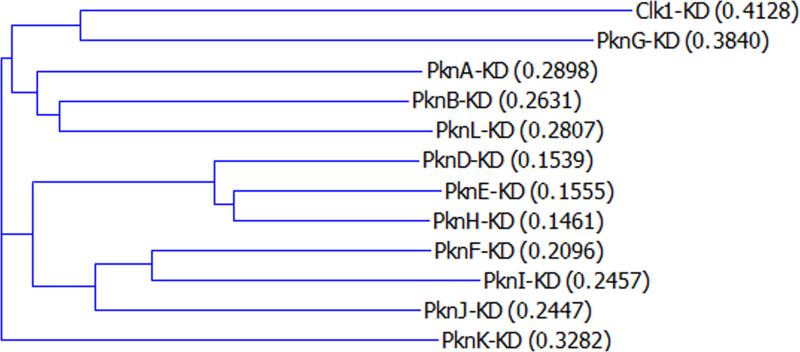
Sequence alignment of the M. tuberculosis STPK KDs shows that the 11 STPKs can be grouped into three clusters of three kinases and two KDs that are less similar to any of the nine clustered domains (Fig. 3). The unclustered outliers, PknG and PknK, are also the two kinases that lack a transmembrane domain. This observation suggests that the genes encoding the nine receptor-type kinases may be derived from a single common ancestral gene via gene duplication, whereas PknG and PknK may have been acquired separately. In contrast to the intracellular KDs, the extracellular domains of the nine transmembrane STPKs show no sequence similarity, indicating that they likely bind to and respond to distinct extracytoplasmic molecular signals. As discussed below for each kinase, motifs are present within the protein sequence of some of the extracellular domains and in the non-KD regions for PknG and PknK. For a few of the extracellular domains, candidate ligands have been identified.
FIGURE 3.
Dendrogram of KDs of M. tuberculosis STPKs. KDs identified by the SMART algorithm were aligned and grouped using the AlignX software (Life Technologies). Human Clk1 kinase is also included for comparison. Distance scores as given by AlignX are shown in parentheses. doi:10.1128/microbiolspec.MGM2-0006-2013.f3
As noted above, STPKs are not evenly distributed among different bacterial phyla. Within the mycobacteria and closely related actinomycetes, their distribution is also uneven. Table 1, which compares the genomes of the pathogens M. tuberculosis and Mycobacterium leprae, the opportunistic pathogens Mycobacterium avium and M. avium subspecies paratuberculosis, the nonpathogen Mycobacterium smegmatis, and the more distantly related nonpathogenic actinomycete Corynebacterium glutamicum, shows these differences in the distribution of the STPKs. The presence of PknA, PknB, PknG, and PknL in all species suggests that these kinases play important roles in regulating key aspects of mycobacterial physiology, though only PknA and PknB are essential in M. tuberculosis (11). The other STPKs likely have more specialized regulatory roles corresponding to the niches occupied by these different species.
TABLE 1.
Closest orthologs of M. tuberculosis STPKa
| Species | PknA | PknB | Pkdd | PkdE | PknF | PknG | PknH | PknI | PknJ | PknK | PknL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mtb | Rv0015c | Rv0014c | Rv0931c | Rvl743 | Rvl746 | Rv0410c | Rvl266c | Rv2914c | Rv2088 | Rv3080c | Rv2176 |
| Mmar | MMAR_0017 | MMAR_0016 | MMAR_4577 | MMAR_2581 | MMAR_2606 | MMAR_0713 | MMAR_1982 | MMAR_1794 | MMAR_1423 | MMAR_2576 | MMAR_32U |
| MMAR_4174 | MMAR_2444 | MMAR_2408 | |||||||||
| MMAR_4156 | MMAR_2941 | ||||||||||
| MMAR_4171 | |||||||||||
| Mav | MAV_0019 | MAV_0017 | MAV_4238 | MAV_3145 | MAV_4751 | MAV_1417 | MAV_2318 | ||||
| MAV_2158 | |||||||||||
| Map | MAP0018C | MAP0016C | MAP3387C | MAP1332 | MAP3893C | MAP2026 | MAP1914 | ||||
| MAP2031C | |||||||||||
| MAP2504 | |||||||||||
| Msmeg | MSMEG_0030 | MSMEG_0028 | MSMEG_0886 | MSMEG_0786 | MSMEG_4366 | MSMEG_5513 | MSMEG_0529 | MSMEG_4243 | |||
| MSMEG_3677 | |||||||||||
| Mle | ML0017 | ML0016 | ML0304 | ML0897 | |||||||
| Cgl | cg0059 | cg0057 | cg3046 | cg2388 |
Selected bacteria are shown: Mtb, M. tuberculosis H37Rv; Mmar ATCC 13032., M. marinum; Mav, M. avium 104; Map, M. avium paratuberculosis klO; Msmeg, M. smeg
matis MC2-155; Mle, M. leprae TN; Cgl, C. glutamicum
STRUCTURAL ANALYSIS OF STPK KINASE DOMAINS
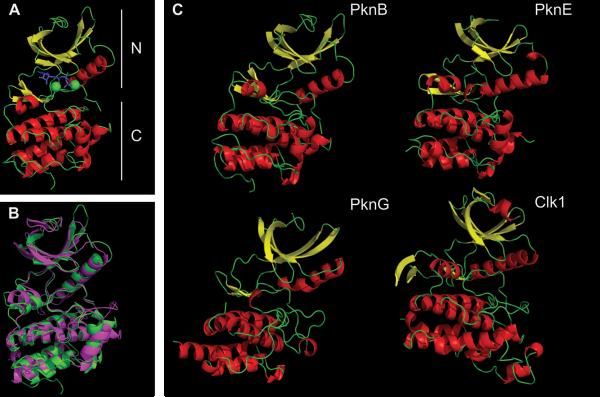
Structures of three KDs from M. tuberculosis STPKs (PknB, PknE, and PknG) have been solved (Table 2). These structures have provided important insights into mechanisms by which the M. tuberculosis STPKs are activated and regulated. The PknB KD was the first bacterial kinase structure described and was published by two groups independently in 2003 (12, 13). Though they were cocrystallized with different ATP analogs, the two PknB KD structures are virtually identical. As in other members of the eukaryotic protein kinase super-family, the PknB KD (as shown in the 1MRU crystal structure) has amino (N)- and carboxy (C)-terminal lobes with a nucleotide/Mg2+ binding site in the cleft between them (Fig. 4A) (13). The N-terminal lobe is mostly composed of β-sheets with a single long α-helix, designated helix C. In contrast, the C-terminal lobe contains only α-helices. Even with very low sequence conservation, this structure closely matches the overall structure of eukaryotic KDs. For example, human Clk1 KD has less than 20% sequence identity with PknB KD but has extensive overlap in structure (Fig. 4B). The KDs from PknE and the more divergent PknG also share this highly conserved kinase fold (Fig. 4C).
TABLE 2.
Structures of M. tuberculosis STPKs available in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Database (PDB)
| Kinase | PDB ID | Notes | Reference |
|---|---|---|---|
| PknB | 1MRU | KD + short linker (1-307) in complex with ATPyS, “back-to-back” dimer | 13 |
| 1O6Y | KD (1-279) in complex with AMP-PCP, monomer | 12 | |
| 2FUM | KD (1-279) in complex with mitoxantrone, “back-to-back” dimer | 25 | |
| 3F69 | L33D/M145L/M155V mutant (PknD surrogate) with Kt5720 “front-to-front” dimer | 21 | |
| 3F61 | L33D / V222D double mutant | ||
| 2KUI | Extracellular sensor (PASTA) | 28 | |
| 2KUE | Extracellular sensor (PASTA)-NMR structures | ||
| 2KUF | |||
| 2KUD | |||
| 3ORI | L33D mutant, various conformations | 22 | |
| 3ORK | |||
| 3ORL | |||
| 3ORO | L33D mutant, no metal ion bound | ||
| 3ORP | |||
| 3ORT | |||
| 3ORM | D76A mutant | ||
| PknD | 1RWI 1RWL | Extracellular sensor domain | 53 |
| PknE | 2H34 | Apo-KD | 14 |
| PknG | 2PZI | Rubredoxin + KD + TPR domain (69 aa truncated from N-terminus) in complex with Ax20017, dimer | 29 |
| PknH | 4ESQ | Extracellular sensor domain, dimer | 70 |
FIGURE 4.
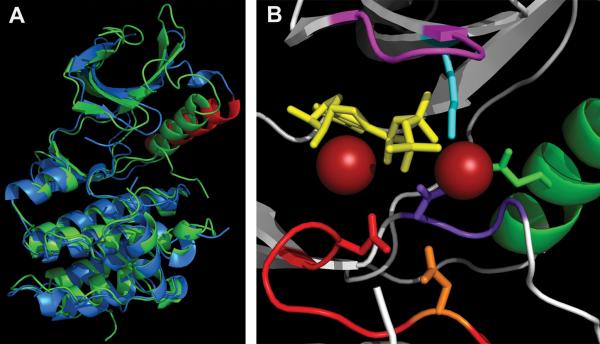
Overview of the M. tuberculosis STPK's KD. (A) Major features of the PknB KD (1MRU_B): N-terminal (upper) and C-terminal (lower) lobes are labeled. The ATP analog is in blue, and two Mg2+ ions are in green. (B) Overlap of PknB (green) and Clk1 (magenta). Clk1 was a top hit when the PknB structure was used to search similar three-dimensional structures using the NCBI VAST program. For clarity, residues 298 to 319 and 395 to 443 in Clk1 that are absent in M. tuberculosis STPKs (see Fig. 2) are truncated in Clk1. (C) PknB (1MRU_B), PknE (2H34_B), PknG (2PZI_A), and Clk1 (1Z57). α-Helix is in red, β-sheet is in yellow. Figures were made using PyMOL (Schrödinger) and POV-Ray (povray.org). doi:10.1128/microbiolspec.MGM2-0006-2013.f4
The PknB KD structures have all of the common elements found in eukaryotic protein kinases, as was predicted from the PknB amino acid sequence (Fig. 2 and 4). Both PknB KD structures (1MRU and 1O6Y) appear to represent the active or “closed” conformation of the kinase (12, 13). In most protein kinases, the conserved C helix is farther from the nucleotide binding cleft in the “open” position, while it is shifted toward this site in the “closed” position. The close proximity of the C loop to the nucleotide binding cleft of PknB in the closed position is shown in Fig. 5. The apo-PknE KD, was crystallized in the open or inactive form (Fig. 5A) (14). Positioning of the C helix away from the active site in the open conformation causes unfavorable placement of the conserved Glu64 (Glu59 in PknB) (Fig. 5A) (14). In contrast, in the PknB KD active conformation Glu59 is close enough to the invariant Lys-40 of the nucleotide binding domain to allow proper positioning toward the α and β phosphates of ATP (Fig. 5B).
FIGURE 5.
Active site of PknB KD. (A) Overlap of “closed” PknB KD (1MRU_B) in green and “open” apo-PknE-KD (2H34) in blue, with the PknE C helix labeled in red. (B) PknB active site (1MRU_B) P loop (GFGGMS), magenta; Mg2+, red balls; ATPγS, yellow; C-helix, green (Glu59-green); Lys40, aqua; catalytic loop, red (Asp138-orange, Asn143-red); DFG motif, purple (Asp156-purple). Figures were made using PyMOL (Schrödinger) and POV-Ray (povray.org). doi:10.1128/microbiolspec.MGM2-0006-2013.f5
Additional conserved features in the PknB structure are the Gly-rich P-loop present in the N-terminal lobe, which interacts with ATP (and ATP analogs) and the highly conserved Asp-Phe-Gly (DFG) residues at the amino-terminal boundary of the activation loop. In particular, Asp156 of the DFG triplet in PknB contributes to positioning of Mg2+ (Fig. 5B). In addition, two important residues in the catalytic loop, Asp138 (catalytic base) and Asn143, are properly paired in this active conformation structure for attack on the substrate (Thr or Ser) hydroxyl group and transfer of γ phosphate from ATP (Fig. 5B). The hydrophobic pocket that binds adenine is also structurally similar to those observed in eukaryotic kinases (12, 13).
In addition to these conserved features, the PknB structure has interesting distinct characteristics. The activation loop sequence between the conserved DFG and APE (SPE in PknB) motifs (Fig. 2) is, surprisingly, disordered in both PknB structures (1MRU and 1O6Y), although it is usually visible in structures of protein kinases in the active form. The activation loop is also missing in the crystal structure of the inactive conformation of apo-PknE KD, indicating that it is also disordered, as is typical in open KD structures (14). There are no reported crystal structures of any M. tuberculosis STPKs in complex with a substrate, but a working model for substrate binding by PknB involves a large portion of the activation loop that likely provides kinase speci-ficity (15). This loop was shown to be phosphorylated on at least four residues in PknB, two of which are important for kinase activation (T171 and T173), while two others may make minor contributions (Ser166 and Ser169) (13, 16). Activation loops of many eukaryotic kinases are phosphorylated in vivo, and most M. tuberculosis kinases have been shown to be autophosphorylated or trans-phosphorylated on activation loop residues in vitro (15, 17–20).
