Abstract
Age-related macular degeneration (AMD) is a progressive degenerative disease which leads to blindness, affecting the quality of life of millions of Americans. More than 1.75 million individuals in the United States are affected by the advanced form of AMD. The etiological pathway of AMD is not yet fully understood, but there is a clear genetic influence on disease risk. To date, the 1q32 (CFH) and 10q26 (PLEKHA1/ARMS2/HTRA1) loci are the most strongly associated with disease; however, the variation in these genomic regions alone is unable to predict disease development with high accuracy. Therefore, current genetic studies are aimed at identifying new genes associated with AMD and their modifiers, with the goal of discovering diagnostic or prognostic biomarkers. Moreover, these studies provide the foundation for further investigation into the pathophysiology of AMD by utilizing a systems-biology-based approach to elucidate underlying mechanistic pathways.
Keywords: ARMS2/HTRA1, CFH, choroidal neovascularization, complement, geographic atrophy, susceptibility
INTRODUCTION
Although much remains elusive about the etiology of age-related macular degeneration (AMD), it has long been clear that genetics plays an important role. Surveys of age- and sex-matched individuals with and without AMD demonstrated that AMD has a tendency to aggregate within families, i.e. patients with AMD were more likely to have relatives with the disease.1,2 Data from the Rotterdam study in the Netherlands demonstrated that first-degree relatives of individuals affected with AMD have a four-fold higher risk of developing advanced AMD than those with unaffected relatives.3 Swaroop and colleagues calculated that having a sibling with AMD increases an individual’s risk 3-6 fold.4 A genetic component is also supported by twin studies that demonstrated a higher degree of AMD concordance between monozygotic twins than dizygotic twins, 37% compared to 19%, respectively.5
AMD is a challenging disease to study from a genetic perspective because, unlike disorders that exhibit Mendelian inheritance patterns (where one gene and one mutation within that gene is responsible for the phenotype observed in a given family), the development and severity of complex diseases like AMD are influenced by many factors. The high prevalence of the disease implies that that there is more than one gene or environmental factor as well as interactions amongst these factors that influence an individual’s susceptibility to disease. In the United States, the Eye Disease Prevalence Group estimates that advanced AMD currently affects more than 1.75 million individuals, a number predicted to rise to near 3 million by the year 2020.6 AMD is also a heterogeneous disease, suggesting that there may be different disease mechanisms and hence different genes responsible for the various clinical subtypes.
Description of AMD
The hallmark of AMD is yellow subretinal deposits called drusen, which are often accompanied by hypoor hyperpigmentary changes of the retinal pigment epithelium (RPE). The number, size, and area of drusen along with the presence or absence of pigmentary changes of the RPE determine the severity of AMD in the early to intermediate stages.7 A relatively small proportion of individuals (10–15%) are affected by the advanced form of AMD, manifested by geographic atrophy or choroidal neovascularization. Geographic atrophy represents an area of loss of the RPE and choriocapillaris with corresponding loss or dysfunction of the overlying photoreceptors. Choroidal neovascularization occurs when abnormal blood vessels develop in the sub-RPE or subretinal space. Although these basic clinical phenotypes have been widely utilized in categorizing various subtypes of AMD, it may be that more precise phenotypic characterization of AMD may be necessary to elucidate significant genotype-phenotype correlations.
Overview of AMD Risk Factors
Previous studies of genetic and epidemiologic factors have not been in agreement as to the exact predictors for AMD risk.4,8,9 The greatest risk factor for development of the disease is age, with individuals over 50 years of age having a greater risk of developing AMD compared to those under age 50.10–12 While body mass index (BMI), hypertension, cardiovascular disease, and alcohol consumption have been implicated in risk of AMD, cigarette smoking is the modifiable epidemiologic risk factor most consistently associated with an increased risk of AMD.13,14 Investigation of the effects of sunlight exposure on AMD risk have yielded inconsistent results. Some studies suggest a positive association between sunlight and AMD,15,16 while others found no correlation.17–19
Recent progress in AMD genetics has established alleles as well as haplotypes (combinations of alleles at a given locus that are inherited together) on chromosome 1 in Complement Factor H (CFH) and on chromosome 10 in Age-Related Maculopathy Susceptibility 2 (ARMS2, formerly LOC387715/HtrA Serine Peptidase 1[HTRA1]), as having large influences on risk for all AMD subtypes in populations of various ethnicities.20–29 In particular, genes that reside on the long arm of chromosome 10 (10q26) have been the most strongly associated with neovascular AMD risk.30–32 Despite the large influence on AMD risk, the combination of these genes alone is insufficient to correctly predict the development and progression of this disease.4,9
While additional genes may be only minor players in terms of their contribution to the total genetic variance of AMD, effect size does not always correlate with the importance to pathogenesis and hence treatment of AMD. In other words, it is important to consider that the proportion of cases attributable to certain genetic variants does not necessarily reflect the role of these genes in pathophysiology of disease and therefore it is important to continue to search for the other loci, which do exist but have yet to be elucidated.30,33–41 One of the best examples of this can be seen in Alzheimer’s disease with the discovery of the presenilin genes. While variation in the presenilin genes accounts for only several thousand Alzheimer’s patients worldwide, the presenilin proteins, whose existence was unknown before the identification of these genes,42–44 are central to the processing of amyloid β, a hallmark of Alzheimer’s pathogenesis and a target for drug therapy.45–47 In ongoing genetic studies of AMD, the priority should be to determine the mechanism, pathways, and networks underlying disease, not just risk factors, so that appropriate avenues of treatment may be identified.
While the functional pathway involving the ARMS2/HTRA1 gene(s) is unknown, the alternative complement pathway where CFH participates is well described and annotated. As a result, further investigation of the entire complement pathway, initially using a candidate gene approach, revealed associations between AMD risk and rare alleles in the Component 2 (C2), Complement Factor B (CFB), Component 3 (C3) genes, and Complement Factor H related genes (CFHR1-5).32,48–55 Furthermore, a recent genome wide association study56 confirmed the majority of these loci as well as an initial report of an association between AMD susceptibility and another complement gene, Complement Factor I (CFI).57 Although these studies illustrate the importance of studying an entire genetic pathway for dissecting genetic susceptibility to a complex disease such as AMD, there are many AMD-free individuals in the population that harbor these disease susceptibility genotypes and a substantial number of AMD patients who lack risk variants at these loci. Moreover, many of the genes that will be discussed in the following review have more than one function or share similar functions, thus underscoring the redundancy of our genome and biological systems in general.
Developing a unifying genetic susceptibility hypothesis, let alone a commercial genetic test, to predict which individuals in the general population will develop AMD prior to any signs of disease remains a challenge.9,58 This is also the case for other diseases of complex etiology such as cardiovascular disease,59–61 diabetes,62,63 Alzheimer’s disease,64–66 and depression,67 where many genes and environmental factors as well interactions amongst them have been implicated.68 In the following review, we discuss the known AMD genetic findings to date and explore future strategies in the search for a unifying risk hypothesis that may refine diagnosis and prognosis and identify new preventative and therapeutic targets.
GENES ASSOCIATED WITH JUVENILE (MENDELIAN) FORMS OF MACULAR DEGENERATION
Genes known to be associated with juvenile onset forms of retinal degeneration were amongst the first to have been assessed for association with AMD (Table 1; for review please see 69). To date, no studies have found an association between mutations in Vitelliform Macular Dystrophy 2 (VMD2; Best disease),70–74 Retinal Degeneration, Slow/Peripherin (RDS; Retinitis Pigmentosa; macular dystrophy),75,76 and Epidermal Growth Factor-containing Fibulin Like Extracellular Matrix Protein 1(EFEMP1; Malattia Leventinese/Doyne honeycomb retinal dystrophy)77–79 and any type of AMD. Studies investigating the ATP-binding Cassette Transporter gene, subfamily A, member 4 (ABCA4; autosomal recessive Stargardt disease), the Elongation of Very Long Chain Fatty Acids gene (ELOVL4; Stargardt disease Type 3 or autosomal dominant Stargardt-like macular dystrophy), and TIMP Metallopeptidase Inhibitor 3 (TIMP3; Sorsby fundus dystrophy), are inconsistent with respect to whether or not variations in these genes are associated with AMD.
TABLE 1.
Candidate genes for AMD derived from hereditary retinal degenerations.
| Gene | Function | Disease Association |
|---|---|---|
| RDS | Protein binding | Retinitis pigmentosa |
| TIMP3* | Metalloendopeptidase inhibitor activity | Sorsby fundus dystrophy |
| ABCA4* | ATP binding, ATPase activity, lipid transport, and metabolism |
Stargardt disease |
| EFEMP1 | Maintenance of extracellular matrix integrity | Malattia leventinese (ML) / Doyne honeycomb retinal dystrophy (DHRD) |
| VMD2 | Chloride ion binding and ion channel | activity Best disease |
| ELOVL4* | Lipid metabolism and transport | Stargardt disease, type III |
Genes positively associated with age-related macular degeneration
ATP-binding Cassette Transporter, Subfamily A, Member 4 (ABCA4)
ABCA4 is located on the short arm of chromosome 1 (1p22.1–p21) and functions in lipid metabolism and transport. Mutations in ABCA4, also known as ABCR, cause Stargardt disease, a juvenile form of macular degeneration characterized by deposits of lipofuscin in and beneath the retinal pigment epithelium. In 1997, Allikmets et al. found an association between heterozygous ABCA4 variation and AMD.80 In particular, G1961E (rs1800553) and D2177N (rs1800555), the two most common variants, along with other rare variants in ABCA4 were reported to be associated with ~4% of AMD cases (mostly the dry form). Further, in a larger screen of more than 1200 patients with both dry and neovascular AMD, Allikmets et al. later reported that ABCA4 variants could probably explain up to 8% of AMD.81 However, attempts to replicate these findings failed to confirm that variants in ABCA4 were associated with either the early or more advanced stages of AMD.82–89
Elongation of Very Long Chain Fatty Acids (ELOVL4)
Like ABCA4, the protein product of ELOVL4 is involved in lipid transport and metabolism. Unlike ABCA4, ELOVL4 is located in a region (6q24–q14) previously identified from genome-wide scans to harbor AMD susceptibility genes.35,38 A five base pair deletion in this gene has been found to be associated with an autosomal dominant Stargardt-like macular dystrophy, which has phenotypic similarity to the dry or atrophic form of AMD. The M299V (rs3812153) variant of ELOVL4 has been demonstrated to increase risk of neovascular AMD,90 although an earlier study did not find this association for either the early or more advanced stages of AMD.91 Similarly, a study of a Finnish population also did not find an association between the M299V variant in ELOVL4 and patients with large drusen, neovascular, or atrophic AMD.92
TIMP Metallopeptidase Inhibitor 3 (TIMP3)/Synapsin III (SYN3)
The genes TIMP3 and SYN3 are located on 22q12.3, near an AMD-associated locus identified by genome wide scans.36,93 TIMP3 is encoded within an intronic region of the gene SYN3 on the opposite strand. SYN3 belongs to a family of synaptic vesicle proteins involved in the calcium dependent regulation of neurotransmitters.94 A member of a family of extracellular matrix remodeling proteins, TIMP3 has been shown to accumulate in aged eyes95 and to inhibit Vascular Endothelial Growth Factor (VEGF).96 Based on its association with the autosomal dominant disorder, Sorsby fundus dystrophy, TIMP3 was proposed as a candidate gene in the study of AMD. Initial investigations did not find an association of TIMP3 with AMD;97,98 however, a recent genome wide association study (GWAS) with a combined sample size of 10,049 cases and 7,148 controls showed significant association between the SNP rs9621532 that lies in the SYN3/TIMP3 locus and AMD.56 As a result of the location of TIMP3 within SYN3, it is not yet possible to distinguish which, if not both, genes are involved in AMD susceptibility.
GENOME WIDE SCANS
Two approaches for genome wide scans which are complementary and overlap are genome wide linkage studies (using family-based cohorts) and genome wide association studies (GWAS, using case-control cohorts). The purpose of both is to search the 23 pairs of chromosomes with the goal of identifying regions significantly harboring disease susceptibility genes or variants. In the last 12 years, 17 genome wide scans have been conducted. Only three of these studies have been GWAS. GWAS may be better for identifying highly frequent and/or common variation associated with a disease, while linkage may be better for identifying rare disease-associated alleles.
