Abstract
RGMs (repulsive guidance molecules) comprise a recently discovered family of GPI (glycosylphosphatidylinositol)-linked cell-membrane-associated proteins found in most vertebrate species. The three proteins, RGMa, RGMb and RGMc, products of distinct single-copy genes that arose early in vertebrate evolution, are ~ 40–50%identical to each other in primary amino acid sequence, and share similarities in predicted protein domains and overall structure, as inferred by ab initio molecular modelling; yet the respective proteins appear to undergo distinct biosynthetic and processing steps, whose regulation has not been characterized to date. Each RGM also displays a discrete tissue-specific pattern of gene and protein expression, and each is proposed to have unique biological functions, ranging from axonal guidance during development (RGMa) to regulation of systemic iron metabolism (RGMc). All three RGM proteins appear capable of binding selected BMPs (bone morphogenetic proteins), and interactions with BMPs mediate at least some of the biological effects of RGMc on iron metabolism, but to date no role for BMPs has been defined in the actions of RGMa or RGMb. RGMa and RGMc have been shown to bind to the transmembrane protein neogenin, which acts as a critical receptor to mediate the biological effects of RGMa on repulsive axonal guidance and on neuronal survival, but its role in the actions of RGMc remains to be elucidated. Similarly, the full spectrum of biological functions of the three RGMs has not been completely characterized yet, and will remain an active topic of ongoing investigation.
Keywords: axon guidance, gene evolution, gene structure, iron metabolism, protein modelling, repulsive guidance molecule (RGM)
INTRODUCTION
The RGM (repulsive guidance molecule) gene family consists of three members, RGMa, RGMb, and RGMc [1–6]. Each gene encodes a protein whose expression is restricted to a small number of tissues and is hypothesized to be involved in distinct biological functions ranging from control of iron metabolism to regulation of axonal guidance and neuronal survival in the developing nervous system. The RGM family receives its name from the axonal guidance molecule RGMa [2], a protein found primarily in the developing and adult central nervous system [1–3,7]. A second member, RGMb (or Dragon [4]) is also detected in the nervous system, but in a different expression pattern than RGMa [4,8]. The biological actions of RGMb are poorly characterized to date. The third member of the family is RGMc [also called HJV (haemojuvelin), HFE2 (HLA-like protein involved in iron (Fe) homoeostasis) and DL-M (Dragon-like muscle)]. Unlike RGMa or RGMb, RGMc is not expressed in the nervous system, but rather is produced by striated muscle and the liver [3,5,8,9].RGMc surprisingly regulates iron metabolism, as inactivating mutations cause juvenile haemochromatosis, a severe systemic iron overload disorder in humans [6]. To date, there has been no comprehensive assessment of the most fundamental aspects of the biology of the RGM family, including regulation of gene expression, control of protein biosynthesis, the relationship of protein structure to function, or mechanisms of action of each of the RGM proteins. In the present review we address the molecular biology and biochemistry of the RGM family, attempt to define and critically evaluate what is known, and identify new areas for future investigation.
RGMa
Chromosomal organization and gene structure
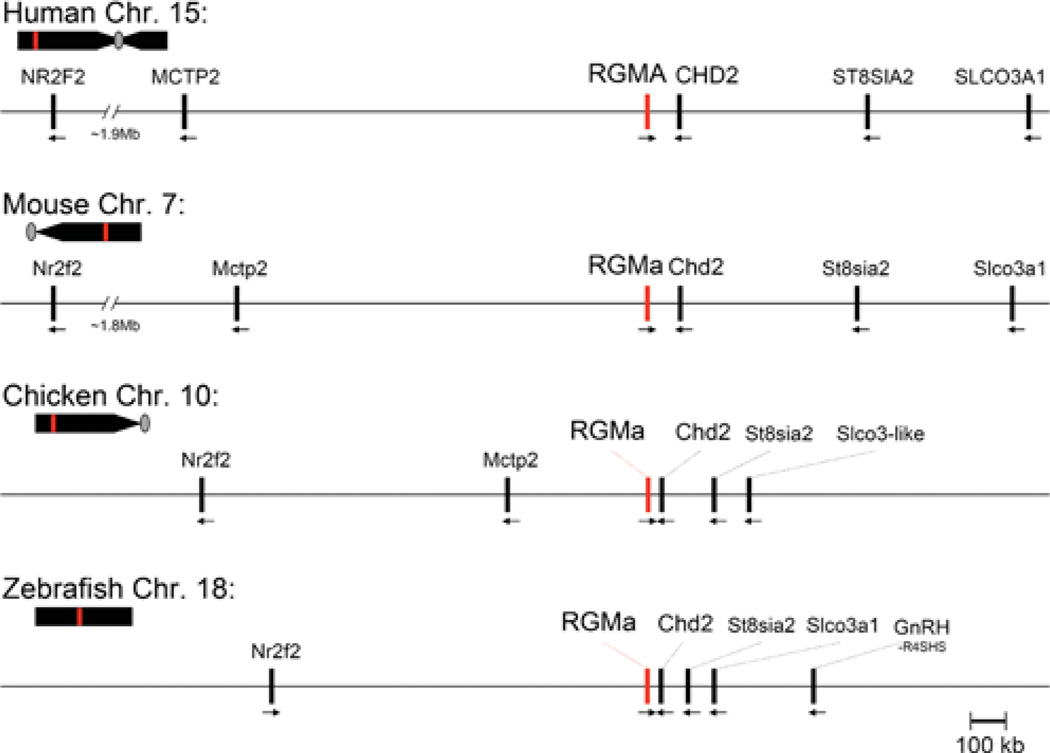
RGMa has been identified in ten mammalian and eight non-mammalian vertebrates, where it is a single-copy gene (Table 1). A single RGM gene also has been described in several invertebrate species, including urochordates, echinoderms, molluscs and nematodes [10], as will be discussed in the molecular evolution section below. In vertebrates, RGMa comprises one of six conserved genes in a syntenic locus [11], as can be assessed by analysis of the corresponding parts of the human, mouse and chicken genomes (Figure 1). In these three species, RGMa is positioned in the opposite transcriptional orientation from the other nearby genes. The locus is also conserved in zebrafish (Figure 1). Within the cluster of six conserved genes near RGMc in human, mouse and chick, Mctp2 (multiple C2 domains, transmembrane 2) is found 5′ to RGMa, and Chd2 (chromodomain helicase DNA-binding protein 2), St8sia2 (ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 2), and Slco3a1 (solute carrier organic anion transporter family member 3A1) are located 3′. The latter three genes also are positioned downstream of RGMa in the zebrafish genome, but only upstream Mctp2 is absent (Figure 1). In addition, in all four species, Nr2f2 (nuclear receptor subfamily 2, group F, member 2) is located upstream of RGMa, although both the relative orientation and the distance among species varies (~ 2 Mb in human and mouse genomes and ~ 830 kb in zebrafish, where the transcriptional direction is reversed) (Figure 1).