Although phosphorylation of the activation loop is required for PknB to be fully activated, “back-to-back” interaction of the unphosphorylated kinase is thought to be the first step in activation (21–24) (Fig. 6A). Although inferred previously from the PknB KD structural data (1MRU and 2FUM), this mechanism of M. tuberculosis kinase activation was first demonstrated in PknD KD fusion constructs that could be brought into proximity by rapamycin binding tags (23). Although PknD KD structures were not obtained, this study clearly showed that bringing KDs together stimulated auto- and transphosphorylation and that this activation depended on residues in a predicted dimer interface homologous to the interface observed in PknB KD crystal structures (13, 23, 25). Subsequent studies of PknB KD back-to-back dimerization provided additional insights into this activation mechanism (21, 22). Conserved residues at the interface of the back of the two N-terminal lobes adjacent to the C terminus of the C helix, such as the conserved Leu33 and the Arg10/Asp76 salt bridge, were demonstrated to be essential for this allosteric activation. When those contact sites were mutated, monomeric structures were obtained in multiple conformational states showing greater flexibility in the N-terminal lobe and misplacement of the C helix and its essential Glu59, resulting in lower enzymatic activity (Fig. 6B,C) (21, 22). Though not typical of many eukaryotic kinases, examples of back-to-back dimerization-induced activation are well described, e.g., for the human kinase PKR (26). The PknE KD was also shown to form a dimer with a similar, although not identical, interface (14).
FIGURE 6.
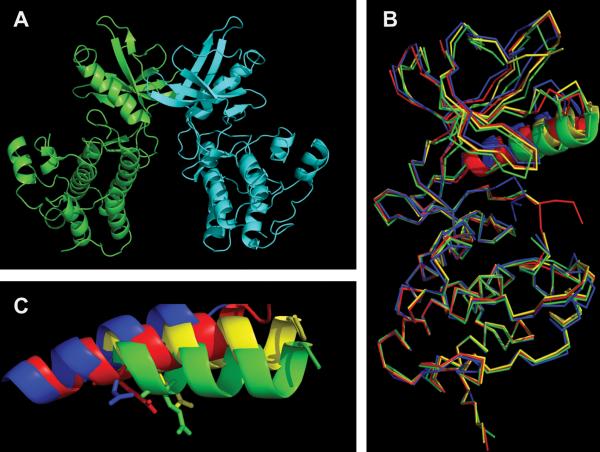
Back-to-back dimerization of KDs. (A) PknB-KD dimer showing “back-to-back” interaction. (B) Overlap of PknB-KD in active form (1MRU_B-blue) and conformations of the PknB-KD L33D mutant that perturbs the dimer interface (3ORK, yellow; 3ORI_A, red; 3ORL, green). The C helix is shown in ribbon, while the rest of the structure is shown in wire. (C) C helix from the PknB structures in panel B magnified to highlight differences in the position of Glu59. Figures were made using PyMOL (Schrödinger) and POV-Ray (povray.org). doi:10.1128/microbiolspec.MGM2-0006-2013.f6
It is important to note that the dimers observed in the PknB crystal structures do not appear to be formed with high affinity in solution (22). Also, the PknB and PknE extracellular domains do not dimerize on their own, so ligand binding is likely required, directly or indirectly, to facilitate dimerization of the KDs (22, 27, 28).
To examine subsequent steps of PknB activation, structural analysis was performed on a form of the PknB KD with substitutions at two Met residues that allow the KD to accommodate the inhibitor Kt5720, plus a single mutation that prevents back-to-back dimerization. This structure revealed an asymmetric “front-to-front” dimer (3f69) (Fig. 7A) (21). The interface between the two PknB KD monomers in this structure was mostly comprised of conserved G helix contacts. The activation loop of one KD monomer was ordered and in contact with the second monomer, while this second monomer had only a partially ordered activation loop with a small portion entering the active site of the first monomer, demonstrating a mechanism for activation by transphosphorylation (Fig. 7A). Mutational analysis of G helix residues that provide the contact surface in the front-to-front dimer showed their requirement for activation loop phosphorylation and demonstrated that they act synergistically with back-to-back contacts to allow full activation (21). Importantly, once activated by the allosteric conformational change to allow autophosphorylation, the primed KD with a fully phosphorylated activation loop can then remain active as a monomer, allowing continuing phosphorylation of substrate proteins even after the initial signal causing dimerization is gone (21–23).
FIGURE 7.
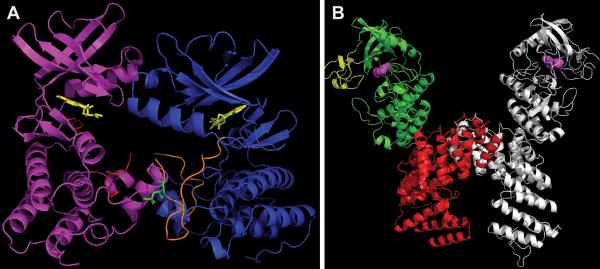
Distinct modes of monomer interaction in dimers of PknB versus PknG (A) “Front-to-front” dimer of mutant PknB KD (3F69) in complex with Kt5720 inhibitor (yellow). The “substrate” subunit (magenta) has most of its activation loop disordered (red), while the “enzyme” subunit (blue) has a well-defined activation loop (orange) with visible phosphor-ylated Thr171 (green). (B) Structure of PknG (2PZI) in complex with inhibitor Ax20017 (magenta). Three domains: rubredoxin (yellow), KD (green), and TPR domain (red) are shown only in one subunit. The second subunit is depicted in gray. Figures were made in PyMOL (Schrödinger) and POV-Ray (povray.org). doi:10.1128/microbiolspec.MGM2-0006-2013.f7
In addition to the KD, the intracellular portion of PknB includes a short juxtamembrane linker that connects the KD to the membrane-spanning segment (Fig. 1). Crystallization of the complete intracellular region of PknB including this region has been unsuccessful, indicating that the linker is disordered. The linker has been found to be phosphorylated in several M. tuberculosis STPKs, however, suggesting that it may have a regulatory role (12, 13, 15, 16, 18, 20).
The cytoplasmic protein kinase PknG is the only M. tuberculosis kinase that has been successfully crystallized as a nearly full-length protein (69 residues were removed from the N-terminus to allow crystallization) (Fig. 7B) (29). This construct includes both kinase and sensor domains: amino-terminal to the PknG KD is an iron binding rubredoxin domain, and carboxy-terminal to the KD is a tetratricopeptide repeat (TPR)–containing domain. TPRs are short repeats that are usually present in multiple copies and are involved in protein-protein interactions (Fig. 7B). The PknG dimer is formed through extensive contacts between the TPR domains of each monomer and not through N-lobe interactions, as was seen in the PknB KD and PknE KD dimer structures. Another difference compared to these other two kinases is that the PknG activation loop is ordered and stabilized, although not phosphorylated, suggesting a distinct mechanism of activation. Further, PknG is the only M. tuberculosis STPK that lacks Arg in front of the invariant Asp in the catalytic loop, and it was suggested that this difference results in the unique conformation of the activation loop in the absence of phosphorylation (Fig. 2) (29). PknG was found to be phosphorylated at the N-terminus in front of the rubredoxin domain, both in vivo and in vitro (15, 19). Although this modification is not required for PknG kinase activity, it was suggested that it might aid in substrate recognition (19). The rubredoxin domain, which contains two conserved Cys-X-X-Cys-Gly iron binding motifs, was shown to interact with both N- and C-terminal lobes of the PknG KD. Importantly, mutational analysis of the Cys residues showed that they are required for PknG kinase activity, suggesting that rubredoxin might sense the redox status of the environment to regulate PknG function (29–31).
These structural data for PknB, PknE, and PknG have provided key insights into mechanisms of STPK activation and regulation. Structures of the extracellular regions of PknB, PknD, and PknH have also been determined and are discussed below in the sections for each of these individual kinases. Going forward, structures of these domains bound to ligands and structures of KDs interacting with substrates may provide further insights into kinase regulation and substrate specificity.
THE M. TUBERCULOSIS PHOSPHOPROTEOME
In eukaryotes, STPKs and Tyr protein kinases are highly abundant. The human genome encodes over 500 of these protein kinases, and it is currently estimated that over two-thirds of human proteins are modified by this posttranslational modification, often at multiple sites (32–34). In contrast, M. tuberculosis encodes 11 STPKs and no typical Tyr kinases, though a protein with Tyr kinase activity has been identified (35). The number of proteins that are phosphorylated within the mycobacterial cell is unknown. In 2010, Prisic et al. published a large-scale analysis of the M. tuberculosis phosphoproteome, identifying over 500 phosphorylation sites in over 300 M. tuberculosis proteins using state-ofthe-art tandem mass-spectrometry methods (15). These data represent the minimal M. tuberculosis phosphoproteome, and many examples have been published of in vivo phosphorylation of additional M. tuberculosis proteins that were not identified in this phosphoproteomic study (Table 3), suggesting that the total M. tuberculosis phosphoproteome, when examined under multiple environmental conditions, is likely to include at least several hundred proteins.
TABLE 3.
Phosphorylated proteins in M. tuberculosis in addition to those identified in the phosphoproteomic study of Prisic et al. (15)a
| Name | Function | Phosphorylation, candidate cognate kinase, and effect | References |
| DacBl | Cell wall synthesis | In vitro (PknH) | (116) |
| DosR | Dormancy | In vitro (PknH), enhances DNA binding | (77) |
| EF-Tu | Protein synthesis | In vitro (PknB), in vivo; reduces interaction with GTP | (15, 117) |
| EmbR | Cell wall synthesis (arabinan) | In vitro (multiple kinases), in vivo (M. smegmatis); FHA domain required for phosphorylation; phosphorylation activates ATPase activity and enhances binding to embCAB promoter | (71, 74, 79) |
| EmbR2 | Cell wall synthesis? | In vitro (PknE, PknF), inhibits PknH | (118) |
| FabD | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases), in vivo (BCG) | (49, 119) |
| FabH | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases), decreases activity | (51) |
| FhaA | Cell wall synthesis | In vitro (multiple kinases), in vivo, FHA interacts with phosphorylated juxtamembrane region of PknB | (15,120,121) |
| FipA | Cell division | In vitro (PknA, PknB), in vivo, depends on FHA, required for activity under oxidative stress | (122) |
| FtsZ | Cell division | In vitro (PknA) (and in E. coli when coexpressed); impairs GTP hydrolysis and polymerization | (123) |
| GarA | Central metabolism | In vitro (PknB, PknG), in vivo; N-terminal tail binds to FHA, regulates interaction with TCA enzymes | (15,19, 69,124) |
| GlgE | Cell wall synthesis | In vitro (PknB), in vivo (BCG), decreases maltosyltransferase activity | (125) |
| GlmU | Cell wall synthesis | In vitro (PknB), decreases acetyltransferase activity | (126) |
| GroELl | Heat shock protein | In vitro (multiple kinases) | (122) |
| InhA | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases) (also when coexpressed in E. coli) in vivo (M. smegmatis, BCG), decreases activity | (47,48) |
| KasA | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases), in vivo (BCG), decreases activity | (49) |
| KasB | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases), in vivo (BCG); increases activity | (49) |
| MabA | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases), in vivo (BCG); decreases activity | (50) |
| MmA4 | Cell wall synthesis (mycolic acid) | In vitro (PknJ) | (79) |
| MmpL7 | Transporter?Virulence factor | Possibly PknD substrate, in vivo | (128) |
| MurD | Cell wall synthesis | In vitro (PknA) (and in E. coli when coexpressed) | (129) |
| MviN | Cell wall synthesis | In vitro (PknB), in vivo, enhances binding to FhaA | (15, 37) |
| PapA5 | Cell wall synthesis | In vitro (PknB) | (130) |
| PbpA | Cell wall synthesis | In vitro (PknB), possibly regulates localization | (36) |
| PepE | Dipeptidase | In vitro (PknJ) | (79) |
| PcaA | Cell wall synthesis (mycolic acid) | In vitro (multiple kinases), decreases activity | (131) |
| PstP | Dephosphorylation | In vitro (PknA, PknB) (and in E. coli when coexpressed), activates | (132) |
| PykA | Glycolysis | In vitro (PknJ) | (133) |
| RshA | Stress response | In vitro (PknB), in vivo, prevents interaction with SigH | (134) |
| Rv0516c | Anti-anti-sigma factor? | In vitro (PknD), in vivo, inhibits protein interactions | (55) |
| Rv0681 | Transcriptional regulator | In vitro (PknH) | (116) |
| Rv1422 | Cell wall synthesis? | In vitro (PknA and PknB), in vivo | (17) |
| Rvl747 | ABC-transporter? Virulence factor | In vitro (multiple kinases), in vivo, FHA domain interacts with kinases, activates | (15, 60, 61, 63, 120) |
| Rv2175c | DNA binding? | In vitro (PknL), in vivo (M. smegmatis), inhibits DNA binding | (85,135) |
| SigH | Stress response | In vitro (PknB), in vivo | (134) |
| VirS | Cell wall synthesis | In vitro (PknK) (and by coexpression in E. coli) increases binding to mym promoter | (81) |
| Wag31 | Cell division | In vitro (PknA), in vivo (PknA, PknB), enhances activity | (17, 42) |
Whether phosphorylation was shown in vitro or in M. tuberculosis (in vivo) is indicated, as are the kinases shown to phosphorylate the protein. Where known, the effects of phosphorylation on protein function are listed.
