Abstract
Detecting somatic single nucleotide variants (SNVs) is an essential component of cancer research with next generation sequencing data. This protocol describes how to run the SomaticSniper somatic SNV detector and then filter the output to eliminate most false positives. It also includes support protocols detailing the compilation of the software.
Keywords: next-generation sequencing, somatic variant calling, somaticsniper
INTRODUCTION
In the last 5 years, the advent of massively parallel sequencing has revolutionized cancer genomics by allowing for the detection of a wide spectrum of tumor-specific variant types including copy number, small insertions and deletions, larger structural variants, and single nucleotide variants (SNVs). The simplest of these variants to detect are the SNVs both due to their small size and abundance relative to the numbers of the other types of variants. Despite this fact, detection of somatic SNVs is far from a solved problem and there remain myriad different approaches for their identification (Cibulskis et al., 2013; Koboldt et al., 2012; Larson et al., 2012; Saunders et al., 2012). In this protocol, we outline the recommended usage for one of the earliest of these methods, SomaticSniper (Larson et al., 2012), and discuss its strengths and weaknesses.
BASIC PROTOCOL 1
Calling Variants Using SomaticSniper
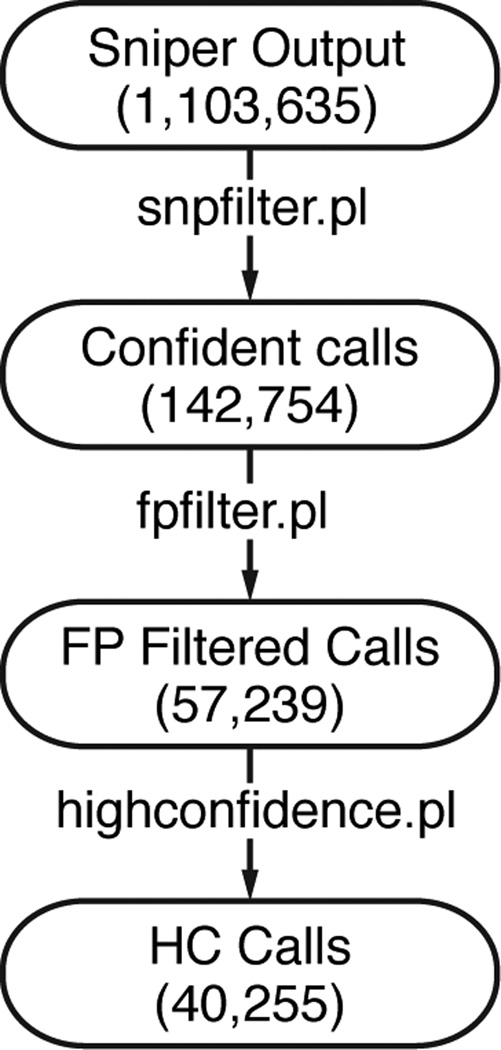
SomaticSniper has a single purpose: to detect somatic single nucleotide variants. This section describes the execution of SomaticSniper and the filtering of its results (Figure 1).
Figure 1. SomaticSniper filtering workflow.
SomaticSniper calls many variants and filtering is essential to eliminate false positives. The numbers in each step indicate the number of calls generated using SomaticSniper version 1.0.2 on the COLO-829 cell line data generated in (Pleasance et al., 2010). In the diagram, FP stands for false positive and HC stands for high confidence.
Necessary Resources
Hardware
A computer running Linux or Mac OS X with at least 1GB of RAM.
Software
The SomaticSniper program (see Support Protocol 1 for information on compiling from source) and the bam-readcount program (see Support Protocol 2 for information on compiling from source). Samtools is also utilized for filtering. An older version is provided (see Support Protocol 1), but versions up to 0.1.16 are supported by the filtering scripts.
Files
A pair of BAM files (tumor and normal), as well as the FASTA file containing the reference sequence that the BAM files are aligned to. Note that both BAM files must be aligned to the same reference sequence and should be indexed by Picard or Samtools. SomaticSniper does not require any special flags produced by specific aligners, but the mapping quality produced by the aligner is utilized to make the calls.
SomaticSniper is typically run on BAM files that have neither been base quality recalibrated nor locally realigned around indels (such as with the GATK). Either of these preprocessing steps can be optionally performed before running SomaticSniper, but we have observed some decrease in sensitivity after base quality recalibration.