Table 1. Species in which more than one RGM has been identified.
| Species (abbreviation) | RGMa | RGMb | RGMc | |
|---|---|---|---|---|
| Mammals | ||||
| Human | Homo sapiens (Hsa) | AK074910 AL136826 | BC067736 | AK223575 AK092682 |
| Chimpanzee | Pan troglodytes (Ptr) | + | + | + |
| Rhesus macaque | Macaca mulatta (Mmul) | + | + | + |
| Pig | Sus scrofa (Sscr) | + | − | − |
| Dog | Canis familiaris (Cfa) | + | − | + |
| Cow | Bos taurus (Bta) | + | + | + |
| Elephant | Loxodonta africana (Laf) | + | + | − |
| Mouse | Mus musculus (Mmu) | BC059072 BC023870 | AK047390 BC096024 | AJ557515 |
| Rat | Rattus norvegicus (Rno) | + | − | BC089203 |
| Armadillo | Dasypus novemcinctus (Dno) | − | + | + |
| Opossum | Monodelphis domestica (Mdo) | + | + | + |
| Non-mammalian vertebrates | ||||
| Chicken | Gallus gallus (Gga) | AY128507 | + | − |
| Frog | Xenopus tropicalis (Xtrop) | BC061329 | BC061325 | + |
| Zebrafish | Danio rerio (Dre) | BC091800 AY613931a | AY613929 | BC134888 BC112964 |
| Salmon | Salmo salar (Ssa) | BT045779 | − | − |
| Japanese Pufferfish | Takifugu rubripes (Tru) | + | + | + |
| Green-spotted Puffer | Tetraodon nigroviridis (Tni) | + | + | + |
| Stickleback | Gasterosteus aculeatus (Gac) | + | + | + |
| Medaka (Killer fish) | Oryzias latipes (Ola) | + | + | + |
Accession numbers for cDNAs are listed. All others have been identified through homology mapping in their respective genomes (+). −, not found.
Mis-labeled in GenBank as DL-M (muscle RGMc).
Figure 1. Comparative structures of RGMa genomic loci.
The relative position of the RGMa gene (red line) is indicated on each chromosome (Chr.; human 15, mouse 7, chicken 10, zebrafish 18) in relation to the centromere (grey oval, if information available) and telomere. Presented below each chromosome is a higher resolution view of the RGMa locus for each species. Neighbouring genes are indicated, with the transcriptional direction represented by an arrow. Gene names corresponding to the abbreviations may be found in Table 4.
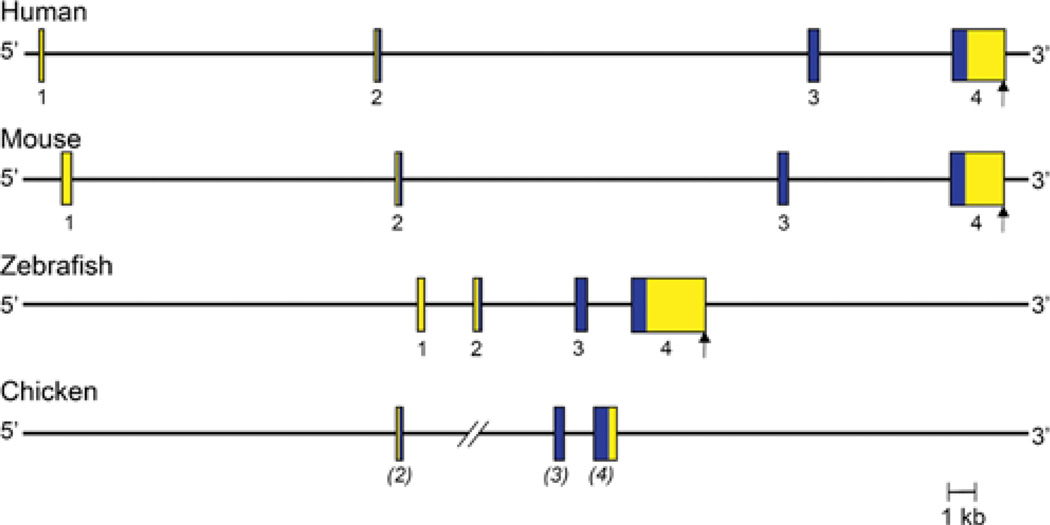
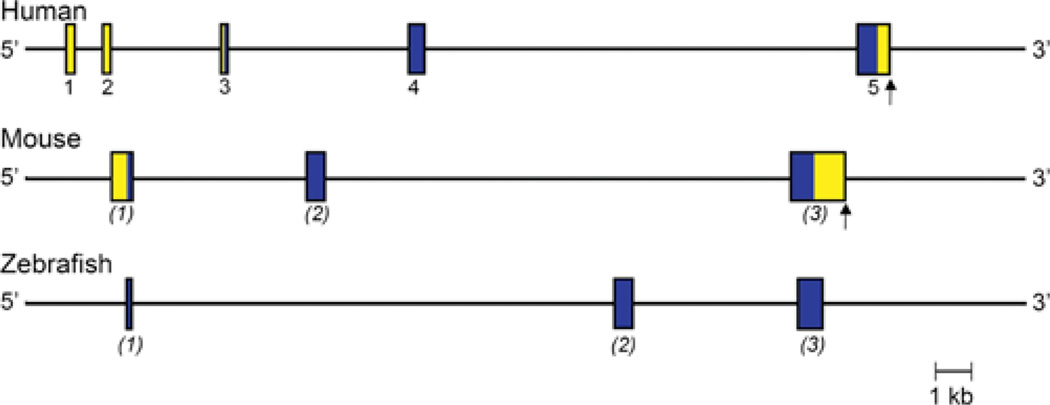
Human and mouse RGMa genes are of comparable size, ~ 46 and ~ 44 kb respectively, and have a similar organization, being composed of four exons separated by three variably-sized introns, although the precise 5′ end of exon 1 has not been defined in either species (Figure 2 and Table 2). In both genes, exon 1 is non-coding, and consists of most of the 5′ UTR (untranslated region) of RGMa mRNA. Exon 2 contains the remaining 35 nucleotides of the 5′ UTR and the first 26 codons of the RGMa protein, whereas exon 3 encodes the next 72 codons (73 in mice), and exon 4 the remaining 328 codons (321 in mice), plus a 3′ UTR of ~ 1800 nucleotides and a single polyadenylation signal (Figure 2). The four exons are well conserved between human and mouse RGMa, with nucleotide identity ranging from a low of 64% for exon 1 to a high of 99% for exon 2 (calculated using data in [12–15]). The three introns are not as conserved as the exons (<30%compared with ~ 60% identity respectively), although their lengths are similar between the two species (Figure 2). Although four exons have been identified in the zebrafish RGMa gene, the nucleotide sequence of exon 1 is not similar to its mammalian counterparts [14–16]. In the chicken, the 5′ end of the largest RGMa cDNA could not be mapped to the RGMa locus, possibly because the genomic sequence is incomplete in this region [17], and its DNA sequence also differs markedly from the other species. Thus only three exons have been identified definitively in chicken RGMa, corresponding to mammalian exons 2–4 (Figure 2).