Sequence data from the human genome project has enabled investigators not only to look at microsatellite markers or short tandem repeat polymorphisms (STRPs), as is characteristic for family-based linkage studies, but also to examine differences in other types of common DNA variation between those with disease and those without disease. Microsatellites or STRPs are variable numbers of tandemly repeated sequences of 2-4 nucleotides generally located in an intronic region. The number of repeats identifies different alleles. Polymerase chain reaction (PCR) probes labeled with a fluorescent tag can be applied to known STRPs/microsatellites in specific genomic regions, yielding products of differing lengths based on the number of repeats. It can then be determined if specific alleles segregate with specific phenotypes, in other words, if there is linkage between a particular genomic region and the phenotype of interest.
Another type of variation that can be used to find genes or regions harboring disease- associated genes are single nucleotide polymorphisms (SNPs). SNPs are inucleotide changes occurring about every 100–300 base pairs and are generally biallelic. SNPs account for 80% of the variation seen in our genome. Because SNPs are so common in the genome, they may pinpoint an actual gene rather than a region harboring perhaps hundreds of genes as is identified when microsatellites/STRPs are used in linkage studies. While a single SNP (whether intonic or exonic) may serve as a marker indicating a gene of importance, it may not actually play a causative role in disease.
Recently, it has been speculated that structural changes in our DNA not related to the portions that encode proteins, such as insertions, deletions, and duplications, may not only influence disease susceptibility but may be causal. Copy number variation (CNV) is a type of such structural change, generally located in intronic regions. A CNV or copy number polymorphism (CNP) can be an insertion or deletion of tandemly repeated blocks of nucleotides (two or more) ranging from 200 base pairs to 2 megabases (for reviews please see 99–101). CNVs can be spontaneous or inherited and may help explain human phenotypic variability, complex behavioral traits, and the presentation of disease. While structural variation may involve only 20% of our genome, it is believed to encompass 70% of the nucleotide variation observed.102 The methodology to statistically analyze this type of variation and its contribution to disease is still under development but represents an active area of research,102,103 which may eventually shed additional light on AMD pathogenesis.
Chromosomal regions found by more than one genome wide scan to be significantly linked to all subtypes of AMD include 1q23.3– q31.1,21,33,34,37,93,104,105 2p25.3– p12,36,37 6q14,35,38 9q31,36,37 10q26,34,37,39,93,104,106 12q23– q24.31,35,3715q13– q15, 35,37 16p12– p12.1, 35,37,38 17q25,104,105 and 22q12.1.36,93 Pooled analysis of several of these genome wide linkage studies and examining all types of AMD confirmed two of the most consistently reported chromosomal regions (1q23.3– q31.1 and 10q26) while identifying others that were not initially significant in the separate studies alone.30 This pooled analysis or meta-analysis also confirmed two regions that had been reported to be significant by only individual studies: 3p14.1–p25.330,40 and 4q32.30,34
Many of the AMD genome wide scans have resulted in the identification of various chromosomal regions that have not been replicated by other studies. Differences across studies with respect to phenotypic definition may make it difficult to compare and interpret findings. For example, sibling pairs concordant for advanced AMD from the Family Age-related Maculopathy Study (FARMS) showed significant linkage to 5q33.3, 14q32.33, 16p13.13, 19q13.31, 21q21.2,40 while other investigators who also studied populations characterized by advanced AMD did not report significant linkage to these same regions.34,37,104,105 Similarly, an association between advanced AMD and the 16p12 region has been observed to be dependent on factors such as body mass index (BMI) and hypertension in some studies38, but not others.37 In summary, genome wide scans employing both linkage and genome wide association methods have helped to identify candidate genes for AMD and candidate regions in need of further investigation.
CANDIDATE GENES
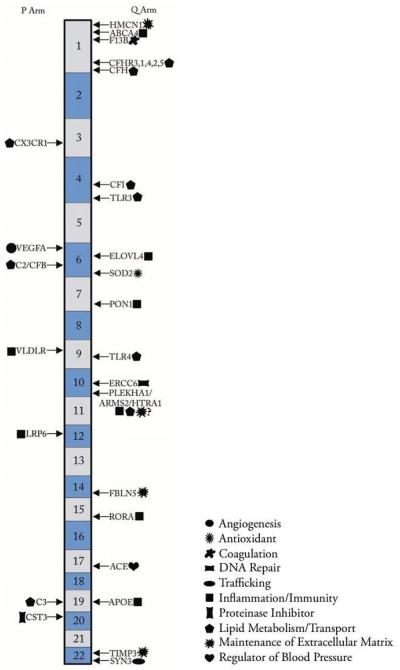
A variety of candidate genes have been evaluated either because of their roles in disease that share phenotypic similarity to AMD and/or because of their location in areas identified in genome wide scans as harboring AMD susceptibility genes. Here we discuss genes that have been found by at least one study to contain variation statistically associated with risk of AMD (Figure 1) and are further supported by additional lines of evidence including biological function, gene expression, or location within a region previously associated with AMD.
FIGURE 1.
Chromosomal location of AMD-associated genes. The schematic represents the location of AMD associated genes on the 22 autosomal chromosomes. The following genes are listed on either side of the cartoon, thereby depicting their location on the p or q arm: Hemicentin 1 (HMCN1), ATP-binding Cassette Transporter, subfamily A, member 4 (ABCA4), Coagulation Factor XIII, B Polypeptide (F13B), Complement Factor H Related Genes (CFHR1-5), Complement Factor H (CFH), Chemokine (C-X3-C Motif) Receptor 1 (CX3CR1), Complement Factor I (CFI), Toll-like Receptor 3 (TLR3), Vascular Endothelial Growth Factor A (VEGFA), Elongation of Very Long Chain Fatty Acids (ELOVL4), Complement Component 2 (C2)/Complement Factor B (CFB), Manganese Superoxide Dismutase 2 (SOD-2), Paraoxonase 1 (PON1), Very Low Density Lipoprotein Receptor (VLDLR), Toll-like Receptor 4 (TLR4), Excision Repair Cross-complementation Group 6 (ERCC6), Pleckstrin Homology Domain Containing, Family A (Phosphoinositide Binding Specific) Member 1 (PLEKHA1)/Age-Related Maculopathy Susceptibility 2 (ARMS2, formerly LOC387715)/HTRA Serine Peptidase 1 (HTRA1), Low Density Lipoprotein Receptor Related Protein 6 (LRP6), Fibulin 5 (FBLN5), RAR-related orphan receptor alpha (RORA), Angiotensin I Converting Enzyme (ACE), Complement Component 3 (C3), Apolipoprotein (APOE), Cystatin C (CST3), TIMP Metallopeptidase Inhibitor 3 (TIMP3), Synapsin III (SYN3). Next to each gene name is a symbol listed in the legend representing the function/cellular process most commonly associated with the gene product.
The 10q26 Region
Pleckstrin Homology Domain Containing, Family A (Phosphoinositide Binding Specific) Member 1 (PLEKHA1), Age-Related Maculopathy Susceptibility 2 (ARMS2), and HTRA Serine Peptidase 1 (HTRA1)
Meta-analysis of findings from six genome wide scans showed that the 10q26 locus demonstrated the greatest significance for linkage compared to other significant regions, such as the 1q25–q32 region.30,32,34,37,39,93,104–106 This region contains the genes PLEKHA1, ARMS2 (formerly LOC387715), and HTRA1.
PLEKHA1 is believed to participate in phospholipid binding and more indirectly in the immune response. Using a combination of linkage and association, Jakobsdottir et al. demonstrated that while both PLEKHA1 and ARMS2 were significantly associated with both neovascular and atrophic AMD, the association was stronger for PLEKHA1. More than 1400 affected individuals comprised of both familial and sporadic cases of AMD were examined.20 However, this finding was not replicated after examination of a cohort that consisted of both early and advanced cases of AMD25 or a cohort that consisted of intermediate and advanced cases, including large drusen (≥125 μm), geographic atrophy, and neovascularization.107 Instead, these two studies concluded that ARMS2, and not PLEKHA1, was associated with AMD.
The ARMS2 gene is nestled between PLEKHA1 and HTRA1 in the 10q26 locus. ARMS2 has been shown to be expressed in multiple tissues, including the retina;108,109 however, its function remains unknown and its cellular location is disputed. Two groups have reported mitochondrial localization,108,110 with conflicting reports finding ARMS2 in the cytosol.109,111 Within ARMS2, rs10490924, encoding the A69S change, has been associated with severe AMD phenotypes including early onset of disease and larger choroidal neovascularization lesions.112–114 This variation has been consistently associated with AMD risk across various ethnicities and has been hypothesized to affect the interaction of ARMS2 with other proteins.108 Moreover, A69S has been shown to significantly interact with smoking in both familial and unrelated case-control cohorts.107 However, this finding of an interaction between smoking and ARMS2 was not replicated in several independent analyses involving cohorts from the Cardiovascular Health Study (CHS) and the Age-Related Eye Disease Study (AREDS),115 the Blue Mountain Eye Study (BMES),116 and a retrospective study of sibling pairs.117
In addition to the A69S variant, a complex insertion/deletion (indel) in the 3′ untranslated region of ARMS2 has also been significantly associated with AMD risk.110,118 The variation is hypothesized to cause a decrease in transcript stability, and in homozygous cases of this indel the protein was not expressed in human cell lines.110 However, a recent publication reported that the indel did not destabilize the mRNA transcript in human cell lines and further suggested that the association with AMD might be attributable to the high linkage disequilibrium between the indel and rs10490924.109
HTRA1, the third gene in the 10q26, is highly conserved among species and has several variants that have consistently been found to be associated with AMD.25,28,32,117,119–123 One of the most consistently AMD-associated variants is the SNP rs11200638, located within the putative HTRA1 promoter.120,122–126This variation has been shown to result in higher levels of HTRA1 expression in lymphocytes and well as RPE cells;28,127 however, a second group found no difference in the expression of a luciferase reporter gene driven by wild type HTRA1 promoter compared to the variant.108 Most recently, a study reported a significant association between AMD and a haplotype containing both the ARMS2 indel and the HTRA1 promoter rs11200638 variant.123 This is not surprising given that variants in both HTRA1 and ARMS2 are in high linkage disequilibrium.110,117
Given that these three genes within the 10q26 are in high linkage disequilibrium, it is difficult to distinguish between the individual contributions of each of these genes to overall AMD susceptibility. As evidenced by varying reports, teasing out precisely which gene(s) in this region is associated with AMD will require further investigation.
Complement Genes
Complement Factor H (CFH)
CFH is an inhibitor that functions in regulation of the alternative-complement-pathway as well as innate immunity. CFH is localized to 1q32, a region found by both linkage21,30,34,37,104,105 and by GWAS21,56 to be associated with all subtypes of AMD. Specifically, several independent reports have shown the functional polymorphism Y402H (rs1061170) in CFH, where a tyrosine is substituted by a histidine, to be associated with increased risk of both early21–24,128 and late stages of AMD (both neovascular and geographic atrophy).21,22,24,90,129,130 These analyses further suggest that the risk allele contributes to almost half of all cases of AMD in the population. Two studies have found no association of the CFH Y402H variant with AMD in Japanese cohorts with neovascular AMD.122,131 This negative finding may be attributed to the low frequency of the CFH Y402H (rs1061170) in the Japanese population. Some groups have reported other common CFH variation, besides Y402H, to be significantly associated with both early and advanced forms of AMD.24,27,32 Interestingly, some of these same variants (or a combination of them, including Y402H) are associated with increased risk for other diseases such as myocardial infarction,132 hemolytic uremic syndrome,133 and membranoproliferative glomerulonephrtis (MPGN).134
Complement Factor H-Related 1-5 (CFHR1-5) and Coagulation Factor XIII, B polypeptide (F13B)
The CFHR genes, along with F13B, are located in tandem with CFH on chromosome 1q32 in the “regulators of complement activation” locus and are likely gene duplications of CFH.135 The CFHR genes contain several short consensus repeats, many of which are homologous to those in CFH. Like CFH, deletion of/variation within several CFHR genes has been associated with hemolytic uremic syndrome and membranoproliferative glomerulonephritis type II134,136–139 as well as AMD. A large deletion encompassing both CFHR1 and CFHR3 has been associated with AMD.50,140 Single SNPs within CFHR2, 4, 5, and F13B were also significantly associated with risk of subtypes of AMD.32,135Similar to the 10q26 region where PLEKHA1, ARMS2, and HTRA1 reside, dissecting the individual and joint contribution of CFH and the CFHR genes on 1q25 to AMD susceptibility requires further investigation.
