Abstract
Coupled translocation of mRNA and tRNA through the ribosome, a process catalyzed by elongation factor EF-G, is a crucial step in protein synthesis. The crystal structure of a translocation complex describes the binding states of two tRNAs trapped in mid-translocation. The deacylated P-site tRNA has moved into a partly-translocated pe/E chimeric hybrid state. The anticodon stem-loop of the peptidyl A-site tRNA is captured in transition toward the 30S P site, while its 3′ acceptor end contacts both the A and P loops of the 50S subunit, forming an ap/ap chimeric hybrid state. The structure shows how features of ribosomal RNA rearrange to hand off the A-site tRNA to the P site, revealing an active role for rRNA in the translocation process.
During the translocation step of the elongation phase of protein synthesis, the mRNA is advanced by one codon, coupled to movement of the tRNAs from the ribosomal A (aminoacyl) to P (peptidyl) and P to E (exit) sites, in a process catalyzed by elongation factor EF-G (1). First, the tRNAs move on the 50S subunit into P/E and A/P hybrid states, followed by movement of the tRNA anticodon stem-loops (ASLs) from the 30S subunit A and P sites to the P and E sites, respectively, coupled to movement of their associated mRNA codons (2). The first step is accompanied by intersubunit rotation (3–7), while the second step requires EF-G·GTP, and involves rotation of the 30S subunit head domain (8–11). Although much has recently been learned about the structural basis of P-tRNA movement to the E site (9, 10, 12, 13), translocation intermediates containing A-tRNA are more difficult to trap. Thus, much of our thinking about the structural basis of A-tRNA and mRNA movement has been based on crystal structures of EF-G bound to vacant (14) or P-tRNA-containing ribosome complexes trapped in classical (15) or hybrid states (10, 12, 13) and two cryo-EM structures of 70S ribosome-EF-G complexes containing two tRNAs bound in P/E and A/P* hybrid states (16), or in ap/P and pe/E chimeric hybrid states (11). [We use the term “chimeric” to indicate binding of a tRNA to elements of two different binding sites within the same subunit.]
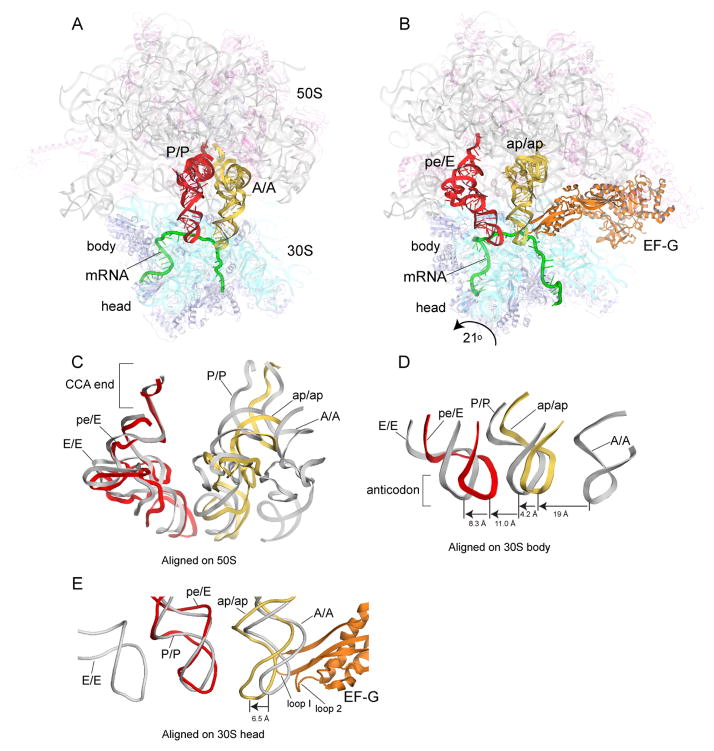
Here we report the crystal structure of a 70S ribosome translocation intermediate containing EF-G, mRNA, and two tRNAs - a deacylated tRNA bound in the pe/E state and a peptidyl-tRNA trapped in an ap/ap chimeric hybrid state. The complex was formed with T. thermophilus 70S ribosomes, a 39-nucleotide mRNA, elongator tRNAMet in the P site and N-acetyl-Val-tRNAVal in the A site. To trap the translocation intermediate, we added neomycin to block completion of translocation, and fusidic acid to prevent release of EF-G (Supplemental Methods; Fig. S1). The structure was solved using diffraction data to 3.8 Å obtained from a single crystal (Table S1). Examples of electron density are shown in supplementary materials (Figs. S2–S13). Relative to the classical-state ribosome (17), the 30S subunit head undergoes a large 21° counterclockwise rotation, and the 30S body a 2.7° rotation relative to the 50S subunit (Fig. 1, Fig 2A, B). The P-tRNA anticodon stem-loop (ASL) moves with the 30S head into a position between the P site of the 30S head and the E site of the 30S body (pe chimeric state; Fig 1D, E), while its acceptor end moves fully into the 50S E site (Fig 1C), forming a pe/E chimeric hybrid state (9–11). The A-tRNA ASL moves to within ~4Å of the P-site elements of the 30S body (Fig. 1D); its elbow rotates towards the classical 50S P site, but its acceptor end is bound between the 50S subunit A and P sites (Fig. 1C), forming an ap/ap chimeric hybrid state. The large, EF-G-dependent rotation of the 30S head in our structure repositions helix H38 of 23S rRNA, allowing the A-tRNA elbow to reach the position of the P-site tRNA elbow (Fig. S14). Domain IV of EF-G is wedged into the site of convergence of the A-site mRNA codon, the anticodon loop of the ap/ap tRNA, and 16S and 23S rRNAs at intersubunit bridge B2a, simultaneously contacting all four RNAs (Fig. S15).
