Abstract
Statins are effective cholesterol-lowering drugs to treat CVDs. Bile acids (BAs), the end products of cholesterol metabolism in the liver, are important nutrient and energy regulators. The present study aims to investigate how statins affect BA homeostasis in the enterohepatic circulation. Male C57BL/6 mice were treated with atorvastatin (100 mg/kg/day po) for 1 week, followed by BA profiling by ultra-performance LC-MS/MS. Atorvastatin decreased BA pool size, mainly due to less BA in the intestine. Surprisingly, atorvastatin did not alter total BAs in the serum or liver. Atorvastatin increased the ratio of 12α-OH/non12α-OH BAs. Atorvastatin increased the mRNAs of the BA-synthetic enzymes cholesterol 7α-hydroxylase (Cyp7a1) (over 10-fold) and cytochrome P450 27a1, the BA uptake transporters Na+/taurocholate cotransporting polypeptide and organic anion transporting polypeptide 1b2, and the efflux transporter multidrug resistance-associated protein 2 in the liver. Noticeably, atorvastatin suppressed the expression of BA nuclear receptor farnesoid X receptor (FXR) target genes, namely small heterodimer partner (liver) and fibroblast growth factor 15 (ileum). Furthermore, atorvastatin increased the mRNAs of the organic cation uptake transporter 1 and cholesterol efflux transporters Abcg5 and Abcg8 in the liver. The increased expression of BA-synthetic enzymes and BA transporters appear to be a compensatory response to maintain BA homeostasis after atorvastatin treatment. The Cyp7a1 induction by atorvastatin appears to be due to suppressed FXR signaling in both the liver and intestine.
Keywords: ultra-performance liquid chromatography-tandem mass spectrometry, cholesterol 7α-hydroxylase, farnesoid X receptor signaling
Statins are a class of drugs that are used to lower cholesterol levels by specifically inhibiting HMG-CoA reductase, which is the rate-limiting enzyme in cholesterol biosynthesis in the liver. Statins reduce LDL (bad cholesterol) as much as 20–60%, lower triglycerides, and slightly increase HDL (good cholesterol) (1, 2). Therefore, statins are used as the most effective drugs for treating CVDs. Commercially available statins include atorvastatin (Lipitor), fluvastatin (Lescol), lovastatin (Mevacor), pitavastatin (Livalo), pravastatin (Pravachol), rosuvastatin (Crestor), and simvastatin (Zocor). Atorvastatin, the best-selling statin, is very potent in lowering LDL levels in patients with CVD (3).
The most important statin uptake transporters in the liver are the organic anion transporting polypeptides (OATP/SLCO1B1 and 1B3 in humans, Oatp/Slco1b2 in mice) (4–6). The Na+/taurocholate cotransporting polypeptide (NTCP/SLC10A1) also plays a role in statin uptake into hepatocytes (5, 7). Moreover, several efflux transporters are important for transporting statins out of hepatocytes, such as the multidrug resistance-associated protein 2 (Mrp2/Abcc2) (8), breast cancer resistant protein (Bcrp/Abcg2) (9), and bile salt export pump (Bsep/Abcb11) (10). The genetic polymorphisms of uptake (OATP1B1, 1B3) and efflux transporters (MRP2, BCRP) that are responsible for inter-individual differences in the pharmacokinetics and pharmacodynamics of statins are well-known in humans (4). The effect of statins on the expression of several transporters has been investigated in cell lines (11). The development of transporter-deficient rodent models provides evidence that Oatp1b2 (6), Mrp2 (12), and Bcrp (13) play important roles in statin transport in the livers of intact animals. However, the in vivo data on the effect of statin treatment on the expression of various transporters in the liver are relatively limited in laboratory animals and in humans.
Another knowledge gap is the role of statins in regulating the homeostasis of bile acids (BAs), which are end products from cholesterol metabolism in the liver. In addition to their roles in cholesterol elimination and nutrient absorption, BAs are known to be signaling molecules that regulate nutrient and energy metabolism (14). BAs are synthesized in the liver (called primary BAs), stored in the gallbladder (GB), secreted into the intestine, reabsorbed at the end of the small intestine (SI), and subsequently returned to the liver. This cycle is called the enterohepatic circulation (EHC) of BAs. The BA reabsorption from the SI is very efficient, which is more than 95% efficient in humans. The rest (∼5%) of BAs are eliminated into feces, the amount of which is often used to estimate the production of BAs in the liver.
Primary BAs are synthesized in the liver and conjugated with taurine and glycine on the side chain, or sulfate on the steroid nucleus (15). Cholic acid (CA) and chenodeoxycholic acid (CDCA), as well as α- and β-muricholic acids (MCAs), are the primary BAs in mice. These primary BAs are converted by bacterial enzymes in the intestine through deconjugation, dehydroxylation, epimerization, and oxidation to form secondary BAs, such as deoxycholic acid (DCA), lithocholic acid (LCA), and ursodeoxycholic acid (UDCA), as well as ωMCA, murideoxycholic acid (MDCA), and hyodeoxycholic acid (HDCA), as previously summarized (16).
Several enzymes catalyze BA synthesis in the liver, either through the classic or alternative pathways. Cholesterol 7α-hydroxylase (Cyp7a1) is the rate-limiting enzyme in the classic pathway (17), which results in the formation of both CA and CDCA. Cytochrome P450 (Cyp)8b1 is responsible for the 12α-hydroxylation of BAs, or the formation of CA. Mitochondrial Cyp27a1 catalyzes the side-chain oxidation of cholesterol. The alternative pathway is initiated by Cyp27a1 and followed by Cyp7b1 and Cyp39a1 to produce CDCA. The expression of key BA-synthetic enzymes (Cyp7a1 and Cyp8b1) can be regulated transcriptionally by BA-feedback inhibition, mainly through two mechanisms (18). One is the BA nuclear receptor farnesoid X receptor (FXR/Nr1b4) and its target gene small heterodimer partner (SHP/Nr0b2) pathway in the liver (19). The other is activation of FXR in the intestine and the induction of its target gene fibroblast growth factor (Fgf)15, which is an intestinal hormone that travels to the liver and also represses the transcription of BA-synthetic enzymes (20).
BA transporters play pivotal roles in maintaining BA homeostasis. Ntcp mediates the uptake of conjugated BAs, whereas Oatp1b2 transports unconjugated BAs (21). Bsep, which is generally accepted as the main BA efflux transporter in the liver, mediates the biliary excretion of monovalent conjugated BAs (such as TCA) and has a relatively poor affinity for unconjugated BAs (22). The finding that Mrp2 mediates the biliary excretion of divalent conjugated BAs (such as tauro-BA-sulfates, TCDCA-S and TLCA-S) (23) suggests that Mrp2 is a promising candidate to be an alternative canalicular BA transporter. It is debatable whether Bcrp transports BAs, because of contradictory findings (24, 25).
Because cholesterol is the substrate for de novo synthesis of BAs in the liver, we hypothesized that the cholesterol-lowering drugs could also affect BA homeostasis, and the changes would be mediated by the coordinated regulation of BA-synthetic enzymes and/or transporters. Therefore, the present study utilized a highly sensitive and accurate ultra-performance (UP)LC-MS/MS method recently developed in our laboratory (26) to characterize BA profiles in various compartments of the BA EHC in mice treated with atorvastatin, and determined the expression of BA-related genes in the liver and intestine to reveal possible regulatory mechanisms. This study also aims to provide the first in vivo evidence for the effect of statins on the expression of transporters in the liver.
EXPERIMENTAL PROCEDURE
Chemicals and reagents
Atorvastatin was purchased from Cayman Chemical (Ann Arbor, MI). The sources of internal standards (ISs) and various taurine-conjugated and unconjugated BA standards were the same as previously reported (26). All other chemicals, unless indicated, were purchased from Sigma-Aldrich (St. Louis, MO).
Animals and treatments
The animal study was approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center. Eight-week-old male C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA). Mice were acclimated for 1 month and fed Teklad Rodent Diet #5053 (Harlan Laboratories, Madison, WI) ad libitum according to the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). Mice (n = 5) were administered vehicle (5 mg/ml β-cyclodextrin in saline) or atorvastatin (100 mg/kg/day) po for seven consecutive days. The dose was chosen based on previous publications (27, 28). Mice were anesthetized 24 h after the final dose, blood was collected by orbital bleeding, and serum was collected by centrifuging blood at 8,000 rpm for 15 min. The liver and GB (tissue with bile inside) were removed, frozen separately in liquid nitrogen, and stored at −80°C. The SI was flushed into 10 ml of saline to collect SI contents, and the large intestine (LI) was flushed into another 10 ml of saline to collect LI contents. The three SI segments, namely duodenum, jejunum, and ileum, as well as the LI, were stored separately at −80°C.