Figure 2. Comparative organization of RGMa genes.
The anatomy of human, mouse, zebrafish and chicken RGMa genes is shown. Exons are indicated by boxes, with coding regions in blue and non-coding regions in yellow. The assignment of exon numbers is based on comparison with mouse RGMa. The polyadenylation site, when known, is depicted by a vertical arrow. The location of zebrafish exon 1 is based on mapping available EST (expressed sequence tag) data taken from GenBank® (accession numbers AL911518 and EH589480). The length of one of the introns of chicken RGMa is not known (shown as two angled lines), as the putative exon 2 cannot be mapped to the genomic DNA sequence, which appears to be incomplete in this region. Chicken exon assignments are in parentheses because the putative exon 1 cannot be mapped to the genome.
Table 2. Characteristics of RGM genes.
| Species | Gene size (kb) | Number of exons | mRNA (kb) | |
|---|---|---|---|---|
| RGMa | Human | 45.8 | 4 | 3.2 |
| Mouse | 44.4 | 4 | 3.6 | |
| Zebrafish | 12.5 | 4 | 4.5 | |
| RGMb | Human | 24.8 | 5 | 2.2 |
| Mouse | 20.3 | 3 | 4.2 | |
| Zebrafish | 18.3 | 3 | >1.3 | |
| RGMc | Human | 4.3 | 4 | 2.1 |
| Mouse | 4.0 | 4 | 2.0 | |
| Zebrafish | 11.4 | 4 | 1.7 |
Gene expression
RGMa was cloned initially from mRNA isolated from chick embryonic optic tectum [2]. Subsequently, RGMa transcripts were shown to be expressed at highest levels in both the adult and developing central nervous system in chicken, mouse and zebrafish [1–4,7,18]. RGMa mRNA also has been detected at lower levels in peripheral tissues, including heart, lung, liver, skin, kidney and testis, at least in the adult rat [19]. By Northern blotting, the major RGMa transcript has been shown to be ~ 3.6 kb in length in the mouse [19], which is consistent with the aggregate size of the four RGMa exons [13,20].Other minor transcripts have been seen by Northern blotting, but their exact relationship with the RGMa gene has not been established to date [19,21].
In the developing mouse embryo, RGMa mRNA has been detected as early as E (embryonic day) 8.5 in the neural folds of the central nervous system [1]. Later in development, RGMa transcripts are found in several brain regions, including hippocampus, midbrain, the ventricular zone of the cortex, and parts of the brainstem and spinal cord [1,8,21]. Similar observations have been reported in the developing chicken [2,7] and zebrafish [4]. The biochemical processes responsible for these distinct patterns of RGMa gene expression in the central nervous system have not been elucidated to date, in large part because nearly nothing is known about the organization or function of the RGMa gene promoter, about mechanisms of regulation of RGMa gene transcription, or about RGMa mRNA turnover. Similarly, the signalling pathways that govern RGMa gene expression in different tissues and in response to physiological and pathological stimuli have not been characterized.
Protein sequence and expression
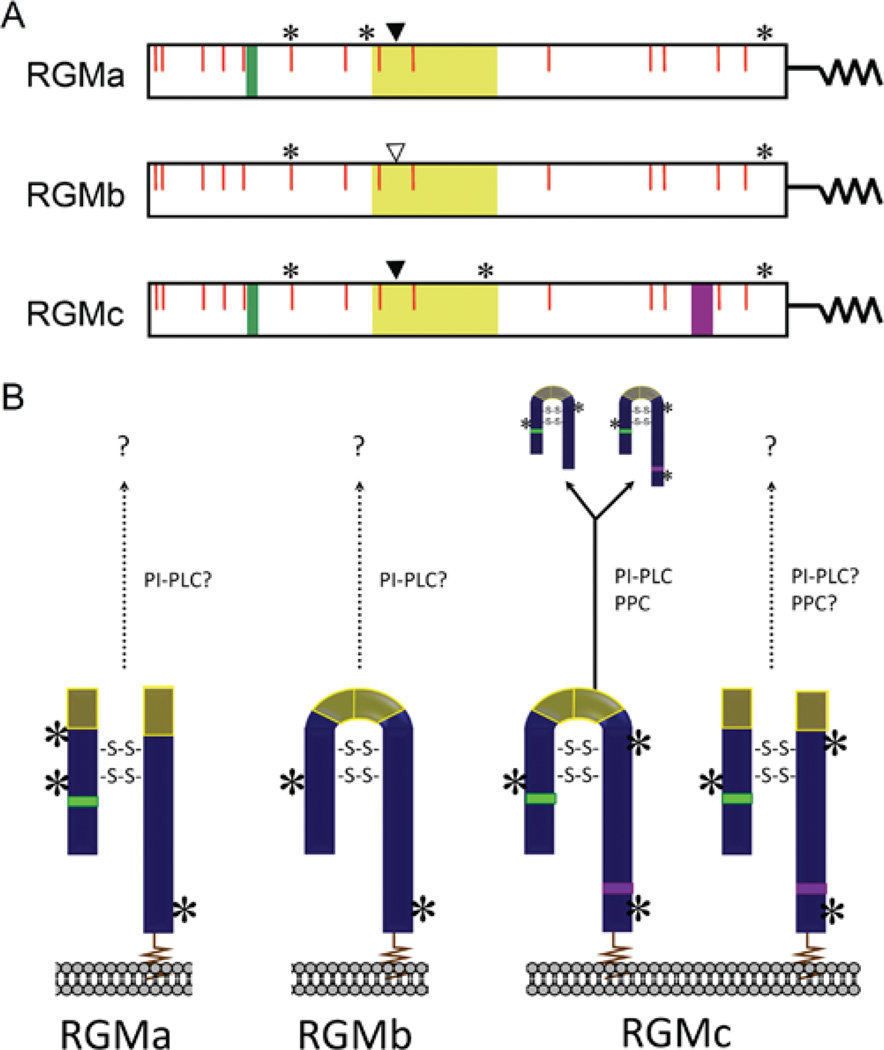
The initial identification of chick RGMa after its cDNA cloning revealed it to be a cell membrane-associated GPI (glycosylphosphatidylinositol)-linked two-chain protein that was derived from a primary translation product of 432 amino acids [2]. Subsequent cloning of human and mouse RGMa cDNAs predicted similarly sized proteins of 434 and 438 residues [1], respectively, that were 91% identical to each other and 80% identical to chick RGMa (Table 3). In all three species and in zebrafish RGMa, the N-terminal signal peptide is estimated to be ~ 30 residues, although the first amino acid of the mature protein has not been characterized experimentally. The RGMa precursor also contains a conserved GPI attachment signal at its C-terminus of ~ 45 amino acids. This segment is removed in the endoplasmic reticulum during RGMa biosynthesis when the GPI anchor is added to the nascent protein [2,22]. Other recognizable protein elements in RGMa include an RGD motif (arginine-glycine-aspartic acid; a potential integrin-binding site [2,23]), and a partial vWD (von Willebrand type-D) domain [2,24] that contains the site of internal cleavage to generate two-chain RGMa (Figure 3) (these domains and other aspects of the biochemistry of RGM proteins will be discussed in the section on structure–function relationships below). The mechanism of intramolecular cleavage of RGMa has not been established, although it appears to occur during its biosynthesis, leading to a mature RGMa that is a disulfide-bonded two-chain protein composed of an N-terminal fragment of ~ 123 residues, and a C-terminal segment of ~ 238 residues [2,25], and that is linked to the outer face of the plasma membrane by its C-terminal GPI anchor [2,26,27] (Figure 3B). The number and pattern of disulfide bonds has not been established yet for the 14 cysteines found in mature RGMa (a molecular model is discussed in the section on structure–function relationships below). RGMa also appears to be a glycoprotein, with three potential asparagine-linked glycosylation sites in mammals and two in the chicken (Figure 3A) [2,26]. At present it is not known if other RGMa isoforms exist, such as single-chain species, or whether soluble forms of the protein are found in the extracellular fluid.