Several interesting findings emerged from this work. First, in contrast to human Ser/Thr phosphorylation, where phosphorylation on Ser and Thr account for 90% and 10% of the identified phosphorylation sites on human proteins, respectively, in M. tuberculosis Thr phosphorylation is predominant, with a 60:40 ratio of phosphorylation on Thr versus Ser. Second, phosphor-ylation was identified on proteins involved in all aspects of M. tuberculosis physiology (Fig. 8). Third, based on in vitro phosphorylation of peptides corresponding to in vivo phosphorylation sites, a conserved Thr-centered phosphorylation motif was identified in which acidic residues are prominent amino-terminal to the phosphoacceptor, particularly at the −2 and −3 positions, and hydrophobic residues are dominant at the +3 and to a lesser extent at the +5 positions. The core components of this motif are shared by six of the M. tuberculosis STPKs (PknA, PknB, PknD, PknE, PknF, and PknH), with less prominent differences among the motifs of each STPK that may provide additional substrate specificity.
FIGURE 8.
M. tuberculosis phosphoproteome. Phosphoproteins were identified in all functional categories of M. tuberculosis proteins (15). doi:10.1128/microbiolspec.MGM2 -0006-2013.f8
While these data have provided an important resource for further investigation, for most of these phosphoproteins the kinase(s) responsible for their phosphorylation and the effects of this posttranslational modification on protein function are not known. Identifying cognate kinase-substrate pairs and determining the functional effects of protein phosphorylation of individual protein substrates are essential for understanding the regulatory role of specific M. tuberculosis kinases. For M. tuberculosis STPKs, as for eukaryotic kinases, where well-characterized kinase-substrate pairs have been characterized and functional effects of phosphorylation have been elucidated for only a tiny fraction of phosphoproteins, these goals are experimentally challenging. In the following sections, we will summarize the current state of knowledge for each of the M. tuberculosis STPKs, including candidate substrates and physiologic pathways potentially regulated by each kinase.
PknA AND PknB
These two STPKs are encoded by adjacent genes in an operon that includes genes for the cell wall synthesis enzyme PBPA and the cell-shape-determining protein RodA, as well as the protein phosphatase PspA. Both pknA and pknB are essential for growth based on transposon mutagenesis experiments (11). Based on their linkage to rodA and pbpA, PknA and PknB were predicted to regulate cell shape and cell wall synthesis. An early study by Kang et al. confirmed this prediction, showing that overexpression of these kinases had marked effects on cell shape, including branching, elongation, and incomplete septation (17). Further support for the role of these kinases in cell wall synthesis and cell morphology were provided by the characterization of proteins involved in cell wall synthesis and its regulation that are likely substrates of one or both of these STPKs. Examples of these phosphoproteins include PBPA, a bifunctional penicillin binding protein that is a possible substrate of PknB; the DivIVA homologue Wag31, a substrate of PknA that has been shown to be required for peptidoglycan (PGN) synthesis in mycobacteria at the growing cell pole; and MviN, an essential protein required for late stages of PGN synthesis that is a likely substrate of PknB (17, 36, 37). In addition to these examples, several other proteins involved in PGN synthesis have been identified as phosphoproteins, in some cases with functional effects of phosphorylation shown (Table 3) (15). Further supporting a role for PknB in regulating cell division and cell wall synthesis, Mir et al. demonstrated that PknB is localized to the cell poles and the mid-cell, the sites of PGN turnover and assembly of the divisome (27). Consistent with PknB regulating cell wall synthesis during M. tuberculosis growth and with data showing 10-fold decreased expression of the pknA/pknB operon in stationary phase compared to log phase, a model was recently proposed in which PknB activity is decreased in hypoxia-induced stasis and PknB activity is required for oxygen-induced re-growth (17, 38).
The sequence of the extracellular region of PknB also suggested a role for this kinase in regulating PGN turnover. This region comprises four penicillin binding protein and serine/threonine kinase associated (PASTA) domains. These domains were first identified bioinformatically and predicted to bind PGN fragments (39). The incorporation of PASTA domains in some PBPs and in PknB-like STPKs led to the prediction that PknB and its homologues, which are widely distributed in Gram-positive bacteria, would regulate PGN synthesis. Evidence that the PASTA domains bind PGN fragments was first obtained in Bacillus subtilis, where spore germination was potently stimulated by muropeptides, PGN fragments derived from cell wall hydrolysis that contain a three- to five-residue stem peptide linked to the N-acetylglucosamine-N-acetymuramic acid disaccharide (40).
In M. tuberculosis, in vitro binding assays to the extracellular domain of PknB, using a comprehensive library of synthetic muropeptides, demonstrated that specific residues at the second and third positions of the stem peptide were required for binding (27). These residues, D-isoglutamine (versus D-isoglutamate) at position 2 and diaminopimelic acid (versus lysine) at position 3 are predominant in M. tuberculosis PGN, indicating that the PknB extracellular domain is adapted to recognize autologous muropeptides. A muropeptide that bound the M. tuberculosis PASTA domain in vitro was able to resuscitate dormant M. tuberculosis cells, albeit with less potency than spent medium, suggesting a link between PASTA binding, PknB activation, and cell growth.
Structural analysis of the PknB extracellular domain using nuclear magnetic resonance showed it to be an elongated structure, with rigid links between each of the four PASTA domains (28). This structure suggests that the PASTA domains protrude from the external surface of the cytoplasmic membrane into the PGN layer, where they may encounter muropeptides produced by cell wall hydrolases. This finding led the authors to propose a model of how ligand binding might activate PknB, in which muropeptide binding serves to cross-link the extracellular domains of two PknB proteins, leading directly to PknB dimerization and activation. In an alternative model, binding of muropeptides to extracellular domains of individual PknB molecules localizes PknB to sites of cell wall turnover, i.e., the septum and cell poles, resulting in high local PknB concentrations allowing dimerization of the intracellular KDs and their activation (27). Data from B. subtilis PrkC show that the PASTA-containing extracellular region does not dimerize in vitro, whether or not muropeptides are present (41). This result, together with the demonstrated ability of the KDs alone to dimerize and activate, favors the latter model (21), though it is possible that additional proteins might play a role in dimerization of the extra-cellular domain in a manner that would support the model of extracellular domain dimerization.
Though phosphorylation by PknA and PknB of several proteins involved in cell shape and cell wall synthesis has been shown or suggested (Table 3), here we will highlight two that play key roles in PGN synthesis and localization: Wag31 and MviN. Wag31, the M. tuberculosis homologue of the cell division protein DivIVA, was shown to be phosphorylated in M. tuberculosis cells and, through a combination of in vivo and in vitro experiments, to be phosphorylated by PknA (17). Subsequent analysis showed that Wag31 localizes to the cell poles, with preference for the old cell pole, the site of new PGN synthesis in mycobacteria and other actinomycetes (42, 43). Depletion of Wag31 led to delocalized PGN synthesis, asymmetric bulging, and ultimately lysis of the cells, indicating a critical role for this protein in proper localization of PGN synthesis. Through the use of phosphomimetic (T73E) or phosphoablative (T73A) substitutions in Wag31, phosphorylation of Wag31 appears to positively affect growth rate, though no clear morphologic differences were observed with expression of the different alleles (42).
MviN, a widely conserved multipass membrane protein, has been shown in Escherichia coli to be the protein that flips the lipid-linked PGN precursor lipid II from the cytoplasm to the periplasmic space, where the muropeptide can be incorporated into PGN (44). The M. tuberculosis MviN homologue, Rv3910, has an essential amino-terminal domain similar to MviN proteins in other bacteria but also has a nonessential carboxy-terminal region with sequence similarities to protein kinases (37). In the M. tuberculosis phosphoproteome study, this protein was found to be phosphorylated on as many as five distinct residues, including one well-localized site at Thr947 in the kinase homology domain (15). Gee et al. demonstrated that although this domain folds and dimerizes in a manner similar to protein kinases such as PknB, it lacks critical residues and motifs required for enzyme activity and is a pseudokinase that lacks both ATP binding and catalytic activities (37). The Thr947 residue was found to be phosphorylated in vitro by PknB and to a lesser extent by PknA, PknD, PknE, and PknH. Phosphorylated MviN was found to bind FhaA tightly; though the direct effect of this binding on MviN function was not shown, analysis of MviN and FhaA depletion strains suggested a model in which PknB phosphorylation of the kinase homology domain of MviN recruits FhaA, leading to inhibition of the terminal steps of PGN synthesis. This negative regulation of PGN synthesis by phosphorylation of MviN, in contrast to positive regulation of cell growth by PknA-mediated phosphorylation of Wag31, suggests a complex regulatory role of phosphorylation, likely by multiple kinases, in controlling M. tuberculosis growth and PGN synthesis.
In addition to cell wall synthesis, PknA and PknB have been linked, directly or indirectly, to regulation of several additional cellular processes, including lipid synthesis, cell division, and transcription regulation, among others (Table 3). The regulation of multiple processes by these kinases suggests that they may function to broadly coordinate cell physiology with cell growth. Linking a specific kinase to a validated in vivo phosphorylation event is challenging, however, and in many cases more than one STPK may target the same protein substrate. For many of the phosphoproteins involved in the processes noted above, whether PknA and/or PknB is the primary cognate kinase is uncertain. In other cases, rather than starting from well-defined in vivo phosphorylation in mycobacteria, investigators have started with a protein (enzyme) of interest, demonstrated in vitro phosphorylation by one or more M. tuberculosis STPKs in vitro or in E. coli, and then characterized effects of phosphorylation on enzyme function in vitro. In some cases the role of phosphorylation of the protein of interest in vivo remains unknown, while in other cases, additional work was undertaken to demonstrate in vivo phosphorylation and effects of phosphorylation on bacterial phenotypes.
An interesting example of a phosphoprotein first characterized in vitro and then shown to be phosphorylated in vivo with important functional effects of phosphorylation on lipid synthesis is the enoyl-acyl carrier protein reductase InhA, an essential component of the FASII fatty acid synthesis pathway in mycobacteria and the primary target of the first-line antitubercular, isoniazid (45, 46). Two groups independently examined in vitro phosphorylation of InhA and then pursued functional effects of phosphorylation in vitro and in vivo (47, 48). InhA was found to be phosphorylated by multiple kinases, including PknA and PknB. Elucidation of the sites of phosphorylation by mutagenesis and mass spectrometry yielded similar results, with one group observing phosphorylation at the carboxy-terminus of the protein at Thr253, Thr254, and Thr266, while the other group identified Thr266 as a unique phosphorylation site. The Thr254 and Thr266 sites have features of the preferred phosphorylation site motifs of PknA and PknB (15, 17). In vivo phosphorylation of InhA was investigated in M. smegmatis wild type and an InhA overexpression strain (47) and in Mycobacterium bovis BCG over-expressing InhA (48), with both studies confirming Thr266 as the primary in vivo site of phosphorylation. Using phosphorylated InhA and phosphomimetic (T266D or T266E) or phosphoablative (T266A) substituted proteins, enzyme activity of InhA was shown to be markedly decreased by phosphorylation. Structural analysis showed that these effects were not the result of gross disruption of InhA folding, and in combination with binding and kinetic analyses suggested a mechanism by which the effects on enzyme activity are the result of decreased affinity of the phosphomimetic-substituted InhA for NADH (48).
In vivo, it was observed that expression of the phosphoablative form of InhA was well tolerated in M. smegmatis but that the phosphomimetic form led to severe growth inhibition. Similarly, inhA conditional expression strains were complemented by inhA expressing the native or phosphoablative proteins but not the phosphomimetic form. These data provide strong evidence for regulation of mycolic acid synthesis by Ser/Thr phosphorylation, though it remains uncertain which specific STPK(s) target InhA. Consistent with this observation, other FASII enzymes, including KasA, KasB, and MabA, have been shown to be phosphorylated in vitro and in mycobacteria with direct effects of phosphorylation on enzyme activity (49, 50); FabD and FabH have also been shown to be phosphorylated in vitro, though not yet in M. tuberculosis (49, 51).
