Abstract
Sphingosine 1-phosphate receptor 1 (S1P1), an abundantly-expressed G protein-coupled receptor which regulates key vascular and immune responses, is a therapeutic target in autoimmune diseases. Fingolimod/Gilenya (FTY720), an oral medication for relapsing-remitting multiple sclerosis, targets S1P1 receptors on immune and neural cells to suppress neuroinflammation. However, suppression of endothelial S1P1 receptors is associated with cardiac and vascular adverse effects. Here we report the genetic variations of the S1P1 coding region from exon sequencing of >12,000 individuals and their functional consequences. We conducted functional analyses of 14 nonsynonymous single nucleotide polymorphisms (SNPs) of the S1PR1 gene. One SNP mutant (Arg120 to Pro) failed to transmit sphingosine 1-phosphate (S1P)-induced intracellular signals such as calcium increase and activation of p44/42 MAPK and Akt. Two other mutants (Ile45 to Thr and Gly305 to Cys) showed normal intracellular signals but impaired S1P-induced endocytosis, which made the receptor resistant to FTY720-induced degradation. Another SNP mutant (Arg13 to Gly) demonstrated protection from coronary artery disease in a high cardiovascular risk population. Individuals with this mutation showed a significantly lower percentage of multi-vessel coronary obstruction in a risk factor-matched case-control study. This study suggests that individual genetic variations of S1P1 can influence receptor function and, therefore, infer differential disease risks and interaction with S1P1-targeted therapeutics.
Keywords: sphingosine phosphate, drug therapy, cell signaling, endocytosis, genetics, sphingosine 1-phosphate receptor, single nucleotide polymorphism
Sphingosine 1-phosphate receptor 1 (S1P1) is a G protein-coupled receptor (GPCR) for a versatile lysophospholipid mediator sphingosine 1-phosphate (S1P) (1, 2). S1P levels are high in blood and lymph, but low in the interstitial tissue fluid, thereby forming a gradient important for immune cell trafficking (3). S1P, which is poorly soluble in aqueous solutions, is bound by specific chaperones. For example, plasma S1P is bound by albumin (4) or apoM (5), a specific apolipoprotein found in HDL. The S1P-S1P1 signaling system is critical for sprouting angiogenesis (6–8) and vascular permeability (2, 9–11). In addition, part of the vasoprotective, anti-inflammatory, and anti-atherosclerotic effects associated with HDL is attributed to its cargo, apoM-S1P (12–15). The S1P-S1P1 signaling system also plays an important role in immune cell trafficking (3). The gradient of S1P concentration between interstitial fluids and lymph is a basis that allows lymphocytes to egress from secondary lymphoid organs to the circulation via the chemotactic activity of S1P1 (3, 16).
Previous studies have accumulated significant functional and structural information on S1P1 (1, 2). The recent crystal structure determination of S1P1 has also enhanced our understanding of ligand interactions with the receptor (17, 18). Upon S1P binding, S1P1 activates heterotrimeric Gαi protein and transmits signals via the p44/42 MAPK, PI3K/Akt, and Rac pathways (19–21). Subsequently, S1P1 gets phosphorylated by GRK2 protein in the carboxyl terminal region (22) and undergoes clathrin-dependent endocytosis, then recycles back to the plasma membrane (23). This endocytosis/recycling pathway is essential for the regulation of S1P1 activity. Impairment of the receptor endocytosis results in enhanced signaling and hyper-immune consequences, whereas vascular endothelial function is enhanced (24–26).
The study of the S1P-S1P1 signaling system has entered the therapeutic era with the successful launch of fingolimod/Gilenya (FTY720) in the treatment of relapsing-remitting multiple sclerosis (27, 28). Indeed, many novel agonists and antagonists for S1P1 have been developed and some of them are undergoing clinical trials to test their efficacy in the control of many diseases, including relapsing-remitting multiple sclerosis, the secondary progressive form of multiple sclerosis, psoriasis, and amyotrophic lateral sclerosis (29). Among them, FTY720, which gets phosphorylated into FTY720-P by sphingosine kinase-2 (10), induces irreversible ubiquitin-dependent degradation of S1P1 and blocks lymphocyte egress from secondary lymphoid organs (25, 30). Genetic variations represented by single nucleotide polymorphisms (SNPs) confer individual variation in terms of susceptibility to certain diseases, development and severity of diseases, and efficacy and adverse effects of therapeutic drugs. However, our knowledge is quite limited on the genetic variations of S1P1 and their effects on the receptor functions and disease development (31). Individual genetic variations of the S1PR1 gene in various disease cohorts and their functional consequences may lead to a better understanding of S1P1 biology in disease outcomes and in targeted therapeutics.
The National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project (NHLBI GO ESP) sequenced protein-coding regions of the human genome across diverse, richly phenotyped populations with the ultimate aim to discover novel genes and mechanisms contributing to heart, lung, and blood disorders (32). The project has identified 14 nonsynonymous SNPs in the S1PR1 gene out of more than 10,000 patients across many different clinical trials, with variations of the SNP prevalence from 1 to 0.001%. In this study, we determined the functional consequences of such nonsynonymous S1P1 mutations. We also conducted a sequencing analysis of the S1PR1 gene in a cardiovascular disease cohort with more than 1,800 patients in which we had access to clinical data including coronary angiograms. We report the identification of one loss-of-function mutation and two mutations that result in impaired receptor endocytosis. We also report one mutation that is associated with a significantly higher incidence in the cardiovascular disease cohort.
MATERIALS AND METHODS
Data collection from NHLBI GO ESP Exome Variant Server
Data for genetic variations of the S1PR1 gene was collected from NHLBI GO ESP Exome Variant Server (32) according to the guidelines of the data usage. The data set was not linked to clinical phenotypes.
Cell culture
CHO-K1 and 293T cells were maintained in Ham’s F-12 medium and DMEM (Sigma-Aldrich), respectively, containing 10% FBS. Human umbilical vein cord endothelial cells (HUVECs) were maintained on fibronectin-coated dishes in EGM2 medium (Lonza). Cells were cultured in a humidified 5% CO2 incubator at 37°C.
Site-directed mutagenesis
cDNAs for S1P1 mutants were generated using a QuikChange site-directed mutagenesis kit (Agilent). Primer information used for the mutagenesis is available in the supplementary material.
Lentivirus vectors and cell transduction
cDNAs for green fluorescent protein (GFP)-tagged WT and mutant S1P1 were cloned in a pCDH-CMV-MCS-EF1-Puro vector (System Biosciences). A lentivirus vector for shRNA-mediated S1P1 suppression was purchased from Sigma-Aldrich (the RNA Interference Consortium clone TRCN0000011359). Pseudoviral particles were made by transfecting these vectors together with packaging (pMDLg/pRRE and pRSV-Rev) and envelope (pMD2.G) plasmids into 293T producer cells. The culture supernatant containing pseudoviral particles was recovered and used to transduce cells. Transduced cells were selected by treatment with puromycin overnight.
Receptor endocytosis assay
Endocytosis of S1P1 upon stimulation with S1P or FTY720-P was observed as described (33).
Western blot analysis
Cells were lysed by sonication in lysis buffer containing 50 mM Tris (pH 7.4), 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% Fos-Choline, 1 mM Na3VO4, 1 mM NaF, 10 mM β-glycerophosphate, and protease inhibitor cocktail (Sigma-Aldrich). Protein concentrations were determined by using a BCA protein assay kit (Thermo Scientific). Equal amounts of proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and then probed with primary antibodies against total- and phospho-p44/42 MAPK (Cell Signaling #9102 and #9106, respectively), total- and phospho-Akt (Cell Signaling #9272 and #9271, respectively), S1P1 (Santa Cruz #sc-25489), β-actin (Sigma-Aldrich #A5316), NA+/K+-ATPase (Developmental Studies Hybridoma Bank #α6F), and HRP-conjugated secondary antibodies. Signals were visualized by using Immobilon Western chemiluminescent HRP substrate (Millipore).