Table 3. Amino acid identity among RGM proteins.
| Mouse | |||||
|---|---|---|---|---|---|
| Species | Size (aa) | RGMa | RGMb | RGMc | |
| RGMa | Human | 434 | 91% | 50% | 48% |
| Mouse | 438 | 100% | 49% | 48% | |
| Zebrafish | 433 | 68% | 48% | 43% | |
| Chicken | 432 | 80% | 53% | 45% | |
| RGMb | Human | 437 | 52% | 89% | 42% |
| Mouse | 438 | 49% | 100% | 42% | |
| Zebrafish | 436 | 46% | 65% | 42% | |
| RGMc | Human | 426 | 49% | 44% | 88% |
| Mouse | 420 | 48% | 42% | 100% | |
| Zebrafish | 410 | 44% | 41% | 46% | |
Calculations are based on Smith-Waterman (local) alignment using Blosum62 matrix, gap open penalty of 10.0, and gap extend penalty of 0.5. The protein sequences are derived from the cDNAs whose accession numbers are listed in Table 1.
Figure 3. Characteristics of RGM proteins.
(A) The linear maps of mature RGMa, RGMb and RGMc contain the following features: RGD motif (RGMa and RGMc, green); vWD, partial vWD domain (yellow); PPC, PPC recognition and cleavage site (RGMc only, purple); *location of asparagine-linked glycosylation sites; solid arrowhead, site of intra-molecular proteolytic cleavage to generate two-chain RGMa and RGMc; vertical open arrowhead, possible site of intra-molecular proteolytic cleavage in RGMb; red vertical lines, conserved cysteine residues. The squiggle at the C-terminus of each protein represents the GPI anchor. (B) Schematic of mature RGMa, RGMb and RGMc on the cell surface, as well as the secreted forms of RGMc. Based on published studies, RGMa appears to be primarily a two-chain molecule, and RGMb a single-chain protein, whereas RGMc appears to be represented by both single- and two-chain species. Experimental data supports at least one disulfide bond between the N- and C-termini [9,45,47], and ab inito molecular modelling (see Figure 9) predicts one or two disulfide bonds connecting the two-chain RGM isoforms (shown as -S-S-), though the exact number is currently unknown. Single chain RGMc is released from the cell surface, and is found in extracellular fluid and in blood [9,43–48], potentially through the actions of a furin-like PPC and/or a PI-PLC. It is not known if RGMa, RGMb or two-chain RGMc are released from the membrane (as indicated by arrows with question marks). Locations of asparagine-linked glycosylation sites are indicated by asterisks, and the GPI anchor is depicted as a squiggle.
Physiological functions and mechanisms of action
RGMa was identified as a factor involved in guiding axons by repulsion from the temporal half of the developing chicken retina toward the anterior optic tectum in the brain, and membranes derived from cells expressing chick RGMa were shown to inhibit temporal retinal growth cones, but had little effect on nasal growth cones [2]. Perhaps surprisingly given these initial observations, genetic knockout of RGMa in mice did not alter retinal axonal patterning, but rather caused defects in neural tube closure [1]. Thus the exact in vivo functions of RGMa in mammals remain to be determined.
It has been shown that RGMa regulates repulsive guidance of retinal axons via binding to neogenin [7,28], a transmembrane protein that is also a receptor for netrins, a family of secreted molecules involved in neuronal development and cell survival (reviewed in [29]). Unlike netrins, RGMa does not bind to proteins related to neogenin, such as DCC (deleted in colorectal cancer) or members of the Unc (unco-ordinated) sub-family [28], although recent observations suggest an indirect association with Unc5b [31]. In addition to regulating retinal axonal guidance, the interaction between RGMa and neogenin has been found to promote neuronal survival [7]. Initial studies of the early events triggered after RGMa binds to neogenin have suggested the involvement of several signal transduction intermediates, including protein kinase C, the small GTPase RhoA, RhoA kinase [27,30], and focal adhesion kinase [31,32], as well as the putative transcriptional co-activator, LIM-only protein 4 [33], but the full spectrum of biochemical mechanisms responsible for mediating the biological effects of RGMa by neogenin has not been established.
Similar to other members of the RGM family, RGMa also has been found to bind to selected BMPs (bone morphogenetic proteins) [19,34], which belong to the TGF (transforming growth factor)-β growth factor family [35]. In initial biochemical studies, a fusion protein composed of human RGMa linked to the IgG Fc fragment was shown to bind radio labelled BMP-2 and BMP-4 but not BMP-7 or TGF-β1 in cross-linking experiments [34]. In cell-based studies, over-expression of RGMa was found to increase activity of a co-transfected promoter-reporter gene containing a BRE (BMP-response element), whereas knockdown of endogenous RGMa led to a reduction in reporter gene expression [34]. Although these preliminary observations are intriguing, a role for BMPs in the biological actions of RGMa has not been defined.