PknD
PknD is encoded by a nonessential gene at a chromosomal locus encoding multiple genes involved in phosphate transport (11, 52). The KD of PknD is most similar to the KDs of PknH and PknE and appears to be activated by allosteric effects of dimerization of the KDs as described above (23). The PknD extracellular domain has been shown to form a highly symmetrical six-bladed β-propeller structure, with variation in the blades concentrated in the membrane-distal “cup” region that is the likely site of ligand binding (53).
Though the ligands of PknD and its function are not known, the linkage of pknD to phosphate transport genes suggests a role in phosphate uptake, and survival of a pknD deletion mutant has been shown to be compromised in a phosphate-deficient growth medium (54). PknD has also been shown to phosphorylate the amino-terminal extension of Rv0516c, a putative regulator of the sigma factor SigF (55), and recent data have linked Rv0516c, SigF, and PknD in an osmosensory signaling pathway (56).
Interestingly, pknD has been linked to central nervous system tuberculosis. A screen for genes required for central nervous system infection by M. tuberculosis identified a pknD deletion strain as defective for central nervous system invasion (57). PknD was shown to be required for invasion of brain endothelial cells in vitro, and the extracytoplasmic domain of the molecule was sufficient to stimulate invasion of these cells. The mechanism by which PknD may contribute to central nervous system invasion and the protein substrates of PknD that may contribute to this phenotype are not known.
PknE
Relatively little is known regarding the substrates or function of PknE. It appears to be a receptor-type kinase, with an extracellular domain, transmembrane domain, and intracellular KD. The extracytoplasmic domain of PknE has not been characterized and does not contain known protein motifs based on amino acid sequence. The gene encoding this STPK is adjacent to genes of unknown function and does not appear to be part of an operon. Interestingly, pknE is separated from pknF by just two genes, suggesting a possible functional link between these two STPKs. A possible link to regulation of lipid synthesis by the type II fatty acid synthase system has been suggested, based on functional effects of phosphorylation or substitutions of phosphoacceptors and in vitro phosphorylation of some of these enzymes by PknE, as well as other kinases (49). PknE has also been suggested to regulate apoptosis in M. tuberculosis– infected macrophages (58).
PknF
As noted above, pknF is separated from pknE by less than 2 kb in the M. tuberculosis chromosome. Immediately 3′ of pknF is Rv1747, which encodes an ABC transporter that contains two forkhead associated (FHA) domains in addition to the transmembrane transporter and ATP binding domains (52, 59). Several studies point to functional and physical interactions between PknF and Rv1747, in which autophosphorylated PknF recruits Rv1747 via its amino-terminal FHA domain, leading to phosphorylation of Rv1747 by PknF (60, 61). Though the transport role of Rv1747 and the effects of phosphorylation on transport were not determined, an Rv1747 deletion mutant was shown to be attenuated for growth in macrophages and in lungs and spleens of mice (61). In a study using pknF overexpression and antisense expressing strains, marked effects on growth, morphology and septum placement were observed, suggesting a role for pknF in regulating growth and cell shape (62).
More recently, it was shown that pknF and Rv1747 are cotranscribed in an operon and that PknF phosphorylates Rv1747 at two sites, Thr150 and Thr208, located between the two FHA domains of this protein (63). In addition to confirming that the Rv1747 deletion strain is attenuated in mice and macrophages, this study demonstrated that phosphoablative mutants of Rv1747 did not complement the virulence defect, indicating a role for phosphorylation in Rv1747 function. Supporting this interpretation, a construct expressing Rv1747 with a mutation in the first FHA domain that is required for phosphothreonine binding failed to complement the growth defect of the Rv1747 deletion strain in macrophages. Interestingly, however, a pknF deletion strain was not clearly defective for growth in macrophages. Given the requirement for phosphorylation in complementing the intracellular growth defects, this result suggests that other kinases may also phosphorylate Rv1747, consistent with the strong similarity between the phosphorylation motifs of PknF and several other STPKs (15, 63). Despite substantial molecular and pathogenic insights into the interactions of PknF and Rv1747, the transport activity of Rv1747 and the effect of phosphorylation on this activity remain unknown. In addition, the extracellular domain of PknF has not been characterized to date.
PknG
PknG is one of the two M. tuberculosis STPKs, PknK being the other, that lack a transmembrane region. Thus, PknG does not have the structure of a typical receptor type kinase that functions in transmembrane signal transduction (Fig. 7B). In addition to its KD, which does not cluster with any of the other M. tuberculosis STPKs based on amino acid sequence (Fig. 2 and 3), PknG has amino and carboxy-terminal sequences that are important for its function. The amino-terminal region contains a rubredoxin redox-sensing domain, and the carboxy-terminus contains a tetratricopeptide (TPR) repeat domain, which may function in protein-protein interactions and regulation of kinase activity (30, 31, 64). As described in detail above, the PknG KD has the characteristic two-lobed structure of Ser/Thr protein kinases, though with differences from the PknB structure including absence of phosphorylation of the activation loop, absence of the highly conserved Arg preceding the catalytic Asp in the catalytic loop, and unusual residues in the ATP-binding pocket (29). Mutagenesis and activity studies based on this structure indicate that the amino-terminal rubredoxin motif likely regulates PknG activity. Deletion of the rubredoxin domain resulted in decreased kinase activity and substitution of Cys residues in the iron binding motif caused reduction or complete elimination of kinase activity, depending on the nature of the substitution (30, 31). PknG is encoded by a gene that is linked in an operon to glnH, a gluta-mine-binding lipoprotein, and a conserved membrane protein of unknown function (52).
An early publication, using M. bovis BCG and M. smegmatis, suggested that PknG was a virulence factor that was secreted into the macrophage phagosome and functioned by modulating host signaling to prevent phagosome-lysosome fusion (65). In the first publication that analyzed a pknG deletion mutant, Cowley et al. found that the ΔpknG strain grew poorly in liquid and on solid media, with decreased growth compared to wild type that was most notable after mid-log phase (66). These investigators also demonstrated that the ΔpknG strain was attenuated in SCID mice and had elevated intracellular levels of glutamate plus glutamine, associated with decreased de novo glutamine synthase.
Further insight regarding the functions of PknG emerged with the identification of its primary substrate, the FHA domain-containing protein GarA (Rv1827), and the role of GarA in regulating central metabolic pathways (67). O'Hare and colleagues first demonstrated three phosphorylation sites in the amino-terminal region of PknG (19). In contrast to other M. tuberculosis STPKs, there was no evidence of phosphorylation of the activation or catalytic loops of the KD, and autophosphorylation was not required for kinase activity of PknG. Building on results obtained in C. glutamicum (68), a related actinomycete, these authors identified GarA as a substrate of PknG that is phosphorylated at Thr22, adjacent to the Thr21 residue of this protein that is phosphorylated by PknB.
In this and subsequent work by Nott et al., an elegant intramolecular phospho-switch mechanism was elucidated as the mechanism by which the phosphorylation state of GarA regulates central carbon metabolism (19, 69). In this research, it was shown that unphosphorylated GarA binds to three enzymes: α-ketoglutarate dehydrogenase (KGD, Rv1248c), NAD-dependent glutamate dehydrogenase (GDH, Rv2476c), and the α-subunit of the glutamate synthase complex (GltB, Rv3859c). In each case, binding regulates their enzymatic activity. Phosphorylation of GarA by either PknG (shown for KGD and GDH) (19) or PknB (shown for KGD, GDH, and GltB) (69) abolished the binding of GarA to these enzymes. Though the amino-terminal region containing the Thr21 or Thr22 phosphoacceptors was not required for binding of GarA to the enzyme ligands, these residues were essential for the phosphor-ylation-mediated abrogation of binding. Structural and mutagenesis analysis demonstrated that the FHA domain of GarA was required for binding of each of the enzyme ligands. Further, phosphorylation of GarA on Thr21 resulted in binding of the amino-terminal region of GarA to the FHA domain, blocking interaction of the FHA domain with the enzymes and thereby increasing their activity in the cell. Unlike most other M. tubercu losis STPKs, PknG has limited in vitro kinase activity toward a wide range of peptide and protein substrates, suggesting that this regulation of metabolic activity by phosphorylation of GarA may be its primary role and/or that additional regulatory inputs are required for its activity.
PknH
PknH is encoded 3′ of the gene encoding the transcriptional regulator EmbR. Sequence analysis indicates that PknH is a typical receptor-type kinase with an intracellular KD and an extracellular receptor domain (52). The extracytoplasmic domain contains no previously described motifs, and its ligand(s) are not known. A recent structural analysis, however, identified unusual features of the structure of this region. The crystal structure revealed the presence of two disulfide bonds that contribute rigidity to the structure and the presence of a deep cleft, the likely site of ligand binding, lined by a mix of hydrophobic and polar residues (70). The presumed ligand binding cleft is highly conserved among PknH orthologues and bears some similarity to binding sites of lipoproteins LprG and LppX. However, the greater polarity of the PknH cleft was thought to make binding of hydrophobic glycolipids unlikely.
The physical proximity between the genes encoding PknH and the transcriptional regulator EmbR suggests a functional interaction between these proteins. The presence of an FHA domain in EmbR, in addition to winged helix-turn-helix and bacterial activation domains that are characteristic of this family of transcription factors led to investigation of the role of phosphorylation, and PknH specifically, in regulating EmbR activity. In an initial study, Molle and colleagues demonstrated Thr phosphorylation of EmbR by PknH in vitro (71). Using substitutions of conserved residues in the EmbR FHA domain, these authors also demonstrated that this domain is essential for EmbR phosphorylation. Singh and colleagues subsequently identified the embCAB operon, which encodes proteins required for arabinosylation of lipoarabinomannan (embC) and arabinogalactan (embA and embB) (72, 73), as a target of EmbR, with apparent binding sites 5′ of embC, embA, and embB (74). EmbR phosphorylation by PknH enhanced its binding to each of these regions. Expression of M. tuberculosis pknH in M. smegmatis resulted in increased phosphor-ylation of M. smegmatis EmbR. In a strain expressing native PknH, but not a kinase inactive form, semiquantitative reverse transcription PCR (RT-PCR) showed increased transcription of embC, embA, and embB. Consistent with increased expression of these enzymes, the ratio of lipoarabinomannan to lipomannan was increased in the pknH overexpression strain. Though expression of M. tuberculosis PknH in M. smegmatis, and the absence of evidence that the same sites on EmbR are phosphorylated in M. tuberculosis and M. smegmatis, complicates interpretation of these data, taken together they do suggest that phosphorylation of EmbR by PknH positively regulates expression of the embA, embB, and embC genes. Decreased expression of embC and embB in a pknH deletion mutant (ΔpknH) further supports this regulatory mechanism (75).
In addition to its regulation of cell envelope glycolipids, proteomic and lipid analysis of a ΔpknH strain compared to wild type suggests a possible role for PknH in regulating lipids in the M. tuberculosis cell envelope (76). Specific phthiocerol dimycoserosates (PDIMs) were found to be decreased in the ΔpknH strain. Surprisingly, enzymes involved in PDIM synthesis were expressed at similar or higher levels in the mutant strain. In contrast to M. smegmatis, where overexpression of PknH led to higher lipoarabinomannan:lipomannan ratios, in this study higher lipoarabinomannan:lipomannan ratios were observed in the M. tuberculosis pknH deletion strain. The mechanism by which PDIM production is affected by pknH deletion, and how lower levels of a virulence-associated surface lipid would lead to hyper-virulence, remain unknown.
The roles of pknH in response to stress and during infection are complex. The ΔpknH strain was found to be more susceptible to peroxide and superoxide (paraquat) stresses but more resistant to nitric oxide (acidified nitrite) stresses (75). The mutant showed decreased survival in THP-1 macrophages but, surprisingly, was hypervirulent in mice, with greater replication in the ΔpknH strain starting at 4 weeks post-infection, reaching 1 to 2 log greater bacterial burden of the deletion strain in both lungs and spleen of Balb/C mice after 4 months of infection.
Further insight into PknH signaling emerged from a proteomic study comparing protein levels in wild type and the ΔpknH strain, untreated or after 48 hours of acidified nitrite treatment (77). The striking finding of this study was the identification of several DosR regulon proteins that were increased in response to acidified nitrite treatment in the pknH mutant. Following this lead, Chao et al. (77) then demonstrated that PknH can phosphorylate DosR in vitro on two Thr residues and that this phosphorylation enhances the binding of DosR to DNA containing a DosR recognition sequence. Subsequent analysis of transcription of DosR regulon genes in response to hypoxia and nitric oxide stress indicated decreased induction of these genes in the ΔpknH strain, though this effect was modest. These data suggest a role for PknH phosphorylation as a mechanism for modulating the M. tuberculosis dormancy response, though in vivo confirmation of DosR phosphorylation and the mechanism by which this affects DosR activity remain to be shown.