BA extraction from serum
2H4-G-CDCA and 2H4-CDCA (40 μg/ml and 20 μg/ml, respectively) were used as ISs. Simple protein precipitation with methanol (MeOH) was used for BA extraction from serum. One milliliter of MeOH was added to 50 μl of serum-spiked with 5 μl of IS, vortexed, and centrifuged at 12,000 g for 10 min. The supernatant was aspirated, evaporated under vacuum, and reconstituted in 50 μl of 50% MeOH. Samples were centrifuged at 20,000 g for 10 min before injection.
BA extraction from the liver
A piece of liver (∼120 mg) was homogenized in 5 vol of water, from which 600 μl of homogenate was taken and mixed with 10 μl of IS. After 10 min equilibration on ice, the homogenate was mixed with 3 ml of ice-cold alkaline acetonitrile (5% ammonia), vortexed vigorously, and shaken for 1 h at room temperature. The mixture was centrifuged at 12,000 g for 10 min, and the supernatant was collected. The pellet was extracted with 1 ml of MeOH, sonicated for 5 min, and centrifuged at 12,000 g for 10 min. The two supernatants were pooled, evaporated under vacuum, and reconstituted in 100 μl of 50% MeOH. The suspension was transferred into a 0.2 μm Costar Spin-X HPLC microcentrifuge filter (purchased from Corning Inc., Corning, NY), and centrifuged at 20,000 g for 10 min. The supernatant was then ready for injection.
BA extraction from the GB
One milliliter of MeOH was added to each GB, which was broken to release the bile inside and premixed with 100 μl of IS. After vigorous vortexing and 10 min sonication, the mixture was centrifuged at 16,000 g for 10 min, and the supernatant was collected. The pellet was extracted with another 2 ml of MeOH. The two supernatants were combined, evaporated under vacuum, and reconstituted in 1 ml of 50% MeOH.
BA extraction from intestinal contents
Intestinal contents were mixed with 100 μl of IS and centrifuged at 12,000 g for 10 min to collect the supernatant. The pellet was extracted with 3 ml of MeOH twice. After shaking for 30 min at room temperature, the mixture was centrifuged at 12,000 g for 20 min to collect the supernatant. The three supernatants were pooled, evaporated under vacuum, and reconstituted in 1 ml of 50% MeOH. The suspension was filtered before injection.
BA extraction from feces
Mice (n = 5) were acclimated to wire-bottomed metabolic cages for 48 h (housed individually), and feces were collected over a 24 h period. Mouse feces were dried under vacuum and ground to powder. Fifty milligrams of feces were mixed with 10 μl IS and 3 ml of MeOH was added. After shaking for 1 h at room temperature, the mixture was centrifuged at 20,000 g for 10 min to collect the supernatant. The pellet was extracted with another 2 ml of MeOH. The two supernatants were pooled, evaporated under vacuum, and reconstituted in 100 μl of 50% MeOH. The suspension was filtered before injection.
BA quantification
BA concentrations were quantified by a highly sensitive and accurate method established in our laboratory using UPLC-MS/MS (26). The conditions of LC and MS were the same as previously reported (26). Major individual BAs quantified include TCA, TCDCA, TαMCA, TβMCA, TDCA, TLCA, TUDCA, TMDCA, TωMCA, THDCA, CA, CDCA, αMCA, βMCA, DCA, LCA, UDCA, MDCA, ωMCA, and HDCA. The concentrations of individual BAs were summed to derive the concentration of conjugated, unconjugated, and total BAs. Primary BAs include (T)CA, (T)CDCA, (T)αMCA, and (T)βMCA, and secondary BAs include (T)DCA, (T)LCA, (T)UDCA, (T)MDCA, (T)ωMCA, and (T)HDCA. The 12α-OH BAs include (T)CA and (T)DCA, and non12α-OH BAs refer to all the remaining BAs.
Total RNA isolation
Total RNA was isolated using RNA Bee reagent (Tel-Test Inc., Friendswood, TX) per the manufacturer’s protocol. RNA concentrations were quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) at a wavelength of 260 nm.
Multiplex suspension assay
The mRNA expression of BA-synthetic enzymes and transporters in control and statin-treated mouse livers were determined with Panomics 2.0 QuantiGene Plex technology (Panomics/Affymetrix, Fremont, CA), following the manufacturer’s protocol. Briefly, individual bead-based oligonucleotide probe sets specific for each gene examined were developed by Panomics Inc. Genes that encode BA-synthetic enzymes and uptake and efflux transporters in the liver can be found on Panomics website under panel number 21021. Samples were analyzed using a Bio-Plex 200 system array reader with Luminex 100 X-MAP technology, and data were acquired using Bio-Plex data manager software version 5.0 (Bio-Rad, Hercules, CA). Assays were performed according to the manufacturer’s protocol. The mRNA expression of target genes was expressed as relative light units (RLU) and normalized to the housekeeping gene, Gapdh.
Reverse transcription and real-time PCR analysis
Total RNA was transcribed to single-stranded cDNA using high capacity cDNA reverse transcription kit 1001073 (Applied Biosystems, Foster City, CA), and then the cDNA products were amplified by PCR, using Power SYBR Green PCR Master Mix in a 7900HT Fast real-time PCR system (Applied Biosystems). The mRNAs of genes encoding BA conjugating enzymes [bile acid-CoA ligase (BAL) and bile acid CoA:amino acid N-acyl-transferase (BAT)], BA transporters in the ileum [apical Na+-dependent bile salt transporter (Asbt), organic solute tranporter (Ost)α, Ostβ, and Mrp3], ileal BA binding protein (Ibabp), and proteins involved in Cyp7a1 regulation (FXR, SHP, Fgf15, and Fgfr4) were quantified, and normalized to β-actin. The sequences of real-time PCR primers (Integrated DNA Technologies, Coralville, IA) are listed in supplementary Table I.
Statistical analysis
Data are presented as mean ± SEM in bar charts, and mean in pie charts. Data were analyzed by Student’s t-test (two-tail), differences being considered significant at P < 0.05. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05).
RESULTS
BA pool size and BA distribution in atorvastatin-treated mice
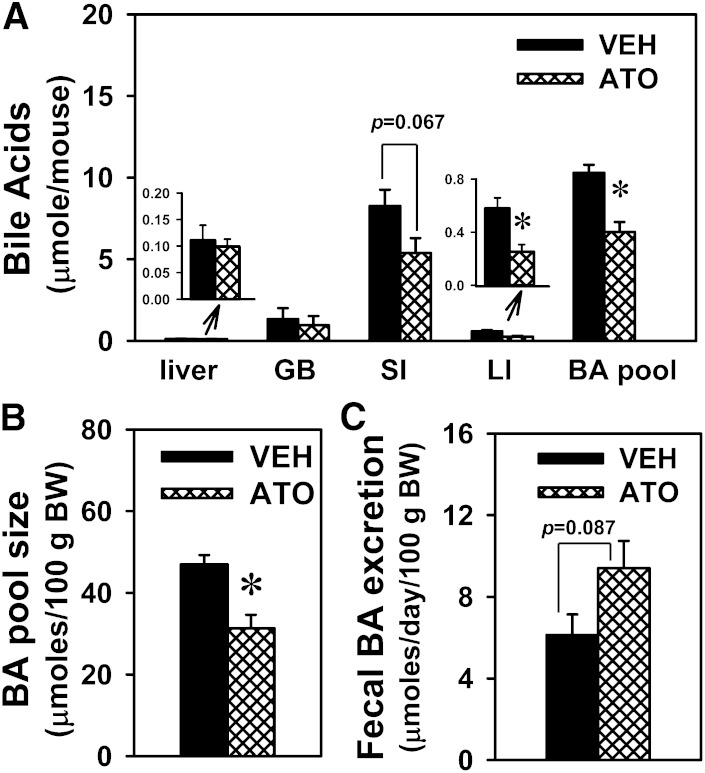
Statins lower cholesterol in the blood by inhibiting cholesterol synthesis, and because BAs are the end products of cholesterol metabolism, statins might also decrease BA synthesis and BAs in the body. Therefore, it was of interest to determine whether atorvastatin alters BA homeostasis in mice. In this study, the dose of atorvastatin (100 mg/kg/day for 7 days) did not cause hepatotoxicity, as evidenced by normal levels of alanine aminotransferase in serum (data not shown). To reveal the overall effects of atorvastatin on BA homeostasis, BA pool size was estimated by adding total BAs in the main compartments of the EHC, namely the liver, GB, and intestine (SI and LI), as published previously (29). As shown in Fig. 1A, B, atorvastatin decreased BA pool size (35.1%), which was primarily due to the decrease in total BAs in the SI (35%) and LI (56.4%). Atorvastatin also tended to increase fecal BA excretion (53.3%) (Fig. 1C). The BA changes in the SI, as well as the fecal excretion, were not statistically significant in the control and statin-treated mice. Furthermore, there were no differences in the body weight or the daily fecal excretion between the control and statin-treated mice (data not shown).