RGMb
Chromosomal organization and gene structure
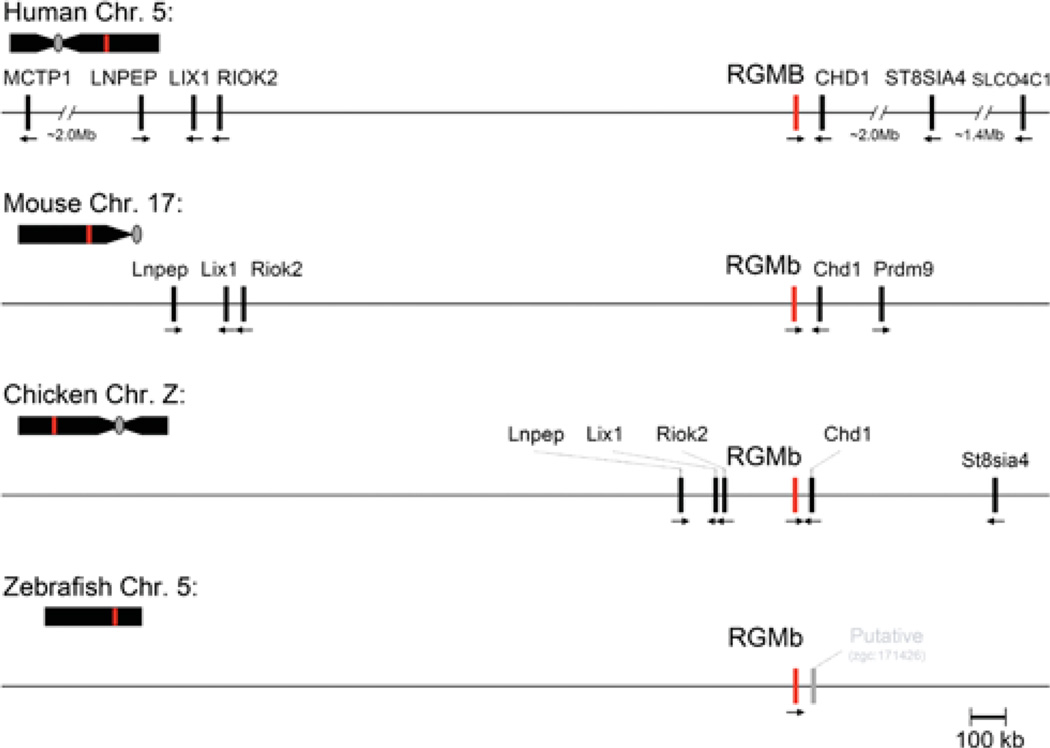
RGMb is a single-copy gene in the eight mammalian and seven non-mammalian vertebrates in which it has been identified (Table 1). Similar to RGMa, RGMb resides within a conserved chromosomal locus, and comprises one of five linked genes that are found in the same relative orientation to each other in the human, mouse and chicken genomes (Figure 4). In each of these species, RGMb is located in a tail-to-tail transcriptional orientation with Chd1, in a relationship similar to that of RGMa and Chd2 (compare Figures 1 and 4). This suggests that a duplication event involving this chromosomal region occurred during evolution prior to the emergence of mammals. Further away and upstream of RGMb are Riok2 (right open reading frame kinase 2), Lix1 (Limb expression 1) and Lnpep (leucyl/cystinyl aminopeptidase) (Figure 4). In contrast, to date very little is known about the chromosomal environment of RGMb in the zebrafish genome (Figure 4).
Figure 4. Comparative structures of RGMb genomic loci.
The relative position of the RGMb gene (red line) is indicated on each chromosome (Chr.; human 5, mouse 17, chicken Z, zebrafish 5) in relation to the centromere (grey oval, if information available) and telomere. Presented below each chromosome is a higher resolution view of the RGMb locus for each species. Neighbouring genes are indicated, with their transcriptional direction represented by an arrow. For the zebrafish RGMb locus, a nearby provisional gene is shown in grey; to date no other genes have been mapped to this region. Gene names corresponding to the abbreviations may be found in Table 4.
The human RGMb gene is ~ 25 kb in length, and contains 5 exons (Figure 5 and Table 2), including two 5′ non-coding exons (1 and 2), which include ~ 406 nucleotides of a ~ 524 nucleotide 5′ UTR of RGMb mRNA. The 5′ end of exon 1 has not been mapped. The remaining 118 nucleotides of the 5′ UTR are found in exon 3, which also includes the first 45 codons of the coding region. Exon 4 encodes the next 170 codons, and exon 5 the remaining 222 codons plus a 3′ UTR of 308 nucleotides that includes a single polyadenylation signal (Figure 5). In the mouse genome, only three RGMb exons have been identified to date, and these correspond to exons 3–5 of the human RGMb gene (Figure 5). The 3′ UTR of mouse RGMb mRNA encoded by exon 3 is longer than its human counterpart, being ~ 2.5 kb in length. In zebrafish, only the coding region for RGMb has been mapped to its genome [4], and is found within three distinct exons (Figure 5).
Figure 5. Comparative organization of RGMb genes.
The anatomy of human, mouse and zebrafish RGMb genes is shown. The assignment of exon numbers is based on comparison with human RGMb, and is provisional for mouse and zebrafish, as indicated by the parentheses. Exons are indicated by boxes, with coding regions in blue and non-coding regions in yellow. The polyadenylation site, when known, is depicted by a vertical arrow. Only coding information is available for zebrafish RGMb.
Gene expression
RGMb was discovered by an informatics-based search for genes related to RGMa [1], and was independently cloned as a gene whose putative promoter was bound by the homeodomain transcription factor, DRG11, which is expressed in DRG (dorsal root ganglia) of the sympathetic nervous system [4,36,37]. RGMb (DRG-‘ON’ or Dragon) was co-localized with DRG11 mRNA in dorsal root ganglia and in the spinal cord. RGMb mRNA also was detected in the developing neural tube prior to the onset of expression of DRG11, and has been found in other areas of the nervous system where DRG11 is not produced [4]. This latter result suggests that RGMb gene expression is controlled by additional regulatory factors besides DRG11. Results of in situ hybridization experiments have found that RGMb mRNA is expressed in the DRG, in the spinal cord excluding the ventricular zone, in the retina, in the optic nerve, and in other distinct regions of the brain, including the developing mouse midbrain, hindbrain and forebrain [1,4,8,38], although the pattern of RGMb gene expression does not overlap appreciably with that of RGMa [1]. RGMb mRNA also has been detected in the nervous system of the developing zebrafish [4], and has been found in the reproductive tract of rodents [39]. Based on results of Northern blotting studies, there appears to be a single RGMb transcript in mice of ~ 4.2 kb [1,4], which is approximately the same size as the three mouse RGMb exons (Table 2). As with RGMa, the mechanisms responsible for RGMb gene expression in different tissues or under different physiological or pathological conditions have not been characterized, and virtually nothing is known about the structure or function of the RGMb gene promoter.