PknI
Relatively little is known regarding the substrates or function of PknI. This STPK has the domain organization of a receptor-type kinase, with an extracellular domain, transmembrane domain, and intracellular KD. The extracytoplasmic domain of PknI has not been characterized and is not similar to known protein domains based on amino acid sequence. Immediately 3′ of the gene encoding PknI (Rv2914c) is a gene that encodes a predicted D-amino acid aminohydrolase (Rv2913c) followed by a transcriptional regulator (Rv2912c) and dacB2 (Rv2911), predicted to encode a D-alanyl-D-alanine hydrolase, raising the possibility of a role for PknI in regulating cell wall turnover. Immediately 5′ of pknI is a gene of unknown function (Rv2915c) and a gene for a predicted signal recognition particle (SRP) protein (52). No well-documented substrates of PknI have been identified, but a pknI deletion strain was found to be hypervirulent for replication in macrophages and for morbidity in SCID mice (78).
PknJ
Like PknI, relatively little is known regarding the substrates or function of PknJ. Jang et al. performed an initial characterization of this protein (79). They demonstrated that the KD can dimerize and that it auto-phosphorylates on three Thr residues in the activation loop (79), consistent with the mode of activation of the other M. tuberculosis STPKs that have been studied. PknJ is a receptor-type kinase and was shown to have a single transmembrane region, an intracellular KD, and an extracellular domain (79). The extracellular domain has not been characterized and has no recognizable motifs. Several M. tuberculosis proteins were shown to be phosphorylated by PknJ in vitro, but in vivo phosphorylation was not demonstrated (79). The several genes 5′ of pknJ encode conserved hypothetical proteins; a gene encoding a predicted dipeptidase is located immediately 3′ of pknJ in the opposite orientation. Thus, the chromosomal locus provides few clues as to a likely function of PknJ. A pknJ transposon insertion mutant did not have a growth phenotype in macrophages or in BALB/c mice (79).
PknK
PknK is one of two M. tuberculosis STPKs that lack a transmembrane segment, the other being PknG, and is therefore predicted to be a cytoplasmic protein. PknK is a large (119 kDa) protein with the KD in the amino-terminal region and a long carboxy-terminal region that shows similarity to the MalT family of ATP-dependent transcription regulators. Within this region are a P-loop ATP binding motif characteristic of AAA+ ATPases, a PDZ domain, and a single tetratricopeptide (TPR) repeat sequence (64). PDZ domains and TPR domains are often involved in protein-protein interactions. TPR sequences typically occur in clusters of three or more repeats and mediate protein-protein interactions (80). The function and interaction partners of the PDZ domain and the TPR motif in PknK are not known.
Kumar et al. demonstrated that, like most other M. tuberculosis STPKs, PknK autophosphorylates on two Thr residues in a TXT motif in the activation loop, and this autophosphorylation is required for kinase activity (81). In addition, they provided evidence for phosphorylation of the carboxy-terminal region of the protein and suggest a positive regulatory role for this region. Despite its lack of a predicted transmembrane domain, probing of subcellular fractions of M. tuberculosis lysates showed PknK to be present in the cell wall/membrane fraction rather than in the cytosol. These investigators demonstrated the ability of PknK to phosphorylate in vitro VirS, a transcription factor encoded by a gene that is separated from pknK by a single gene in the M. tuberculosis chromosome. PknK also phosphorylated several proteins encoded in the mym operon that is regulated by VirS and that is adjacent to virS. Phosphorylation of VirS increased binding to the intergenic region that likely contains the virS and mym promoters, and increased transcriptional activity from a mym promoter-luciferase transcriptional fusion construct when active PknK and VirS were coexpressed in M. smegmatis. Though these data suggest a role for PknK-mediated phosphorylation in regulating VirS activity, whether VirS is phosphorylated in M. tuberculosis and whether PknK might be a cognate kinase for VirS were not shown.
Malhotra et al. have investigated the expression of pknK and phenotypes of a pknK deletion strain (ΔpknK) (82). Expression of pknK was found to be greater in virulent M. tuberculosis H37Rv compared to avirulent H37Ra, and PknK protein was shown to be much more abundant in stationary versus log phase cells grown in broth culture in vitro. The ΔpknK strain was found to grow to higher levels during stationary phase, and the cells of the mutant strain were shorter than wild type. The ΔpknK strain was slightly more resistant to acidic, oxidative, and hypoxic stress, and in a low-dose aerosol infection of C57BL/6 the mutant strain showed a transient early growth defect. In a follow-up report the same group showed striking effects on the expression of several tRNAs in the ΔpknK strain compared to wild type, with a large subset of tRNAs expressed at lower levels in the mutant in logarithmic growth and at higher levels during stationary phase (83). These phenotypes and expression data suggest a role for PknK in regulating translation in relation to growth and environmental conditions. These investigators also demonstrated dimerization of the PknK KD and, based on indirect evidence of effects on kinase activity, proposed a model in which the carboxy terminal domain negatively regulates the activity of the amino-terminal KD through steric hindrance.
PknL
PknL is encoded by a gene adjacent to a transposase and a putative DNA binding protein (52). Additional genes in the region encode several conserved hypothetical proteins and biosynthetic enzymes for aromatic amino acids, mannan, and lipid biosynthesis. Located further 5′ is a division and cell wall cluster (dcw) that includes several genes that encode enzymes for PGN precursor synthesis, a penicillin binding protein, and several cell division proteins, suggesting the possibility that PknL regulates cell wall synthesis. Though the M. tuberculosis PknL lacks an extracellular domain, the PknL orthologue in C. glutamicum has an extracellular domain comprised of three PASTA domains, further supporting a role for this STPK in regulating the mycobacterial cell wall (84).
There has been relatively little investigation of PknL in mycobacteria. The intracellular domain has been shown to autophosphorylate on two Thr residues in the activation loop, similar to the phosphorylation required for activation of PknB and several other M. tuberculosis STPKs (16, 85). Rv2175c, a putative transcription factor encoded by the gene adjacent to pknL in M. tuberculosis, was shown to be phosphorylated in vitro by recombinant PknL. Overexpression of Rv2175c in M. smegmatis allowed detection of three isoforms of the protein with different pIs, consistent with one and two phosphorylations, but direct evidence of Rv2175c phosphorylation in M. tuberculosis, and whether PknL is the cognate kinase, has not been shown.
TYROSINE PHOSPHORYLATION IN M. TUBERCULOSIS
No tyrosine kinases were annotated in the M. tuberculosis H37Rv genome sequence or in other sequenced strains. Speculating that a Tyr kinase might be linked to a Tyr phosphatase, Bach et al. examined Rv2232, a protein of unknown function encoded by the gene adjacent to the gene encoding the Tyr phosphatase PtpA, for kinase activity. These investigators determined that recombinant Rv2232, which they renamed PtkA, could autophosphorylate and that the site of phosphorylation was a Tyr residue (35). They further demonstrated that PtkA interacted with PtpA and phosphorylated PtpA in vitro on two Tyr residues. Surprisingly, PtpA did not have phosphatase activity toward PtkA. PtkA has no characteristic Tyr kinase motifs and thus would be an atypical Tyr kinase, whose role in M. tuberculosis remains to be defined.
M. TUBERCULOSIS PROTEIN PHOSPHATASES
Phosphorylation of Ser, Thr, or Tyr is a relatively stable modification, so that reversal of phosphorylation requires the enzymatic activity of a phosphatase that can bind to and act on specific phosphorylated residues on proteins. In most organisms, the number of protein kinases substantially exceeds the number of protein phosphatases. In M. tuberculosis there is one annotated phospho-Ser/phospho-Thr phosphatase, PstP, encoded by the first gene in the operon that includes pknA and pknB. There are two annotated Tyr phosphatases, the low-molecular-weight phosphatase PtpA and the high-molecular-weight phosphatase PtpB.
PstP is a member of the PPM family of Ser/Thr phosphatases, which utilize Mg2+ or Mn2+ to catalyze dephosphorylation (86). PstP was initially shown to dephosphorylate Thr residues that had been phosphor-ylated by PknB (16) and has since been shown to have broad activity in dephosphorylating Ser and Thr residues targeted by several M. tuberculosis kinases. The X-ray crystal structure of PstP was determined and found to be highly similar to the structure of PP2Cα, the prototype of the PPM phosphatases, with additional features of interest (87). In addition to the two Mn2+ ions present in PP2Cα, a third Mn2+ was found to be present in the PstP active site, with positioning of a large flap domain to bring it in contact with this metal ion. Given the broad reactivity of PstP, how dephosphorylation by this enzyme may contribute to regulating specific Ser/Thr signaling pathways is not clear and remains to be elucidated.
The two Tyr phosphatases have attracted a great deal of interest based on the hypothesis that one or both may target host phosphotyrosine signaling pathways. In an early paper, Koul et al. demonstrated that these enzymes specifically target phospho-Tyr and not phospho-Ser or phospho-Thr (88). Examination of subcellular fractions by this group indicated that these proteins were present in culture supernatants, suggesting that they are secreted proteins. These proteins lack SecI or TAT secretion signals, however, and the mechanism by which they are secreted is not known.
Both ptpA and ptpB are widely distributed among pathogenic and nonpathogenic mycobacteria. Using a combination of bioinformatic and experimental approaches, Beresford et al. identified features of the conserved active site motif of PtpB, suggesting that it may be a dual-specificity phosphatase (89). While confirming greater activity against phospho-Tyr-containing substrates, these investigators demonstrated phosphatase activity against phospho-Ser and phospho-Thr residues as well. Importantly, they also demonstrated potent phosphatase activity against phosphoinositides, demonstrating that PtpB may be a triple specificity phosphatase that could target multiple host signaling pathways to alter macrophage activity against M. tuberculosis.
Crystal and solution structures of PtpA have been determined and demonstrate that PtpA has a structure and active site characteristic of eukaryotic low-molecular-protein phosphatases despite limited sequence similarity (90, 91). Specific differences in residues of the substrate binding site suggested differences in specificity. The d-loop in the apo form of the protein was found to be in a more open orientation compared to ligand bound PtpA, and phosphorylation of two adjacent Tyr residues that are likely to play regulatory roles was demonstrated.
In the case of PtpB, crystal structures demonstrated preservation of the typical phosphatase fold plus two unique features: a disordered loop that may play a role in substrate binding or regulation and a mobile lid over the active site that may serve to protect this site from oxidative damage encountered in the phagosome of macrophages or neutrophils that have ingested M. tuberculosis (92, 93).
Supporting the hypothesis that these Tyr phosphatases act on host Tyr signaling pathways, PtpA has been implicated in modulating phagosome acidification. In an initial report Bach et al. demonstrated that a ptpA deletion mutant was defective for survival in macrophages (94). Using a substrate trap form of PtpA, vacuolar protein sorting 33B (VPS33B) was identified as a candidate substrate of PtpA. This protein was found to autophosphorylate on Tyr residues and to be bound and dephosphorylated by PtpA. PtpA and VPS33B were found to colocalize in M. tuberculosis macrophages. In a subsequent study from the same group, Wong et al. demonstrated that PtpA binds to subunit H of the host vacuolar ATPase (V-ATPase) in the phagosome membrane and that the macrophage class C sorting complex associates with the V-ATPase during phagosome maturation (95). These interactions are required for the dephosphorylation of VPS33B and result in exclusion of V-ATPase from the M. tuberculosis–containing phagosomes, thereby preventing acidification.
PtpB has also been implicated in modulating host responses, apparently by targeting specific Tyr signaling pathways to alter host immune function (96). Zhou et al. demonstrated decreased interferon-gamma-stimulated production of IL-6 from a macrophage cell line stably transfected with ptpB compared to nontransfected cells and demonstrated decreased phosphorylation of Erk2 and p38 in the MptB-expressing cells, but not in cells expressing an inactive form of PtpB. These investigators also noted decreased apoptosis in PtpB-expressing cells and an associated decrease in Caspase 3 activity. Though these data are indirect, i.e., experiments were not performed using cells infected with M. tuberculosis, they suggest a mechanism that may explain the decreased viability of ptpB deletion strains in activated macrophages and guinea pigs (97).
A lipid phosphatase of M. tuberculosis, SapM, has also been identified and shown to be a secreted protein (98). In macrophages this protein appears to target phosphatidyl inositol-3-phosphate in a manner that prevents phagosome maturation (99).
M. TUBERCULOSIS STPKS AND PHOSPHATASES AS POTENTIAL DRUG TARGETS
Inhibition of protein kinases and phosphatases has been one the most active areas of current pharmaceutical research and drug development to target human diseases. The critical role of protein phosphorylation in controlling key physiologies that are dysregulated in diseases such as cancer and autoimmune diseases, the extensive detailed structural information on proteins and complexes involved in signal transduction, and the ability to develop potent inhibitors of these enzymes has led to the development of several approved drugs targeting protein phosphorylation, with many more in development (100). The importance of protein phosphorylation in regulating key processes for M. tuberculosis viability and virulence, together with substantial structural information on M. tuberculosis kinases and phosphatases and the extensive expertise in design and development of small molecule inhibitors of these enzymes, make them highly attractive targets for the development of new antituberculars.