Complement Component 2 (C2) and Complement Factor B (CFB)
C2 and CFB are adjacent genes on 6p21.3; therefore, as in the case of variants on 1q25 and 10q26, it is difficult to discern the individual contribution of each gene to AMD risk. Gold et al. postulated that because variation in CFH had been significantly associated with AMD, other genes in the alternative complement-pathway may also play a role in the pathophysiology of AMD.48 This study demonstrated that combinations of rare alleles in both C2 and CFB decreased an individual’s risk of AMD. Additionally, the protective effect for an individual with variation in these genes was most significant for those who were homozygous for the Y402H variation of CFH. These findings were confirmed in a report that examined simultaneously CFH, C2, CFB, and ARMS2 on risk of advanced AMD.54 Recently, a genome wide scan validated the candidate gene association of C2/CFB results in a separate population.56
Complement Component 3 (C3)
C3 is an important component of all three complement cascades: the alternative, the classical, and the lectin pathways. Numerous studies have demonstrated an association of rare alleles in C3 with AMD risk.51,52,125,141,142 The most significantly associated SNP, rs2230199, is non-synonymous and results in an amino acid change from arginine to glycine with functional consequences illustrated by a shift in electrophoretic mobility and a change in the ability of C3 to bind monocyte complement receptors.143 This SNP has been shown to be significantly associated with late stage AMD in diverse populations.51,141,144,145 However, this and other C3 SNPs have not been associated with AMD in all populations.142,146,147 A role for C3 in AMD is not only supported by genetics studies but also by the presence of the protein in drusen.24,51,135,148
The association of CFH and its related pathway genes with AMD susceptibility and pathophysiology underscores the importance of examining common variation in multiple susceptibility genes as well as contributing environmental factors simultaneously to get a better estimate of an individual’s risk of AMD. Several groups have begun to do this with respect to the contribution of CFH along with other reported genetic and epidemiological risk factors for AMD. Some of these studies include investigations of CFH, ARMS2 and smoking,107,117,149,150 CFH and smoking,29,130 CFH, Complement Component 2 Gene (C2), and Complement Factor B Gene (CFB),48 and CFH, C2, CFB, and ARMS2.151
Other Genes Involved in Immune Regulation
Chemokine (C-X3-C Motif) Receptor 1 (CX3CR1)
CX3CR1 functions as an acute phase regulator of the immune response. CX3CR1 is localized to the short arm of chromosome 3 (3p21.3), a region found to be significantly linked to the advanced stages of AMD by two independent genome-wide scans34,40 Two polymorphisms in CX3CR1, V249I (rs3732379) and T280M (rs3732378), were found to be associated with cases of advanced AMD for both homozygous and heterozygous states.152 This study also demonstrated decreased level of expression of this gene in the maculae of a patient homozygous for the T280M variation when compared to the maculae from a control individual.
Toll-like Receptor 3 (TLR3)
TLR3 is located on 4q35. The protein product acts as a viral sensor by recognizing double-stranded RNA and is thus important to cellular immunity.153 Variation within TLR3, specifically in the SNP rs3775291, has been associated with protection against geographic atrophy;154 however, studies done using several independent case-control cohorts including those from the National Eye Institute Clinical Center, the Age-Related Eye Disease Study, Columbia University, the University of Iowa, and the Blue Mountains Eye Study were unable to replicate association of this particular SNP with geographic atrophy as well as all other AMD subtypes.155–158
Toll-like Receptor 4 (TLR4)
TLR4 is localized to the long arm of chromosome 9 (9q32–q33), a region previously linked to patients with the advanced forms of AMD.34 A variant in this gene, which functions in innate immunity, lipid transport, and metabolism, has been previously shown to be associated with a decreased risk of arteriosclerosis.159,160 This same variant, D299G (rs4986790), was found to be associated with a 2.6-fold increased risk of AMD.161 However, this same group, as well as other groups, have not been able to replicate the association of TLR4 in larger populations representing all AMD subtypes.162,163
Lipid Metabolism Genes
Apolipoprotein (APOE)
Like ELOVL4, APOE functions in lipid transport and metabolism. APOE was the first candidate gene investigated for a role in AMD outside of the genes responsible for the juvenile forms of macular degenerations. The role of APOE in susceptibility to Alzheimer’s disease is well established; i.e., APOE is a component of the amyloid plaques that are a hallmark of this degenerative disease of the central nervous system. Three alleles, E2, E3, E4 (E3 is the most frequent allele in the general population), were determined by the last exon, exon 4, of APOE influence disease risk for Alzheimer’s. The E4 allele, and in particular the E4/E4 genotype, is thought to be associated with increased risk of an earlier onset of Alzheimer’s disease while the E2 allele is thought to be protective for development of Alzheimer’s disease (as reviewed in 164). The E4 allele of APOE was first reported by the Rotterdam group in 1998 to be associated with a decreased risk of AMD.165 A trend toward increased risk of development of AMD was observed with the E2 allele, but this finding was not statistically significant. This study further histologically demonstrated the presence of APOE in large soft drusen from eyes of patients with AMD as well as the absence of this protein from small hard drusen. Many other studies have supported the finding that the E4 allele is associated with a decreased risk of both early and more advanced stages of AMD.166–171 Although several of these studies have shown a trend toward an association of the E2 allele with increased risk of AMD,165,168,169 this finding only reached statistical significance in one study.171
However, several studies have also failed to confirm an association between APOE variants and AMD risk.172–175 Further supporting the lack of association between APOE and AMD is the failure of any genome wide scan to show the 19q13.2 region containing APOE to be linked to AMD. Other data suggest that while APOE genotype alone might not significantly influence AMD risk, it could modify the effect of smoking as an AMD risk factor.175 Specifically, smoking was found to greatly increase the risk of neovascular AMD in individuals with the E2 allele compared to non-smokers with the E3/E3 genotype. This finding suggests that if APOE is involved in AMD pathogenesis, it may function as a weak modifier of risk.
Paraoxonase 1 (PON1)
PON1, like APOE, encodes for a protein that regulates both lipid metabolism and transport. The three studies to date that have been published are not in agreement as to the contribution of PON1 in the etiology of AMD. Specifically, the M55L (rs854560) and Q192R (rs662) variants were reported to be associated with neovascular AMD in 72 individuals from Japan.176 The affected individuals also had significantly higher levels of low density lipoprotein in the plasma.177 Two other studies in patients of English, Scottish, and northern Irish ancestry, however, found no association between these variants and advanced AMD.178,179
Very Low Density Lipoprotein Receptor (VLDLR)
There is a lack of agreement between the two studies conducted to date as to the association of VLDLR and AMD. VLDLR is located on the short arm of chromosome 9 (9p24), a region found to be significantly associated with late AMD in one genome-wide scan.37 While one study found a significant association of SNPs within this gene with intermediate and late AMD in Caucasian patients,180 another large study on Caucasian patients with similar AMD severity did not find a significant association with VLDRL.90 In support of a possible role for VLDLR in AMD are data showing that the VLDLR knockout mouse develops subretinal neovascularization characteristic of retinal angiomatous proliferation seen in humans with AMD.181–183
Low Density Lipoprotein Receptor Related Protein 6 (LRP6)
Using a combination of linkage and association analysis on family-based and case-control populations, respectively, Haines et al. demonstrated that LRP6 was associated with advanced AMD in Caucasian patients.180 Although LRP6 is located in a region, 12p11– p13, not identified by genome wide scans as significant, its recent identification as a gene of interest with respect to AMD risk warrants further study.
Extracellular Matrix
Hemicentin1 (HMCN1, FIBULIN 6, FIBL6)
In 1998, Klein and colleagues described a family with AMD linked to the long arm of chromosome 1 (1q25–1q32).33 Subsequently, this same region has been found by many studies to be linked or associated with both early and advanced stages of AMD.21,30,34,37,93,104,105 In 2003, Schultz et al. analyzed this same multigenerational family and found that a variant in HMCN1 was associated with early/intermediate AMD.184 HMCN1 has been shown to be involved in the maintenance of extracellular matrix integrity. Specifically, this group found that a Q5345R change in exon 104 (rs35856562) segregated with the disease. Based on these results, it was suggested that HMCN1 was the gene responsible for the phenotype observed in this family.184 Since this time, there have been no other confirmatory reports that the Q5345R variant in HMCN1 is associated with any subtype of AMD.90,92,185,186 However, Iyengar et al. reported that this gene could be involved in the etiology of AMD because linkage was observed for four other SNPs in this gene, concluding that other variants in HMCN1, as yet to be discovered, could be associated with AMD.37
Fibulin 5 (FBLN5)
FBLN5, located on the long arm of chromosome 14 (14q32.1), was chosen as a candidate gene because of its similarity to Fibulin 3, which is associated with Doyne’s honeycomb dystrophy, a disease with some phenotypic similarities to AMD. Additionally, the 14q32 region was reported by one genome wide scan to be linked to advanced AMD.40 Similar to HMCN1, FBLN5 is an extracellular matrix protein that functions in maintaining the integrity of elastic lamina, such as that found in Bruch’s membrane. Several variants in FIBLN5 were reported to be associated with AMD in 7 out of 402 AMD patients (1.7%) in a case-control study.187 Phenotypically, the seven patients all had the basal laminar or cuticular form of drusen. Three out of the seven patients also had neovascularization. None of these variants were found in unaffected individuals. This same group, while replicating their initial finding, also demonstrated that two additional novel variants in FIBLN5 were associated with other subtypes of AMD.188 This study further demonstrated in vitro that there is a reduction in secretion of FIBLN5 in COS7 cells transfected with constructs containing these variants when compared to wild type cells.
Other Genes
Manganese Superoxide Dismutase 2 (SOD-2)
SOD2, localized to 6q25.3, encodes an enzyme with anti-oxidant properties and is also expressed in the retina. A homozygous variation in exon 2 of this gene, resulting in the substitution of an alanine for a valine, was found to be associated with a 10-fold increased risk of neovascular AMD in 102 Japanese patients when compared to 200 ethnically matched controls.189 However, the variant in SOD2 was found to have an opposite effect in a separate Japanese population190 and could not be found in patients with neovascular AMD at a statistically significant level when compared to controls in additional studies from northern Ireland as well as Japan.179,191
Cystatin C (CST3)
CST3, similar to TIMP3 (Sorsby’s fundus dystrophy), is a protease inhibitor. CST3 is localized to 20p11.21, a region as yet to be reported as being linked to any type of AMD. Homozygosity for a variant in the 5′ untranslated region of CST3 was found in almost 7% with neovascular AMD in a German cohort.192
Angiotensin I Converting Enzyme (ACE)
ACE is localized to the long arm of chromosome 17 (17q23.3) and its role as a regulator of blood pressure is well established. An Alu sequence (a short or long interspersed sequence of nucleotides of around 300 base pairs repeated within the gene, not usually in tandem, originally characterized by the action of the Alu restriction endonuclease), was found to confer a five-fold decreased risk of early or advanced dry AMD when present homozygously.193 However, another study that examined a similar spectrum of AMD phenotypes concluded that there was no association of ACE with risk of AMD.90
Vascular Endothelial Growth Factor A (VEGFA)
VEGFA is localized to the short arm of chromosome 6 (6p12), a region not highlighted by any genome wide scan. The role of VEGFA as a regulator of angiogenesis is well established. Further, VEGFA is the target for current therapeutic interventions for neovascular AMD, such as ranibizumab. Common variation in VEGFA has been found to be significantly associated with both early and advanced forms of AMD in studies of various populations, including both family-based and case-control cohorts.180,194,195 Recently, Francis et al. showed increased risk of advanced disease for those homozygous for the risk allele of VEGFA SNP rs833070 in two separate unrelated AMD case-control cohorts.196 However, this finding could not be replicated in a family-based AMD cohort in the same study. Others have found no association between variation in VEGFA and neovascular AMD197 or any AMD subtype in a prospectively ascertained cohort of 4,228 individuals.198
Excision Repair Cross-complementation Group 6 (ERCC6)
ERCC6 is localized to 10q11.23 and functions in the repair of DNA. A SNP in the 5′-untranslated region of this gene was reported to be significantly associated in the homozygous state with advanced AMD.154 Further, increased expression of this gene was observed in lymphocytes from individuals that were homozygous for this variation. To date this is the only study showing an association of this gene with AMD.