It is important to have good coverage for detection of somatic variants. SomaticSniper was designed to work on whole genomes sequenced to 30X depth of coverage. Higher depths of coverage will improve variant calling in most cases, but will also increase some types of SomaticSniper false positives. Example files for the sections below can be obtained from https://github.com/genome/somatic-snv-test-data.
Run SomaticSniper and filter its output
- Call SomaticSniper from the command line
bam-somaticsniper –f ref.fa tumor.bam normal.bam output.txt
The main options for altering calling by SomaticSniper are the mapping quality threshold and somatic score threshold. The -q option can be used to adjust the minimum mapping quality of reads used for calling variants and the -Q option can be used to adjust the minimum somatic score for reporting variants. By default, there is no minimum mapping quality and the minimum somatic score for reporting a variant is 15. To increase stringency further, the -s option may be used to take into account a global expected somatic mutation rate (in mutations per base). A realistic value for this parameter would be 0.000001, but this will reduce sensitivity substantially for typical 30X genomes (Larson et al., 2012). Other options should be left as their defaults and a full outline of all options is available by running the tool without any options. - Examine the output available in output.txt.By default, SomaticSniper produces output in the so-called “classic” format, which contains a number of tab-delimited output fields. See Table 1 for a full description of the file format. This format is maintained in subsequent filtering steps.
- Generate a samtools pileup indel file for indel filtering
samtools pileup –cvi –f ref.fa tumor.bam indel.pileup
- Filter out low quality indels called by samtools.
perl samtools/misc/samtools.pl varFilter indel.pileup > indel.pileup.filtered
Note that the samtools directory listed above is the place where you have installed samtools. - Filter SomaticSniper’s predictions by quality and proximity to indels using the provided filtering script.
perl snpfilter.pl --snp-file output.txt --indel-file indel.pileup.filtered --out-file output.SNPfilter.txt
These scripts are located within the SomaticSniper source code at somatic-sniper/src/scripts/. Users may want to adjust the mapping quality threshold from the default of 40 (--min-mapping-quality) if an aligner other than BWA is utilized. Users may also wish to filter out reads with excess depth (--max-read-depth), but this setting will depend on the experimental coverage and by default is set extremely high. Users desiring higher stringency may also wish to raise the minimum coverage as well (--min-read-depth). Other parameters should generally be left to their defaults. - Adapt the remaining unfiltered SNVs for use with bam-readcount.
perl prepare_for_readcount.pl --snp-file output.SNPfilter.txt --out-file output.SNPfilter.pos
- Run bam-readcount using the same mapping quality (-q) setting as used for SomaticSniper.
bam-readcount -b 15 -f ref.fa –l output.SNPfilter.pos tumor.bam > output.SNPfilter.rc
Make certain that your BAM file has been sorted and indexed by Samtools or Picard before using bam-readcount. - Run the false positive filter to eliminate likely incorrect calls.
perl fpfilter.pl --snp-file output.SNPfilter.txt -- readcount-file output.SNPfilter.rc
This will generate a file containing those sites that passed the filter called output.SNPfilter.txt.fp_pass. Note that this filter is highly optimized for 100 bp paired end Illumina reads and may be too stringent for other technologies. - Lastly, run the "high confidence" filter which filters based on the somatic score and mapping quality.
perl highconfidence.pl --snp-file output.SNPfilter.txt.fp_pass --out-file output.SNPfilter.txt.fp_pass.hc
Users may want to adjust the mapping quality threshold (--min-mapping-quality) if an aligner other than BWA is utilized. The value for the somatic score threshold (--min-somatic-score) may be adjusted to increase the stringency of the calls.
Table 1. SomaticSniper output file columns.
A description of each column of SomaticSniper’s output file is below.