Cell-surface protein biotinylation assay
Cells were treated with a cell nonpermeable biotinylation reagent, EZ-link Sulfo-NHS-SS-Biotin (Thermo Scientific), and lysed in the lysis buffer. Biotinylated proteins were purified by using NeutrAvidin agarose beads (Thermo Scientific).
Measurement of intracellular calcium concentration
Changes in intracellular calcium concentrations upon ligand stimulation were monitored with a FLEX-station scanning fluorometer system (Molecular Devices) as described (34).
Collection of patient samples and sequencing analysis of the S1PR1 gene
Patients were recruited from Dartmouth-Hitchcock Medical Center under a protocol approved by the Institutional Review Board (35). Genomic DNA was extracted from blood using a PuregeneH system (Gentra Systems). The coding sequence of the human S1PR1 gene was amplified by PCR using the human genomic DNAs as templates and the following primers: 5′-tctccagccaaggaaaagc-3′ and 5′-agtaaagagcgcttccgg-3′. The amplified DNAs were purified using a QIAquick PCR purification kit (Qiagen) and subjected to Sanger DNA sequencing analyses at the Genomics Facility of the Biotechnology Resource Center, Cornell University.
Human HDL preparation
Lipoprotein particles with the density ranging from 1.066 to 1.21 were collected from human plasma by repeated ultracentrifugation as described (36), and dialyzed against PBS. S1P concentration contained in the HDL fraction was determined by a LC-MS/MS analysis in the Lipidomics Shared Resources at the Medical University of South Carolina (37).
Measurement of trans-endothelial electric resistance
HUVECs were seeded onto gelatin-coated gold electrodes (8W10E plates, Applied BioPhysics), serum starved in EBM2 medium (Lonza) for 1 h, and stimulated with S1P. Endothelial barrier function was assessed by monitoring electric resistance of the electrodes with an ECIS θ system (Applied BioPhysics).
RESULTS
Genetic variations of the S1PR1 gene found in the NHLBI GO ESP database
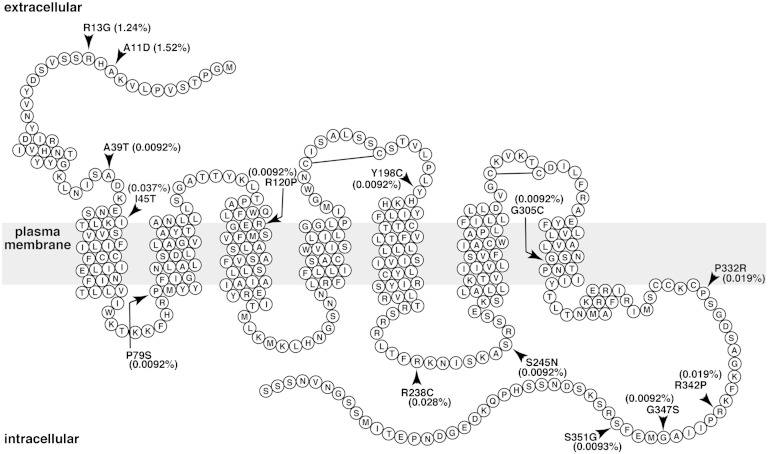
To examine the genetic variations of the S1PR1 gene, we performed a database search in the NHLBI GO ESP database (32). As of June 2012, we found 33 SNPs in the S1PR1 gene, consisting of 14 nonsynonymous mutations (Table 1), 14 synonymous mutations, 4 mutations in the 5′-untranslated region (UTR), and 1 mutation in the 3′-UTR. The positions of the amino acids changed in the 14 nonsynonymous mutations are illustrated in the secondary structure of S1P1 (Fig. 1). Those include Ala11 to Asp (A11D), R13G, A39T, I45T, P79S, R120P, Y198C, R238C, S245N, G305C, P332R, R342P, G347S, and S351G. Among them, A11D and R13G show relatively higher prevalence (>1%) in the African American and European American populations, respectively (Table 1).
TABLE 1.
Non-synonymous mutations of the human S1PR1 gene found in the NHLBI GO ESP database
| SNP Position | Identification Number (rs) | Alleles | EA Allele Number | AA Allele Number | Total Allele Number | cDNA Position | Protein Position | Amino Acid |
| 1:101704572 | rs61734752 | A/C | A = 1/C = 7,019 (0.014%) | A = 163/C = 3,575 (4.36%) | A = 164/C = 10,594 (1.52%) | 32 | 11 | Asp/Ala |
| 1:101704577 | rs41287280 | G/C | G = 116/C = 6,904 (1.65%) | G = 17/C = 3,721 (0.45%) | G = 133/C = 10,625 (1.24%) | 37 | 13 | Gly/Arg |
| 1:101704655 | rs140298731 | A/G | A = 0/G = 7,020 (0%) | A = 1/G = 3,737 (0.027%) | A = 1/G = 10,757 (0.0092%) | 115 | 39 | Thr/Ala |
| 1:101704674 | rs148977042 | C/T | C = 2/T = 7,018 (0.028%) | C = 2/T = 3,736 (0.054%) | C = 4/T = 10,754 (0.037%) | 134 | 45 | Thr/Ile |
| 1:101704775 | rs370003570 | T/C | T = 0/C = 7,020 (0%) | T = 1/C = 3,737 (0.027%) | T = 1/C = 10,757 (0.0092%) | 235 | 79 | Ser/Pro |
| 1:101704899 | rs149198314 | C/G | C = 1/G = 7,019 (0.014%) | C = 0/G = 3,738 (0%) | C = 1/G = 10,757 (0.0092%) | 359 | 120 | Pro/Arg |
| 1:101705133 | rs370425078 | G/A | G = 0/A = 7,020 (0%) | G = 1/A = 3,737 (0.027%) | G = 1/A = 10,757 (0.0092%) | 593 | 198 | Cys/Tyr |
| 1:101705252 | rs149041375 | T/C | T = 2/C = 7,018 (0.028%) | T = 1/C = 3,737 (0.027%) | T = 3/C = 10,755 (0.028%) | 712 | 238 | Cys/Arg |
| 1:101705274 | rs151135641 | A/G | A = 0/G = 7,020 (0%) | A = 1/G = 3,737 (0.027%) | A = 1/G = 10,757 (0.0092%) | 734 | 245 | Asn/Ser |
| 1:101705453 | rs146890331 | T/G | T = 1/G = 7,019 (0.014%) | T = 0/G = 3,738 (0%) | T = 1/G = 10,757 (0.0092%) | 913 | 305 | Cys/Gly |
| 1:101705535 | rsrs7549921 | G/C | G = 0/C = 7,020 (0%) | G = 2/C = 3,736 (0.054%) | G = 2/C = 10,756 (0.019%) | 995 | 332 | Arg/Pro |
| 1:101705565 | rs138199836 | C/G | C = 2/G = 7,018 (0.028%) | C = 0/G = 3,738 (0%) | C = 2/G = 10,756 (0.019%) | 1,025 | 342 | Pro/Arg |
| 1:101705579 | rs375635245 | A/G | A = 1/G = 7,019 (0.014%) | A = 0/G = 3,734 (0%) | A = 1/G = 10,753 (0.0092%) | 1,039 | 347 | Ser/Gly |
| 1:101705591 | rs149581242 | G/A | G = 0/A = 7,010 (0%) | G = 1/A = 3,735 (0.027%) | G = 1/A = 10,745 (0.0093%) | 1,051 | 351 | Cly/Ser |
The allele numbers in the European American (EA allele) and African American (AA allele) population are shown, as well as the total allele number as of June 2012.