Figure 1. Structure of trapped translocation intermediate containing EF-G, mRNA, and two partially-translocated tRNAs.
(A) 70S ribosome with tRNAs bound in classical A/A and P/P states (17); (B) Translocation intermediate complex, showing tRNAs trapped in intermediate chimeric hybrid ap/ap and pe/E states. (C) Comparison of positions of tRNA in classical states and in the translocation intermediate, aligned on the 50S subunit; (D) relative positions of tRNA ASLs, aligned on the 30S subunit body or (E) head. Molecular components are colored throughout as: 16S rRNA, cyan; 30S proteins, blue; 23S rRNA, grey; 5S rRNA, light blue; 50S proteins, magenta; mRNA, green; A/A and ap/ap tRNAs, yellow; P/P and pe/E tRNAs, red; EF-G, orange.
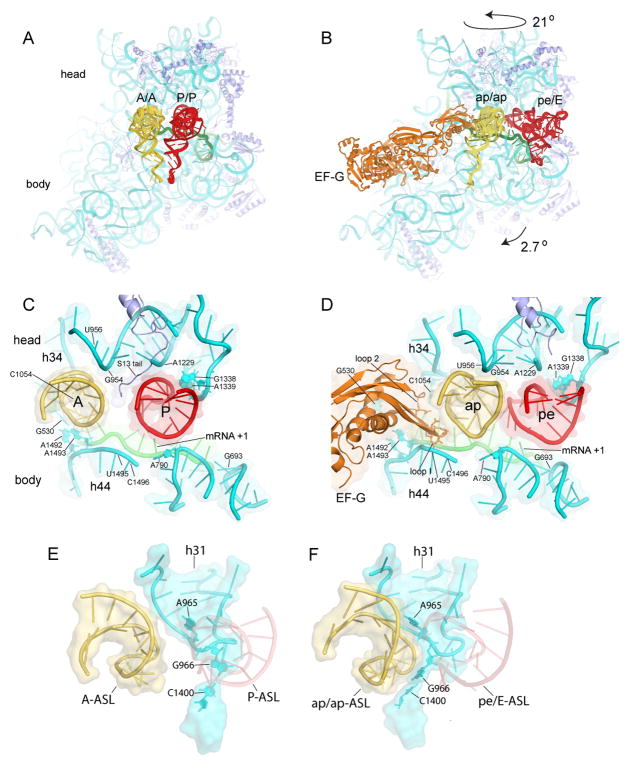
Figure 2. Movement of tRNA ASLs on the 30S subunit, and capture of translocating A-tRNA by P-site elements of the 30S subunit head.
(A–B) Positions of tRNAs in the (A) classical-state ribosome and (B) translocation intermediate. (C–D) Interactions of the tRNA ASLs and mRNA as they move from the (C) A and P classical states to the (D) ap and pe chimeric hybrid states. (E–F) Rearrangement of the 966 loop (h31) of 16S rRNA in the 30S subunit head from its (E) classical P-tRNA-binding position to (F) forms a surface-fitting pocket around the ap/ap tRNA ASL.
Although 30S head rotation clearly facilitates P-site ASL translocation (9–11), the A-site ASL is translocated by a different mechanism. The P-site ASL moves precisely with 30S head rotation into the pe/E state, whereas the A-site ASL has moved further than the rotational movement of the head into the ap state on the 30S subunit (Fig. 1E). Movement of the A-site ASL precisely with head rotation would result in severe clash with domain IV of EF-G as it is positioned in our complex, which, together with the contact formed between the tip of domain IV and the codon-anticodon helix of the ap/ap tRNA (Fig S16), suggests that movement of mRNA and ASL are coupled to that of domain IV. The additional displacement of the ap/ap ASL brings it close to the pe/E ASL (Fig. 2C, D) (11). The position of the head may be stabilized by interaction between phosphate 1210 of 16S rRNA with domain IV of EF-G at Gly531.