RAR-related Orphan Receptor Alpha (RORA)
RORA is located on chromosome 15q and encodes an intracellular receptor that has been shown to be involved in several cellular processes implicated in AMD pathogenesis including oxidative stress, inflammation, and cholesterol/lipid metabolism.90,199–206 Our group recently showed a genetic association between neovascular AMD and RORA, which was first suggested by converging data from linkage analysis, gene expression microarray analysis, and association analysis.41 The association of both single SNPs and haplotypes in intron 1 of RORA was initially demonstrated in a cohort of 150 extremely discordant sibling pairs of 300 individuals, where one sibling had neovascular AMD and the other had normal maculae. Significant findings from this initial cohort were replicated in an unrelated case-control cohort from central Greece and prospectively validated in nested case-control cohorts.207 Analysis of sibling pairs as well as unrelated cases and controls for comparison of individuals with neovascular AMD to those with the early dry form (AREDS Category 2) demonstrated that variation within RORA is specifically associated with neovascular disease. Additionally, a gene-gene interaction between RORA and ARMS2/HTRA1 was demonstrated and was recently replicated in prospective nested case control cohorts.207 This gene-gene interaction suggests that RORA may be functioning in the same or related pathway to ARMS2/HTRA1. Functional studies in our laboratory are underway to validate these statistical findings.
FUTURE STUDIES
The differences in association of the aforementioned genes with AMD can potentially be attributed to variation in study design including differing cohort compositions (case control versus family based, ethnic heterogeneity), disease severities (all AMD versus advanced cases only), and statistical methodological approaches.48,54,126,208 Population stratification (differences in allele frequencies between cases and controls occurring by chance alone due to different ethnic origins), phenotypic heterogeneity (different presentations of the same disease), and diagnostic heterogeneity (inter-observer differences; different grading scales e.g., AREDS; International Classification System; Wisconsin Grading Scale) can confound results. Therefore, a challenge in the study of AMD is to standardize phenotypic characterization. One means of achieving this may be to develop better clinical diagnostic techniques in order to ascertain truly unaffected individuals.
Inconsistencies seen with more modest risk associations may occur due to the existence of multiple susceptibility genes for AMD, which do not necessarily contribute to disease in every patient with AMD. However, it is likely that numerous variations could influence common pathways underlying the disease. Therefore, it is important to study the pathways and mechanisms underlying AMD via several methods. By employing a systems biology approach utilizing information from various lines of investigation involving diverse scientific techniques in tandem with genetic studies, the functional consequences of genetic variation and the pathophysiology of the disease can be better understood.
With few exceptions, the biological implications of the genetic variation associated with AMD have yet to be fully explored and understood in terms of both normal function and pathophysiology. In cases where there is no apparent functional change, such as synonymous substitutions in exons or intronic variation, it is interesting to speculate that there may still be a change in the structure of a transcript and/or the sequence of unidentified modifying elements (e.g. silencers or enhancers).102,103 In turn, these variations could theoretically influence gene expression resulting in differential total expression, isoform expression, and/or transcript stability.209–211
Proteomics in Age Related Macular Degeneration
As discussed earlier in this review, a genomic convergence approach, or systems biology approach, where information is combined from several sources including robust phenotyping (as well as epidemiologic data), genetic data, expression data, and protein data, is key to the design of appropriate in vitro and in vivo experiments with the goal of elucidating true disease causality. Much work has been done in human donor eyes to identify genes that are differentially expressed between those with and without all subtypes of age related macular degeneration (for a recent review please see 135). However, not all genes will be involved in disease pathogenesis at the mRNA level but rather will function at the protein level to influence disease onset and progression. Studying genes at the protein level may confirm or uncover novel pathways underlying AMD pathophysiology and also help to resolve inconsistencies in genetic studies. Proteomics, or the evaluation of the structure and function of proteins in an organism, can be used to not only understand AMD pathogenesis but to enhance diagnostic and prognostic tools in the clinical setting. Early work in this field focused mainly on evaluating risk factors in serum and plasma for cardiovascular disease such as C-reactive protein and homocysteine. The measurement of these and additional biomarkers of inflammation/innate immunity, lipid metabolism, and nutrition has been conducted in serum and plasma from subjects with and without AMD, but results have been inconsistent and await larger confirmatory studies.212–223
With current more sensitive and higher throughput techniques, it has become possible to examine hundreds of proteins at one time. This provides an unbiased avenue to evaluate protein levels. However, similar to gene expression studies, access to appropriate tissues remains a challenge for large-scale proteomic studies. Since there is not yet an ideal animal model recapitulating AMD, fresh human donor eye tissue from those with and without AMD represents the best platform to conduct proteomic studies, but challenges remain with respect to timely collection of these tissues.
Nevertheless, use of human donor eye tissue has provided some important insights by showing additional pathways that may underlie AMD pathogenesis.224–226 For example, a study by Crabb and colleagues demonstrated that the protein crystallin was more common in drusen from AMD eyes compared to normal eyes and that more carboxyethyl pyrrole (CEP) adducts were present in Bruch’s membrane from AMD eyes vs. normal eyes.224 Subsequently, it has been shown that CEP adduct and autoantibodies are elevated in the plasma of AMD patients vs. controls and may serve as a biomarker for AMD.227
Additionally, proteomic studies of the mitochondria from the RPE in donor eyes with varying stages of AMD progression revealed AMD-associated changes in 8 proteins, including ATP synthase subunits and the mitochondrial translation factor Tu, implicating mitochondrial function in disease onset and progression.226 These proteomic studies complement genetic studies also supporting a role for mitochondria in AMD pathogenesis.228, 229
CONCLUSIONS
While the findings to date have begun to provide insights into AMD etiology, there are still many questions about AMD risk and pathogenesis that cannot be explained by the known AMD-related genes. Models incorporating the existing disease risk factors cannot fully account for the observed degree of heritability, nor can the combined assessment of the known genetic and environmental risk factors adequately predict the risk of disease (For review, please see Swaroop, et al., 20074 and 20098). These observations, coupled with data from the numerous linkage studies that included all AMD subtypes,30,33,41,93 suggest that important loci remain to be identified and characterized. To this end, the AMD GWAS consortium, a meta-analysis of cohorts throughout the world should help to refine regions and validate or uncover novel common variation associated with AMD risk.
However, complete characterization of both common and rare alleles that influence AMD risk will be necessary for optimal and accurate determination of an individual’s overall genetic risk of developing advanced disease as well as identification of new targets for therapeutic intervention.54,230 Therefore, efforts are underway by our group and others to perform whole exome sequencing on families enriched for advanced disease to uncover rare alleles that may contribute to disease pathophysiology. Additionally, the genetic evaluation of both common and rare variants in diverse ethnic populations throughout the world with varying prevalence of advanced AMD should help to pinpoint disease causality so that appropriate functional studies can follow.
With respect to clinical applications, although the currently recognized genes and environmental factors may help to identify those most susceptible to AMD, the lack of specific, efficacious preventative treatments severely limits the utility of genetic testing for individuals without any signs of disease. For those already affected by some degree of AMD, retinal findings remain the strongest predictor of advanced AMD, with genotype adding minimally to risk prediction.231 Finally, for those who develop advanced AMD, no consistent correlations have been made with respect to treatment response and genotype. Therefore, while genetic studies should be an integral aspect of clinical studies of AMD, routine genotyping of AMD patients may not yet be indicated.
ACkNOWLEDGMENT
The authors alone are responsible for the content and writing of the paper.
Footnotes
Declaration of interest: The authors report no conflicts of interest.
REFERENCES
- 1.Silvestri G, Johnston PB, Hughes AE. Is genetic predisposition an important risk factor in age-related macular degeneration? Eye (Lond) 1994;8(Pt 5):564–568. doi: 10.1038/eye.1994.138. [DOI] [PubMed] [Google Scholar]
- 2.Seddon JM, Ajani UA, Mitchell BD. Familial aggregation of age-related maculopathy. Am. J. Ophthalmol. 1997;123(2):199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 3.Klaver CC, Wolfs RC, Assink JJ, et al. Genetic risk of age-related maculopathy. Population-based familial aggregation study. Arch. Ophthalmol. 1998;116(12):1646–1651. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 4.Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: A paradigm for dissecting complex disease traits. Hum. Mol. Genet. 2007;16(Spec No. 2):R174–182. doi: 10.1093/hmg/ddm212. [DOI] [PubMed] [Google Scholar]
- 5.Hammond CJ, Webster AR, Snieder H, et al. Genetic influence on early age-related maculopathy: A twin study. Ophthalmology. 2002;109(4):730–736. doi: 10.1016/s0161-6420(01)01049-1. [DOI] [PubMed] [Google Scholar]
- 6.Friedman DS, O’Colmain BJ, Muñoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004;122(4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 7.Ferris FL, Davis MD, Clemons TE, et al. A simplified severity scale for age-related macular degeneration: AREDS Report No. 18. Arch. Ophthalmol. 2005;123(11):1570–1574. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: From genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: Markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman DS, Katz J, Bressler NM, Rahmani B, Tielsch JM. Racial differences in the prevalence of age-related macular degeneration: The Baltimore Eye Survey. Ophthalmology. 1999;106(6):1049–1055. doi: 10.1016/S0161-6420(99)90267-1. [DOI] [PubMed] [Google Scholar]
- 11.VanNewkirk MR, Nanjan MB, Wang JJ, et al. The prevalence of age-related maculopathy: The visual impairment project. Ophthalmology. 2000;107(8):1593–1600. doi: 10.1016/s0161-6420(00)00175-5. [DOI] [PubMed] [Google Scholar]
- 12.Muñoz B, Klein R, Rodriguez J, Snyder R, West SK. Prevalence of age-related macular degeneration in a population-based sample of Hispanic people in Arizona: Proyecto VER. Arch. Ophthalmol. 2005;123(11):1575–1580. doi: 10.1001/archopht.123.11.1575. [DOI] [PubMed] [Google Scholar]