Fig. 1.
Serpentine diagram of the secondary structure of human S1P1. Each circle represents one amino acid residue with a one letter amino acid code. The gray rectangle represents the plasma membrane. Positions of α-helixes were determined by referring the crystal structure of S1P1 (Protein Data Bank number 3V2Y) and by using a hydrophobicity-calculation program, SOSUI. Arrowheads denote the positions of amino acids changed in the 14 nonsynonymous mutations in the NHLBI GO ESP database. Prevalence of the 14 mutations is shown as percent of total sample numbers. Two extracellular disulfide bonds are also shown.
Functional analysis of the S1P1 SNP mutants
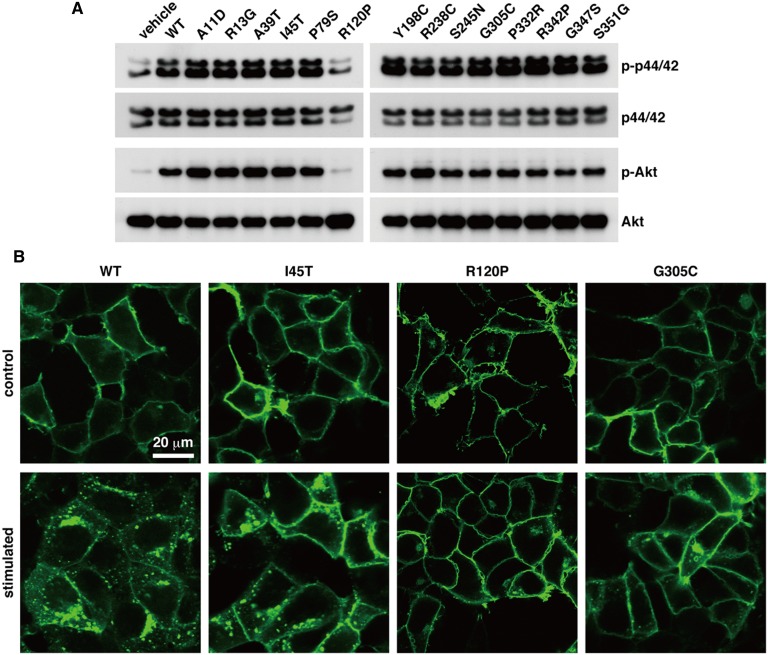
We expressed S1P1 harboring each mutation in CHO-K1 cells, and stimulated the cells with the ligand S1P to test the ability of the SNP mutations to influence the receptor-mediated activation of p44/42 MAPK and Akt signaling proteins. As shown in Fig. 2A, R120P mutant failed to mediate the activation of p44/42 MAPK and Akt in response to 100 nM S1P, while the other mutants did not show any change compared with WT S1P1. Arg120 is a critical residue for the binding of S1P through the interaction of its basic side chain with the phosphate group of S1P (17, 38). S1P binding ability of the receptor was largely impaired by R120A mutation (38). Thus, the defect of the R120P mutant in S1P-induced signaling is likely due to impaired S1P binding ability.
Fig. 2.
Characterization of 14 SNP mutants found in the NHLBI GO ESP database. CHO-K1 (A) or 293T (B) cells were transduced with lentivirus vectors for the expression of GFP-tagged S1P1 mutants, starved overnight, and then stimulated with 100 nM S1P for 5 min (A) or 200 nM S1P for 1 h (B). A: Total cell lysates were prepared and subjected to Western blot analyses for the activation of p44/42 MAPK and Akt. B: The cells were fixed with paraformaldehyde, and the GFP signal was observed for the localization of the S1P1 mutants under a confocal microscope system. Data are representative of at least three independent experiments with the same tendency.
We also examined ligand-induced receptor endocytosis that is an important step for the receptor desensitization processes (23). All of the mutants showed cell surface localization after serum starvation in the same way as WT S1P1 (Fig. 2B, supplementary Fig. I), which indicates that none of the mutations affect receptor translation and trafficking to the plasma membrane in the basal condition. Upon S1P stimulation, R120P mutant showed no receptor endocytosis (Fig. 2B), which confirms that activation of the receptor by S1P is impaired in the R120P mutant. Surprisingly, I45T showed diminished endocytosis and G305C failed to undergo endocytosis upon S1P stimulation (Fig. 2B). All other mutants responded to S1P and endocytosed in an indistinguishable manner from the WT (supplementary Fig. I).
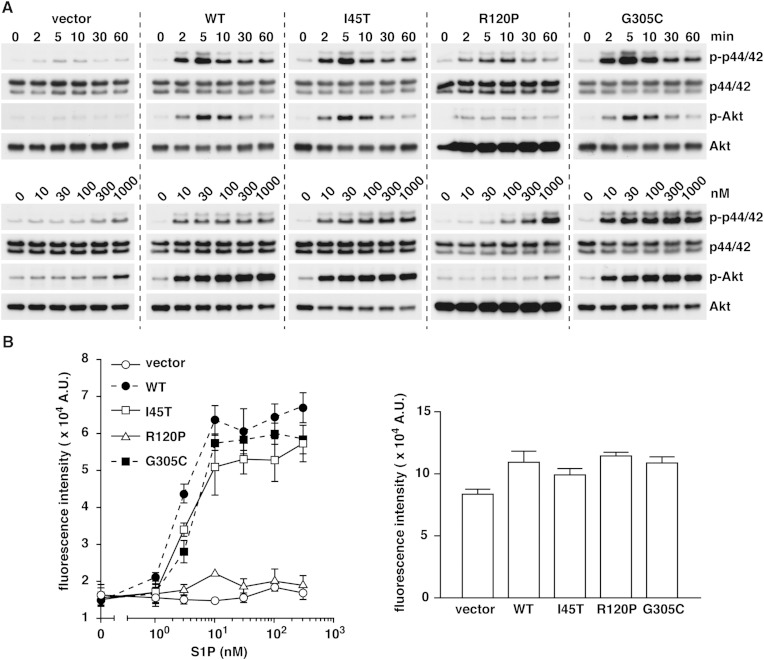
We further explored the time course and ligand-dose dependency of p44/42 and Akt activation in CHO-K1 cells overexpressing WT S1P1, I45T, R120P, and G305C mutants. We confirmed a similar level of expression of each receptor (supplementary Fig. II). As shown in Fig. 3A, I45T did not show any differences compared with WT S1P1 both in the time course and dose dependency. G305C had overall similar, but slightly enhanced, signaling compared with WT S1P1, which could be due to the defect in endocytosis-mediated desensitization processes (Fig. 2B). A higher dose of S1P (>100 nM) did induce modest activation of p44/42 MAPK in cells expressing the R120P mutant. This indicates that the R120P mutant is not completely insensitive to S1P, suggesting that several other residues, such as Tyr29, Lys34, Asn101, and Glu121, also contribute to the recognition of the head group of S1P (17). We also measured the increase in intracellular calcium concentration that is a proximal downstream event of the receptor activation. I45T and G305C mutants did not show any differences from WT S1P1, but the R120P mutant failed to mediate the calcium response (Fig. 3B). These results show that I45T and G305C mutants maintain the normal properties in the receptor activation but the receptor desensitization processes are impaired, while the R120P mutant shows largely impaired receptor activation.
Fig. 3.