Fig. 1.
BA pool size and BA distribution in atorvastatin-treated mice. Amounts of BAs in major EHC compartments (liver, GB, SI, and LI) (A), BA pool size per 100 g body weight (BW) (B), and fecal BA excretion per 100 g body weight (C) in mice treated with vehicle (VEH) or atorvastatin (ATO) (100 mg/kg po for 7 days). Data are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05).
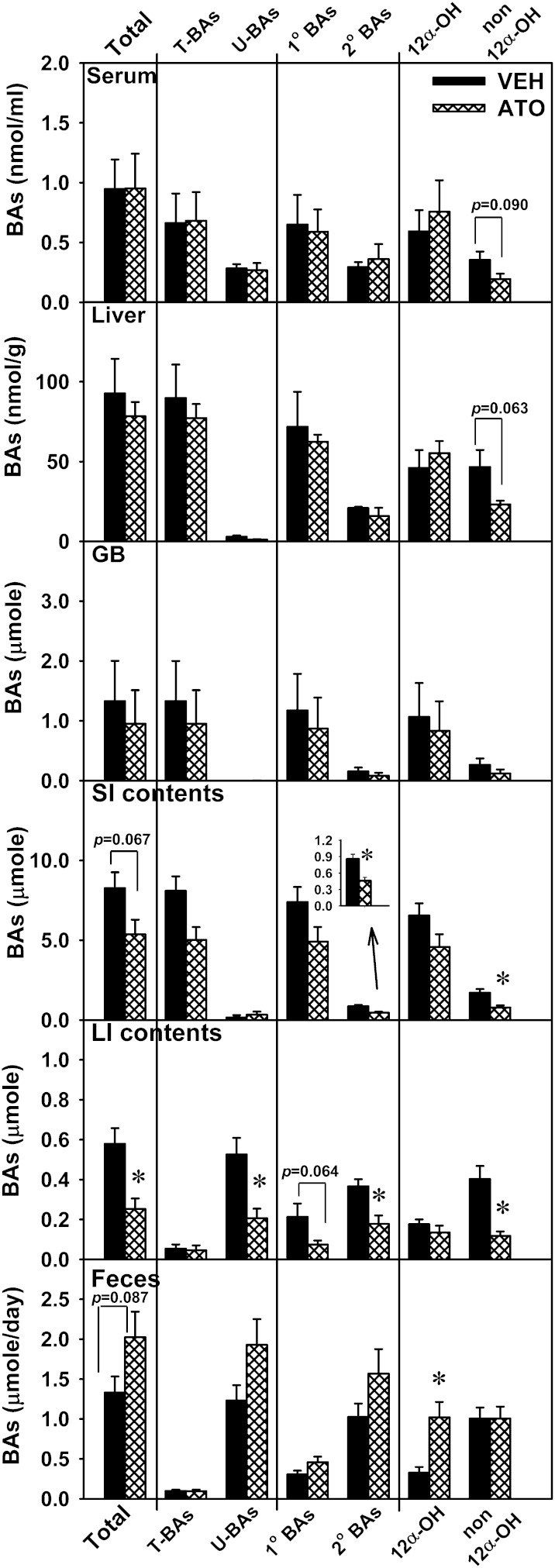
In addition, atorvastatin decreased the amounts of unconjugated BAs in the LI (60.7%) and secondary BAs in the SI (46.5%) and LI (51.3%), and tended to decrease the amounts of primary BAs in the LI (65%) (Fig. 2). CA and DCA, which have 12α-OH groups, are referred to as 12α-OH BAs, whereas other BAs are called non12α-OH BAs. Atorvastatin tended to decrease non12α-OH BAs in serum (45.3%) and the liver (50.4%), and decreased non12α-OH BAs in the SI (54.2%) and LI (70.7%) contents (Fig. 2).
Fig. 2.
Total, taurine-conjugated (T-), unconjugated (U-), primary (1°), secondary (2°), 12α-OH, and non12α-OH BAs in serum, liver, GB, intestinal contents (small and large), and feces of atorvastatin-treated mice. Data are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05). VEH, vehicle; ATO, atorvastatin.
Individual BA concentrations/amounts in atorvastatin-treated mice
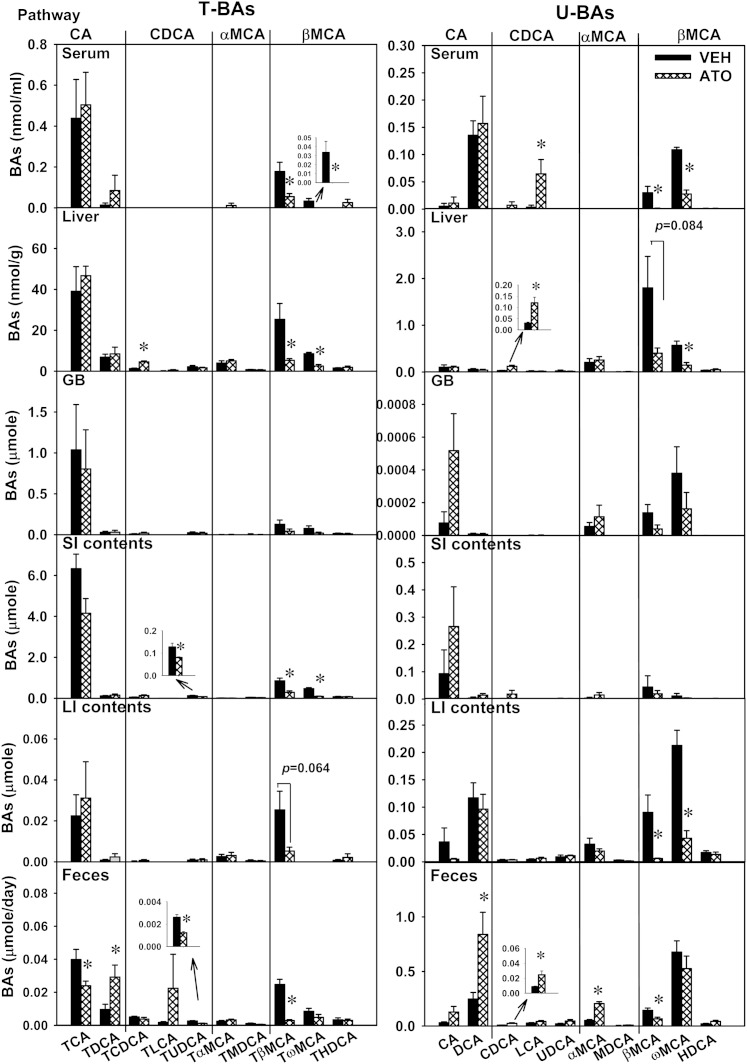
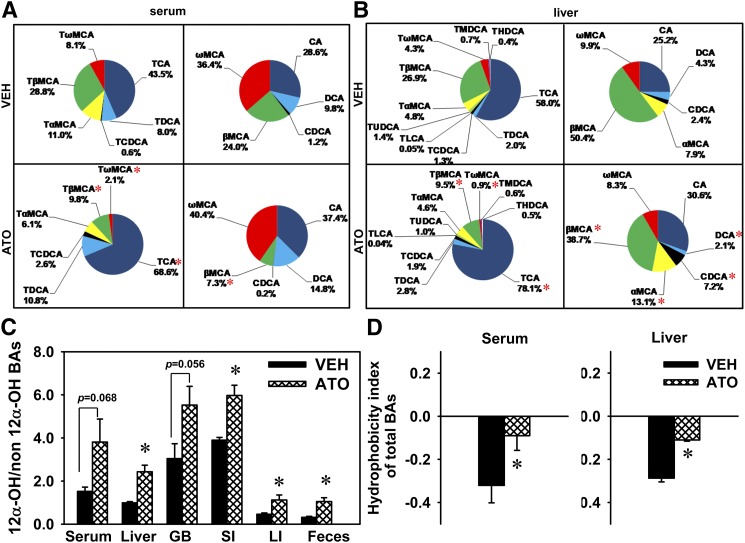
In order to provide detailed information on the regulation of BA homeostasis by statins, concentrations of individual BAs were quantified in serum, liver, GB, SI, LI, and feces, as shown in Fig. 3. In serum, atorvastatin did not change concentrations of BAs in the CA or αMCA pathway, did not change most BAs in the CDCA pathway, except increasing CDCA, and decreased several BAs in the βMCA pathway, including TβMCA (69%) and its secondary BA, TωMCA (100%), as well as βMCA (96.6%) and ωMCA (74.6%). In the liver, atorvastatin did not change the amount of BAs in the CA or αMCA pathways, did not change most BAs in the CDCA pathway, except increasing TCDCA (250%) and CDCA (303%), and decreased several BAs in the βMCA pathway, including TβMCA (79.1%), TωMCA (70.7%), βMCA (77.6%, not statistically significant), and ωMCA (74.3%). In the GB, atorvastatin did not alter the amount of BAs in any pathway. In the SI, atorvastatin did not alter the amount of BAs in the CA or αMCA pathways, did not change most BAs in the CDCA pathway, except decreasing TUDCA (36.7%), and decreased TβMCA (65.8%) and TωMCA (80.3%), but did not change other BAs in the βMCA pathway. In the LI, atorvastatin did not alter the amount of BAs in the CA, CDCA, or αMCA pathways, and decreased several BAs in the βMCA pathway, including TβMCA (79.3%), βMCA (93.4%), and ωMCA (79.7%). In feces, atorvastatin decreased TCA (39.9%), increased TDCA (203%) and DCA (239%), but did not alter the amount of CA. Atorvastatin did not alter most BAs in the CDCA pathway in feces, except decreasing TUDCA (52.8%) and increasing CDCA (187%). Atorvastatin did not alter the amount of most BAs in the αMCA pathway in feces, except increasing αMCA (300%). Atorvastatin decreased TβMCA (88.2%) and βMCA (53.7%), but did not alter other BAs in the βMCA pathway in feces.