The wealth of experience in targeting human Ser/Thr and Tyr kinases provides a huge advantage in developing inhibitors of M. tuberculosis kinases and phosphatases. The similarity of the mycobacterial proteins to their eukaryotic counterparts, however, has led to concern that inhibitors of M. tuberculosis enzymes will not be sufficiently selective relative to human kinases and phosphatases to avoid important toxicities. While this is a valid concern, the success in developing specific inhibitors of individual human kinases suggests that this challenge can be overcome, even with ATP-competitive inhibitors that target the conserved ATP-binding site of kinases. Both type I inhibitors that bind to the active conformation through interactions with the purine binding site and adjacent hydrophobic pockets and type 2 inhibitors that bind to the inactive conformation, in which an additional binding site is exposed, have been developed as FDA-approved selective inhibitors of human kinases (100). Other classes of kinase inhibitors, e.g., allosteric inhibitors that bind at sites separate from the ATP binding, are of great interest in that they may target unique features of a kinase and offer the potential for very high selectivity.
While the conservation of the KDs of M. tuberculosis kinases offers advantages and disadvantages for the inhibitor identification and candidate drug development described above, other regions of the M. tuberculosis kinases may also serve as targets for inhibitor development. The extracellular domains of the receptor-type M. tuberculosis kinases, and the regulatory and protein interaction domains of PknG and PknK, may provide targets that are unrelated to human proteins and thus allow development of highly selective inhibitors of M. tuberculosis kinases.
A number of groups have undertaken screens to identify inhibitors of M. tuberculosis kinases, with most efforts focusing on the essential kinases PknA and PknB and on PknG, which is required for virulence and regulates central metabolism (29, 65, 101–103). A number of M. tuberculosis kinase inhibitors have advanced through medicinal chemistry efforts to optimize their activity. In some cases enzyme inhibitory activity in the low nanomolar range has been achieved. Some compounds also show substantial antibacterial activity, with MICs in the low micromolar range (102–104), but none has achieved potency sufficient to lead to evaluation in clinical trials.
The protein phosphatases also present important opportunities for antitubercular drug development (105). Phosphatases have advantages and challenges as targets for drug development that are similar to those of the M. tuberculosis kinases; i.e., the presence of similar enzymes in humans provides a wealth of knowledge but raises concerns regarding selectivity and potential for toxicity (106). The smaller number of protein phosphatases relative to kinases suggests that these phosphatases will be less specific than protein kinases, and lack of selectivity has been a challenge in developing phosphatase inhibitors to treat human diseases. In addition, the natural substrate includes the charged phosphate residue; active site inhibitors that share this charge are unlikely to enter the host cell, a problem that has slowed phosphatase inhibitor development for human diseases and will be a challenge in targeting the secreted protein phosphatases of M. tuberculosis. Potentially offsetting these challenges, the fact that PtpA and PtpB are both secreted proteins means that a major impediment to developing antituberculars that target proteins in the bacterial cytoplasm, passage across the relatively impermeable mycobacterial cell envelope, does not have to be overcome. The PtpA structure is highly similar to that of human protein phosphatases, so that development of highly selective inhibitors may be difficult. In contrast, though PtpB retains the phosphatase active site signature sequence, it is otherwise highly divergent from known human phosphatases and thus may be especially attractive as a target for inhibitors. Further, as described above, both PtpA and PtpB have unique structural features that may allow more specific targeting of these proteins.
Several groups have identified compounds that inhibit PtpA. Manger et al. identified two classes of natural compounds with IC50s in the low micromolar range and also screened a fragment-based library to identify synthetic molecules that inhibit PtpA activity, with the most potent having Kis of 1 to 2 μM (107). The compounds that were tested, however, also had significant activity against one or more human phosphatases. Chiradia et al. identified synthetic chalcones as inhi bitors of PtpA, and in follow-up work elucidated structure-activity relationships and demonstrated inhibition by one compound of M. bovis BCG in human macrophages but not in axenic culture (108–110). A group that had previously identified PtpB inhibitors using a fragment-based approach applied this method to identify inhibitors of PtpA (111). The most potent of these (Ki = 1.4 μM) was also selective relative to human Tyr and dual-specificity kinases.
In the case of PtpB, a number of groups have identified potent inhibitory compounds. Grundner et al., using scaffolds from which inhibitors of eukaryotic Tyr phosphatases had been developed, identified a competitive inhibitor of PtpB with IC50 of 0.44 μM, with good selectivity versus several human tyrosine phosphatases (112). Crystal structure analysis of this compound in complex with PtpB demonstrated conformational changes and key residues that may guide further inhibitor development. Soellner et al. subsequently used a fragment-based approach to identify a more potent inhibitor that maintained good selectivity (113). Beresford et al. identified inhibitors with IC50s in the low micromolar range with good selectivity (114). One of these compounds was shown to inhibit growth of M. bovis BCG in resting murine macrophages, but not in axenic culture, consistent with an effect on macrophage function rather than a direct antibacterial effect.
SUMMARY AND FUTURE RESEARCH
Since the identification of 11 STPKs in the M. tuberculosis genome sequence in 1998 (115), substantial progress has been made in gaining insight into the role of Ser/Thr phosphorylation in regulating M. tuberculosis physiology. The effect of phosphorylation on protein function has been elucidated for several M. tuberculosis proteins, a number of which have been highlighted in this article. In particular, important insights have been gained into the roles of protein phosphorylation in regulating the synthesis of several components of the mycobacterial cell envelope, including PGN, glycans, and lipids. To a lesser extent, knowledge has been acquired regarding the cognate STPKs that target specific enzymes to control these and other cell functions. Structural analyses have also provided key insights into STPK function, first demonstrating the structural and mechanistic similarity between the mycobacterial and eukaryotic protein kinases, and then providing important and novel insights regarding activation and regulation of M. tuberculosis kinases and phosphatases, as well as potential ways to target these molecules with inhibitors.
Despite these advances, in many ways we have barely scratched the surface of the regulatory roles of protein phosphorylation in M. tuberculosis. Going forward, we expect that many new phosphoproteins will be identi-fied, and the effects of phosphorylation on protein function will be discerned for many of these. Identification of cognate kinases for individual phosphoproteins, though challenging, is critical for deepening our understanding of the regulatory roles of Ser/Thr phosphorylation in M. tuberculosis. Identifying ligands for the receptor-type STPKs and how ligand binding affects kinase function will provide key new information on how these signaling molecules sense the external environment. Ultimately, integration of these data to construct functional signal transduction networks will provide deep insights into the mechanisms by which M. tuberculosis adapts during the course of infection of the mammalian host. Finally, we hope to see these insights translated into novel and potent antituberculars to improve the treatment of drug-susceptible and drug-resistant tuberculosis.
ACKNOWLEDGMENTS
The authors thank Benoit Smagghe for help with the protein structure figures. The authors’ work on the M. tuberculosis kinases has been supported by grants from the National Institute of Allergy and Infectious Diseases and from Vertex Pharmaceuticals Incorporated.
REFERENCES
- 1.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staroń A, Sofia H, Dietrich S, Ulrich L, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 4.Lonetto M, Brown K, Rudd K, Buttner M. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song T, Dove SL, Lee KH, Husson RN. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol Microbiol. 2003;50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 6.Hahn MY, Raman S, Anaya M, Husson RN. The Mycobacterium tuberculosis ECF sigma factor SigL regulates polyketide synthases and membrane/secreted proteins, and is required for virulence. J Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez J, Castaneda-Garcia A, Jenke-Kodama H, Muller R, Munoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci USA. 2008;105:15950–15955. doi: 10.1073/pnas.0806851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munoz-Dorado J, Inouye S, Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell. 1991;67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 9.Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 2000;8:238–244. doi: 10.1016/s0966-842x(00)01734-0. [DOI] [PubMed] [Google Scholar]
- 10.Hanks S, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 11.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 12.Ortiz-Lombardia M, Pompeo F, Boitel B, Alzari PM. Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:13094–13100. doi: 10.1074/jbc.M300660200. [DOI] [PubMed] [Google Scholar]
- 13.Young TA, Delagoutte B, Endrizzi JA, Falick AM, Alber T. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Biol. 2003;10:168–174. doi: 10.1038/nsb897. [DOI] [PubMed] [Google Scholar]
- 14.Gay LM, Ng HL, Alber T. A conserved dimer and global conformational changes in the structure of apo-PknE Ser/Thr protein kinase from Mycobacterium tuberculosis. J Mol Biol. 2006;360:409–420. doi: 10.1016/j.jmb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang CM, Bemis G, Church GM, Steen H, Husson RN. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci USA. 2010;107:7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boitel B, Ortiz-Lombardia M, Duran R, Pompeo F, Cole ST, Cervenansky C, Alzari PM. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol Microbiol. 2003;49:1493–1508. doi: 10.1046/j.1365-2958.2003.03657.x. [DOI] [PubMed] [Google Scholar]
- 17.Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molle V, Zanella-Cleon I, Robin JP, Mallejac S, Cozzone AJ, Becchi M. Characterization of the phosphorylation sites of Mycobacterium tuberculosis serine/threonine protein kinases, PknA, PknD, PknE, and PknH by mass spectrometry. Proteomics. 2006;6:3754–3766. doi: 10.1002/pmic.200500900. [DOI] [PubMed] [Google Scholar]
- 19.O'Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, Pritsch O, Obal G, Baumgartner J, Vialaret J, Johnsson K, Alzari PM. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70:1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 20.Duran R, Villarino A, Bellinzoni M, Wehenkel A, Fernandez P, Boitel B, Cole ST, Alzari PM, Cervenansky C. Conserved autophosphorylation pattern in activation loops and juxtamembrane regions of Mycobacterium tuberculosis Ser/Thr protein kinases. Biochem Biophys Res Comm. 2005;333:858–867. doi: 10.1016/j.bbrc.2005.05.173. [DOI] [PubMed] [Google Scholar]
- 21.Mieczkowski C, Iavarone AT, Alber T. Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. Embo J. 2008;27:3186–3197. doi: 10.1038/emboj.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lombana TN, Echols N, Good MC, Thomsen ND, Ng H-L, Greenstein AE, Falick AM, King DS, Alber T. Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB. Structure. 2010;18:1667–1677. doi: 10.1016/j.str.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenstein AE, Echols N, Lombana TN, King DS, Alber T. Allosteric activation by dimerization of the PknD receptor Ser/Thr protein kinase from Mycobacterium tuberculosis. J Biol Chem. 2007;282:11427–11435. doi: 10.1074/jbc.M610193200. [DOI] [PubMed] [Google Scholar]