Comparison of WT S1P1 with I45T, R120P, and G305C mutants. A: The CHO-K1 cells expressing each receptor were serum starved overnight and stimulated with 100 nM S1P for the indicated times (upper panel) or with the indicated concentrations of S1P for 5 min (lower panel). Data are the representative of two independent experiments with the same tendency. B (left): The cells were loaded with a calcium-sensitive fluorescent dye, fluo-8 AM. The changes in intracellular calcium concentration upon exposure to S1P were analyzed by a FlexStation system. Data represent the mean ± SEM (n = 3). B (right): The cells were also stimulated with a positive control ATP (10 μM) to check the similar fluo-8 loading efficiency and the cell viability. Data are representative of three independent experiments with the same tendency.
Diminished efficacy of FTY720 on I45T and G305C mutants
Endocytosis of S1P1 plays an important role in the receptor desensitization. It is a crucial process in the regulation of lymphocyte trafficking. In circulating lymphocytes, S1P1 is internalized in response to high concentrations of S1P in blood and lymph. In secondary lymphoid organs, S1P concentrations are maintained low, thus S1P1 recovers its cell surface localization (3). This is the basis for enabling lymphocytes to egress from lymphoid organs toward circulation along the gradient of S1P concentration. FTY720 inhibits the egress of lymphocytes from secondary lymphoid organs by inducing internalization and irreversible S1P1 degradation in the proteasome (25, 30). Recently, our group showed that mutant mice harboring the S1PR1 gene encoding phosphorylation-deficient, and as such endocytosis-deficient, receptors [S1P1 (S5A)] developed severe experimental autoimmune encephalomyelitis due to T helper 17 cell-mediated autoimmunity, both in the peripheral immune system and the nervous system (26). This S1P1 (S5A) receptor shows resistance to FTY720-induced degradation both in vivo and in vitro (25, 30).
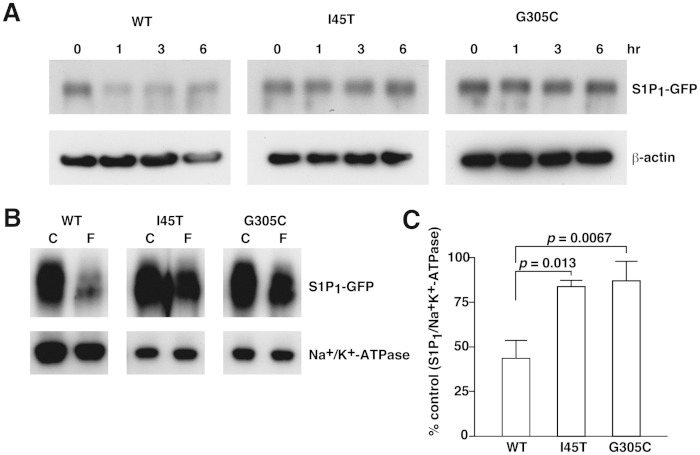
The defect in the endocytosis response found in I45T and G305C mutants phenocopies the S1P1 (S5A) receptor and may also lead to the resistance to FTY720-induced endocytosis and degradation. We examined the efficacy of FTY720-P, a phosphorylated form of FTY720, on I45T and G305C mutants, and found that these mutants showed much weaker endocytosis in response to FTY720-P treatment compared with the strong internalization of WT S1P1 (supplementary Fig. III). As a consequence of diminished endocytosis, I45T and G305C mutants showed much less FTY720-induced degradation than WT S1P1 (Fig. 4A). To confirm quantitatively that the endocytosis response is impaired in I45T and G305C mutants, we measured cell surface S1P1 before and after FTY720-P treatment. As shown in Fig. 4B, C, WT S1P1 showed a 56% decrease in the cell surface amount after 1 h of FTY720-P treatment, while I45T and G305C showed only 16 and 13% decreases, respectively. These results suggest that the efficacy of FTY720 treatment could be impaired in individuals who carry these genetic variations in the S1PR1 gene.
Fig. 4.
Diminished efficacy of FTY720-P on I45T and G305C mutants. CHO-K1 cells were transduced with lentivirus vectors for the expression of GFP-tagged S1P1, I45T, and G305C, starved overnight, and then stimulated with 10 nM FTY720-P for the indicated time (A) or for 1 h (B). A: Total cell lysates were prepared and subjected to Western blot analyses for the degradation of S1P1. B: Cell-surface proteins were biotinylated, purified, and subjected to Western blot analyses to check the amount of S1P1 remaining on the cell surface after FTY720-P treatment. C: Densitometric analysis of (B) using the National Institutes of Health ImageJ program is shown. Data represent the mean ± SD (n = 3).
Sequencing analysis and phenotype correlation of the S1PR1 gene in a cardiovascular cohort
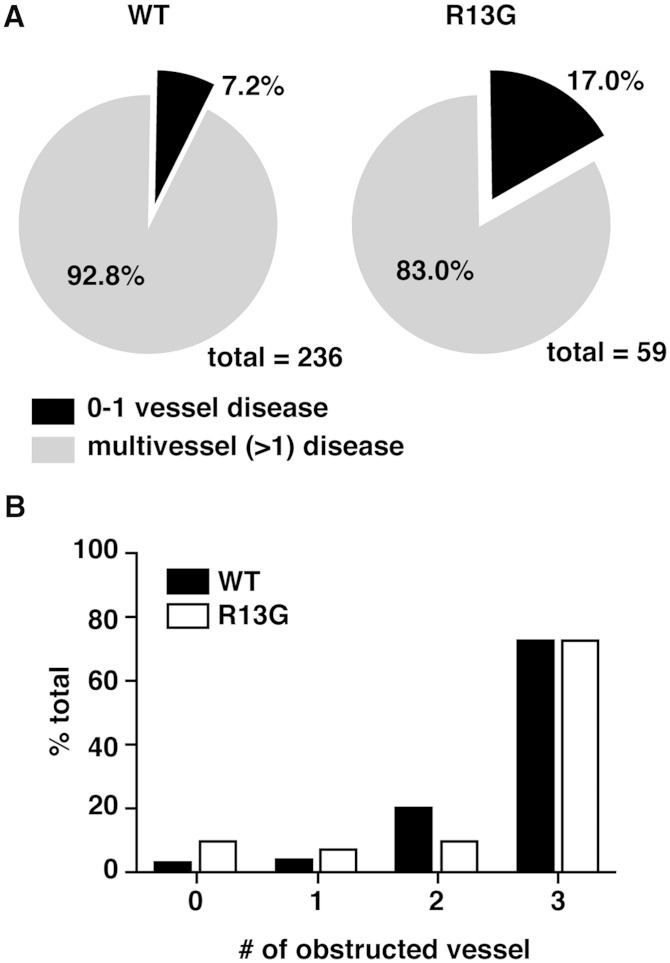
The S1P-S1P1 signaling system also plays an important role in vascular development and homeostasis. Dysfunction of this system leads to increased vascular permeability and hypersensitivity to anaphylactic stimulus (2, 9–11). Anti-inflammatory effects of HDL on the endothelium are partially attributed to its cargo S1P (12–15). Moreover, one of the adverse effects of FTY720 in therapeutics for multiple sclerosis is bradycardia, which is associated with the agonistic effects of FTY720 on S1P1 and subsequent activation of the G protein-coupled inwardly rectifying potassium channel in cardiomyocytes (39). Therefore, it is of clinical importance to examine the prevalence of nonsynonymous mutations in the S1PR1 gene in a cardiovascular disease cohort and its relationship with disease severity or with sensitivity to FTY720. Toward this end, we performed a sequencing analysis of the S1PR1 gene in 1,814 patients in a high cardiovascular risk population (35). As summarized in Table 2, we found five nonsynonymous mutations that resulted in amino acid changes of A11D, R13G, V20I, R238C, and S333R. The mutations for V20I and S333R have not been reported before. R13G showed a significantly higher incidence than that observed in the NHLBI GO ESP (3.58% versus 1.24%). The other detected mutations were rare (0.0055%). Because coronary angiograms (for detection of coronary obstruction) were available on all the patients, we compared the R13G patients (n = 59) to age- and risk factor-matched control patients (n = 236 in a 4:1 match). R13G appeared to protect from coronary artery disease (CAD); significantly more patients had no CAD and significantly fewer patients had multiple vessel obstruction compared with the control patients (Table 3, Fig. 5).