| Column # | Description |
|---|---|
| 1 | Chromosome name |
| 2 | Chromosomal location (1-based) |
| 3 | Reference base at this position |
| 4 | Genotype of tumor (as IUB code) |
| 5 | Genotype of normal (as IUB code) |
| 6 | Somatic Score |
| 7 | Tumor consensus quality |
| 8 | Tumor variant allele quality |
| 9 | Tumor mean mapping quality |
| 10 | Normal consensus quality |
| 11 | Normal variant allele quality |
| 12 | Normal mean mapping quality |
| 13 | Depth in tumor |
| 14 | Depth in normal |
| 15 | Mean base quality of reads supporting reference in tumor |
| 16 | Mean mapping quality of reads supporting reference in tumor |
| 17 | Depth of reads supporting reference in tumor |
| 18 | Mean base quality of reads supporting variant(s) in tumor |
| 19 | Mean mapping quality of reads supporting variant(s) in tumor |
| 20 | Depth of reads supporting variant(s) in tumor |
| 21 | Mean base quality of reads supporting reference in normal |
| 22 | Mean mapping quality of reads supporting reference in normal |
| 23 | Depth of reads supporting reference in normal |
| 24 | Mean base quality of reads supporting variant(s) in normal |
| 25 | Mean mapping quality of reads supporting variant(s) in normal |
| 26 | Depth of reads supporting variant(s) in normal |
SUPPORT PROTOCOL 1
Compiling SomaticSniper from Source Code
SomaticSniper requires several dependencies be installed in order to compile successfully. Most of these dependencies can be installed via package managers, e.g., Apt or Yum on Linux platforms.
Necessary Resources
Hardware
A computer running Linux or Mac OS X with at least 1GB of RAM.
Software
The following programs are required to compile SomaticSniper: git, cmake (v2.8 or greater), the make utility, and a C compiler such as gcc. For Debian based distributions, these can be installed using:
sudo apt-get install cmake build-essential zlib1g-dev libncurses-dev git-core
For RedHat based distributions, this can be done using:
sudo yum groupinstall "Development tools" sudo yum install zlib-devel ncurses-devel cmake.
Note that RHEL and CentOS 6.4 both ship with cmake versions earlier than 2.8 and cmake will need to be installed from a source tarball obtained from http://www.cmake.org/.
- Clone a copy of the source code using git.
git clone --recursive git://github.com/genome/somaticsniper. git
Failing to include the --recursive option to this command or attempting to avoid using git and instead downloading the compressed archive available through github will cause compilation errors. See the SomaticSniper compilation FAQ section at github (https://github.com/genome/somatic-sniper). - Create a directory within the somatic-sniper directory just created by git in which to compile.
mkdir somatic-sniper/build
- Change to the new directory.
cd somatic-sniper/build
- Run cmake.
cmake ..
- Run make to compile dependencies and SomaticSniper.
make deps make
The SomaticSniper executable (bam-somaticsniper) is now compiled within the bin subdirectory. You can see help by running the command with no options. bin/bam-somaticsniper
SUPPORT PROTOCOL 2
Compiling bam-readcount from Source Code
The bam-readcount program is essential for properly filtering somatic mutations called by SomaticSniper and its compilation is similar.
Necessary Resources
Hardware
A computer running Linux or Mac OS X with at least 1GB of RAM.
Software
The following programs are required to compile bam-readcount: git, cmake (v2.8 or greater), the make utility and a C compiler such as gcc. The same packages as described in Support Protocol 1 can be used to meet these dependencies.
- Clone a copy of the source code using git.
git clone --recursive git://github.com/genome/bamreadcount. git
Failing to include the --recursive option to this command or attempting to avoid using git and instead downloading the compressed archive available through github will cause compilation errors. See the bam-readcount compilation FAQ section at github (https://github.com/genome/bam-readcount). - Create a directory within the bam-readcount directory just created by git in which to compile.
mkdir bam-readcount/build
- Change to the new directory.
cd bam-readcount/build
- Run cmake.
cmake ..
-
Run make to compile dependencies and the program itself.
make deps make
The bam-readcount executable (bam-readcount) is now compiled within the bin subdirectory. You can see help by running the command with no options: bin/bam-readcount
GUIDELINES FOR UNDERSTANDING RESULTS
SomaticSniper predicts somatic single nucleotide variants by calculating a somatic score indicating the confidence that the two samples have different genotypes at a position. This score is the chief metric reporting SomaticSniper’s confidence in a call and ranges from 0 to 255 with higher scores indicating higher confidence. The program uses a custom tab-delimited format (see Table 1) and the somatic score is located in column 6. Since SomaticSniper only compares that the two genotypes are different, this means that it will call variants resulting from loss of heterozygosity as well as variants overlapping germline SNPs. While the former are not strictly somatic, they are frequently important in cancer if the variant allele is retained in the tumor. The latter are relatively rare and the expectation would be that roughly 1/1000 true somatic SNVs would overlap a germline SNP. The called genotype of the normal is included in the output file (in column 5), along with the tumor genotype (in column 4) and examining these genotypes allows the user to distinguish between the situations. Large numbers of predicted somatic SNVs overlapping dbSNPs or where a homozygous SNP site acquires the reference allele in the tumor are indicative of a potential contamination problem or, in some cases, a patient that received an allotransplant.