Fig. 3.
Individual BAs in atorvastatin-treated mice. Individual BAs in serum, liver, GB, intestine (small and large), and feces of mice treated with vehicle (VEH) or atorvastatin (ATO). Data in bar charts are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05). Four BA pathways are defined to aid in reading the figure, each of which involves the primary BA and its secondary BA, as well as their taurine conjugates. For example, CA pathway involves TCA, TDCA, CA, and DCA.
BA composition in atorvastatin-treated mice
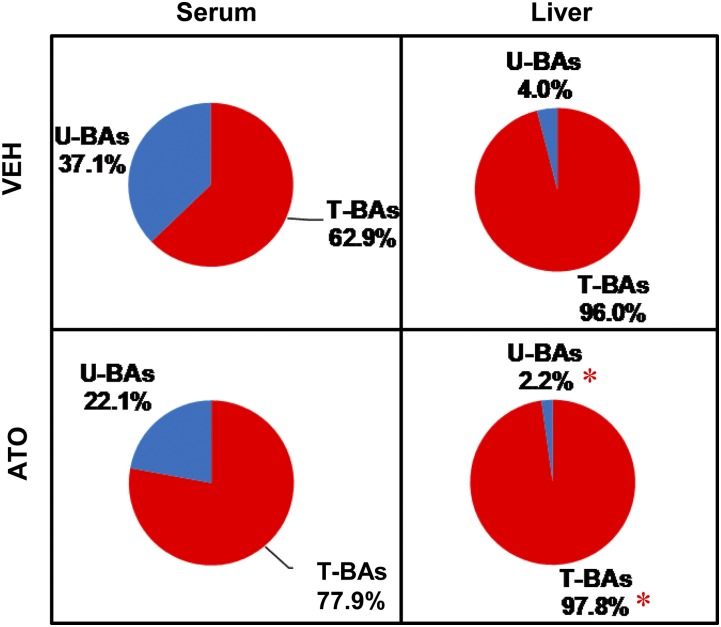
In addition to BA concentration, BA composition is another important factor that leads to different biological effects. For instance, individual BAs have various potencies to activate their receptors, FXR (CDCA > LCA, DCA > CA) and TGR5 (LCA > DCA > CDCA > CA) (30). Therefore, it is important to analyze BA composition in statin-treated mice. Atorvastatin did not alter the proportions of total taurine-conjugated or total unconjugated BAs in serum (Fig. 4). In regard to taurine-conjugated BAs in serum, atorvastatin increased the proportion of TCA (43.5→68.6%), and decreased the proportions of TβMCA (28.8→9.8%) and TωMCA (8.1→2.1%). In regard to unconjugated BAs in serum, atorvastatin decreased the proportion of βMCA (24.0→7.3%), but not other unconjugated BAs (Fig. 5A).
Fig. 4.
The proportions of taurine-conjugated (T-) and unconjugated (U-) BAs in serum and the livers of atorvastatin-treated mice. Data are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05). VEH, vehicle; ATO, atorvastatin.
Fig. 5.
BA composition in atorvastatin-treated mice. BA composition in total taurine-conjugated or unconjugated BAs in serum (A) or livers (B), the ratio of 12α-OH versus non12α-OH BAs in serum, liver, GB, intestine (small and large), and feces (C), and the HI of total BAs in serum and liver (D) of mice treated with vehicle (VEH) or atorvastatin (ATO). Data in bar charts are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05).
In the liver, atorvastatin increased the proportion of total taurine-conjugated BAs (96→97.8%) and decreased the proportion of total unconjugated BAs (4→2.2%) (Fig. 4). Atorvastatin increased the proportion of TCA and decreased the proportions of TβMCA and TωMCA in total taurine-conjugated BAs in the liver. Atorvastatin also increased the proportions of CDCA and αMCA and decreased the proportions of DCA and βMCA in total unconjugated BAs in liver (Fig. 5B).
The ratio of 12α-OH versus non12α-OH BAs, which is an important aspect of BA composition, was analyzed in this study. Atorvastatin increased the ratio of 12α-OH versus non12α-OH BAs in the liver (146%), SI (53.1%), LI (143%), and feces (233%), and tended to increase this ratio in serum (151%) and the GB (81.5%) (Fig. 5C).
Because BAs in serum determine the effects of BAs on peripheral tissues and statins inhibit cholesterol synthesis in the liver, we focused on the changes of BA composition by statins in the serum and liver. Individual BAs have different hydrophobicity, and thus changes in BA composition could lead to altered hydrophobicity of total BAs. The hydrophobicity index (HI) of individual BAs and their proportions in biological samples are used to predict the HI of total BAs at physiological pH, as published previously (29). Atorvastatin increased the HI of total BAs in the serum and liver (Fig. 5D). However, total BAs remained hydrophilic in the serum and liver after atorvastatin treatment.
mRNA expression of BA-synthetic enzymes in the livers of atorvastatin-treated mice
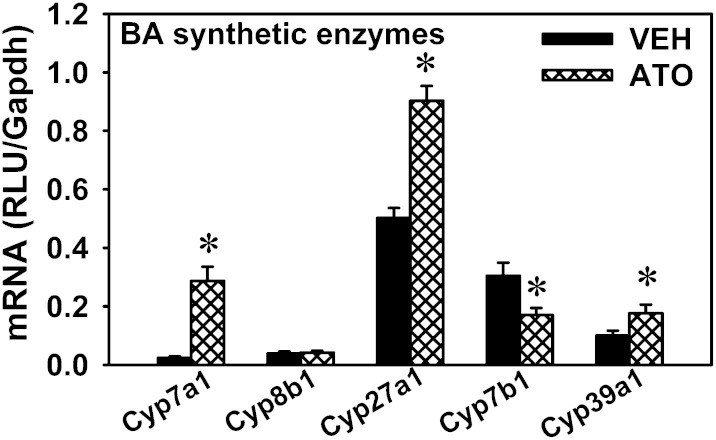
The present observation that statins decrease cholesterol (the substrate for BA synthesis) but do not decrease total BAs in either the liver or serum, suggests that there are compensatory mechanisms to maintain BA homeostasis. The mRNA of Cyp7a1, which encodes the rate-limiting enzyme for the classic pathway of BA synthesis in the liver, was markedly increased by atorvastatin (11.1-fold) (Fig. 6), whereas the mRNA of Cyp8b1, which is responsible for CA formation, remained unchanged after atorvastatin treatment. For the alternative pathway of BA synthesis, the mRNA of Cyp27a1 in the liver was increased by atorvastatin (79.9%). The mRNA of Cyp7b1 in the liver was decreased 44.1% by atorvastatin. The mRNA of Cyp39a1 in the liver was increased by atorvastatin (76.8%). In summary, the mRNAs of the rate-limiting enzyme in the classic pathway (Cyp7a1) and the initial enzyme in the alternative pathway (Cyp27a1) were both increased by atorvastatin treatment, whereas other genes for BA synthesis were only moderately altered (Cyp7b1 and 39a1) or remained unchanged (Cyp8b1).