Protein sequence and expression
Cloning of mouse RGMb cDNA revealed a predicted protein of 438 amino acids [1,4], which is 89% identical to human RGMb (437 amino acids) and 65%identical to zebrafish RGMb (436 amino acids) (Table 3). The primary RGMb translation product is predicted to contain an N-terminal signal peptide of ~ 50 residues, although this has not been verified experimentally, and a C-terminal GPI attachment signal of ~ 35 amino acids [1,4]. Other identifiable motifs in RGMb include a partial vWD element. After forced expression of mouse RGMb in HEK-293 and COS-7 cells, only a single protein band of ~ 50 kDa could be detected in cell extracts by immunoblotting, and a similarly sized protein was released into the culture medium after incubation of cells with PI-PLC (phosphoinositide-specific phospholipase C), which cleaves the GPI anchor [1,4]. These latter results indicate that only a single-chain RGMb species is attached to the outer face of the cell membrane [4,40] (Figure 3B), although the protein contains a putative internal proteolytic cleavage site similar to that in RGMa. RGMb also appears to be a glycoprotein, and is predicted to encode up to two asparagine-linked glycosylation sites (Figure 3A). As with RGMa, mature RGMb contains 14 cysteines whose potential organization into disulfide bonded residues has not been established (but see discussion of potential molecular models in the section on structure–function relationships below).
Potential physiological functions
No biological functions of RGMb have been elucidated, except for its possible ability to promote cell–cell adhesion by homophilic interactions [1,4], and its capability to bind selected BMPs [40,41]. As with RGMa, overexpressed full-length RGMb has been found to increase the activity of a promoter–reporter gene containing a BMP-responsive transcriptional control element in cell culture systems [39,40], but unlike RGMa, RGMb has not been shown to bind to neogenin.
RGMc/HAEMOJUVELIN
Chromosomal organization and gene structure
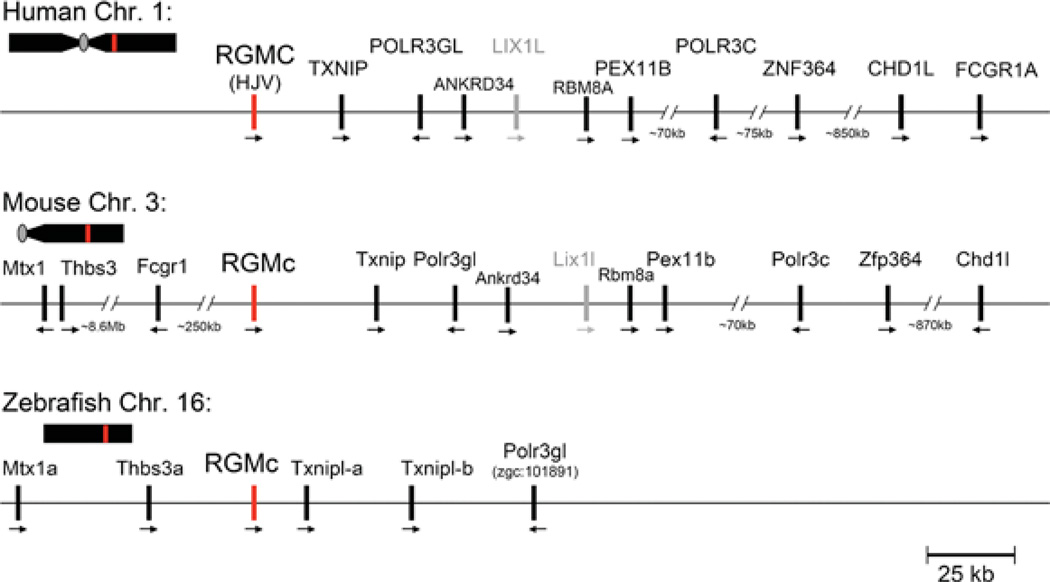
RGMc is a single-copy gene in the nine mammalian and six non-mammalian vertebrates in which it has been identified (Table 1). Unlike RGMa and RGMb, RGMc has not been found to date in the chicken or other avian species. In human and mouse genomes, RGMc comprises one of 10 linked genes in a syntenic locus that includes among others, Txnip (thioredoxin interacting protein), Polr3gl [polymerase (RNA) III (DNA directed) polypeptide G-like], Ankrd34 (ankyrin repeat domain 34), Lix1l (related to Lix1, which maps near RGMb), and Chd1l [related to Chd1 and Chd2, which are located near RGMb and RGMa respectively (compare Figures 1, 4 and 6)]. Of note, however, the relative transcriptional orientation of RGMc and Chd1l (tail-to-head) differs from that of RGMa–Chd2 and RGMb–Chd1 (tail-to-tail). Moreover, in zebrafish, the RGMc chromosomal environment differs from mammals (Figure 6). Although the location of two Txnip-like genes and Polr3gl are adjacent to RGMc, and is similar to what is seen in mammals, Mtx1 and Thbs3a are just upstream of zebrafish RGMc, but are located at a distance of more than 8 Mb from mouse RGMc. Furthermore, there is no Chd homologue present on the zebrafish RGMc locus.
Figure 6. Comparative structures of RGMc genomic loci.
The relative position of the RGMc gene (red line) is indicated on each chromosome (Chr.; human 1, mouse 3, zebrafish 16) in relation to the centromere (grey oval, if information available) and telomere. Presented below each chromosome is a higher resolution view of the RGMc locus for each species. Neighbouring genes are indicated, with their transcriptional direction represented by an arrow. Lix1-like, shown in grey, is a putative pseudo-gene (Lix1l), as there is no known transcript available in GenBank®. Gene names corresponding to the abbreviations may be found in Table 4.
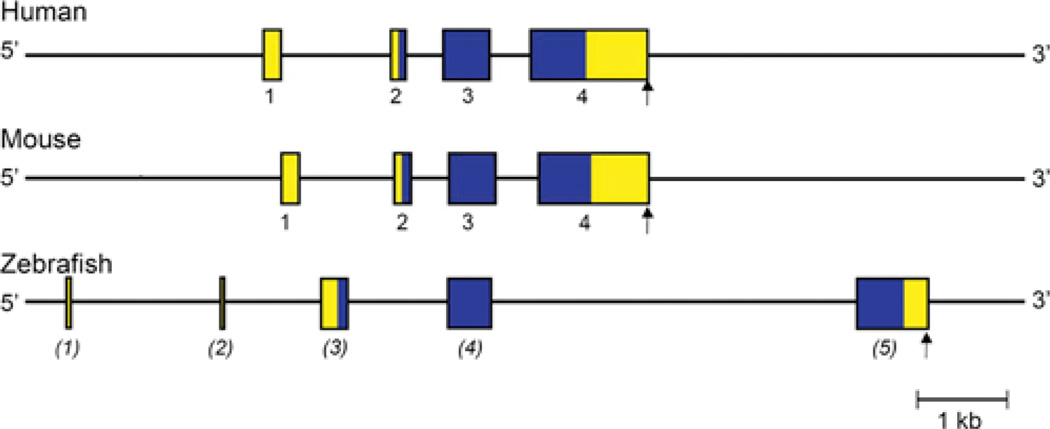
Human and mouse RGMc genes are similar in size (~ 4.3 and ~ 4.0 kb respectively, Table 2) and organization, being composed of four exons separated by three introns (Figure 7), and are considerably smaller than mammalian RGMa or RGMb (Table 2). In both species, exon 1 is ~ 160 nucleotides in length, although the 5′ end has not been identified, and contains most of the 5′ UTR of RGMc mRNA. The remaining 90 nucleotides of the 5′ UTR are found in exon 2, along with the first 31 codons of the RGMc protein (28 in mouse). Exon 3 encodes the next 173 codons (169 in mouse), and exon 4 the remaining 222 codons (223 in mouse), plus a 3′ UTR of ~ 1150 nucleotides with a single polyadenylation signal (Figure 7). The four RGMc exons are well-conserved between the mouse and human genes, with nucleotide sequence identity ranging from 73 to 83% (calculated using references [12–15]). The three introns are less conserved, although their lengths are similar between mouse and human (Figure 7). The zebrafish RGMc gene is larger than its mammalian counterparts, and contains 5 exons distributed over ~ 11.4 kb (Figure 7). Exons 1 and 2 are non-coding but are not similar in DNA sequence to mammalian RGMc exon 1. In contrast, zebrafish exons 3–5 correspond to mammalian RGMc exons 2–4, with nucleotide sequence identity ranging from 50 to 59%.