TABLE 2.
Non-synonymous mutations of the human S1PR1 gene found in the sequence analysis of 1,814 patients of the cardiovascular cohort
| Alleles | Allele Number | cDNA Position | Protein Position | Amino Acid |
| A/C | A = 1/C = 1,813 (0.055%) | 32 | 11 | Asp/Ala |
| G/C | G = 65/C = 1,749 (3.58%) | 37 | 13 | Gly/Arg |
| A/G | A = 1/G = 1,813 (0.055%) | 58 | 20 | Ile/Val |
| T/C | T = 1/C = 1,813 (0.055%) | 712 | 238 | Cys/Arg |
| A/C | A = 1/C = 1,813 (0.055%) | 999 | 333 | Arg/Ser |
Sample number analyzed = 1,814.
TABLE 3.
Age-, gender-, and risk factor-matched case-control comparison between R13G and WT S1P1
| Factor | R13G (n = 59) | WT (n = 236) | Significance |
| Age (± SEM)a | 66.49 ± 1.51 | 66.80 ± 0.83 | NS |
| Male (% total)a | 41 (69.5%) | 165 (69.9%) | NS |
| Female (% total)a | 18 (30.5%) | 71 (30.1%) | NS |
| BMI (± SEM)a | 29.37 ± 0.76 | 29.03 ± 0.42 | NS |
| HTN (% total)a | 40 (67.8%) | 163 (69.1%) | NS |
| Chol (% total)a | 46 (77.9%) | 179 (75.8%) | NS |
| FHx (% total)a | 26 (44.1%) | 103 (43.6%) | NS |
| DM (% total)a | 23 (38.9%) | 98 (41.5%) | NS |
| Smoking (% total)a | 9 (15.2%) | 33 (14.0%) | NS |
| LM (% total)b | 26 (44.1%) | 105 (44.5%) | NS |
| LAD (% total)b | 50 (84.7%) | 212 (89.8%) | NS |
| LCx (% total)b | 46 (77.9%) | 200 (84.7%) | NS |
| RCA (% total)b | 49 (83.0%) | 207 (87.7%) | NS |
| Multivessel disease (% total)b | 49 (83.0%) | 219 (93.2%) | P < 0.05 |
| Number of diseased vessels (± SEM)b | 2.98 ± 0.17 | 3.07 ± 0.06 | NS |
| Number of cardiovascular events (± SEM)b | 1.89 ± 0.20 | 1.78 ± 0.78 | NS |
| Number of post interventions (± SEM)b | 0.57 ± 0.07 | 0.62 ± 0.04 | NS |
HTN, hypertension; Chol, dyslipidemia; FHx, family history of cardiovascular diseases; DM, diabetes mellitus; LM, left main coronary artery; LAD, left anterior descending coronary artery; LCx, left circumflex coronary artery; RCA, right coronary artery.
Age, gender and risk factors were matched between the case (R13G mutant, n = 59) and the control (WT S1P1, n = 236).
Numbers of patients with no CAD and with multivessel disease (obstruction in >1 major coronary artery) are shown.
Fig. 5.
Lower percentage of multi-vessel obstructions in the patients with R13G mutation. A: A pie chart shows the percentages of the patients with zero or one (black) or with two or more (gray) obstructed vessels observed in coronary angiography in the case-control study between the R13G mutant and WT S1P1. B: Distribution of the obstructed vessel number is shown.
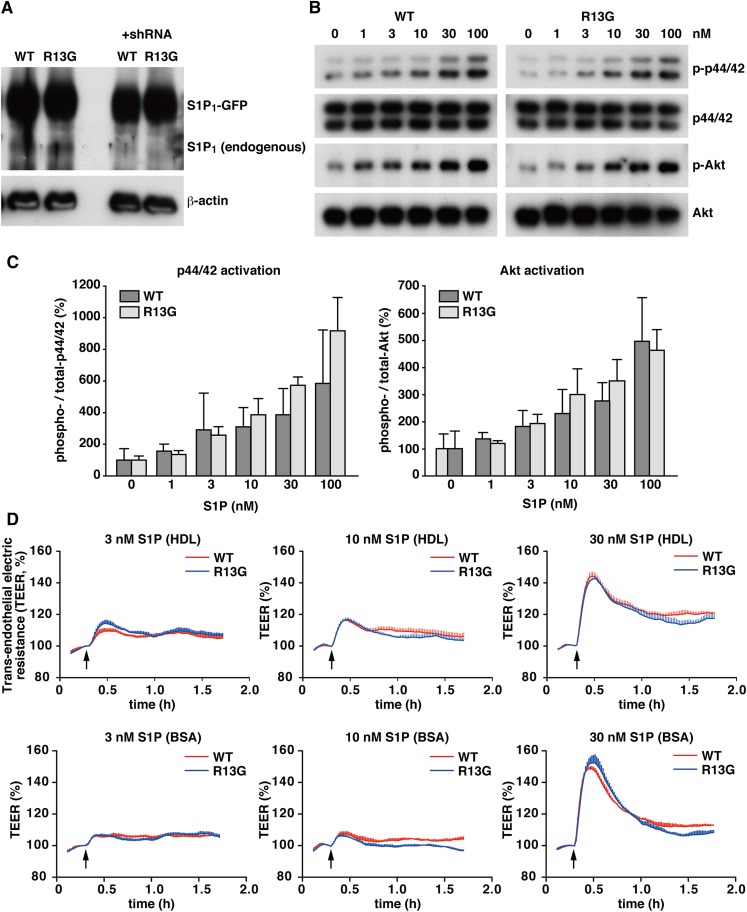
Functional analysis of the R13G mutation of S1P1 in endothelial cells
R13G mutation was enriched in the high cardiovascular risk population, but appeared to protect from CAD. To understand this apparent discrepancy, we examined functional differences of the R13G mutant in endothelial cells. GFP-tagged WT S1P1 or R13G mutant were overexpressed in HUVECs with endogenous S1P1 knocked-down by specific shRNA against 3′-UTR of S1P1 (Fig. 6A). These cells were stimulated with an increasing dose of HDL prepared from human plasma (S1P concentration from 1 to 100 nM) to check activation of p44/42 MAPK and Akt. As shown in Fig. 6B, C, R13G mutant did not show any significant differences from WT S1P1. We also measured changes in trans-endothelial electric resistance (TEER) upon S1P stimulation in these HUVECs, because one of the important functions of S1P1 in endothelial cells is strengthening adherens junctions to maintain vascular barrier integrity (9, 40). HDL-bound S1P increased the TEER in a dose-dependent manner, and the R13G mutant did not show any difference from WT S1P1 (Fig. 6D, upper). The same tendency was observed when these cells were stimulated with BSA-bound S1P (Fig. 6D, lower). Thus, we did not find detectable difference of the R13G mutant in endothelial cells, at least in the assay systems we performed, though the cohort analysis of cardiovascular patients showed that patients with the R13G mutant had a significantly lower incidence of multi-vessel obstructions. We also examined the effects of I45T and G305C mutations on the S1P-induced increases in TEER, but did not find any differences compared with the WT receptor (supplementary Fig. IV).
Fig. 6.