- 24.Wehenkel A, Bellinzoni M, Graña M, Duran R. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim Biophys Acta. 2008;1784:193–202. doi: 10.1016/j.bbapap.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Wehenkel A, Fernandez P, Bellinzoni M, Catherinot V, Barilone N, Labesse G, Jackson M, Alzari PM. The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett. 2006;580:3018–3022. doi: 10.1016/j.febslet.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 26.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 27.Mir M, Asong J, Li X, Cardot J, Boons GJ, Husson RN. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barthe P, Mukamolova GV, Roumestand C, Cohen-Gonsaud M. The structure of PknB extracellular PASTA domain from Mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure. 2010;18:606–615. doi: 10.1016/j.str.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Scherr N, Honnappa S, Kunz G, Mueller P, Jayachandran R, Winkler F, Pieters J, Steinmetz MO. Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2007;104:12151–12156. doi: 10.1073/pnas.0702842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiwari D, Singh RK, Goswami K, Verma SK, Prakash B, Nandicoori VK. Key residues in Mycobacterium tuberculosis protein kinase G play a role in regulating kinase activity and survival in the host. Journal of Biological Chemistry. 2009;284:27467–27479. doi: 10.1074/jbc.M109.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gil M, Graña M, Schopfer FJ, Wagner T, Denicola A, Freeman BA, Alzari PM, Batthyány C, Duran R. Inhibition of Mycobacterium tuberculosis PknG by non-catalytic rubredoxin domain specific modification: reaction of an electrophilic nitro-fatty acid with the Fe-S center. Free Radic. Biol. Med. 2013;65:150–161. doi: 10.1016/j.freeradbiomed.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braconi Quintaje S, Orchard S. The annotation of both human and mouse kinomes in UniProtKB/Swiss-Prot: one small step in manual annotation, one giant leap for full comprehension of genomes. Mol Cell Proteomics. 2008;7:1409–1419. doi: 10.1074/mcp.R700001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 34.Kinexus Phosphonet: Human Phospho-Site KnowledgeBase. 2013 Mar 1; 2013, posting date. [Online.] [Google Scholar]
- 35.Bach H, Wong D, Av-Gay Y. Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem J. 2009;420:155–160. doi: 10.1042/BJ20090478. [DOI] [PubMed] [Google Scholar]
- 36.Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152:493–504. doi: 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- 37.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei JR, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortega C, Liao R, Anderson LN, Rustad T, Ollodart AR, Wright AT, Sherman DR, Grundner C. Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch. PLoS Biol. 2014;12:e1001746. doi: 10.1371/journal.pbio.1001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeats C, Finn RD, Bateman A. The PASTA domain: a beta-lactam-binding domain. Trends Biochem Sci. 2002;27:438–440. doi: 10.1016/s0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 40.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Squeglia F, Marchetti R, Ruggiero A, Lanzetta R, Marasco D, Dworkin J, Petoukhov M, Molinaro A, Berisio R, Silipo A. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J Am Chem Soc. 2011;133:20676–20679. doi: 10.1021/ja208080r. [DOI] [PubMed] [Google Scholar]
- 42.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 43.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen MH, Vilchèze C, Kremer L, Besra GS, Parsons L, Salfinger M, Heifets L, Hazbon MH, Alland D, Sacchettini JC, Jacobs WR. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol Microbiol. 2002;46:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 46.Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC, Jacobs WR. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med. 2006;12:1027–1029. doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 47.Khan S, Nagarajan SN, Parikh A, Samantaray S, Singh A, Kumar D, Roy RP, Bhatt A, Nandicoori VK. Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J Biol Chem. 2010;285:37860–37871. doi: 10.1074/jbc.M110.143131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molle V, Gulten G, Vilchèze C, Veyron-Churlet R, Zanella-Cleon I, Sacchettini JC, Jacobs WR, Kremer L. Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol Microbiol. 2010;78:1591–1605. doi: 10.1111/j.1365-2958.2010.07446.x. [DOI] [PubMed] [Google Scholar]
- 49.Molle V, Brown AK, Besra GS, Cozzone AJ, Kremer L. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J Biol Chem. 2006;281:30094–30103. doi: 10.1074/jbc.M601691200. [DOI] [PubMed] [Google Scholar]
- 50.Veyron-Churlet R, Zanella-Cleon I, Cohen-Gonsaud M, Molle V, Kremer L. Phosphorylation of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J Biol Chem. 2010;285:12714–12725. doi: 10.1074/jbc.M110.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veyron-Churlet R, Molle V, Taylor RC, Brown AK, Besra GS, Zanella-Cleon I, Fütterer K, Kremer L. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J Biol Chem. 2009;284:6414–6424. doi: 10.1074/jbc.M806537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 53.Good MC, Greenstein AE, Young TA, Ng HL, Alber T. Sensor domain of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknD, forms a highly symmetric beta propeller. J Mol Biol. 2004;339:459–469. doi: 10.1016/j.jmb.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 54.Vanzembergh F, Peirs P, Lefèvre P, Celio N, Mathys V, Content J, Kalai M. Effect of PstS sub-units or PknD deficiency on the survival of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2010;90:338–345. doi: 10.1016/j.tube.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Greenstein AE, Macgurn JA, Baer CE, Falick AM, Cox JS, Alber T. M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog. 2007;3:e49. doi: 10.1371/journal.ppat.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatzios SK, Baer CE, Rustad TR, Siegrist MS, Pang JM, Ortega C, Alber T, Grundner C, Sherman DR, Bertozzi CR. Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proceedings of the National Academy of Sciences. 2013;110:E5069–5077. doi: 10.1073/pnas.1321205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Be NA, Bishai WR, Jain SK. Role of Mycobacterium tuberculosis pknD in the pathogenesis of central nervous system tuberculosis. BMC Microbiol. 2012;12:7. doi: 10.1186/1471-2180-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar D, Narayanan S. PknE, a serine/threonine kinase of Mycobacterium tuberculosis modulates multiple apoptotic paradigms. INFECTION, GENETICS AND EVOLUTION. 2011:1–11. doi: 10.1016/j.meegid.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 60.Molle V, Soulat D, Jault JM, Grangeasse C, Cozzone AJ, Prost JF. Two FHA domains on an ABC transporter, Rv1747, mediate its phosphorylation by PknF, a Ser/Thr protein kinase from Mycobacterium tuberculosis. FEMS Microbiol Lett. 2004;234:215–223. doi: 10.1016/j.femsle.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 61.Curry JM, Whalan R, Hunt DM, Gohil K, Strom M, Rickman L, Colston MJ, Smerdon SJ, Buxton RS. An ABC transporter containing a forkhead-associated domain interacts with a serine-threonine protein kinase and is required for growth of Mycobacterium tuberculosis in mice. Infect Immun. 2005;73:4471–4477. doi: 10.1128/IAI.73.8.4471-4477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deol P, Vohra R, Saini AK, Singh A, Chandra H, Chopra P, Das TK, Tyagi AK, Singh Y. Role of Mycobacterium tuberculosis Ser/Thr kinase PknF: implications in glucose transport and cell division. J Bacteriol. 2005;187:3415–3420. doi: 10.1128/JB.187.10.3415-3420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spivey VL, Molle V, Whalan RH, Rodgers A, Leiba J, Stach L, Walker KB, Smerdon SJ, Buxton RS. Forkhead-associated (FHA) domain containing ABC transporter Rv1747 is positively regulated by Ser/Thr phosphorylation in Mycobacterium tuberculosis. J Biol Chem. 2011;286:26198–26209. doi: 10.1074/jbc.M111.246132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 66.Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, Betts JC, Mizrahi V, Smith DA, Stokes RW, Av-Gay Y. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. 2004;52:1691–1702. doi: 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 67.Ventura M, Rieck B, Boldrin F, Degiacomi G, Bellinzoni M, Barilone N, Alzaidi F, Alzari PM, Manganelli R, O'Hare HM. GarA is an essential regulator of metabolism in Mycobacterium tuberculosis. Mol Microbiol. 2013;90:356–366. doi: 10.1111/mmi.12368. [DOI] [PubMed] [Google Scholar]
- 68.Niebisch A, Kabus A, Schultz C, Weil B, Bott M. Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J Biol Chem. 2006;281:12300–12307. doi: 10.1074/jbc.M512515200. [DOI] [PubMed] [Google Scholar]
- 69.Nott TJ, Kelly G, Stach L, Li J, Westcott S, Patel D, Hunt DM, Howell S, Buxton RS, O'Hare HM, Smerdon SJ. An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis. Sci Signaling. 2009;2:1–9. doi: 10.1126/scisignal.2000212. [DOI] [PubMed] [Google Scholar]
- 70.Cavazos A, Prigozhin DM, Alber T. Structure of the sensor domain of Mycobacterium tuberculosis PknH receptor kinase reveals a conserved binding cleft. J Mol Biol. 2012;422:488–494. doi: 10.1016/j.jmb.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molle V, Kremer L, Girard-Blanc C, Besra GS, Cozzone AJ, Prost JF. An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry. 2003;42:15300–15309. doi: 10.1021/bi035150b. [DOI] [PubMed] [Google Scholar]
- 72.Zhang N, Torrelles JB, McNeil MR, Escuyer VE, Khoo K-H, Brennan PJ, Chatterjee D. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol Microbiol. 2003;50:69–76. doi: 10.1046/j.1365-2958.2003.03681.x. [DOI] [PubMed] [Google Scholar]
- 73.Escuyer VE, Lety MA, Torrelles JB, Khoo KH, Tang JB, Rithner CD, Frehel C, Mcneil MR, Brennan PJ, Chatterjee D. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J Biol Chem. 2001;276:48854–48862. doi: 10.1074/jbc.M102272200. [DOI] [PubMed] [Google Scholar]
- 74.Sharma K, Gupta M, Pathak M, Gupta N, Koul A, Sarangi S, Baweja R, Singh Y. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J Bacteriol. 2006;188:2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papavinasasundaram KG, Chan B, Chung JH, Colston MJ, Davis EO, Av-Gay Y. Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J Bacteriol. 2005;187:5751–5760. doi: 10.1128/JB.187.16.5751-5760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gomez-Velasco A, Bach H, Rana AK, Cox LR, Bhatt A, Besra GS, Av-Gay Y. Disruption of the serine/threonine protein kinase H affects phthiocerol dimycocerosates synthesis in Mycobacterium tuberculosis. Microbiology. 2013;159:726–736. doi: 10.1099/mic.0.062067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chao JD, Papavinasasundaram KG, Zheng X, Chávez-Steenbock A, Wang X, Lee GQ, Av-Gay Y. Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J Biol Chem. 2010;285:29239–29246. doi: 10.1074/jbc.M110.132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gopalaswamy R, Narayanan S, Chen B, Jacobs WR, Av-Gay Y. The serine/threonine protein kinase PknI controls the growth of Mycobacterium tuberculosis upon infection. FEMS Microbiol Lett. 2009;295:23–29. doi: 10.1111/j.1574-6968.2009.01570.x. [DOI] [PubMed] [Google Scholar]
- 79.Jang J, Stella A, Boudou F, Levillain F, Darthuy E, Vaubourgeix J, Wang C, Bardou F, Puzo G, Gilleron M, Burlet-Schiltz O, Monsarrat B, Brodin P, Gicquel B, Neyrolles O. Functional characterization of the Mycobacterium tuberculosis serine/threonine kinase PknJ. Microbiology. 2010;156:1619–1631. doi: 10.1099/mic.0.038133-0. [DOI] [PubMed] [Google Scholar]
- 80.Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 81.Kumar P, Kumar D, Parikh A, Rananaware D, Gupta M, Singh Y, Nandicoori VK. The Mycobacterium tuberculosis protein kinase K modulates activation of transcription from the promoter of mycobacterial monooxygenase operon through phosphorylation of the transcriptional regulator VirS. J Biol Chem. 2009;284:11090–11099. doi: 10.1074/jbc.M808705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malhotra V, Arteaga-Cortés LT, Clay G, Clark-Curtiss JE. Mycobacterium tuberculosis protein kinase K confers survival advantage during early infection in mice and regulates growth in culture and during persistent infection: implications for immune modulation. Microbiology. 2010;156:2829–2841. doi: 10.1099/mic.0.040675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malhotra V, Okon BP, Clark-Curtiss JE. Mycobacterium tuberculosis protein kinase K enables growth adaptation through translation control. J Bacteriol. 2012;194:4184–4196. doi: 10.1128/JB.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narayan A, Sachdeva P, Sharma K, Saini AK, Tyagi AK, Singh Y. Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol Genomics. 2007;29:66–75. doi: 10.1152/physiolgenomics.00221.2006. [DOI] [PubMed] [Google Scholar]
- 85.Canova MJ, Veyron-Churlet R, Zanella-Cleon I, Cohen-Gonsaud M, Cozzone AJ, Becchi M, Kremer L, Molle V. The Mycobacterium tuberculosis serine/threonine kinase PknL phosphorylates Rv2175c: mass spectrometric profiling of the activation loop phosphorylation sites and their role in the recruitment of Rv2175c. Proteomics. 2008;8:521–533. doi: 10.1002/pmic.200700442. [DOI] [PubMed] [Google Scholar]