- 13.Thornton J, Edwards R, Mitchell P, et al. Smoking and age-related macular degeneration: A review of association. Eye (Lond) 2005;19(9):935–944. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- 14.Klein R. Overview of progress in the epidemiology of age-related macular degeneration. Ophthalmic Epidemiol. 2007;14(4):184–187. doi: 10.1080/09286580701344381. [DOI] [PubMed] [Google Scholar]
- 15.Cruickshanks KJ, Klein R, Klein BE, Nondahl DM. Sunlight and the 5-year incidence of early age-related maculopathy: The beaver dam eye study. Arch. Ophthalmol. 2001;119(2):246–250. [PubMed] [Google Scholar]
- 16.Taylor HR, Muñoz B, West S, et al. Visible light and risk of age-related macular degeneration. Trans Am Ophthalmol Soc. 1990;88:163–173. discussion 173–178. [PMC free article] [PubMed] [Google Scholar]
- 17.Delcourt C, Carrière I, Ponton-Sanchez A, et al. Light exposure and the risk of age-related macular degeneration: The Pathologies Oculaires Liées à l’Age (POLA) study. Arch. Ophthalmol. 2001;119(10):1463–1468. doi: 10.1001/archopht.119.10.1463. [DOI] [PubMed] [Google Scholar]
- 18.Hyman LG, Lilienfeld AM, Ferris FL, Fine SL. Senile macular degeneration: A case-control study. Am. J. Epidemiol. 1983;118(2):213–227. doi: 10.1093/oxfordjournals.aje.a113629. [DOI] [PubMed] [Google Scholar]
- 19.West SK, Duncan DD, Muñoz B, et al. Sunlight exposure and risk of lens opacities in a population-based study: The Salisbury Eye Evaluation project. JAMA. 1998;280(8):714–718. doi: 10.1001/jama.280.8.714. [DOI] [PubMed] [Google Scholar]
- 20.Jakobsdottir J, Conley YP, Weeks DE, et al. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am. J. Hum. Genet. 2005;77(3):389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 23.Edwards AO, Ritter R, Abel KJ, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 24.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 2005;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet. 2005;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 26.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314(5801):989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Atmaca-Sonmez P, Othman M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat. Genet. 2006;38(9):1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314(5801):992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 29.DeAngelis MM, Ji F, Kim IK, et al. Cigarette smoking, CFH, APOE, ELOVL4, and risk of neovascular age-related macular degeneration. Arch. Ophthalmol. 2007;125(1):49–54. doi: 10.1001/archopht.125.1.49. [DOI] [PubMed] [Google Scholar]
- 30.Fisher SA, Abecasis GR, Yashar BM, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum. Mol. Genet. 2005;14(15):2257–2264. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 31.Shuler RK, Hauser MA, Caldwell J, et al. Neovascular age-related macular degeneration and its association with LOC387715 and complement factor H polymorphism. Arch. Ophthalmol. 2007;125(1):63–67. doi: 10.1001/archopht.125.1.63. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Morrison MA, Dewan A, et al. The NEI/NCBI dbGAP database: Genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med. Genet. 2008;9:51. doi: 10.1186/1471-2350-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein ML, Schultz DW, Edwards A, et al. Age-related macular degeneration. Clinical features in a large family and linkage to chromosome 1q. Arch. Ophthalmol. 1998;116(8):1082–1088. doi: 10.1001/archopht.116.8.1082. [DOI] [PubMed] [Google Scholar]
- 34.Majewski J, Schultz DW, Weleber RG, et al. Age-related macular degeneration--A genome scan in extended families. Am. J. Hum. Genet. 2003;73(3):540–550. doi: 10.1086/377701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schick JH, Iyengar SK, Klein BE, et al. A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am. J. Hum. Genet. 2003;72(6):1412–1424. doi: 10.1086/375500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis GR, Yashar BM, Zhao Y, et al. Age-related macular degeneration: A high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am. J. Hum. Genet. 2004;74(3):482–494. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyengar SK, Song D, Klein BEK, et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am. J. Hum. Genet. 2004;74(1):20–39. doi: 10.1086/380912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt S, Scott WK, Postel EA, et al. Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12. BMC Genet. 2004;5:18. doi: 10.1186/1471-2156-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenealy SJ, Schmidt S, Agarwal A, et al. Linkage analysis for age-related macular degeneration supports a gene on chromosome 10q26. Mol. Vis. 2004;10:57–61. [PubMed] [Google Scholar]
- 40.Jun G, Klein BEK, Klein R, et al. Genome-wide analyses demonstrate novel loci that predispose to drusen formation. Invest. Ophthalmol. Vis. Sci. 2005;46(9):3081–3088. doi: 10.1167/iovs.04-1360. [DOI] [PubMed] [Google Scholar]
- 41.Silveira AC, Morrison MA, Ji F, et al. Convergence of linkage, gene expression and association data demonstrates the influence of the RAR-related orphan receptor alpha (RORA) gene on neovascular AMD: A systems biology based approach. Vision Res. 2010;50(7):698–715. doi: 10.1016/j.visres.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St George-Hyslop P, Haines J, Rogaev E, et al. Genetic evidence for a novel familial Alzheimer’s disease locus on chromosome 14. Nat. Genet. 1992;2(4):330–334. doi: 10.1038/ng1292-330. [DOI] [PubMed] [Google Scholar]
- 43.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376(6543):775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 44.Kennedy JL, Farrer LA, Andreasen NC, Mayeux R, St George-Hyslop P. The genetics of adult-onset neuropsychiatric disease: Complexities and conundra? Science. 2003;302(5646):822–826. doi: 10.1126/science.1092132. [DOI] [PubMed] [Google Scholar]
- 45.Yu P, Oberto G. Alzheimer’s disease: Transgenic mouse models and drug assessment. Pharmacol. Res. 2000;42(2):107–114. doi: 10.1006/phrs.2000.0670. [DOI] [PubMed] [Google Scholar]
- 46.Ishii K, Lippa C, Tomiyama T, et al. Distinguishable effects of presenilin-1 and APP717 mutations on amyloid plaque deposition. Neurobiol. Aging. 2001;22(3):367–376. doi: 10.1016/s0197-4580(01)00216-0. [DOI] [PubMed] [Google Scholar]
- 47.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 48.Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006;38(4):458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 2006;38(9):1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 50.Hughes AE, Orr N, Esfandiary H, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat. Genet. 2006;38(10):1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 51.Yates ?Sepp T, Matharu BK, et al. Complement C3 variant and the risk of age-related macular degeneration. N. Engl. J. Med. 2007;357(6):553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 52.Maller JB, Fagerness JA, Reynolds RC, et al. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat. Genet. 2007;39(10):1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 53.Spencer KL, Hauser MA, Olson LM, et al. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Hum. Mol. Genet. 2007;16(16):1986–1992. doi: 10.1093/hmg/ddm146. [DOI] [PubMed] [Google Scholar]
- 54.Jakobsdottir J, Conley YP, Weeks DE, Ferrell RE, Gorin MB. C2 and CFB genes in age-related maculopathy and joint action with CFH and LOC387715 genes. PLoS ONE. 2008;3(5):e2199. doi: 10.1371/journal.pone.0002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gehrs KM, Jackson JR, Brown EN, Allikmets R, Hageman GS. Complement, age-related macular degeneration and a vision of the future. Arch. Ophthalmol. 2010;128(3):349–358. doi: 10.1001/archophthalmol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 2010;107(16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fagerness JA, Maller JB, Neale BM, et al. Variation near complement factor I is associated with risk of advanced AMD. Eur. J. Hum. Genet. 2009;17(1):100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moonesinghe R, Liu T, Khoury MJ. Evaluation of the discriminative accuracy of genomic profiling in the prediction of common complex diseases. Eur. J. Hum. Genet. 2010;18(4):485–489. doi: 10.1038/ejhg.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelly M, Semsarian C. Multiple mutations in genetic cardiovascular disease: A marker of disease severity? Circ Cardiovasc Genet. 2009;2(2):182–190. doi: 10.1161/CIRCGENETICS.108.836478. [DOI] [PubMed] [Google Scholar]
- 60.Ioannidis JPA. Prediction of cardiovascular disease outcomes and established cardiovascular risk factors by genome-wide association markers. Circ Cardiovasc Genet. 2009;2(1):7–15. doi: 10.1161/CIRCGENETICS.108.833392. [DOI] [PubMed] [Google Scholar]
- 61.Lusis AJ, Weiss JN. Cardiovascular networks: Systems-based approaches to cardiovascular disease. Circulation. 2010;121(1):157–170. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462(7271):307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 63.Hakonarson H, Grant SFA. Genome-wide association studies in type 1 diabetes, inflammatory bowel disease and other immune-mediated disorders. Semin. Immunol. 2009;21(6):355–362. doi: 10.1016/j.smim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Reitz C, Mayeux R. Use of genetic variation as biomarkers for Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2009;1180:75–96. doi: 10.1111/j.1749-6632.2009.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gubitz AK, Gwinn K. Mining the genome for susceptibility to complex neurological disorders. Curr. Mol. Med. 2009;9(7):801–813. doi: 10.2174/156652409789105534. [DOI] [PubMed] [Google Scholar]
- 66.Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer’s disease. Hum. Mol. Genet. 2009;18(R2):R137–145. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cichon S, Craddock N, Daly M, et al. Genomewide association studies: History, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeAngelis M, Ji F. Albert & Jakobiec’s Principles & Practice of Ophthalmology. 3rd ed Saunders; Philadelphia, PA: 2008. Genetics of age-related macular degeneration; pp. 1881–1900. [Google Scholar]
- 70.Allikmets R, Seddon JM, Bernstein PS, et al. Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies. Hum. Genet. 1999;104(6):449–453. doi: 10.1007/s004390050986. [DOI] [PubMed] [Google Scholar]
- 71.Krämer F, White K, Pauleikhoff D, et al. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur. J. Hum. Genet. 2000;8(4):286–292. doi: 10.1038/sj.ejhg.5200447. [DOI] [PubMed] [Google Scholar]
- 72.Lotery AJ, Munier FL, Fishman GA, et al. Allelic variation in the VMD2 gene in best disease and age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2000;41(6):1291–1296. [PubMed] [Google Scholar]
- 73.Akimoto A, Akimoto M, Kuroiwa S, Kikuchi T, Yoshimura N. Lack of association of mutations of the bestrophin gene with age-related macular degeneration in non-familial Japanese patients. Graefes Arch. Clin. Exp. Ophthalmol. 2001;239(1):66–68. doi: 10.1007/pl00007900. [DOI] [PubMed] [Google Scholar]
- 74.Seddon JM, Afshari MA, Sharma S, et al. Assessment of mutations in the Best macular dystrophy (VMD2) gene in patients with adult-onset foveomacular vitelliform dystrophy, age-related maculopathy, and bull’s-eye maculopathy. Ophthalmology. 2001;108(11):2060–2067. doi: 10.1016/s0161-6420(01)00777-1. [DOI] [PubMed] [Google Scholar]
- 75.Kemp CM, Jacobson SG, Cideciyan AV, et al. RDS gene mutations causing retinitis pigmentosa or macular degeneration lead to the same abnormality in photoreceptor function. Invest. Ophthalmol. Vis. Sci. 1994;35(8):3154–3162. [PubMed] [Google Scholar]
- 76.Gorin MB, Jackson KE, Ferrell RE, et al. A peripherin/retinal degeneration slow mutation (Pro-210-Arg) associated with macular and peripheral retinal degeneration. Ophthalmology. 1995;102(2):246–255. doi: 10.1016/s0161-6420(95)31029-9. [DOI] [PubMed] [Google Scholar]
- 77.Stone EM, Lotery AJ, Munier FL, et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 1999;22(2):199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 78.Guymer RH, McNeil R, Cain M, et al. Analysis of the Arg345Trp disease-associated allele of the EFEMP1 gene in individuals with early onset drusen or familial age-related macular degeneration. Clin. Experiment. Ophthalmol. 2002;30(6):419–423. doi: 10.1046/j.1442-9071.2002.00572.x. [DOI] [PubMed] [Google Scholar]