Functional comparison between WT S1P1 and R13G mutant in HUVECs. HUVECs were transduced with lentivirus vectors for the expression of GFP-tagged S1P1 or R13G mutant together with a vector for endogenous S1P1-specifc shRNA. A: Western blot analysis was performed to check the overexpression of WT S1P1 and R13G and the suppression of endogenous S1P1 by shRNA. B: The cells were starved for 3 h and stimulated with the indicated concentrations of HDL-bound S1P for 5 min. Total cell lysates were prepared and subjected to Western blot analyses for the activation of p44/42 MAPK and Akt. C: Densitometric analysis of (B) by the National Institutes of Health ImageJ program is shown. Data represent the mean + SD (n = 3). D: The cells were starved for 3 h and stimulated with the indicated concentrations of HDL-bound (upper panel) or BSA-bound (lower panel) S1P. The changes in TEER were monitored in an ECIS θ system (Applied Biophysics). Data are the mean ± SEM (n = 3) and are representative of three independent experiments with the same tendency.
DISCUSSION
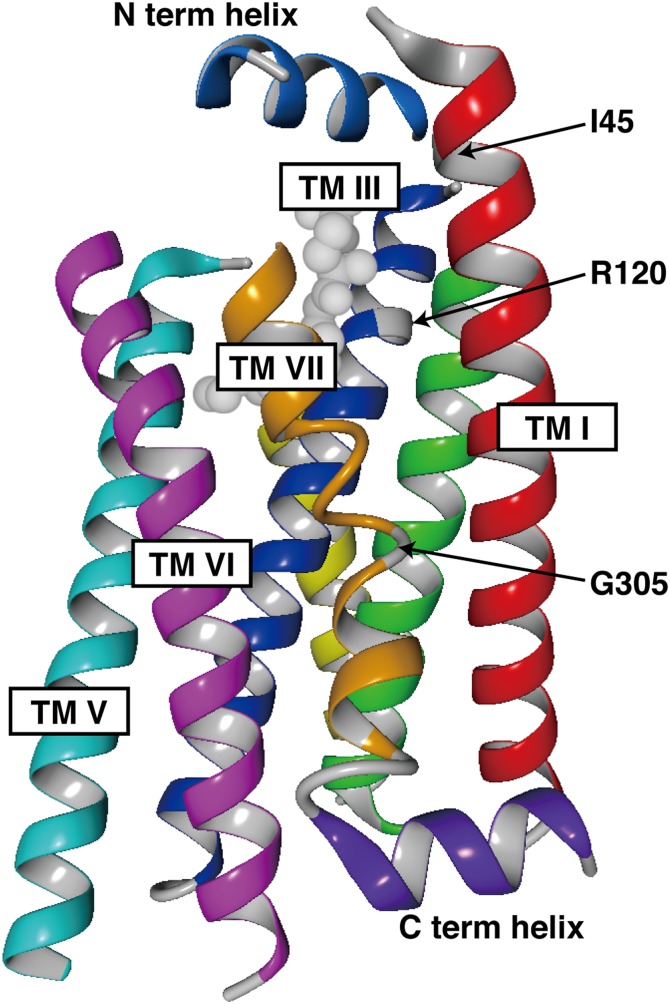
In this study, we characterized 14 nonsynonymous mutations of S1P1 found in the NHLBI GO ESP database. We found three mutations that showed functional differences; R120P showed a defect in both ligand-induced activation and endocytosis, while I45T and G305C only showed a defect in ligand-induced endocytosis. These three residues are all located in the transmembrane helixes in sharp contrast to the other 11 mutations in the extra- or intra-cellular region (Fig. 1, and Fig. 7), which suggests that nonsynonymous mutations in transmembrane helixes of GPCRs tend to result in functional alterations. R120P is a loss-of-function mutation and can be a risk factor for various diseases, considering a pivotal role of the S1P-S1P1 signaling system in cardiovascular and immune systems. Because the S1P1 knockout mouse is embryonic lethal at around E12.5 due to defects in vascular development (41, 42), homozygotes for the R120P mutation are most likely embryonic lethal. Effects of heterozygous loss-of-function of S1P1 should be explored in mouse disease models and/or in various cohort studies.
Fig. 7.
Three-dimensional structure of S1P1. Three-dimensional structure of human S1P1 is illustrated by using a program (YASARA) based on the crystal structure of S1P1 (Protein Data Bank number 3V2Y). Only α-helixes and a cocrystalized antagonist (ML056) are shown. Arrows denote the positions of Ile45, Arg120, and Gly305. TM, transmembrane.
I45T and G305C showed attenuated ligand-induced endocytosis/degradation, though they mediate intracellular signaling normally. I45T and G305C mutants phenocopy a S1P1 (S5A) mutant in which five serine residues in the carboxyl-terminal region are substituted with alanine. Recently, our group reported that S1P1 (S5A) mutant mice showed enhanced autoimmunity and developed severe experimental autoimmune encephalomyelitis (26). Thus, impairment of the receptor internalization in I45T and G305C mutants may result in increased autoimmunity as observed in S1P1 (S5A) mice, raising a possibility that these SNPs can be potential risk factors for autoimmune diseases. In multiple sclerosis patients, these two SNPs would hamper the ability of FTY720 treatment to induce degradation of S1P1. It is of clinical importance to explore the prevalence of I45T and G305C mutations and their involvement in disease development in several autoimmune disease cohorts.
In contrast to the established importance of Arg120 in S1P binding (17, 38), no clear function has been attributed to Ile45 and Gly305 so far. Substitution of Ile45 to Thr results in an amino acid sequence of Asn43Ser44Thr45, which makes a potential N-linked glycosylation site (N-X-S/T). N-linked glycosylation of transmembrane proteins, including GPCRs, occurs in the endoplasmic reticulum and plays roles in proper folding and trafficking to the cell surface (43). Most GPCRs have one or more N-linked glycosylation sites in the extracellular domain (44), though the functions of N-linked glycosylation have not been well-clarified. The I45T mutant of S1P1 showed normal localization on the cell surface and ligand-induced activation of intracellular signaling pathways, which means this mutation does not affect proper folding of the receptor and trafficking to the cell surface. In mouse S1P1, which has one N-linked glycosylation site in the N-terminal region, mutation of this Asn resulted in poor association with caveolae and impaired ligand-induced endocytosis (45). A similar defect in ligand-induced endocytosis by a nonglycosylated mutant was observed in protease-activated receptor 1 (46). Thus, glycosylation of the extracellular region of GPCRs is somehow involved in the regulation of ligand-induced endocytosis in some cases. Human S1P1 has two putative N-linked glycosylation sites (Asp30 and Asp36) in the close vicinity of Asp43, though glycosylation of these residues has not been confirmed. Additional glycosylation of Asp43 might interfere with functions of these putative glycosylated residues. Because Ile45 is located in the beginning of transmembrane helix I and faces the ligand-binding pocket, as illustrated in Figs. 1 and 7, another possibility is that the I45T mutation and/or glycosylation of Asp43 interferes with ligand-induced conformational changes of the receptor necessary for internalization responses.
G305C shows an even more severe defect in the endocytosis than I45T. Gly305 is located in the middle of transmembrane helix VII (Figs. 1, 7). A stretch of polar amino acids with a glycine in the middle (Asn303Ser304Gly305Thr306Asn307) precedes a highly conserved NPXXY motif and makes the helix VII loose, allowing some flexibility of this transmembrane region (Fig. 7). Glycine is energetically unfavorable for the stability of α-helixes and has the second lowest helix propensity next to proline (47). Gly305 to Cys mutation may constrain this flexible loop and interfere with ligand-induced conformational changes required for endocytosis. Another possibility is that Cys305 might create an additional disulfide bond between another Cys residue. There are four cysteine residues within the transmembrane helixes of S1P1 [Cys56, Cys57 (helix I), Cys206 (helix V), and Cys268 (helix VI)]. These cysteine residues could be potential intra-molecule bonding partners. It is also possible that Cys305 could make an inter-molecule disulfide bond with a cysteine residue of a transmembrane domain of another molecule, because many GPCRs are shown to homo/hetero-dimerize (48).