COMMENTARY
Background Information
At this time there exists an increasing number of somatic mutation callers such as Strelka (Saunders et al., 2012), MuTect (Cibulskis et al., 2013), VarScan 2 (Koboldt et al., 2012; UNIT 15.4), EBCall (Shiraishi et al., 2013), and Seurat (Christoforides et al., 2013). Virtually all perform a comparison between a tumor and a matched normal and SomaticSniper calculates its somatic score by performing a comparison of genotype calls between the two samples. These genotypes are calculated using Samtools’ (Li et al., 2009; Li et al., 2008) genotype likelihood model and, by default, there are no constraints or restrictions on the genotype of the tumor or normal. This differs significantly from recent callers such as MuTect (Cibulskis et al., 2013) or Strelka (Saunders et al., 2012) which impose strict penalties on sites overlapping variation in the normal. This restriction greatly improves the specificity of these callers, but also eliminates a small number of true somatic variants. In general, these newer callers are more sensitive to lower variant allele frequencies and more specific. In liquid tumors like AML, however, their tuning to avoid calls with support in the normal may prove detrimental. Circulating tumor cells in the blood frequently contaminate normal biopsy samples and this results in true somatic variation being present in the normal sample. The handling of this type of situation remains an area for further research, as existing callers do not typically do so explicitly.
More so than germline variants, somatic variant calling is prone to false positives due to the rarity of the variants themselves. All of the somatic variant callers discussed here rely on alignments to a reference genome in order to perform their calls, and errors in these alignments may result in erroneous calls. In our experience, the majority of somatic false positives can be attributed to alignment errors (Koboldt et al., 2012; Larson et al., 2012; Li, 2011). To that end, we have developed a thorough filtering strategy (Koboldt et al., 2012) that eliminates most of these errors while only decreasing sensitivity by ~5%. Other callers utilize internal filters, but that means that filtering cannot be generally applied across approaches.
Critical Parameters and Troubleshooting
Depending on the purity, tumor type, and validation plans for a given study, a user may want to increase the precision of SomaticSniper calls by altering key parameters. Raising the mapping quality cutoff (-q) is one way to increase the precision, as less confidently mapped reads are more susceptible to producing false positives due to alignment issues. Raising the minimum somatic score of reported variants (-Q) will also increase precision. Our high confidence filter sets both equal to 40 for BWA aligned reads. To further increase precision, the -s parameter can be used to set the prior probability of a somatic mutation. While this may significantly decrease power at low coverage depths, it will also substantially decrease the false positive rate. A realistic rate for this parameter is 10−6.
In troubleshooting calls, manual review of the alignments and variants with a tool such as IGV (http://www.broadinstitute.org/software/igv/) is recommended to understand the characteristics of the site. Common issues with SomaticSniper include missing low frequency mutations and false positives with significant support in the normal. In the former case, the sensitivity of SomaticSniper is significantly reduced when variants occur in less than 20% of the reads (Larson et al., 2012). In the latter case, these false positives occur due to algorithmic insufficiencies and are generated when both the tumor and the normal have high depth and variant allele frequencies around 20%. These calls can be filtered out, when a pure normal is present, by removing sites with variant reads in the normal (for example by filtering the classic output using a simple awk script awk '{if ($26 < 2) print}' test.txt).
Acknowledgements
This research was funded by the National Human Genome Research Institute, grant number HG003079.
Footnotes
INTERNET RESOURCES
http://gmt.genome.wustl.edu/somatic-sniper
This is the homepage of the SomaticSniper algorithm and contains links to the source code, documentation, and information on how to receive support.
Contributor Information
Travis E. Abbott, Email: tabbott@genome.wustl.edu.
Richard K. Wilson, Email: rwilson@genome.wustl.edu.
LITERATURE CITED
- Christoforides A, Carpten JD, Weiss GJ, Demeure MJ, Von Hoff DD, Craig DW. Identification of somatic mutations in cancer through Bayesian-based analysis of sequenced genome pairs. BMC Genomics. 2013;14:302. doi: 10.1186/1471-2164-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, Sanada M, Ogawa S, Miyano S. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucl eic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]