Most striking is the rearrangement of the 16S rRNA 966 loop in the 30S head, which breaks away from the P site to reach toward the A site, where it initiates contact with the partly translocated ap/ap-ASL (Fig. 2E, F). This rearrangement involves disruption of the packing of m22G966 against ribose 34 of the P-site ASL (18, 19), and formation of a surface-fitting pocket around the ap/ap-ASL by nucleotides A965, m22G966 and C1400. In a crystal structure of the corresponding single-tRNA complex (10) (Fig. S6), the pe/E tRNA ASL was found to maintain all canonical P-site contacts with the 30S head, including the 966 loop, as it rotates towards the E site. However, in the two-tRNA complex reported here, only a sub-set of the P-site 30S head interactions with the pe/E ASL are maintained, involving G1338, A1339, A1340, and the C-terminal tail of protein uS9. The formation of post-translocation interactions simultaneously with disruption of pre-translocation interactions, suggests a mechanism for how the ASLs are handed off between the A and P sites. It may also represent an initial shift in contacts that must be disrupted to release the pe/E tRNA ASL prior to back-rotation of the 30S head in the final stages of translocation (20, 21).
The network of interactions formed between the conserved loop 1 (residues 499-504) in domain IV of EF-G, the ap/ap ASL and its mRNA codon (Fig. S15) (11) are similar to those observed in the post-translocation state (15), suggesting that they are maintained throughout the translocation cycle. Their resemblance to the minor-groove interactions made by A1492 and A1493 with the codon-anticodon helix in the decoding site (22) suggests that they may help to maintain correct codon-anticodon pairing during movement between the A and P sites (15), and are consistent with the proposal that EF-G facilitates translocation by destabilizing interactions in the 30S decoding site (23).
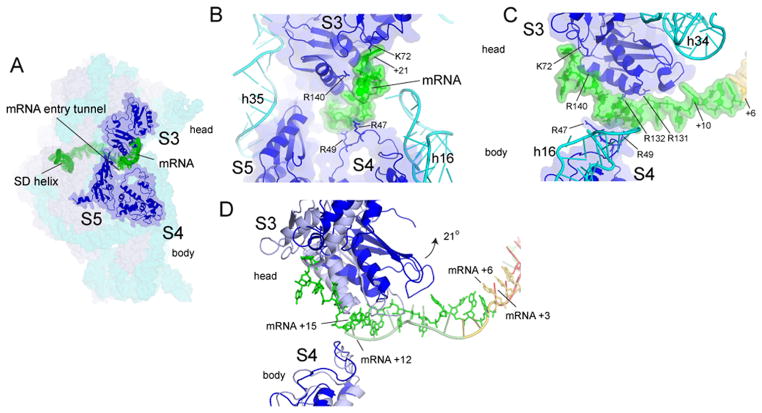
Interactions between the mRNA and elements of the 30S head within the downstream mRNA entry tunnel have been proposed to carry the mRNA forward with the tRNA during head rotation (17). However, it is not clear that net movement of the mRNA could be sustained in this way, because these interactions are disrupted during head rotation. We find that the extended tail of the mRNA, as it emerges from the downstream tunnel, curves upward to bind to the surface of protein uS3 in the head of the subunit (Fig. 3, S17). Thus, forward head rotation would actively move the mRNA in the direction of translocation. Comparison of the position of a reference nucleotide on the mRNA in the rotated and non-rotated states further shows that forward head rotation would advance the mRNA by one codon (Fig. 3D), supporting the possibility that the mRNA is actively moved by the ribosome during translocation. Since the dimensions of the downstream tunnel exclude entry of an RNA helix, disruption of mRNA secondary structure by the ribosomal helicase (24) must occur at or near the entry to the tunnel, by interaction with the ribosome outside the tunnel. Binding of the mRNA tail immediately flanking the entrance to the tunnel exclusively to the 30S head provides such an interaction. The forces created by head rotation could thus destabilize base pairing in helical elements of mRNA as they enter the downstream tunnel.
Figure 3. Contact between the downstream region of mRNA with the head of the 30S subunit.
(A) The entry to the downstream mRNA entry tunnel is surrounded by proteins uS3, uS4, and uS5. Positions +14 to +21 contact a positively charged patch on the surface of protein uS3 in the 30S head (Fig. S17). (B) Path of the downstream mRNA and (C) 90° rotated view. (D) Docking on the 30S body of the classical structure (17) (gray) shows that rotation of the head in the ap/ap intermediate structure translocates the mRNA by one codon. A-codon (yellow) and P-codon (red).