Fig. 6.
mRNA expression of BA-synthetic enzymes in the livers of atorvastatin-treated mice. Data are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05). VEH, vehicle; ATO, atorvastatin.
mRNA expression of regulators for Cyp7a1 transcription in atorvastatin-treated mice
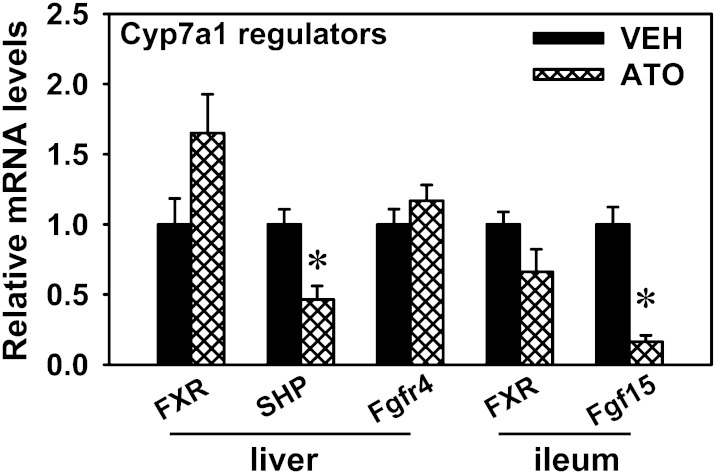
In order to determine the possible regulatory mechanisms for the robust increase in Cyp7a1 mRNA expression by statins, the mRNA expression of several key regulators that are involved in the feedback inhibition of Cyp7a1 were quantified in the liver and ileum. Atorvastatin decreased the mRNAs of SHP (53.6%) in the liver and Fgf15 (83.6%) in the ileum (Fig. 7). Atorvastatin tended to increase FXR mRNA in the liver, but it was not statistically significant. The mRNA levels of Fgfr4 in the liver and FXR in the ileum remained relatively constant after atorvastatin treatment.
Fig. 7.
mRNA expression of regulatory factors for Cyp7a1 transcription in the liver and ileum of atorvastatin-treated mice. The transcription of Cyp7a1 is feedback inhibited by BAs through two proposed mechanisms, namely the hepatic FXR-SHP pathway and ileal Fgf15-hepatic Fgfr4 pathway. Data are presented as mean ± SEM of five mice. The mRNAs of each gene in the control group were set as one. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05). VEH, vehicle; ATO, atorvastatin.
mRNA expression of various transporters in the liver and ileum of atorvastatin-treated mice
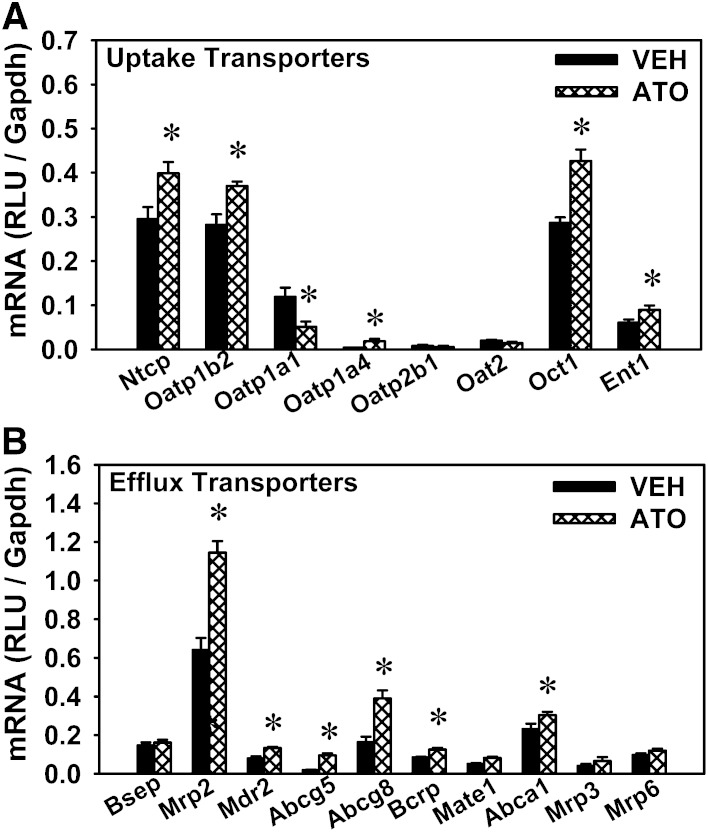
In addition to BA-synthetic enzymes, BA transporters play a crucial role in promoting the EHC of BAs. Thus, the mRNA expression of BA transporters was quantified in the liver and ileum of statin-treated mice, as well as other hepatic transporters that mediate the transport of phospholipids, cholesterol, etc., and promote bile formation. For the uptake transporters in the liver, atorvastatin increased the mRNAs of Ntcp (35.1%), Oatp1b2 (31%), Oatp1a4 (384%), organic cation transporter (Oct)1 (48.9%), and equilibrative nucleoside transporter 1 (Ent1) (47.5%), but decreased Oatp1a1 mRNA (57%) (Fig. 8A). For the efflux transporters in the liver (Fig. 8B), atorvastatin increased the mRNAs of Mrp2 (78.7%), multidrug resistance protein (Mdr)2 (68.1%), Abcg5 (402%), Abcg8 (138%), Bcrp (48.3%), and multidrug and toxin extrusion protein (Mate)1 (60.1%) present on canalicular membranes, as well as Abca1 (31.5%) on the basolateral membrane. Atorvastatin did not alter the mRNA expression of canalicular transporter Bsep, nor the basolateral transporters Mrp3 and Mrp6.
Fig. 8.
mRNA expression of various transporters in the livers of atorvastatin-treated mice. Expression of various uptake transporters (A) as well as basolateral and canalicular efflux transporters (B) in the livers of mice treated with vehicle (VEH) or atorvastatin (ATO). Data are presented as mean ± SEM. Asterisks (*) represent significant differences compared with vehicle-control (P < 0.05).
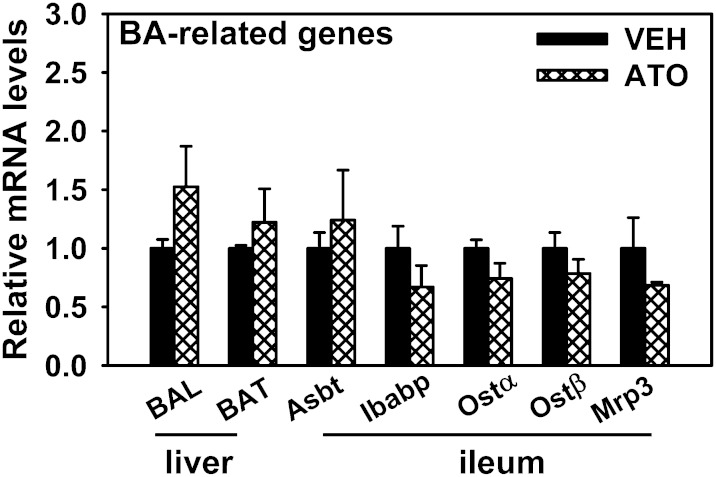
mRNA expression of BA-conjugating enzymes in the liver, as well as proteins responsible for BA reabsorption in the ileum of atorvastatin-treated mice
The mRNA expression of BA-conjugating enzymes (BAL and BAT) was not altered by atorvastatin. In the ileum, none of the transporters or proteins involved in BA reabsorption (Asbt, Ibabp, Ostα, Ostβ, or Mrp3) had altered expression after atorvastatin treatment (Fig. 9).
Fig. 9.
mRNA expression of BA-conjugating enzymes in the liver as well as the proteins responsible for BA reabsorption in ileum of atorvastatin-treated mice. Data are presented as mean ± SEM. VEH, vehicle; ATO, atorvastatin.