- 79.Narendran N, Guymer RH, Cain M, Baird PN. Analysis of the EFEMP1 gene in individuals and families with early onset drusen. Eye (Lond) 2005;19(1):11–15. doi: 10.1038/sj.eye.6701435. [DOI] [PubMed] [Google Scholar]
- 80.Allikmets R, Shroyer NF, Singh N, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277(5333):1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 81.Allikmets R. Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am. J. Hum. Genet. 2000;67(2):487–491. doi: 10.1086/303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stone EM, Webster AR, Vandenburgh K, et al. Allelic variation in ABCR associated with Stargardt disease but not age-related macular degeneration. Nat. Genet. 1998;20(4):328–329. doi: 10.1038/3798. [DOI] [PubMed] [Google Scholar]
- 83.De La Paz MA, Guy VK, Abou-Donia S, et al. Analysis of the Stargardt disease gene (ABCR) in age-related macular degeneration. Ophthalmology. 1999;106(8):1531–1536. doi: 10.1016/S0161-6420(99)90449-9. [DOI] [PubMed] [Google Scholar]
- 84.Souied EH, Ducroq D, Gerber S, et al. Age-related macular degeneration in grandparents of patients with Stargardt disease: Genetic study. Am. J. Ophthalmol. 1999;128(2):173–178. doi: 10.1016/s0002-9394(99)00145-2. [DOI] [PubMed] [Google Scholar]
- 85.Rivera A, White K, Stöhr H, et al. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am. J. Hum. Genet. 2000;67(4):800–813. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guymer RH, Héon E, Lotery AJ, et al. Variation of codons 1961 and 2177 of the Stargardt disease gene is not associated with age-related macular degeneration. Arch. Ophthalmol. 2001;119(5):745–751. doi: 10.1001/archopht.119.5.745. [DOI] [PubMed] [Google Scholar]
- 87.Webster AR, Héon E, Lotery AJ, et al. An analysis of allelic variation in the ABCA4 gene. Invest. Ophthalmol. Vis. Sci. 2001;42(6):1179–1189. [PubMed] [Google Scholar]
- 88.Bernstein PS, Leppert M, Singh N, et al. Genotype-phenotype analysis of ABCR variants in macular degeneration probands and siblings. Invest. Ophthalmol. Vis. Sci. 2002;43(2):466–473. [PubMed] [Google Scholar]
- 89.Schmidt S, Postel EA, Agarwal A, et al. Detailed analysis of allelic variation in the ABCA4 gene in age-related maculopathy. Invest. Ophthalmol. Vis. Sci. 2003;44(7):2868–2875. doi: 10.1167/iovs.02-0957. [DOI] [PubMed] [Google Scholar]
- 90.Conley YP, Thalamuthu A, Jakobsdottir J, et al. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum. Mol. Genet. 2005;14(14):1991–2002. doi: 10.1093/hmg/ddi204. [DOI] [PubMed] [Google Scholar]
- 91.Ayyagari R, Zhang K, Hutchinson A, et al. Evaluation of the ELOVL4 gene in patients with age-related macular degeneration. Ophthalmic Genet. 2001;22(4):233–239. doi: 10.1076/opge.22.4.233.2219. [DOI] [PubMed] [Google Scholar]
- 92.Seitsonen S, Lemmelä S, Holopainen J, et al. Analysis of variants in the complement factor H, the elongation of very long chain fatty acids-like 4 and the hemicentin 1 genes of age-related macular degeneration in the Finnish population. Mol. Vis. 2006;12:796–801. [PubMed] [Google Scholar]
- 93.Seddon JM, Santangelo SL, Book K, Chong S, Cote J. A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am. J. Hum. Genet. 2003;73(4):780–790. doi: 10.1086/378505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hosaka M, Südhof TC. Synapsin III, a novel synapsin with an unusual regulation by Ca2+ J. Biol. Chem. 1998;273(22):13371–13374. doi: 10.1074/jbc.273.22.13371. [DOI] [PubMed] [Google Scholar]
- 95.Macgregor AM, Eberhart CG, Fraig M, Lu J, Halushka MK. Tissue inhibitor of matrix metalloproteinase-3 levels in the extracellular matrix of lung, kidney, and eye increase with age. J. Histochem. Cytochem. 2009;57(3):207–213. doi: 10.1369/jhc.2008.952531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi JH, Ebrahem Q, Moore N, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): Inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 2003;9(4):407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- 97.De La Paz MA, Pericak-Vance MA, Lennon F, Haines JL, Seddon JM. Exclusion of TIMP3 as a candidate locus in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 1997;38(6):1060–1065. [PubMed] [Google Scholar]
- 98.Felbor U, Doepner D, Schneider U, Zrenner E, Weber BH. Evaluation of the gene encoding the tissue inhibitor of metal-loproteinases-3 in various maculopathies. Invest. Ophthalmol. Vis. Sci. 1997;38(6):1054–1059. [PubMed] [Google Scholar]
- 99.Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum. Mol. Genet. 2010;19(R2):R176–187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ku CS, Loy EY, Salim A, Pawitan Y, Chia KS. The discovery of human genetic variations and their use as disease markers: Past, present and future. J. Hum. Genet. 2010;55(7):403–415. doi: 10.1038/jhg.2010.55. [DOI] [PubMed] [Google Scholar]
- 101.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 102.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lupski JR. Genomic rearrangements and sporadic disease. Nat. Genet. 2007;39(7 Suppl):S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 104.Weeks DE, Conley YP, Tsai HJ, et al. Age-related maculopathy: An expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am. J. Ophthalmol. 2001;132(5):682–692. doi: 10.1016/s0002-9394(01)01214-4. [DOI] [PubMed] [Google Scholar]
- 105.Weeks DE, Conley YP, Tsai H, et al. Age-related maculopathy: A genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am. J. Hum. Genet. 2004;75(2):174–189. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weeks DE, Conley YP, Mah TS, et al. A full genome scan for age-related maculopathy. Hum. Mol. Genet. 2000;9(9):1329–1349. doi: 10.1093/hmg/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 107.Schmidt S, Hauser MA, Scott WK, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am. J. Hum. Genet. 2006;78(5):852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 2007;104(41):16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang G, Spencer KL, Scott WK, et al. Analysis of the indel at the ARMS2 3′UTR in age-related macular degeneration. Hum. Genet. 2010;127(5):595–602. doi: 10.1007/s00439-010-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fritsche LG, Loenhardt T, Janssen A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat. Genet. 2008;40(7):892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 111.Wang G, Spencer KL, Court BL, et al. Localization of age-related macular degeneration-associated ARMS2 in cytosol, not mitochondria. Invest. Ophthalmol. Vis. Sci. 2009;50(7):3084–3090. doi: 10.1167/iovs.08-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brantley MA, Fang AM, King JM, et al. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114(12):2168–2173. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 113.Shuler RK, Schmidt S, Gallins P, et al. Phenotype analysis of patients with the risk variant LOC387715 (A69S) in age-related macular degeneration. Am. J. Ophthalmol. 2008;145(2):303–307. doi: 10.1016/j.ajo.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 114.Andreoli MT, Morrison MA, Kim BJ, et al. Comprehensive analysis of complement factor H and LOC387715/ARMS2/HTRA1 variants with respect to phenotype in advanced age-related macular degeneration. Am. J. Ophthalmol. 2009;148(6):869–874. doi: 10.1016/j.ajo.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conley YP, Jakobsdottir J, Mah T, et al. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum. Mol. Genet. 2006;15(21):3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 116.Ross RJ, Bojanowski CM, Wang JJ, et al. The LOC387715 polymorphism and age-related macular degeneration: Replication in three case-control samples. Invest. Ophthalmol. Vis. Sci. 2007;48(3):1128–1132. doi: 10.1167/iovs.06-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deangelis MM, Ji F, Adams S, et al. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;115(7):1209–1215. e7. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gotoh N, Nakanishi H, Hayashi H, et al. ARMS2 (LOC387715) variants in Japanese patients with exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Am. J. Ophthalmol. 2009;147(6):1037–1041. 1041.e1–2. doi: 10.1016/j.ajo.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 119.Gibbs D, Yang Z, Constantine R, et al. Further mapping of 10q26 supports strong association of HTRA1 polymorphisms with age-related macular degeneration. Vision Res. 2008;48(5):685–689. doi: 10.1016/j.visres.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 120.Hadley D, Orlin A, Brown G, et al. Analysis of six genetic risk factors highly associated with AMD in the region surrounding ARMS2 and HTRA1 on chromosome 10, region q26. Invest. Ophthalmol. Vis. Sci. 2010;51(4):2191–2196. doi: 10.1167/iovs.09-3798. [DOI] [PubMed] [Google Scholar]
- 121.Losonczy G, Fekete A, Vokó Z, et al. [Accessed February 28, 2011];Analysis of complement factor H Y402H, LOC387715, HTRA1 polymorphisms and ApoE alleles with susceptibility to age-related macular degeneration in Hungarian patients. Acta Ophthalmol. 2009 doi: 10.1111/j.1755-3768.2009.01687.x. Available at: http://www.ncbi.nlm.nih.gov.ezp-prod1.hul.harvard.edu/pubmed/19845562. [DOI] [PubMed] [Google Scholar]
- 122.Mori K, Horie-Inoue K, Gehlbach PL, et al. Phenotype and genotype characteristics of age-related macular degeneration in a Japanese population. Ophthalmology. 2010;117(5):928–938. doi: 10.1016/j.ophtha.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 123.Yang Z, Tong Z, Chen Y, et al. Genetic and functional dissection of HTRA1 and LOC387715 in age-related macular degeneration. PLoS Genet. 2010;6(2):e1000836. doi: 10.1371/journal.pgen.1000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jiang H, Qu Y, Dang G, et al. Analyses of single nucleotide polymorphisms and haplotype linkage of LOC387715 and the HTRA1 gene in exudative age-related macular degeneration in a Chinese cohort. Retina. 2009;29(7):974–979. doi: 10.1097/IAE.0b013e3181a3b90e. [DOI] [PubMed] [Google Scholar]
- 125.Bergeron-Sawitzke J, Gold B, Olsh A, et al. Multilocus analysis of age-related macular degeneration. Eur. J. Hum. Genet. 2009;17(9):1190–1199. doi: 10.1038/ejhg.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Farwick A, Dasch B, Weber BHF, et al. Variations in five genes and the severity of age-related macular degeneration: results from the Muenster aging and retina study. Eye (Lond) 2009;23(12):2238–2244. doi: 10.1038/eye.2008.426. [DOI] [PubMed] [Google Scholar]
- 127.An E, Sen S, Park SK, Gordish-Dressman H, Hathout Y. Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest. Ophthalmol. Vis. Sci. 2010;51(7):3379–3386. doi: 10.1167/iovs.09-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zareparsi S, Branham KEH, Li M, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am. J. Hum. Genet. 2005;77(1):149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Souied EH, Leveziel N, Richard F, et al. Y402H complement factor H polymorphism associated with exudative age-related macular degeneration in the French population. Mol. Vis. 2005;11:1135–1140. [PubMed] [Google Scholar]
- 130.Sepp T, Khan JC, Thurlby DA, et al. Complement factor H variant Y402H is a major risk determinant for geographic atrophy and choroidal neovascularization in smokers and nonsmokers. Invest. Ophthalmol. Vis. Sci. 2006;47(2):536–540. doi: 10.1167/iovs.05-1143. [DOI] [PubMed] [Google Scholar]
- 131.Gotoh N, Yamada R, Hiratani H, et al. No association between complement factor H gene polymorphism and exu-dative age-related macular degeneration in Japanese. Hum. Genet. 2006;120(1):139–143. doi: 10.1007/s00439-006-0187-0. [DOI] [PubMed] [Google Scholar]