In the classical state, C74 and C75 in the CCA tail of P-site tRNA form base pairs with G2252 and G2251, respectively, in the P loop (H80) of 23S rRNA, while C75 of the A-site tRNA pairs with G2553 in the A loop (H92) (18, 25, 26). These interactions position the peptidyl and aminoacyl moieties for the peptidyl transferase reaction (27), which leaves a deacylated tRNA in the P site, and a peptidyl tRNA in the A site. A critical step in translocation is therefore the subsequent movement of the peptidyl-CCA end from the A loop to the P loop. The structure of the translocation intermediate reveals a chimeric state, distinct from those previously described (Fig. S18), in which the acceptor end of the peptidyl-tRNA interacts simultaneously with both the A and P loops (Fig 4). In this state, base pairing of C75 with G2553 of the A loop is preserved, while movement of its acceptor stem into a position near that of classical P-site tRNA allows G2252 and G2253 in a rearranged P loop to contact the tRNA backbone at positions 72 and 70 (Fig. 4B). Thus, the end of the acceptor stem of the ap/ap-tRNA is anchored on the P loop, facilitating transfer of its adjacent C74 and C75 residues from the A loop to the P loop. A76 of the ap/ap tRNA is oriented similarly to that observed for tRNA bound in the P/P classical state (17), with its N-acetyl-valine peptidyl moiety positioned at the entrance to the peptide exit tunnel, whereas C74 and C75 maintain their interactions with the A loop of 23S rRNA (Figs. 4, S19).
Figure 4. The acceptor end of ap/ap tRNA simultaneously contacts the A and P loops of 23S rRNA.

During translocation, the 3′ acceptor end of peptidyl-tRNA moves from the (A) classical A/A state to the (B) intermediate ap/ap state to the (C) classical P/P states, facilitated by conformational changes in the A and P loops.
The ap/ap-tRNA may correspond to intermediate states that have been characterized kinetically. Fluorescence quenching kinetic studies identified an EF-G-dependent intermediate (INT) in which the peptidyl-tRNA elbow moves from the A site towards the P site, but remains only partially reactive with the aminoacyl-tRNA mimic puromycin, suggesting that its CCA end is not fully accommodated into the P site (28). Kinetic studies in which a probe was attached directly to the peptidyl-tRNA CCA end, identified EF-G-dependent translocation intermediates in which the acceptor end of peptidyl-tRNA moves towards the P site, but remains puromycin-unreactive (29). The properties of these intermediates are compatible with the trapped ap/ap state observed here. The structure of this trapped translocation intermediate begins to describe how tRNA moves between the ribosomal A and P sites. The tRNA binding sites themselves are dynamic, forming simultaneous contacts between the A-tRNA and elements of both the A and P sites, resembling the passing of the baton in a relay race (30). This is seen for both the 30S and 50S subunits. In the 30S subunit, rearrangement of the 966 loop of 16S rRNA releases G966 from the P-site ASL to contact the A-site ASL as it moves into the 30S P site (Fig. 2E, F). In the 50S subunit, the A and P loops of 23S rRNA rearrange to allow both loops to contact the 3′-acceptor end of the A-site tRNA as it begins its transition into the 50S P site (Fig. 4). These findings show that the structural dynamics of ribosomal RNA play an active role in the mechanism of translocation.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM-17129 and GM59140. We thank the beamline staffs at the ALS, SSRL and APS for their expert support. Structures have been deposited with PDB accession numbers 4QS0, 4QS1, 4QS2, 4QS3.
References
- 1.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Noller HF. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 3.Frank J, Agrawal RK. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 4.Valle M, et al. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 5.Agirrezabala X, et al. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julian P, et al. Proc Natl Acad Sci U S A. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ermolenko DN, et al. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 8.Schuwirth BS, et al. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 9.Ratje AH, et al. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Lancaster L, Donohue JP, Noller HF. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramrath DJ, et al. Proc Natl Acad Sci U S A. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Nat Struct Mol Biol. 2013;20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 13.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pulk A, Cate JH. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao YG, et al. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Proc Natl Acad Sci U S A. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 18.Selmer M, et al. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 19.Berk V, Zhang W, Pai RD, Cate JH. Proc Natl Acad Sci U S A. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunha BRCE, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV. Translation. 2013;1 doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, Noller HF. Proc Natl Acad Sci U S A. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogle JM, et al. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 23.Khade PK, Joseph S. Nat Struct Mol Biol. 2011;18:1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takyar S, Hickerson RP, Noller HF. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 25.Samaha RR, Green R, Noller HF. Nature. 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 26.Kim DF, Green R. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 27.Hansen JL, Schmeing TM, Moore PB, Steitz TA. Proc Natl Acad Sci U S A. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan D, Kirillov SV, Cooperman BS. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtkamp W, et al. EMBO J. 2014 [Google Scholar]
- 30.Bock LV, et al. Nat Struct Mol Biol. 2013;20:1390–1396. doi: 10.1038/nsmb.2690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.