Figure 7. Comparative organization of RGMc genes.
The anatomy of human, mouse and zebrafish RGMc genes is shown. Exons are indicated by boxes, with coding regions in blue and non-coding regions in yellow. The assignment of exon numbers is based on comparison with mouse RGMc, and is provisional for zebrafish (in parentheses). The polyadenylation site is represented by a vertical arrow.
Gene expression
RGMc was independently discovered as a gene within a locus linked to the human iron overload disorder juvenile haemochromatosis [6], as an mRNA related to RGMa and RGMb [1,3,4,8], and as a novel transcript expressed during skeletal muscle differentiation [5]. In addition to skeletal muscle, RGMc mRNA has been detected in the heart and in the liver [1,5,8]. During mouse development, RGMc transcripts are found first in the somites, precursors of skeletal muscle, as early as E11.5, which is before muscle can be identified morphologically [5]. Similar observations have been made in zebrafish [4,16]. In the mouse, RGMc mRNA is detected by E13.5 in the heart and liver [5,42].
Very little is known about RGMc gene regulation. In mice, RGMc mRNA levels were shown to be increased in the liver but not in skeletal or cardiac muscle after systemic injection of bacterial lipopolysaccharide [42], but as with RGMa and RGMb, the biochemical mechanisms responsible for controlling RGMc gene transcription or mRNA stability in different tissues or under different physiological or pathological conditions have not been established, and virtually nothing is known about the structure or function of the RGMc gene promoter.
Protein sequence, processing and expression
The initial cloning of human and mouse RGMc cDNAs revealed primary translation products of 426 and 420 amino acids respectively, with a predicted N-terminal signal peptide of ~ 31 residues and a C-terminal GPI-attachment signal of ~ 45 amino acids [1,3,9], although as in other RGM molecules, the precise boundaries have not been determined experimentally. Mouse and human RGMc precursor proteins are 88% identical to each other (Table 3). Similar to RGMa, RGMc contains up to three asparagine-linked glycosylation sites, and similar to its paralogues, has several shared protein motifs, including an RGD sequence and a partial vWD domain with a conserved proteolytic cleavage site (Figure 3A). In addition, and unlike RGMa or RGMb, mammalian RGMc proteins encode a furin-like PPC (pro-protein convertase) recognition and cleavage sequence near the C-terminus (Figure 3A), and the protein has been shown to be cleaved by furin at this site [43–45]. As a consequence, RGMc appears to undergo a complex series of biosynthetic and processing steps, leading to the production of four distinct protein isoforms in skeletal muscle and after expression of the recombinant protein in heterologous mammalian cells [9,43,45,46]. Two of the RGMc proteins, a disulfide-bonded two-chain species that is similar to RGMa, and a single-chain isoform similar to RGMb, are attached to the extracellular face of the plasma membrane by a GPI linkage [9,43,45,47] (Figure 3B). In addition, single-chain RGMc species have been detected in the extracellular fluid of cultured cells, and in blood [9,43–48] (Figure 3B). These latter two proteins differ at their C-termini, with the smaller species being derived from the larger by PPC-mediated proteolytic cleavage [9,43,45]. Results of biosynthesis experiments additionally support the idea that the two soluble single-chain RGMc proteins originate from the single-chain cell-associated molecule [9,43]. Analogous studies have not been reported for RGMa or RGMb. As in RGMa and RGMb, the disulfide bonding pattern of the 14 cysteines found in mature full-length RGMc has not been experimentally defined, but a possible model is discussed below.
Physiological functions and mechanisms of action
A role for RGMc in systemic iron metabolism was first inferred when mutations in the human gene were linked to the severe iron overload disorder, juvenile haemochromatosis [6]. This relationship was strengthened when mice engineered to lack RGMc were found to have excessive accumulation of iron in multiple tissues [42,49]. It has been postulated that the normal biological actions of RGMc lead to induction of expression of the secreted hepatic peptide hepcidin [6,42], which functions as a negative regulator of the uptake of dietary iron from the duodenum and of the release of stored iron from macrophages [6,50].Humans with juvenile haemochromatosis and mice with RGMc deficiency have low levels of serum or urinary hepcidin [51,52], and mice lacking RGMc also have diminished expression of hepcidin mRNA in the liver [42,49]. The mechanism of regulation of hepcidin by RGMc is currently under active investigation, with the leading hypothesis being that cell-membrane associated RGMc facilitates signalling by BMPs through its receptors to promote hepcidin gene expression [41,53–55]. In this model, soluble RGMc has been proposed to act as an inhibitor, presumably by sequestering BMPs away from cell-surface receptors [45,48].
Similar to RGMa, RGMc binds to the extracellular portion of neogenin [46,47,56], although the role of neogenin in the biological actions of RGMc has not been established. One report has demonstrated preferential binding of two-chain RGMc to neogenin [46], and mouse versions of two juvenile haemochromatosis-associated RGMc amino acid substitution mutants, D172E and G320V, which did not form a two-chain species [9,46], were unable to bind [46]. Similar results were observed with the human G320V juvenile haemochromatosis-associated protein [9,43,45,47]. In other experiments, neogenin was unable to alter BMP-mediated hepcidin gene expression [55], although it is unclear which RGMc protein isoforms were used in these studies. Further studies will be needed to elucidate the biochemical mechanisms by which RGMc regulates systemic iron metabolism under different physiological conditions, to determine if there is a role for neogenin in the biological actions of RGMc, and to characterize the functions of different RGMc species in normal physiology and in disease.