- 132.Kardys I, Klaver CCW, Despriet DDG, et al. A common polymorphism in the complement factor H gene is associated with increased risk of myocardial infarction: The Rotterdam Study. J. Am. Coll. Cardiol. 2006;47(8):1568–1575. doi: 10.1016/j.jacc.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 133.Fremeaux-Bacchi V, Kemp EJ, Goodship JA, et al. The development of atypical haemolytic-uraemic syndrome is influenced by susceptibility factors in factor H and membrane cofactor protein: Evidence from two independent cohorts. J. Med. Genet. 2005;42(11):852–856. doi: 10.1136/jmg.2005.030783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abrera-Abeleda MA, Nishimura C, Smith JLH, et al. Variations in the complement regulatory genes factor H (CFH) and factor H related 5 (CFHR5) are associated with membranoproliferative glomerulonephritis type II (dense deposit disease) J. Med. Genet. 2006;43(7):582–589. doi: 10.1136/jmg.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Anderson DH, Radeke MJ, Gallo NB, et al. The pivotal role of the complement system in aging and age-related macular degeneration: Hypothesis re-visited. Prog Retin Eye Res. 2010;29(2):95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Abarrategui-Garrido C, Martínez-Barricarte R, López-Trascasa M, de Có,rdoba SR, Sánchez-Corral P. Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood. 2009;114(19):4261–4271. doi: 10.1182/blood-2009-05-223834. [DOI] [PubMed] [Google Scholar]
- 137.Józsi M, Licht C, Strobel S, et al. Factor H autoantibodies in atypical hemolytic uremic syndrome correlate with CFHR1/CFHR3 deficiency. Blood. 2008;111(3):1512–1514. doi: 10.1182/blood-2007-09-109876. [DOI] [PubMed] [Google Scholar]
- 138.Moore I, Strain L, Pappworth I, et al. Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood. 2010;115(2):379–387. doi: 10.1182/blood-2009-05-221549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zipfel PF, Mache C, Müller D, et al. DEAP-HUS: Deficiency of CFHR plasma proteins and autoantibody-positive form of hemolytic uremic syndrome. Pediatr. Nephrol. 2010;25(10):2009–2019. doi: 10.1007/s00467-010-1446-9. [DOI] [PubMed] [Google Scholar]
- 140.Hageman GS, Hancox LS, Taiber AJ, et al. Extended haplo-types in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: Characterization, ethnic distribution and evolutionary implications. Ann. Med. 2006;38(8):592–604. [PMC free article] [PubMed] [Google Scholar]
- 141.Spencer KL, Olson LM, Anderson BM, et al. C3 R102G polymorphism increases risk of age-related macular degeneration. Hum. Mol. Genet. 2008;17(12):1821–1824. doi: 10.1093/hmg/ddn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pei X, Li X, Bao Y, et al. Association of c3 gene polymorphisms with neovascular age-related macular degeneration in a chinese population. Curr. Eye Res. 2009;34(8):615–622. doi: 10.1080/02713680903003484. [DOI] [PubMed] [Google Scholar]
- 143.Arvilommi H. Capacity of complement c3 phenotypes to bind on to mononuclear cells in man. Nature. 1974;251(5477):740–741. doi: 10.1038/251740a0. [DOI] [PubMed] [Google Scholar]
- 144.Park KH, Fridley BL, Ryu E, Tosakulwong N, Edwards AO. Complement component 3 (C3) haplotypes and risk of advanced age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2009;50(7):3386–3393. doi: 10.1167/iovs.08-3231. [DOI] [PubMed] [Google Scholar]
- 145.McKay GJ, Dasari S, Patterson CC, Chakravarthy U, Silvestri G. Complement component 3: an assessment of association with AMD and analysis of gene-gene and gene-environment interactions in a Northern Irish cohort. Mol. Vis. 2010;16:194–199. [PMC free article] [PubMed] [Google Scholar]
- 146.Cui L, Zhou H, Yu J, et al. Noncoding variant in the complement factor H gene and risk of exudative age-related macular degeneration in a Chinese population. Invest. Ophthalmol. Vis. Sci. 2010;51(2):1116–1120. doi: 10.1167/iovs.09-4265. [DOI] [PubMed] [Google Scholar]
- 147.Farwick A, Wellmann J, Stoll M, Pauleikhoff D, Hense H. Susceptibility genes and progression in age-related maculopathy: A study of single eyes. Invest. Ophthalmol. Vis. Sci. 2010;51(2):731–736. doi: 10.1167/iovs.09-3953. [DOI] [PubMed] [Google Scholar]
- 148.Donoso LA, Kim D, Frost A, Callahan A, Hageman G. The role of inflammation in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2006;51(2):137–152. doi: 10.1016/j.survophthal.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Francis PJ, Zhang H, Dewan A, Hoh J, Klein ML. Joint effects of polymorphisms in the HTRA1, LOC387715/ARMS2, and CFH genes on AMD in a Caucasian population. Mol. Vis. 2008;14:1395–1400. [PMC free article] [PubMed] [Google Scholar]
- 150.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch. Ophthalmol. 2007;125(1):55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 151.Seddon JM, Reynolds R. Rosner B. Peripheral retinal drusen and reticular pigment: association with CFHY402H and CFHrs1410996 genotypes in family and twin studies. Invest. Ophthalmol. Vis. Sci. 2009;50(2):586–591. doi: 10.1167/iovs.08-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tuo J, Smith BC, Bojanowski CM, et al. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J. 2004;18(11):1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matsumoto M, Seya T. TLR3: Interferon induction by double-stranded RNA including poly(I:C) Adv. Drug Deliv. Rev. 2008;60(7):805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 154.Yang Z, Stratton C, Francis PJ, et al. Toll-like receptor 3 and geographic atrophy in age-related macular degeneration. N. Engl. J. Med. 2008;359(14):1456–1463. doi: 10.1056/NEJMoa0802437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cho Y, Wang JJ, Chew EY, et al. Toll-like receptor polymorphisms and age-related macular degeneration: Replication in three case-control samples. Invest. Ophthalmol. Vis. Sci. 2009;50(12):5614–5618. doi: 10.1167/iovs.09-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Edwards AO, Swaroop A, Seddon JM. Geographic atrophy in age-related macular degeneration and TLR3. N. Engl. J. Med. 2009;360(21):2254–2255. author reply 2255–2256. [PubMed] [Google Scholar]
- 157.Allikmets R, Bergen AA, Dean M, et al. Geographic atrophy in age-related macular degeneration and TLR3. N. Engl. J. Med. 2009;360(21):2252–2254. author reply 2255–2256. [PMC free article] [PubMed] [Google Scholar]
- 158.Lewin AS. Geographic atrophy in age-related macular degeneration and TLR3. N. Engl. J. Med. 2009;360(21):2251. doi: 10.1056/NEJMc082233. author reply 2255–2256. [DOI] [PubMed] [Google Scholar]
- 159.Kiechl S, Lorenz E, Reindl M, et al. Toll-like receptor 4 polymorphisms and atherogenesis. N. Engl. J. Med. 2002;347(3):185–192. doi: 10.1056/NEJMoa012673. [DOI] [PubMed] [Google Scholar]
- 160.Boekholdt SM, Agema WRP, Peters RJG, et al. Variants of toll-like receptor 4 modify the efficacy of statin therapy and the risk of cardiovascular events. Circulation. 2003;107(19):2416–2421. doi: 10.1161/01.CIR.0000068311.40161.28. [DOI] [PubMed] [Google Scholar]
- 161.Zareparsi S, Buraczynska M, Branham KEH, et al. Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum. Mol. Genet. 2005;14(11):1449–1455. doi: 10.1093/hmg/ddi154. [DOI] [PubMed] [Google Scholar]
- 162.Despriet DDG, Bergen AAB, Merriam JE, et al. Comprehensive analysis of the candidate genes CCL2, CCR2, and TLR4 in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2008;49(1):364–371. doi: 10.1167/iovs.07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Edwards AO, Chen D, Fridley BL, et al. Toll-like receptor polymorphisms and age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2008;49(4):1652–1659. doi: 10.1167/iovs.07-1378. [DOI] [PubMed] [Google Scholar]
- 164.Saunders AM. Apolipoprotein E and Alzheimer disease: An update on genetic and functional analyses. J. Neuropathol. Exp. Neurol. 2000;59(9):751–758. doi: 10.1093/jnen/59.9.751. [DOI] [PubMed] [Google Scholar]
- 165.Klaver CC, Kliffen M, van Duijn CM, et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am. J. Hum. Genet. 1998;63(1):200–206. doi: 10.1086/301901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Souied EH, Benlian P, Amouyel P, et al. The epsilon4 allele of the apolipoprotein E gene as a potential protective factor for exudative age-related macular degeneration. Am. J. Ophthalmol. 1998;125(3):353–359. doi: 10.1016/s0002-9394(99)80146-9. [DOI] [PubMed] [Google Scholar]
- 167.Schmidt S, Saunders AM, De La Paz MA, et al. Association of the apolipoprotein E gene with age-related macular degeneration: Possible effect modification by family history, age, and gender. Mol. Vis. 2000;6:287–293. [PubMed] [Google Scholar]
- 168.Simonelli F, Margaglione M, Testa F, et al. Apolipoprotein E polymorphisms in age-related macular degeneration in an Italian population. Ophthalmic Res. 2001;33(6):325–328. doi: 10.1159/000055688. [DOI] [PubMed] [Google Scholar]
- 169.Schmidt S, Klaver C, Saunders A, et al. A pooled case-control study of the apolipoprotein E (APOE) gene in age-related maculopathy. Ophthalmic Genet. 2002;23(4):209–223. doi: 10.1076/opge.23.4.209.13883. [DOI] [PubMed] [Google Scholar]
- 170.Zareparsi S, Reddick AC, Branham KEH, et al. Association of apolipoprotein E alleles with susceptibility to age-related macular degeneration in a large cohort from a single center. Invest. Ophthalmol. Vis. Sci. 2004;45(5):1306–1310. doi: 10.1167/iovs.03-1253. [DOI] [PubMed] [Google Scholar]
- 171.Baird PN, Guida E, Chu DT, Vu HTV, Guymer RH. The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2004;45(5):1311–1315. doi: 10.1167/iovs.03-1121. [DOI] [PubMed] [Google Scholar]
- 172.Pang CP, Baum L, Chan WM, et al. The apolipoprotein E epsilon4 allele is unlikely to be a major risk factor of age-related macular degeneration in Chinese. Ophthalmologica. 2000;214(4):289–291. doi: 10.1159/000027506. [DOI] [PubMed] [Google Scholar]
- 173.Schultz DW, Klein ML, Humpert A, et al. Lack of an association of apolipoprotein E gene polymorphisms with familial age-related macular degeneration. Arch. Ophthalmol. 2003;121(5):679–683. doi: 10.1001/archopht.121.5.679. [DOI] [PubMed] [Google Scholar]
- 174.Gotoh N, Kuroiwa S, Kikuchi T, et al. Apolipoprotein E polymorphisms in Japanese patients with polypoidal choroidal vasculopathy and exudative age-related macular degeneration. Am. J. Ophthalmol. 2004;138(4):567–573. doi: 10.1016/j.ajo.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 175.Schmidt S, Haines JL, Postel EA, et al. Joint effects of smoking history and APOE genotypes in age-related macular degeneration. Mol. Vis. 2005;11:941–949. [PubMed] [Google Scholar]
- 176.Ikeda T, Obayashi H, Hasegawa G, et al. Paraoxonase gene polymorphisms and plasma oxidized low-density lipoprotein level as possible risk factors for exudative age-related macular degeneration. Am. J. Ophthalmol. 2001;132(2):191–195. doi: 10.1016/s0002-9394(01)00975-8. [DOI] [PubMed] [Google Scholar]
- 177.Hara K, Shiga A, Fukutake T, et al. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N. Engl. J. Med. 2009;360(17):1729–1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- 178.Baird PN, Chu D, Guida E, Vu HTV, Guymer R. Association of the M55L and Q192R paraoxonase gene polymorphisms with age-related macular degeneration. Am. J. Ophthalmol. 2004;138(4):665–666. doi: 10.1016/j.ajo.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 179.Esfandiary H, Chakravarthy U, Patterson C, Young I, Hughes AE. Association study of detoxification genes in age related macular degeneration. Br J Ophthalmol. 2005;89(4):470–474. doi: 10.1136/bjo.2004.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Haines JL, Schnetz-Boutaud N, Schmidt S, et al. Functional candidate genes in age-related macular degeneration: Significant association with VEGF, VLDLR, and LRP6. Invest. Ophthalmol. Vis. Sci. 2006;47(1):329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 181.Frykman PK, Brown MS, Yamamoto T, Goldstein JL, Herz J. Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 1995;92(18):8453–8457. doi: 10.1073/pnas.92.18.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Heckenlively JR, Hawes NL, Friedlander M, et al. Mouse model of subretinal neovascularization with choroidal anastomosis. Retina. 2003;23(4):518–522. doi: 10.1097/00006982-200308000-00012. [DOI] [PubMed] [Google Scholar]