DISCUSSION
The present study revealed a novel role of statins in regulating BA homeostasis. Although statins are well-known to decrease cholesterol, which is the substrate for BA synthesis in the liver, total BA concentrations in the serum and liver were not decreased by atorvastatin (Figs. 1, 2). However, atorvastatin decreases the BA pool size, which is primarily due to lower BAs in the intestine. The fecal BA excretion usually equals the amount of newly biosynthesized BAs in the liver. The total amount of BAs synthesized in the liver is thought to be regulated by Cyp7a1. In this study, atorvastatin increased fecal BA excretion, which is consistent with the upregulation of Cyp7a1 expression in the liver. A marked decrease (over 50%) in unconjugated BAs was observed in the large intestinal contents of mice treated by atorvastatin (Fig. 2), which correspondingly leads to an increased ratio of conjugated versus unconjugated BAs. Conjugated BAs are more hydrophilic, and thus less ready to cross cell membranes by passive diffusion. In contrast to the transporter-mediated active reabsorption that is restricted to the ileum, passive diffusion occurs in both the SI and LI (31). Primary BAs undergo deconjugation catalyzed by bacterial enzymes, which are mainly colonized in the LI lumen. Furthermore, it has been reported that diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiome (32). Therefore, it is possible that atorvastatin may alter the intestinal microbiota composition and decrease the bacterial enzyme-catalyzed deconjugation of primary BAs, and thus passive absorption of BAs in the intestine.
Because individual BAs have different physiochemical properties and biological effects, such as the capability to activate BA receptors, it is pivotal to analyze the concentrations of individual BAs as well as the proportion of each BA. In general, the majority of BAs that have altered concentrations after atorvastatin treatment are βMCA and its secondary BAs (Fig. 3). For instance, atorvastatin decreases TβMCA, TωMCA, and ωMCA in serum and the liver, TβMCA and TωMCA in the SI, and βMCA and ωMCA in the LI, as well as TβMCA and βMCA in feces. Moreover, the ratio of 12α-OH versus non12α-OH BAs is increased by atorvastatin in the liver, SI, LI, and feces. This is consistent with a previous report on the correlation of 12α-OH BAs and improved glucose and lipid parameters (33), considering the lipid-lowering effects of statins.
Interestingly, atorvastatin increases the proportion of (T)CA and decreases the proportion of (T)MCAs in both serum and the liver (Fig. 5A, B). Correspondingly, atorvastatin increases the hydrophobicity of total BAs in serum and the liver (Fig. 5C). However, the BAs whose levels are altered by statins are mainly trihydroxy-BAs, which are considered hydrophilic. In contrast, DCA and LCA are considered hydrophobic BAs and can disrupt cell membranes (34) and mediate mitochondria-derived cell death (35). Therefore, the increase in BA hydrophobicity after atorvastatin treatment is not of concern, which is confirmed by the normal activity of alanine aminotransferase after statin treatment (data not shown).
Atorvastatin has robust effects on regulating the expression of BA-synthetic enzymes in the liver. Interestingly, the mRNA of Cyp7a1, the rate-limiting BA-synthetic enzyme in the classic pathway, is increased over 10-fold by atorvastatin. The mRNA of the initial enzyme in the alternative pathway, Cyp27a1, is also increased by atorvastatin (Fig. 6). These results suggest that the rate of BA de novo synthesis may be enhanced by atorvastatin. The present finding is consistent with a number of previous reports that Cyp7a1 was stimulated by pitavastatin in guinea pigs (36) and crilvastatin in rats (37); 7α-hydroxylase activity was increased by pravastatin in gallstone patients (38); and CYP7A1 promoter polymorphism was associated with poor response to atorvastatin (39). However, there are also some contradicting data indicating that Cyp7a1 mRNA was decreased by atorvastatin and simvastatin in guinea pigs (36) and by pravastatin in bile duct-obstructed rats (40).
The abundant induction of Cyp7a1 transcription by atorvastatin is likely due to decreased FXR signaling in both the liver and intestine. Statins inhibit cholesterol synthesis, thus leading to a decrease in substrates for BA synthesis. This would possibly lead to decreased amounts of BAs in the EHC. Thus, BA-feedback inhibition of Cyp7a1 transcription is removed. Specifically, the FXR signaling activated by BAs in both the liver and intestine is lower after atorvastatin treatment, evidenced by decreased mRNA levels of the FXR target gene, SHP, in the liver, as well as the FXR target gene, Fgf15, in the ileum (Fig. 7). The inhibited FXR signaling in the liver and intestine probably contributes to the abundant induction of Cyp7a1 transcription in atorvastatin-treated mice. The elevated expression of Cyp7a1, the rate-limiting enzyme for BA synthesis, possibly compensates the prior BA changes by suppressed cholesterol synthesis, and helps to maintain BA homeostasis in liver and serum.
In addition to increasing the expression of BA-synthetic enzymes, another pathway to maintain BA concentrations in the liver after statin treatment might be through increasing BA uptake transporters in the liver. Indeed, atorvastatin increases the expression of the unconjugated BA uptake transporter Oatp1b2. Moreover, atorvastatin also increases the expression of the conjugated BA uptake transporter Ntcp (Fig. 8A). The increase in BA uptake transporters in the liver could be a direct effect by atorvastatin or an indirect effect due to decrease in BA recycling from the intestine. Although atorvastatin does not alter the expression of the BA efflux transporter Bsep (Fig. 8B), it increases the mRNA of Mrp2, which mediates the biliary excretion of tauro-BA-sulfates (23) as well as glutathione, which provides the osmotic force that drives BA-independent bile flow (41). This suggests that atorvastatin might increase bile flow. Several reports have shown that BAs seem to cause more changes in SHP and Fgf15 than the BA transporters, which are also FXR targets (42–44). Comparing germ-free and conventional mice, Sayin et al. (43) revealed that gut microbiota primarily affect FXR targets in the ileum and not the liver, and despite the marked increase in Fgf15 and SHP, the expression of other FXR target genes were slightly increased (Ostα), not altered (Ostβ, Ibabp, Ntcp, and Bsep), or even decreased (Mrp2) in the presence of gut microbiota. Consistent with these findings, the present study showed that the expression of SHP and Fgf15 were decreased by atorvastatin, whereas other FXR targets, such as Bsep, Asbt, and Ostα/β, remained relatively constant.
In addition to BA transporters, the present study is the first report on the effect of statins in regulating the expression of many transporters (such as Oatp1a1, Oatp1a4, Oct1, Ent1, Mate1, etc.) in the liver of mice. Atorvastatin increases the hepatic expression of the statin uptake transporter Oatp1b2, as well as statin efflux transporters Mrp2 and Bcrp (Fig. 8). The upregulation of Mrp2 and Bcrp is consistent with previous reports in the HepG2 cell line with atorvastatin (11). In addition, the expression of Oct1 is increased by atorvastatin (Fig. 8A), indicating a possible increase in the uptake of organic cations, such as its model substrate tetraethylammonium (45), as well as the antidiabetic drug metformin (46).
Furthermore, statins may increase the efflux of phospholipids and cholesterol out of the liver. Atorvastatin increases the mRNA of Mdr2 (Fig. 8B), which encodes a phospholipid efflux transporter to facilitate micelle formation and the function of BAs. In addition, atorvastatin increases the expression of Abca1, which is the basolateral efflux transporter for cholesterol, as well as Abcg5 and Abcg8, which function as dimers to mediate the canalicular efflux of cholesterol. This finding is consistent with previous reports that pitavastatin increased the mRNA of Abca1 in rat hepatoma cell lines (31) and simvastatin promoted Abca1 expression in mice with atherogenesis (47). A synergistic hypocholesterolemic effect could be potentially achieved by simultaneously inhibiting cholesterol biosynthesis and promoting Abcg5/Abcg8-mediated cholesterol excretion (48). Therefore, enhanced cholesterol excretion is likely an additional mechanism for atorvastatin to lower cholesterol. Further studies on protein expression and transporter membrane localization, as well as transporting activity, are necessary to draw a definite conclusion.
The findings of the present study are in general agreement with the previous report in mice that atorvastatin increased fecal BA excretion and expression of Cyp7a1, but did not affect BA conjugating enzymes (49). Several reports showed that statins at therapeutic doses had little effect on hepatic BA synthesis or turnover in humans. Specifically, atorvastatin did not alter BA concentrations or composition in patients with primary biliary cirrhosis (50); lovastatin did not alter the synthesis or turn-over of BAs (51); fluvastatin did not alter BA composition in bile (52); and pravastatin did not alter the cholesterol 7α-hydroxylase activity (53) or fecal BAs (54), but was shown to decrease 7α-hydroxy-4-cholesten-3-one level, which reflects BA synthesis (55). In addition, pravastatin was shown to decrease BA synthesis (56) and decrease BA concentrations in the plasma of bile duct-obstructed rats (40). The discrepancies in the literature about BA regulation by statins may be due to species differences, different disease/toxicity models, or different statins used. Further studies are needed to determine the effect of atorvastatin on BA homeostasis in disease status, and whether atorvastatin could be used as a combined treatment with UDCA to treat hepatobilliary diseases.