In addition to the functional analysis of 14 nonsynonymous mutations of S1P1 found in the NHLBI GO ESP database, we performed a sequencing analysis of the S1PR1 gene in the cardiovascular disease cohort and found that R13G shows a significantly higher incidence than that observed in the NHLBI GO ESP (3.58% versus 1.24%). Moreover, the number of patients with multi-vessel obstructions was significantly lower in the R13G group in comparison with the age-, gender-, and risk factor-matched control group, which suggests that the R13G mutation might reduce the severity of coronary atherosclerotic obstructions. Although we tried to clarify functional differences of the R13G mutant in an in vitro study using HUVECs, we did not find any differences compared with WT S1P1, at least in the assay systems we performed. However, atherosclerotic arterial obstructions develop through complex interactions between endothelial cells, smooth muscle cells, macrophages, and lymphocytes. Therefore, it is possible that the R13G mutant might be functionally different in some of these cell types or might affect interaction between these cell types. To establish physiological/clinical significance of the R13G mutation, further case-control studies should be conducted in a cardiovascular disease cohort, as well as other disease cohorts. In vitro characterization of the R13G mutant in various cell types will also be necessary.
In summary, findings in this study suggest that individual variations in S1P1 may influence cardiovascular/immune functions as well as drug sensitivity and, therefore, infer differential risks for cardiovascular and autoimmune diseases as well as response to S1P receptor modulatory drugs. Sufficiently powered prospective cohort studies should be conducted to validate possible relationships between individual variations in the S1P1 receptor and specific diseases/drug sensitivity.
Supplementary Material
Acknowledgments
The authors thank the Lipidomics Shared Resources at the Medical University of South Carolina for the measurement of S1P concentration and Dr. DiLorenzo for her help in the TEER measurements.
Footnotes
Abbreviations:
- CAD
- coronary artery disease
- FTY720
- fingolimod/Gilenya
- GFP
- green fluorescent protein
- GPCR
- G protein-coupled receptor
- HUVEC
- human umbilical vein cord endothelial cell
- NHLBI GO ESP
- National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project
- SNP
- single nucleotide polymorphism
- S1P
- sphingosine 1-phosphate
- S1P1
- sphingosine 1-phosphate receptor 1
- TEER
- trans-endothelial electric resistance
- UTR
- untranslated region
This work was supported by National Institutes of Health Grants HL89934 and HL117798 to T.H., and the National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL102923), the WHI Sequencing Project (HL102924), the Broad GO Sequencing Project (HL102925), the Seattle GO Sequencing Project (HL102926), and the Heart GO Sequencing Project (HL103010).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of text and four figures.
REFERENCES
- 1.Blaho V. A., Hla T. 2011. Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem. Rev. 111: 6299–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obinata H., Hla T. 2012. Sphingosine 1-phosphate in coagulation and inflammation. Semin. Immunopathol. 34: 73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cyster J. G., Schwab S. R. 2012. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 30: 69–94. [DOI] [PubMed] [Google Scholar]
- 4.Murata N., Sato K., Kon J., Tomura H., Yanagita M., Kuwabara A., Ui M., Okajima F. 2000. Interaction of sphingosine 1-phosphate with plasma components, including lipoproteins, regulates the lipid receptor-mediated actions. Biochem. J. 352: 809–815. [PMC free article] [PubMed] [Google Scholar]
- 5.Christoffersen C., Obinata H., Kumaraswamy S. B., Galvani S., Ahnström J., Sevvana M., Egerer-Sieber C., Muller Y. A., Hla T., Nielsen L. B., et al. 2011. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M. Proc. Natl. Acad. Sci. USA. 108: 9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung B., Obinata H., Galvani S., Mendelson K., Ding B-S., Skoura A., Kinzel B., Brinkmann V., Rafii S., Evans T., et al. 2012. Flow-regulated endothelial S1P receptor-1 signaling sustains vascular development. Dev. Cell. 23: 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaengel K., Niaudet C., Hagikura K., Laviña B., Siemsen B. L., Muhl L., Hofmann J. J., Ebarasi L., Nyström S., Rymo S., et al. 2012. The sphingosine-1-phosphate receptor S1PR1 restricts sprouting angiogenesis by regulating the interplay between VE-cadherin and VEGFR2. Dev. Cell. 23: 587–599. [Erratum. 2012. Dev. Cell. 23: 1264.] [DOI] [PubMed] [Google Scholar]
- 8.Ben Shoham A., Malkinson G., Krief S., Shwartz Y., Ely Y., Ferrara N., Yaniv K., Zelzer E. 2012. S1P1 inhibits sprouting angiogenesis during vascular development. Development. 139: 3859–3869. [DOI] [PubMed] [Google Scholar]
- 9.Lee M. J., Thangada S., Claffey K. P., Ancellin N., Liu C. H., Kluk M., Volpi M., Sha’afi R. I., Hla T. 1999. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell. 99: 301–312. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez T., Estrada-Hernandez T., Paik J. H., Wu M. T., Venkataraman K., Brinkmann V., Claffey K., Hla T. 2003. Phosphorylation and action of the immunomodulator FTY720 inhibits vascular endothelial cell growth factor-induced vascular permeability. J. Biol. Chem. 278: 47281–47290. [DOI] [PubMed] [Google Scholar]
- 11.Camerer E., Regard J. B., Cornelissen I., Srinivasan Y., Duong D. N., Palmer D., Pham T. H., Wong J. S., Pappu R., Coughlin S. R. 2009. Sphingosine-1-phosphate in the plasma compartment regulates basal and inflammation-induced vascular leak in mice. J. Clin. Invest. 119: 1871–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nofer J. R., van der Giet M., Tölle M., Wolinska I., von Wnuck Lipinski K., Baba H. A., Tietge U. J., Gödecke A., Ishii I., Kleuser B., et al. 2004. HDL induces NO-dependent vasorelaxation via the lysophospholipid receptor S1P3. J. Clin. Invest. 113: 569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Argraves K. M., Argraves W. S. 2007. HDL serves as a S1P signaling platform mediating a multitude of cardiovascular effects. J. Lipid Res. 48: 2325–2333. [DOI] [PubMed] [Google Scholar]
- 14.Okajima F., Sato K., Kimura T. 2009. Anti-atherogenic actions of high-density lipoprotein through sphingosine 1-phosphate receptors and scavenger receptor class B type I. Endocr. J. 56: 317–334. [DOI] [PubMed] [Google Scholar]
- 15.Potì F., Simoni M., Nofer J. R. 2014. Atheroprotective role of high density lipoprotein (HDL)-associated sphingosine 1-phosphate (S1P). Cardiovasc. Res. 103: 395–404. [DOI] [PubMed] [Google Scholar]
- 16.Hla T., Venkataraman K., Michaud J. 2008. The vascular S1P gradient-cellular sources and biological significance. Biochim. Biophys. Acta. 1781: 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson M. A., Roth C. B., Jo E., Griffith M. T., Scott F. L., Reinhart G., Desale H., Clemons B., Cahalan S. M., Schuerer S. C., et al. 2012. Crystal structure of a lipid G protein-coupled receptor. Science. 335: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosen H., Stevens R. C., Hanson M., Roberts E., Oldstone M. B. A. 2013. Sphingosine-1-phosphate and its receptors: structure, signaling, and influence. Annu. Rev. Biochem. 82: 637–662. [DOI] [PubMed] [Google Scholar]