- 183.Hu W, Jiang A, Liang J, et al. Expression of VLDLR in the retina and evolution of subretinal neovascularization in the knockout mouse model’s retinal angiomatous proliferation. Invest. Ophthalmol. Vis. Sci. 2008;49(1):407–415. doi: 10.1167/iovs.07-0870. [DOI] [PubMed] [Google Scholar]
- 184.Tuo J, Ning B, Bojanowski CM, et al. Synergic effect of polymorphisms in ERCC6 5′ flanking region and complement factor H on age-related macular degeneration predisposition. Proc. Natl. Acad. Sci. U.S.A. 2006;103(24):9256–9261. doi: 10.1073/pnas.0603485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schultz DW, Klein ML, Humpert AJ, et al. Analysis of the ARMD1 locus: Evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum. Mol. Genet. 2003;12(24):3315–3323. doi: 10.1093/hmg/ddg348. [DOI] [PubMed] [Google Scholar]
- 186.Hayashi M, Merriam JE, Klaver CCW, et al. Evaluation of the ARMD1 locus on 1q25–31 in patients with age-related maculopathy: Genetic variation in laminin genes and in exon 104 of HEMICENTIN-1. Ophthalmic Genet. 2004;25(2):111–119. doi: 10.1080/13816810490514342. [DOI] [PubMed] [Google Scholar]
- 187.Stone EM, Braun TA, Russell SR, et al. Missense variations in the fibulin 5 gene and age-related macular degeneration. N. Engl. J. Med. 2004;351(4):346–353. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- 188.Lotery AJ, Baas D, Ridley C, et al. Reduced secretion of fibulin 5 in age-related macular degeneration and cutis laxa. Hum. Mutat. 2006;27(6):568–574. doi: 10.1002/humu.20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kimura K, Isashiki Y, Sonoda S, Kakiuchi-Matsumoto T, Ohba N. Genetic association of manganese superoxide dismutase with exudative age-related macular degeneration. Am. J. Ophthalmol. 2000;130(6):769–773. doi: 10.1016/s0002-9394(00)00552-3. [DOI] [PubMed] [Google Scholar]
- 190.Gotoh N, Yamada R, Matsuda F, Yoshimura N, Iida T. Manganese superoxide dismutase gene (SOD2) polymorphism and exudative age-related macular degeneration in the Japanese population. Am. J. Ophthalmol. 2008;146(1):146. doi: 10.1016/j.ajo.2008.03.017. author reply 146–147. [DOI] [PubMed] [Google Scholar]
- 191.Kondo N, Bessho H, Honda S, Negi A. SOD2 gene polymorphisms in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol. Vis. 2009;15:1819–1826. [PMC free article] [PubMed] [Google Scholar]
- 192.Zurdel J, Finckh U, Menzer G, Nitsch RM, Richard G. CST3 genotype associated with exudative age related macular degeneration. Br J Ophthalmol. 2002;86(2):214–219. doi: 10.1136/bjo.86.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hamdi HK, Reznik J, Castellon R, et al. Alu DNA polymorphism in ACE gene is protective for age-related macular degeneration. Biochem. Biophys. Res. Commun. 2002;295(3):668–672. doi: 10.1016/s0006-291x(02)00728-3. [DOI] [PubMed] [Google Scholar]
- 194.Churchill AJ, Carter JG, Lovell HC, et al. VEGF polymorphisms are associated with neovascular age-related macular degeneration. Hum. Mol. Genet. 2006;15(19):2955–2961. doi: 10.1093/hmg/ddl238. [DOI] [PubMed] [Google Scholar]
- 195.Janik-Papis K, Zaras M, Krzyzanowska A, et al. Association between vascular endothelial growth factor gene polymorphisms and age-related macular degeneration in a Polish population. Exp. Mol. Pathol. 2009;87(3):234–238. doi: 10.1016/j.yexmp.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 196.Francis PJ, Hamon SC, Ott J, Weleber RG, Klein ML. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age related macular degeneration associated with visual loss. J. Med. Genet. 2009;46(5):300–307. doi: 10.1136/jmg.2008.062737. [DOI] [PubMed] [Google Scholar]
- 197.Fang AM, Lee AY, Kulkarni M, Osborn MP, Brantley MA. Polymorphisms in the VEGFA and VEGFR-2 genes and neovascular age-related macular degeneration. Mol. Vis. 2009;15:2710–2719. [PMC free article] [PubMed] [Google Scholar]
- 198.Boekhoorn SS, Isaacs A, Uitterlinden AG, et al. Polymorphisms in the vascular endothelial growth factor gene and risk of age-related macular degeneration: The Rotterdam Study. Ophthalmology. 2008;115(11):1899–1903. doi: 10.1016/j.ophtha.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 199.Besnard S, Silvestre JS, Duriez M, et al. Increased ischemia-induced angiogenesis in the staggerer mouse, a mutant of the nuclear receptor Roralpha. Circ. Res. 2001;89(12):1209–1215. doi: 10.1161/hh2401.101755. [DOI] [PubMed] [Google Scholar]
- 200.Hageman GS, Luthert PJ, Victor Chong NH, et al. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20(6):705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 201.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am. J. Ophthalmol. 2002;134(3):411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 202.Klein R, Klein BEK, Tomany SC, Danforth LG, Cruickshanks KJ. Relation of statin use to the 5-year incidence and progression of age-related maculopathy. Arch. Ophthalmol. 2003;121(8):1151–1155. doi: 10.1001/archopht.121.8.1151. [DOI] [PubMed] [Google Scholar]
- 203.Boukhtouche F, Mariani J, Tedgui A. The “CholesteROR” protective pathway in the vascular system. Arterioscler. Thromb. Vasc. Biol. 2004;24(4):637–643. doi: 10.1161/01.ATV.0000119355.56036.de. [DOI] [PubMed] [Google Scholar]
- 204.Johnson EJ. Obesity, lutein metabolism, and age-related macular degeneration: A web of connections. Nutr. Rev. 2005;63(1):9–15. doi: 10.1111/j.1753-4887.2005.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 205.Zhu Y, McAvoy S, Kuhn R, Smith DI. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25(20):2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- 206.Lau P, Fitzsimmons RL, Raichur S, et al. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: Staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283(26):18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 207.Schaumberg DA, Chasman D, Morrison MA, et al. Prospective study of common variants in the retinoic acid receptor-related orphan receptor α gene and risk of neovascular age-related macular degeneration. Arch. Ophthalmol. 2010;128(11):1462–1471. doi: 10.1001/archophthalmol.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Scholl HPN, Charbel Issa P, Walier M, et al. Systemic complement activation in age-related macular degeneration. PLoS ONE. 2008;3(7):e2593. doi: 10.1371/journal.pone.0002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hollams EM, Giles KM, Thomson AM, Leedman PJ. MRNA stability and the control of gene expression: Implications for human disease. Neurochem. Res. 2002;27(10):957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- 210.Maddox DM, Vessey KA, Yarbrough GL, et al. Allelic variance between GRM6 mutants, Grm6nob3 and Grm6nob4 results in differences in retinal ganglion cell visual responses. J. Physiol. (Lond.) 2008;586(Pt 18):4409–4424. doi: 10.1113/jphysiol.2008.157289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Margulies EH, Birney E. Approaches to comparative sequence analysis: Towards a functional view of vertebrate genomes. Nat. Rev. Genet. 2008;9(4):303–313. doi: 10.1038/nrg2185. [DOI] [PubMed] [Google Scholar]
- 212.Reynolds R, Rosner B, Seddon JM. Serum lipid biomarkers and hepatic lipase gene associations with age-related macular degeneration. Ophthalmology. 2010;117(10):1989–1995. doi: 10.1016/j.ophtha.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gu J, Pauer GJT, Yue X, et al. Proteomic and genomic bio-markers for age-related macular degeneration. Adv Exp Med Biol. 2010;664:411–417. doi: 10.1007/978-1-4419-1399-9_47. [DOI] [PubMed] [Google Scholar]
- 214.Robman L, Baird PN, Dimitrov PN, Richardson AJ, Guymer RH. C-reactive protein levels and complement factor H polymorphism interaction in age-related macular degeneration and its progression. Ophthalmology. 2010;117(10):1982–1988. doi: 10.1016/j.ophtha.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 215.Gu J, Pauer GJT, Yue X, et al. Assessing susceptibility to age-related macular degeneration with proteomic and genomic biomarkers. Mol. Cell Proteomics. 2009;8(6):1338–1349. doi: 10.1074/mcp.M800453-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ni J, Yuan X, Gu J, et al. Plasma protein pentosidine and carboxymethyllysine, biomarkers for age-related macular degeneration. Mol. Cell Proteomics. 2009;8(8):1921–1933. doi: 10.1074/mcp.M900127-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lu L, Gu X, Hong L, et al. Synthesis and structural characterization of carboxyethylpyrrole-modified proteins: Mediators of age-related macular degeneration. Bioorg. Med. Chem. 2009;17(21):7548–7561. doi: 10.1016/j.bmc.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Klein R, Knudtson MD, Lee KE, Klein BEK. Serum cystatin C level, kidney disease markers, and incidence of age-related macular degeneration: The Beaver Dam Eye Study. Arch. Ophthalmol. 2009;127(2):193–199. doi: 10.1001/archophthalmol.2008.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Boekhoorn SS, Vingerling JR, Witteman JCM, Hofman A, de Jong PTVM. C-reactive protein level and risk of aging macula disorder: The Rotterdam Study. Arch. Ophthalmol. 2007;125(10):1396–1401. doi: 10.1001/archopht.125.10.1396. [DOI] [PubMed] [Google Scholar]
- 220.Wang JJ, Ross RJ, Tuo J, et al. The LOC387715 polymorphism, inflammatory markers, smoking, and age-related macular degeneration. A population-based case-control study. Ophthalmology. 2008;115(4):693–699. doi: 10.1016/j.ophtha.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Vine AK, Stader J, Branham K, Musch DC, Swaroop A. Biomarkers of cardiovascular disease as risk factors for age-related macular degeneration. Ophthalmology. 2005;112(12):2076–2080. doi: 10.1016/j.ophtha.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 222.Richer S, Rudy D, Statkute L, Karofty K, Frankowski J. Serum iron, transferrin saturation, ferritin, and dietary data in age-related macular degeneration. Am J Ther. 2002;9(1):25–28. doi: 10.1097/00045391-200201000-00006. [DOI] [PubMed] [Google Scholar]
- 223.Smith W, Mitchell P, Rochester C. Serum beta carotene, alpha tocopherol, and age-related maculopathy: The Blue Mountains Eye Study. Am. J. Ophthalmol. 1997;124(6):838–840. doi: 10.1016/s0002-9394(14)71702-7. [DOI] [PubMed] [Google Scholar]
- 224.Crabb JW, Miyagi M, Gu X, et al. Drusen proteome analysis: An approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 2002;99(23):14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ethen CM, Reilly C, Feng X, Olsen TW, Ferrington DA. The proteome of central and peripheral retina with progression of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2006;47(6):2280–2290. doi: 10.1167/iovs.05-1395. [DOI] [PubMed] [Google Scholar]
- 226.Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2008;49(7):2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gu J, Pauer GJ, Yue X, Narendra U, Sturgill GM, Bena J, Gu X, Peachey NS, Salomon RG, Hagstrom SA, Crabb JW. The Clinical Genomic and Proteomic AMD Study Group. Proteomic and genomic biomarkers for age-related macular degeneration. Adv Exp Med Biol. 2010;664:411–417. doi: 10.1007/978-1-4419-1399-9_47. [DOI] [PubMed] [Google Scholar]
- 228.Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM. Mitochondrial DNA haplogroups and age-related maculopathy. Arch Ophthalmol. 2007 Sep;125(9):1235–40. doi: 10.1001/archopht.125.9.1235. [DOI] [PubMed] [Google Scholar]
- 229.SanGiovanni JP, Arking DE, Iyengar SK, Elashoff M, Clemons TE, Reed GF, Henning AK, Sivakumaran TA, Xu X, DeWan A, Agrón E, Rochtchina E, Sue CM, Wang JJ, Mitchell P, Hoh J, Francis PJ, Klein ML, Chew EY, Chakravarti A. Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS One. 2009;4(5):e5508. doi: 10.1371/journal.pone.0005508. Epub 2009 May 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hamsten A, Eriksson P. Identifying the susceptibility genes for coronary artery disease: from hyperbole through doubt to cautious optimism. J. Intern. Med. 2008;263(5):538–552. doi: 10.1111/j.1365-2796.2008.01958.x. [DOI] [PubMed] [Google Scholar]
- 231.Seddon JM, Reynolds R, Maller J, Fagerness JA, Daly MJ, Rosner B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009 May;50(5):2044–53. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]