In conclusion, the present study demonstrated that the cholesterol-lowering drug atorvastatin maintains total BAs in the liver and serum of mice, and this appears to be due to the compensatory induction of some critical BA-synthetic enzymes (Cyp7a1 and Cyp27a1) and BA uptake transporters (Ntcp and Oatp1b2) in the liver. The marked induction of Cyp7a1 by atorvastatin is likely due to the repression of FXR signaling in both the liver and intestine. The induction of Cyp7a1 could serve as an additional novel mechanism for the cholesterol-lowering effects of statins in addition to the inhibition of HMG-CoA reductase and induction of the LDL receptor. Moreover, this study provides comprehensive information on the regulatory role of statins on the expression of various transporters in the liver in vivo.
Supplementary Material
Footnotes
Abbreviations:
- Asbt
- apical Na+-dependent bile salt transporter
- BA
- bile acid
- BAL
- bile acid-CoA ligase
- BAT
- bile acid CoA:amino acid N-acyl-transferase
- Bcrp
- breast cancer resistant protein
- Bsep
- bile salt export pump
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- Cyp
- cytochrome P450
- Cyp7a1
- cholesterol 7α-hydroxylase
- DCA
- deoxycholic acid
- EHC
- enterohepatic circulation
- Ent1
- equilibrative nucleoside transporter 1
- Fgf
- fibroblast growth factor
- FXR
- farnesoid X receptor
- GB
- gall bladder
- HDCA
- hyodeoxycholic acid
- HI
- hydrophobicity index
- Ibabp
- ileal bile acid binding protein
- IS
- internal standard
- LCA
- lithocholic acid
- LI
- large intestine
- Mate
- multidrug and toxin extrusion
- MCA
- muricholic acid
- MDCA
- murideoxycholic acid
- Mdr
- multidrug resistance protein
- MeOH
- methanol
- Mrp
- multidrug resistance-associated protein
- Ntcp
- Na+/taurocholate cotransporting polypeptide
- Oatp
- organic anion transporting polypeptide
- Oct
- organic cation transporter
- Ost
- organic solute tranporter
- SHP
- small heterodimer partner
- SI
- small intestine
- UDCA
- ursodeoxycholic acid
- UPLC-MS/MS
- ultra-performance LC-MS/MS
This study was supported by National Institutes of Health Grant ES009649.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
REFERENCES
- 1.McTaggart F., Jones P. 2008. Effects of statins on high-density lipoproteins: a potential contribution to cardiovascular benefit. Cardiovasc. Drugs Ther. 22: 321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maron D. J., Fazio S., Linton M. F. 2000. Current perspectives on statins. Circulation. 101: 207–213. [DOI] [PubMed] [Google Scholar]
- 3.Shepherd J., Hunninghake D. B., Barter P., McKenney J. M., Hutchinson H. G. 2003. Guidelines for lowering lipids to reduce coronary artery disease risk: a comparison of rosuvastatin with atorvastatin, pravastatin, and simvastatin for achieving lipid-lowering goals. Am. J. Cardiol. 91: 11C–17C; discussion 17C–19C. [DOI] [PubMed] [Google Scholar]
- 4.Ieiri I., Higuchi S., Sugiyama Y. 2009. Genetic polymorphisms of uptake (OATP1B1, 1B3) and efflux (MRP2, BCRP) transporters: implications for inter-individual differences in the pharmacokinetics and pharmacodynamics of statins and other clinically relevant drugs. Expert Opin. Drug Metab. Toxicol. 5: 703–729. [DOI] [PubMed] [Google Scholar]
- 5.Ho R. H., Tirona R. G., Leake B. F., Glaeser H., Lee W., Lemke C. J., Wang Y., Kim R. B. 2006. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology. 130: 1793–1806. [DOI] [PubMed] [Google Scholar]
- 6.Zaher H., Meyer zu Schwabedissen H. E., Tirona R. G., Cox M. L., Obert L. A., Agrawal N., Palandra J., Stock J. L., Kim R. B., Ware J. A. 2008. Targeted disruption of murine organic anion-transporting polypeptide 1b2 (Oatp1b2/Slco1b2) significantly alters disposition of prototypical drug substrates pravastatin and rifampin. Mol. Pharmacol. 74: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greupink R., Dillen L., Monshouwer M., Huisman M. T., Russel F. G. 2011. Interaction of fluvastatin with the liver-specific Na+ -dependent taurocholate cotransporting polypeptide (NTCP). Eur. J. Pharm. Sci. 44: 487–496. [DOI] [PubMed] [Google Scholar]
- 8.Jemnitz K., Veres Z., Tugyi R., Vereczkey L. 2010. Biliary efflux transporters involved in the clearance of rosuvastatin in sandwich culture of primary rat hepatocytes. Toxicol. In Vitro. 24: 605–610. [DOI] [PubMed] [Google Scholar]
- 9.Huang L., Wang Y., Grimm S. 2006. ATP-dependent transport of rosuvastatin in membrane vesicles expressing breast cancer resistance protein. Drug Metab. Dispos. 34: 738–742. [DOI] [PubMed] [Google Scholar]
- 10.Hirano M., Maeda K., Hayashi H., Kusuhara H., Sugiyama Y. 2005. Bile salt export pump (BSEP/ABCB11) can transport a nonbile acid substrate, pravastatin. J. Pharmacol. Exp. Ther. 314: 876–882. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues A. C., Curi R., Genvigir F. D., Hirata M. H., Hirata R. D. 2009. The expression of efflux and uptake transporters are regulated by statins in Caco-2 and HepG2 cells. Acta Pharmacol. Sin. 30: 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kivistö K. T., Grisk O., Hofmann U., Meissner K., Möritz K. U., Ritter C., Arnold K. A., Lutjöohann D., von Bergmann K., Klöting I., et al. 2005. Disposition of oral and intravenous pravastatin in MRP2-deficient TR- rats. Drug Metab. Dispos. 33: 1593–1596. [DOI] [PubMed] [Google Scholar]
- 13.Hirano M., Maeda K., Matsushima S., Nozaki Y., Kusuhara H., Sugiyama Y. 2005. Involvement of BCRP (ABCG2) in the biliary excretion of pitavastatin. Mol. Pharmacol. 68: 800–807. [DOI] [PubMed] [Google Scholar]
- 14.Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. 2009. Bile acids as regulatory molecules. J. Lipid Res. 50: 1509–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann A. F., Sjovall J., Kurz G., Radominska A., Schteingart C. D., Tint G. S., Vlahcevic Z. R., Setchell K. D. 1992. A proposed nomenclature for bile acids. J. Lipid Res. 33: 599–604. [PubMed] [Google Scholar]
- 16.Fu Z. D., Csanaky I. L., Klaassen C. D. 2012. Gender-divergent profile of bile acid homeostasis during aging of mice. PLoS ONE. 7: e32551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myant N. B., Mitropoulos K. A. 1977. Cholesterol 7 alpha-hydroxylase. J. Lipid Res. 18: 135–153. [PubMed] [Google Scholar]
- 18.Chiang J. Y. 2004. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 40: 539–551. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 21.Csanaky I. L., Lu H., Zhang Y., Ogura K., Choudhuri S., Klaassen C. D. 2011. Organic anion-transporting polypeptide 1b2 (Oatp1b2) is important for the hepatic uptake of unconjugated bile acids: studies in Oatp1b2-null mice. Hepatology. 53: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trauner M., Boyer J. L. 2003. Bile salt transporters: molecular characterization, function, and regulation. Physiol. Rev. 83: 633–671. [DOI] [PubMed] [Google Scholar]
- 23.Akita H., Suzuki H., Ito K., Kinoshita S., Sato N., Takikawa H., Sugiyama Y. 2001. Characterization of bile acid transport mediated by multidrug resistance associated protein 2 and bile salt export pump. Biochim. Biophys. Acta. 1511: 7–16. [DOI] [PubMed] [Google Scholar]
- 24.Imai Y., Asada S., Tsukahara S., Ishikawa E., Tsuruo T., Sugimoto Y. 2003. Breast cancer resistance protein exports sulfated estrogens but not free estrogens. Mol. Pharmacol. 64: 610–618. [DOI] [PubMed] [Google Scholar]