- 86.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 87.Pullen KE, Ng H-L, Sung P-Y, Good MC, Smith SM, Alber T. An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-family Ser/Thr protein phosphatase. Structure. 2004;12:1947–1954. doi: 10.1016/j.str.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 88.Koul A, Choidas A, Treder M, Tyagi AK, Drlica K, Singh Y, Ullrich A. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J Bacteriol. 2000;182:5425–5432. doi: 10.1128/jb.182.19.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beresford N, Patel S, Armstrong J, Szöor B, Fordham-Skelton AP, Tabernero L. MptpB, a virulence factor from Mycobacterium tuberculosis, exhibits triple-specificity phosphatase activity. Biochem J. 2007;406:13–18. doi: 10.1042/BJ20070670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stehle T, Sreeramulu S, Löhr F, Richter C, Saxena K, Jonker HRA, Schwalbe H. The apo-structure of the low molecular weight protein-tyrosine phosphatase A (MptpA) from Mycobacterium tuberculosis allows for better target-specific drug development. J Biol Chem. 2012;287:34569–34582. doi: 10.1074/jbc.M112.399261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Madhurantakam C, Rajakumara E, Mazumdar PA, Saha B, Mitra D, Wiker HG, Sankaranarayanan R, Das AK. Crystal structure of low-molecular-weight protein tyrosine phosphatase from Mycobacterium tuberculosis at 1.9-A resolution. J Bacteriol. 2005;187:2175–2181. doi: 10.1128/JB.187.6.2175-2181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flynn EM, Hanson JA, Alber T, Yang H. Dynamic active-site protection by the M. tuberculosis protein tyrosine phosphatase PtpB lid domain. J Am Chem Soc. 2010;132:4772–4780. doi: 10.1021/ja909968n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grundner C, Ng H-L, Alber T. Mycobacterium tuberculosis protein tyrosine phosphatase PtpB structure reveals a diverged fold and a buried active site. Structure. 2005;13:1625–1634. doi: 10.1016/j.str.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 94.Bach H, Papavinasasundaram KG, Wong D, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe. 2008;3:316–322. doi: 10.1016/j.chom.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 95.Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc Natl Acad Sci USA. 2011;108:19371–19376. doi: 10.1073/pnas.1109201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou B, He Y, Zhang X, Xu J, Luo Y, Wang Y, Franzblau SG, Yang Z, Chan RJ, Liu Y, Zheng J, Zhang Z-Y. Targeting mycobacterium protein tyrosine phosphatase B for antituberculosis agents. Proc Natl Acad Sci USA. 2010;107:4573–4578. doi: 10.1073/pnas.0909133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh R, Rao V, Shakila H, Gupta R, Khera A, Dhar N, Singh A, Koul A, Singh Y, Naseema M, Narayanan PR, Paramasivan CN, Ramanathan VD, Tyagi AK. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol Microbiol. 2003;50:751–762. doi: 10.1046/j.1365-2958.2003.03712.x. [DOI] [PubMed] [Google Scholar]
- 98.Saleh MT, Belisle JT. Secretion of an acid phosphatase (SapM) by Mycobacterium tuberculosis that is similar to eukaryotic acid phosphatases. J Bacteriol. 2000;182:6850–6853. doi: 10.1128/jb.182.23.6850-6853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vergne I, Chua J, Lee H-H, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005;102:4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chapman TM, Bouloc N, Buxton RS, Chugh J, Lougheed KEA, Osborne SA, Saxty B, Smerdon SJ, Taylor DL, Whalley D. Substituted aminopyrimidine protein kinase B (PknB) inhibitors show activity against Mycobacterium tuberculosis. Bioorg Med Chem Lett. 2012;22:3349–3353. doi: 10.1016/j.bmcl.2012.02.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lougheed KEA, Osborne SA, Saxty B, Whalley D, Chapman T, Bouloc N, Chugh J, Nott TJ, Patel D, Spivey VL, Kettleborough CA, Bryans JS, Taylor DL, Smerdon SJ, Buxton RS. Effective inhibitors of the essential kinase PknB and their potential as anti-mycobacterial agents. Tuberculosis (Edinb) 2011;91:277–286. doi: 10.1016/j.tube.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hanzelka BL, Wang T, Bemis G, Zuccola H, Doyle T, Stuver-Moody C, Huang YN, Sears C, Fleming M, Erwin AL, Locher C, Muh U. Evidence that inhibition of the serine-threonine protein kinases of Mycobacterium tuberculosis holds promise for developing novel anti-tuberculosis drugs.. Gordon Research Conference on Tuberculosis Drug Development; Oxford, U.K.: Oxford University; 2009. [Google Scholar]
- 104.Székely R, Wáczek F, Szabadkai I, Németh G, Hegymegi-Barakonyi B, Eros D, Szokol B, Pató J, Hafenbradl D, Satchell J, Saint-Joanis B, Cole ST, Orfi L, Klebl BM, Kéri G. A novel drug discovery concept for tuberculosis: inhibition of bacterial and host cell signalling. Immunol Lett. 2008;116:225–231. doi: 10.1016/j.imlet.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 105.Wong D, Chao JD, Av-Gay Y. Mycobacterium tuberculosis-secreted phosphatases: from pathogenesis to targets for TB drug development. Trends Microbiol. 2013;21:100–109. doi: 10.1016/j.tim.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 106.Vintonyak VV, Antonchick AP, Rauh D, Waldmann H. The therapeutic potential of phosphatase inhibitors. Curr Opin Chem Biol. 2009;13:272–283. doi: 10.1016/j.cbpa.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 107.Manger M, Scheck M, Prinz H, von Kries JP, Langer T, Saxena K, Schwalbe H, Fürstner A, Rademann J, Waldmann H. Discovery of Mycobacterium tuberculosis protein tyrosine phosphatase A (MptpA) inhibitors based on natural products and a fragment-based approach. Chembiochem. 2005;6:1749–1753. doi: 10.1002/cbic.200500171. [DOI] [PubMed] [Google Scholar]
- 108.Chiaradia LD, Martins PGA, Cordeiro MNS, Guido RVC, Ecco G, Andricopulo AD, Yunes RA, Vernal J, Nunes RJ, Terenzi H. Synthesis, biological evaluation, and molecular modeling of chalcone derivatives as potent inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatases (PtpA and PtpB). J Med Chem. 2012;55:390–402. doi: 10.1021/jm2012062. [DOI] [PubMed] [Google Scholar]
- 109.Chiaradia LD, Mascarello A, Purificação M, Vernal J, Cordeiro MNS, Zenteno ME, Villarino A, Nunes RJ, Yunes RA, Terenzi H. Synthetic chalcones as efficient inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Bioorg Med Chem Lett. 2008;18:6227–6230. doi: 10.1016/j.bmcl.2008.09.105. [DOI] [PubMed] [Google Scholar]
- 110.Mascarello A, Chiaradia LD, Vernal J, Villarino A, Guido RVC, Perizzolo P, Poirier V, Wong D, Martins PGA, Nunes RJ, Yunes RA, Andricopulo AD, Av-Gay Y, Terenzi H. Inhibition of Mycobacterium tuberculosis tyrosine phosphatase PtpA by synthetic chalcones: kinetics, molecular modeling, toxicity and effect on growth. Bioorg Med Chem. 2010;18:3783–3789. doi: 10.1016/j.bmc.2010.04.051. [DOI] [PubMed] [Google Scholar]
- 111.Rawls KA, Lang PT, Takeuchi J, Imamura S, Baguley TD, Grundner C, Alber T, Ellman JA. Fragment-based discovery of selective inhibitors of the Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Bioorg Med Chem Lett. 2009;19:6851–6854. doi: 10.1016/j.bmcl.2009.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grundner C, Perrin D, Hooft van Huijsduijnen R, Swinnen D, Gonzalez J, Gee CL, Wells TN, Alber T. Structural basis for selective inhibition of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB. Structure. 2007;15:499–509. doi: 10.1016/j.str.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Soellner MB, Rawls KA, Grundner C, Alber T, Ellman JA. Fragment-based substrate activity screening method for the identification of potent inhibitors of the Mycobacterium tuberculosis phosphatase PtpB. J Am Chem Soc. 2007;129:9613–9615. doi: 10.1021/ja0727520. [DOI] [PubMed] [Google Scholar]
- 114.Beresford NJ, Mulhearn D, Szczepankiewicz B, Liu G, Johnson ME, Fordham-Skelton A, Abad-Zapatero C, Cavet JS, Tabernero L. Inhibition of MptpB phosphatase from Mycobacterium tuberculosis impairs mycobacterial survival in macrophages. J Antimicrob Chemother. 2009;63:928–936. doi: 10.1093/jac/dkp031. [DOI] [PubMed] [Google Scholar]
- 115.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 116.Zheng X, Papavinasasundaram KG, Av-Gay Y. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem Biophys Res Comm. 2007;355:162–168. doi: 10.1016/j.bbrc.2007.01.122. [DOI] [PubMed] [Google Scholar]
- 117.Sajid A, Arora G, Gupta M, Singhal A, Chakraborty K, Nandicoori VK, Singh Y. Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation. J Bacteriol. 2011;193:5347–5358. doi: 10.1128/JB.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Molle V, Reynolds RC, Alderwick LJ, Besra GS, Cozzone AJ, Fütterer K, Kremer L. EmbR2, a structural homologue of EmbR, inhibits the Mycobacterium tuberculosis kinase/substrate pair PknH/EmbR. Biochem J. 2008;410:309–317. doi: 10.1042/BJ20071384. [DOI] [PubMed] [Google Scholar]
- 119.Sinha I, Boon C, Dick T. Apparent growth phase-dependent phosphorylation of malonyl coenzyme A:acyl carrier protein transacylase (MCAT), a major fatty acid synthase II component in Mycobacterium bovis BCG. FEMS Microbiol Lett. 2003;227:141–147. doi: 10.1016/S0378-1097(03)00664-5. [DOI] [PubMed] [Google Scholar]
- 120.Grundner C, Gay LM, Alber T. Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 2005;14:1918–1921. doi: 10.1110/ps.051413405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roumestand C, Leiba J, Galophe N, Margeat E, Padilla A, Bessin Y, Barthe P, Molle V, Cohen-Gonsaud M. Structural insight into the Mycobacterium tuberculosis Rv0020c protein and its interaction with the PknB kinase. Structure. 2011;19:1525–1534. doi: 10.1016/j.str.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 122.Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, Kundu M, Basu J. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One. 2010;5:e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Thakur M, Chakraborti PK. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J Biol Chem. 2006;281:40107–40113. doi: 10.1074/jbc.M607216200. [DOI] [PubMed] [Google Scholar]
- 124.Villarino A, Duran R, Wehenkel A, Fernandez P, England P, Brodin P, Cole ST, Zimny-Arndt U, Jungblut PR, Cervenansky C, Alzari PM. Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J Mol Biol. 2005;350:953–963. doi: 10.1016/j.jmb.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 125.Leiba J, Syson K, Baronian G, Zanella-Cleon I, Kalscheuer R, Kremer L, Bornemann S, Molle V. Mycobacterium tuberculosis maltosyltransferase GlgE, a genetically validated antituberculosis target, is negatively regulated by Ser/Thr phosphorylation. Journal of Biological Chemistry. 2013;288:16546–16556. doi: 10.1074/jbc.M112.398503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J Mol Biol. 2009;386:451–464. doi: 10.1016/j.jmb.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 127.Canova MJ, Kremer L, Molle V. The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J Bacteriol. 2009;191:2876–2883. doi: 10.1128/JB.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perez J, Garcia R, Bach H, de Waard JH, Jacobs WR, Jr, Av-Gay Y, Bubis J, Takiff HE. Mycobacterium tuberculosis transporter MmpL7 is a potential substrate for kinase PknD. Biochem Biophys Res Commun. 2006;348:6–12. doi: 10.1016/j.bbrc.2006.06.164. [DOI] [PubMed] [Google Scholar]
- 129.Thakur M, Chakraborti PK. Ability of PknA, a mycobacterial eukaryotic-type serine/threonine kinase, to transphosphorylate MurD, a ligase involved in the process of peptidoglycan biosynthesis. Biochem J. 2008;415:27–33. doi: 10.1042/BJ20080234. [DOI] [PubMed] [Google Scholar]
- 130.Gupta M, Sajid A, Arora G, Tandon V, Singh Y. Forkhead-associated domain-containing protein Rv0019c and polyketide-associated protein PapA5, from substrates of serine/threonine protein kinase PknB to interacting proteins of Mycobacterium tuberculosis. J Biol Chem. 2009;284:34723–34734. doi: 10.1074/jbc.M109.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Corrales RM, Molle V, Leiba J, Mourey L, de Chastellier C, Kremer L. Phosphorylation of mycobacterial PcaA inhibits mycolic acid cyclopropanation: consequences for intracellular survival and for phago-some maturation block. J Biol Chem. 2012;287:26187–26199. doi: 10.1074/jbc.M112.373209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sajid A, Arora G, Gupta M, Upadhyay S, Nandicoori VK, Singh Y. Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB. PLoS One. 2011;6:e17871. doi: 10.1371/journal.pone.0017871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arora G, Sajid A, Gupta M, Bhaduri A, Kumar P, Basu-Modak S, Singh Y. Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS One. 2010;5:e10772. doi: 10.1371/journal.pone.0010772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park ST, Kang CM, Husson RN. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:13105–13110. doi: 10.1073/pnas.0801143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cohen-Gonsaud M, Barthe P, Canova MJ, Stagier-Simon C, Kremer L, Roumestand C, Molle V. The Mycobacterium tuberculosis Ser/Thr kinase substrate Rv2175c is a DNA-binding protein regulated by phosphorylation. J Biol Chem. 2009;284:19290–19300. doi: 10.1074/jbc.M109.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Parish T. Two-component regulatory systems of mycobacteria. Microbiol Spectrum. 2014;2(1):MGM2–0010-2013. doi: 10.1128/microbiolspec.MGM2-0010-2013. doi:10.1128/microbiolspec.MGM2-0010-2013. [DOI] [PubMed] [Google Scholar]
- 139.Manganelli R. Sigma factors: key molecules in Mycobacterium tuberculosis physiology and virulence. Microbiol Spectrum. 2014;2(1):MGM2–0007-2013. doi: 10.1128/microbiolspec.MGM2-0007-2013. doi:10.1128/microbiolspec.MGM2-0007-2013. [DOI] [PubMed] [Google Scholar]