- 19.Lee M. J., Evans M., Hla T. 1996. The inducible G protein-coupled receptor edg-1 signals via the G(i)/mitogen-activated protein kinase pathway. J. Biol. Chem. 271: 11272–11279. [DOI] [PubMed] [Google Scholar]
- 20.Windh R. T., Lee M. J., Hla T., An S., Barr A. J., Manning D. R. 1999. Differential coupling of the sphingosine 1-phosphate receptors Edg-1, Edg-3, and H218/Edg-5 to the G(i), G(q), and G(12) families of heterotrimeric G proteins. J. Biol. Chem. 274: 27351–27358. [DOI] [PubMed] [Google Scholar]
- 21.Lee M. J., Thangada S., Paik J. H., Sapkota G. P., Ancellin N., Chae S. S., Wu M., Morales-Ruiz M., Sessa W. C., Alessi D. R., et al. 2001. Akt-mediated phosphorylation of the G protein-coupled receptor EDG-1 is required for endothelial cell chemotaxis. Mol. Cell. 8: 693–704. [DOI] [PubMed] [Google Scholar]
- 22.Watterson K. R., Johnston E., Chalmers C., Pronin A., Cook S. J., Benovic J. L., Palmer T. M. 2002. Dual regulation of EDG1/S1P(1) receptor phosphorylation and internalization by protein kinase C and G-protein-coupled receptor kinase 2. J. Biol. Chem. 277: 5767–5777. [DOI] [PubMed] [Google Scholar]
- 23.Liu C. H., Thangada S., Lee M. J., Van Brocklyn J. R., Spiegel S., Hla T. 1999. Ligand-induced trafficking of the sphingosine-1-phosphate receptor EDG-1. Mol. Biol. Cell. 10: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thangada S., Khanna K. M., Blaho V. A., Oo M. L., Im D. S., Guo C., Lefrancois L., Hla T. 2010. Cell-surface residence of sphingosine 1-phosphate receptor 1 on lymphocytes determines lymphocyte egress kinetics. J. Exp. Med. 207: 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oo M. L., Chang S. H., Thangada S., Wu M. T., Rezaul K., Blaho V., Hwang S. I., Han D. K., Hla T. 2011. Engagement of S1P1-degradative mechanisms leads to vascular leak in mice. J. Clin. Invest. 121: 2290–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garris C. S., Wu L., Acharya S., Arac A., Blaho V. A., Huang Y., Moon B. S., Axtell R. C., Ho P. P., Steinberg G. K., et al. 2013. Defective sphingosine 1-phosphate receptor 1 (S1P1) phosphorylation exacerbates TH17-mediated autoimmune neuroinflammation. Nat. Immunol. 14: 1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkmann V., Billich A., Baumruker T., Heining P., Schmouder R., Francis G., Aradhye S., Burtin P. 2010. Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 9: 883–897. [DOI] [PubMed] [Google Scholar]
- 28.Obinata H., Hla T. 2012. Fine-tuning S1P therapeutics. Chem. Biol. 19: 1080–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigaud M., Guerini D., Billich A., Bassilana F., Brinkmann V. 2014. Second generation S1P pathway modulators: research strategies and clinical developments. Biochim. Biophys. Acta. 1841: 745– 758. [DOI] [PubMed] [Google Scholar]
- 30.Oo M. L., Thangada S., Wu M. T., Liu C. H., Macdonald T. L., Lynch K. R., Lin C. Y., Hla T. 2007. Immunosuppressive and anti-angiogenic sphingosine 1-phosphate receptor-1 agonists induce ubiquitinylation and proteasomal degradation of the receptor. J. Biol. Chem. 282: 9082–9089. [DOI] [PubMed] [Google Scholar]
- 31.Sun X., Ma S. F., Wade M. S., Flores C., Pino-Yanes M., Morita J., Ober C., Kittles R., Husain A. N., Ford J. G., et al. 2010. Functional variants of the sphingosine-1-phosphate receptor 1 gene associate with asthma susceptibility. J. Allergy Clin. Immunol. 126: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NHLBI GO Exome Sequencing Project (ESP). Exome Variant Server, Seattle, WA. Accessed June 2012, at http://evs.gs.washington.edu/EVS/.
- 33.Obinata H., Hla T. 2012. Assessment of sphingosine-1-phosphate activity in biological samples by receptor internalization and adherens junction formation. Methods Mol. Biol. 874: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aratake Y., Okuno T., Matsunobu T., Saeki K., Takayanagi R., Furuya S., Yokomizo T. 2012. Helix 8 of leukotriene B4 receptor 1 inhibits ligand-induced internalization. FASEB J. 26: 4068–4078. [DOI] [PubMed] [Google Scholar]
- 35.Arehart E., Stitham J., Asselbergs F. W., Douville K., MacKenzie T., Fetalvero K. M., Gleim S., Kasza Z., Rao Y., Martel L., et al. 2008. Acceleration of cardiovascular disease by a dysfunctional prostacyclin receptor mutation: potential implications for cyclooxygenase-2 inhibition. Circ. Res. 102: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bielawski J., Pierce J. S., Snider J., Rembiesa B., Szulc Z. M., Bielawska A. 2009. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 579: 443–467. [DOI] [PubMed] [Google Scholar]
- 38.Parrill A. L., Wang D., Bautista D. L., Van Brocklyn J. R., Lorincz Z., Fischer D. J., Baker D. L., Liliom K., Spiegel S., Tigyi G. 2000. Identification of Edg1 receptor residues that recognize sphingosine 1-phosphate. J. Biol. Chem. 275: 39379–39384. [DOI] [PubMed] [Google Scholar]
- 39.Koyrakh L., Roman M. I., Brinkmann V., Wickman K. 2005. The heart rate decrease caused by acute FTY720 administration is mediated by the G protein-gated potassium channel IKACh. Am. J. Transplant. 5: 529–536. [DOI] [PubMed] [Google Scholar]
- 40.Garcia J. G., Liu F., Verin A. D., Birukova A., Dechert M. A., Gerthoffer W. T., Bamberg J. R., English D. 2001. Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. J. Clin. Invest. 108: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Wada R., Yamashita T., Mi Y., Deng C. X., Hobson J. P., Rosenfeldt H. M., Nava V. E., Chae S. S., Lee M. J., et al. 2000. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Invest. 106: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allende M. L., Yamashita T., Proia R. L. 2003. G protein-coupled receptor S1P1 acts within endothelial cells to regulate vascular maturation. Blood. 102: 3665–3667. [DOI] [PubMed] [Google Scholar]
- 43.Helenius A. 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell. 5: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheatley M., Wootten D., Conner M. T., Simms J., Kendrick R., Logan R. T., Poyner D. R., Barwell J. 2012. Lifting the lid on GPCRs: the role of extracellular loops. Br. J. Pharmacol. 165: 1688–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohno T., Wada A., Igarashi Y. 2002. N-glycans of sphingosine 1-phosphate receptor Edg-1 regulate ligand-induced receptor internalization. FASEB J. 16: 983–992. [DOI] [PubMed] [Google Scholar]
- 46.Soto A. G., Trejo J. 2010. N-linked glycosylation of protease-activated receptor-1 second extracellular loop: a critical determinant for ligand-induced receptor activaiton and internalization. J. Biol. Chem. 285: 18781–18793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pace C. N., Scholtz J. M. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Audet M., Bouvier M. 2012. Restructuring G protein-coupled receptor activation. Cell. 151: 14–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.