- 25.Janvilisri T., Shahi S., Venter H., Balakrishnan L., van Veen H. W. 2005. Arginine-482 is not essential for transport of antibiotics, primary bile acids and unconjugated sterols by the human breast cancer resistance protein (ABCG2). Biochem. J. 385: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Klaassen C. D. 2010. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51: 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nachtigal P., Pospisilova N., Jamborova G., Pospechova K., Solichova D., Andrys C., Zdansky P., Micuda S., Semecky V. 2008. Atorvastatin has hypolipidemic and anti-inflammatory effects in apoE/LDL receptor-double-knockout mice. Life Sci. 82: 708–717. [DOI] [PubMed] [Google Scholar]
- 28.Dostal L. A., Schardein J. L., Anderson J. A. 1994. Developmental toxicity of the HMG-CoA reductase inhibitor, atorvastatin, in rats and rabbits. Teratology. 50: 387–394. [DOI] [PubMed] [Google Scholar]
- 29.Fu Z. D., Klaassen C. D. 2013. Increased bile acids in enterohepatic circulation by short-term calorie restriction in male mice. Toxicol. Appl. Pharmacol. 273: 680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Lou G., Meng Z., Huang W. 2011. TGR5: a novel target for weight maintenance and glucose metabolism. Exp. Diabetes Res. 2011: 853501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawson P. A., Lan T., Rao A. 2009. Bile acid transporters. J. Lipid Res. 50: 2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez I., Perdicaro D. J., Brown A. W., Hammons S., Carden T. J., Carr T. P., Eskridge K. M., Walter J. 2013. Diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl. Environ. Microbiol. 79: 516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haeusler R. A., Pratt-Hyatt M., Welch C. L., Klaassen C. D., Accili D. 2012. Impaired generation of 12-hydroxylated bile acids links hepatic insulin signaling with dyslipidemia. Cell Metab. 15: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schölmerich J., Becher M. S., Schmidt K., Schubert R., Kremer B., Feldhaus S., Gerok W. 1984. Influence of hydroxylation and conjugation of bile salts on their membrane-damaging properties–studies on isolated hepatocytes and lipid membrane vesicles. Hepatology. 4: 661–666. [DOI] [PubMed] [Google Scholar]
- 35.Rolo A. P., Oliveira P. J., Moreno A. J., Palmeira C. M. 2000. Bile acids affect liver mitochondrial bioenergetics: possible relevance for cholestasis therapy. Toxicol. Sci. 57: 177–185. [DOI] [PubMed] [Google Scholar]
- 36.Aoki T., Yamazaki H., Maejima T., Sato F., Kitahara M., Saito Y. 2003. Effect of pitavastatin on sterol and bile acid excretion in guinea pigs. Arzneimittelforschung. 53: 351–356. [PubMed] [Google Scholar]
- 37.Clerc T., Jomier M., Chautan M., Portugal H., Senft M., Pauli A. M., Laruelle C., Morel O., Lafont H., Chanussot F. 1993. Mechanisms of action in the liver of crilvastatin, a new hydroxymethylglutaryl-coenzyme A reductase inhibitor. Eur. J. Pharmacol. 235: 59–68. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto S., Nakano K., Kosahara K., Kishinaka M., Oda H., Ichimiya H., Chijiiwa K., Kuroki S. 1994. Effects of pravastatin and ursodeoxycholic acid on cholesterol and bile acid metabolism in patients with cholesterol gallstones. J. Gastroenterol. 29: 47–55. [DOI] [PubMed] [Google Scholar]
- 39.Kajinami K., Brousseau M. E., Ordovas J. M., Schaefer E. J. 2005. A promoter polymorphism in cholesterol 7alpha-hydroxylase interacts with apolipoprotein E genotype in the LDL-lowering response to atorvastatin. Atherosclerosis. 180: 407–415. [DOI] [PubMed] [Google Scholar]
- 40.Kolouchova G., Brcakova E., Hirsova P., Sispera L., Tomsik P., Cermanova J., Hyspler R., Slanarova M., Fuksa L., Lotkova H., et al. 2011. Pravastatin modulates liver bile acid and cholesterol homeostasis in rats with chronic cholestasis. J. Gastroenterol. Hepatol. 26: 1544–1551. [DOI] [PubMed] [Google Scholar]
- 41.Johnson D. R., Habeebu S. S., Klaassen C. D. 2002. Increase in bile flow and biliary excretion of glutathione-derived sulfhydryls in rats by drug-metabolizing enzyme inducers is mediated by multidrug resistance protein 2. Toxicol. Sci. 66: 16–26. [DOI] [PubMed] [Google Scholar]
- 42.Klaassen C. D., Song P. 2010. Bile acids regulate bile acid synthetic enzymes (Cyp7a1 and 8b1) but not hepatic transporters (Ntcp or Bsep) (Abstract in the XXI International Bile Acid Meeting. Freiburg, Baden-Württemberg, Germany, October 7–8, 2010).
- 43.Sayin S. I., Wahlstrom A., Felin J., Jantti S., Marschall H. U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., Backhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 44.Hu X., Bonde Y., Eggertsen G., Rudling M. 2014. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J. Intern. Med. 275: 27–38. [DOI] [PubMed] [Google Scholar]
- 45.Jonker J. W., Wagenaar E., Mol C. A., Buitelaar M., Koepsell H., Smit J. W., Schinkel A. H. 2001. Reduced hepatic uptake and intestinal excretion of organic cations in mice with a targeted disruption of the organic cation transporter 1 (Oct1 [Slc22a1]) gene. Mol. Cell. Biol. 21: 5471–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu Y., Sheardown S. A., Brown C., Owen R. P., Zhang S., Castro R. A., Ianculescu A. G., Yue L., Lo J. C., Burchard E. G., et al. 2007. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 117: 1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song G., Liu J., Zhao Z., Yu Y., Tian H., Yao S., Li G., Qin S. 2011. Simvastatin reduces atherogenesis and promotes the expression of hepatic genes associated with reverse cholesterol transport in apoE-knockout mice fed high-fat diet. Lipids Health Dis. 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang W., Ma Y., Yu L. 2006. Plasma cholesterol is hyperresponsive to statin in ABCG5/ABCG8 transgenic mice. Hepatology. 44: 1259–1266. [DOI] [PubMed] [Google Scholar]
- 49.Parker R. A., Garcia R., Ryan C. S., Liu X., Shipkova P., Livanov V., Patel P., Ho S. P. 2013. Bile acid and sterol metabolism with combined HMG-CoA reductase and PCSK9 suppression. J. Lipid Res. 54: 2400–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stojakovic T., Putz-Bankuti C., Fauler G., Scharnagl H., Wagner M., Stadlbauer V., Gurakuqi G., Stauber R. E., Marz W., Trauner M. 2007. Atorvastatin in patients with primary biliary cirrhosis and incomplete biochemical response to ursodeoxycholic acid. Hepatology. 46: 776–784. [DOI] [PubMed] [Google Scholar]
- 51.Duane W. C. 1994. Effects of lovastatin and dietary cholesterol on bile acid kinetics and bile lipid composition in healthy male subjects. J. Lipid Res. 35: 501–509. [PubMed] [Google Scholar]
- 52.Tazuma S., Ohya T., Mizuno T., Takizawa I., Kunita T., Takata K., Hayashi K., Hino F., Tokumo H., Watanabe T., et al. 1995. Effects of fluvastatin on human biliary lipids. Am. J. Cardiol. 76: 110A–113A. [DOI] [PubMed] [Google Scholar]
- 53.Reihnér E., Rudling M., Ståhlberg D., Berglund L., Ewerth S., Björkhem I., Einarsson K., Angelin B. 1990. Influence of pravastatin, a specific inhibitor of HMG-CoA reductase, on hepatic metabolism of cholesterol. N. Engl. J. Med. 323: 224–228. [DOI] [PubMed] [Google Scholar]
- 54.Hoogerbrugge-vd Linden N., de Rooy F. W., Jansen H., van Blankenstein M. 1990. Effect of pravastatin on biliary lipid composition and bile acid synthesis in familial hypercholesterolaemia. Gut. 31: 348–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida T., Honda A., Shoda J., Abei M., Matsuzaki Y., Tanaka N., Miyazaki H., Osuga T. 1997. Short-term effects of 3-hydroxy-3-methylglutaryl-CoA reductase inhibitor on cholesterol and bile acid synthesis in humans. Lipids. 32: 873–878. [DOI] [PubMed] [Google Scholar]
- 56.Scheibner J., Fuchs M., Schiemann M., Tauber G., Hormann E., Stange E. F. 1993. Bile acid synthesis from newly synthesized vs. preformed cholesterol precursor pools in the rat. Hepatology. 17: 1095–1102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.