MOLECULAR EVOLUTION OF THE RGM FAMILY
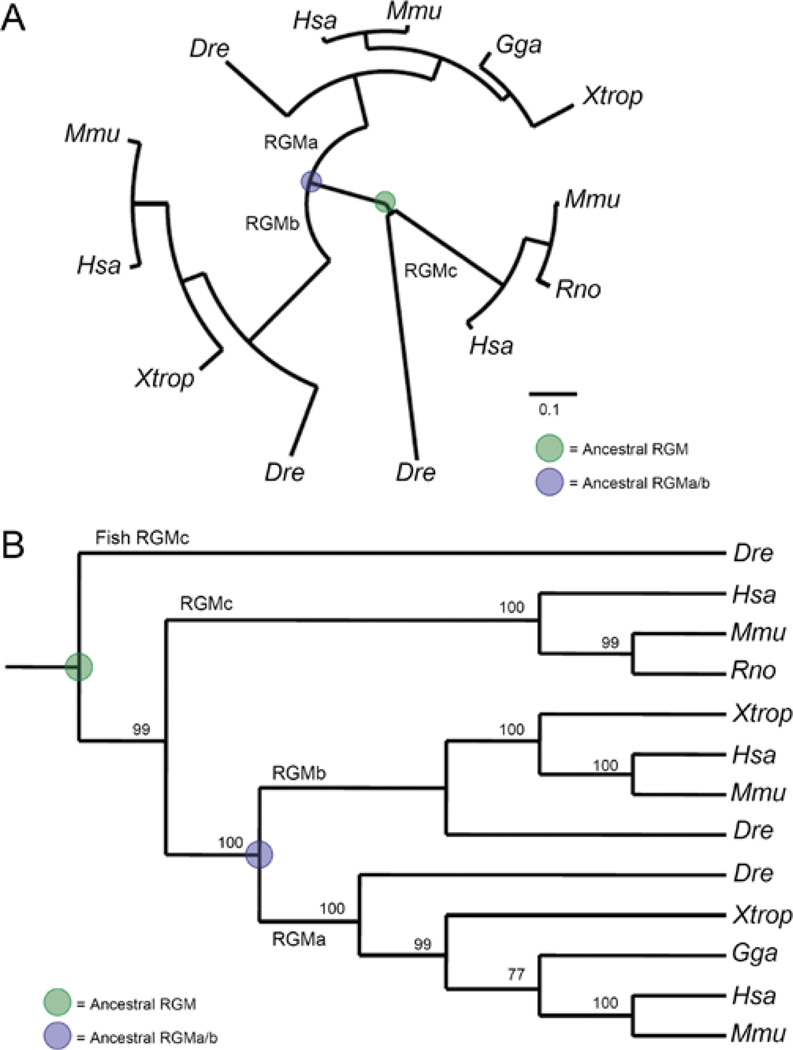
One unresolved question about the RGM family concerns the evolutionary relationships among the three members. To address this issue, we performed a series of phylogenetic analyses by querying multiple sequence alignments of selected RGM proteins after applying the following two criteria: (i) using only well-annotated sequences in which the protein defined by translation from both mRNA and genomic sequences is identical, and (ii) minimizing the level of ‘mammalian bias’ by selecting RGM genes from a diversity of organisms. We found that three out of four assessments supported the hypothesis that RGMc diverged from a common ancestor earlier than did RGMa or RGMb (see legend to Figure 8 for a summary of methods). Two of the phylogenetic trees are presented in Figure 8. Similar conclusions were reached by Schmidtmer and Engelkamp [3], whereas Camus and Lambert [10] have advocated the alternative viewpoint that RGMa and RGMc are more closely related to one another.
Figure 8. Phylogeny of the RGM family.
Evolutionary trees have been derived from the protein translation of well-annotated RGM DNA sequences in which the mRNA and gene agrees. Methods of analysis are as follows: seven separate MSAs (multiple sequence alignments) of full-length RGM proteins were performed with MUSCLE [14], Clustal-W [75] or hand alignment, followed by direct submission or a codon-optimized alignment through PAL2NAL [76]. Either protein MSAs or codon-based alignments were submitted to several phylogenetic methods, including neighbour joining with unrooted and rooted trees (via MacVector), maximum likelihood [77,78] (with and without Bootstrap methods on neighbour joining and maximum likelihood) and Bayesian [79,80] analysis. (A) RGM family phylogeny using an unrooted maximum likelihood method, displaying a distance of 0.1 amino acid substitutions per position (scale bar). (B) RGM family cladogram derived from the neighbour joining method (Poisson-correction with gaps distributed proportionally) rooted with zebrafish (Dre) RGMc, displaying bootstrap values as percentage of 5000 replications supporting that branch on the cladogram. Species abbreviations for (A) and (B) may be found in Table 1. For both (A) and (B), the putative ancestral RGM is highlighted in green and the ancestral gene to RGMa and RGMb is shown in blue. Phylogeny and cladogram created using Pal2NAL [76], Selection Server [81], Phylogeny.fr [78], PhyML 3.0 [77], TreeDyn [82] and MacVector v7.2.3.
Inspection of RGM genomic loci strengthens the view that RGMa and RGMb have a closer relationship to each other than to RGMc. RGMa and RGMb genes are physically linked to Chd2 and Chd1 respectively, in mammalian, chicken, and zebrafish genomes (Figures 1 and 4), and are each part of a more extensive syntenic linkage group that includes in order (at least in the human genome) RGMA - CHD2 - ST8SIA1 - SLCO3a1 and RGMB - CHD1 - ST8SIA4 - SLCO4C1, indicating that the organization of paralogous genes within the duplicated chromosomal regions has been maintained (Figures 1 and 4). In contrast, only a Chd1-related pseudo-gene is found near the same chromosomal locus as RGMc in mammals, but is located at a much greater distance from RGMc than Chd2 or Chd1 are from RGMa or RGMb respectively (compare Figures 1, 4 and 6). Also, in mammals, the pseudo-gene Lix1-like is found near RGMc, but in a different relationship than Lix1 and RGMb (compare Figures 4 and 6).
Single RGM genes have been identified in several invertebrates. The evidence is strongest for existence of an RGM protein in the sea squirt, Ciona intestinalis, where a polyadenylated mRNA has been characterized that corresponds to the four-exon genomic DNA sequence (NCBI accession number AK173741), and encodes a predicted protein of 637 amino acids (calculated using Transeq [15]), with multiple cysteine residues (15 in the putative mature protein compared with 14 in vertebrate RGMs), and overall similarity of 40%, 38% or 27% to mouse RGMa, RGMb or RGMc respectively. Similar to RGMb, Ciona RGM contains no RGD motif, but instead has an RGN sequence [15,57]. Similar to mammalian RGMc, the Ciona RGM has a predicted PPC site near its C-terminus. To date, however, this putative protein has not been characterized.
An RGM gene also has been identified in the purple sea urchin, Strongylocentrotus purpuratus, where it maps near a CHD1-like gene (LOC575959) as seen in RGMa and RGMb loci in vertebrates (Figures 1 and 4). The protein predicted to be encoded by this gene contains an RGD motif and 16 cysteines (14 of which align with the 14 conserved cysteines in mammalian RGMs), and is ~ 40%identical to mammalian RGMa or RGMb, and ~ 35% identical to RGMc [58]. In the nematode, Caenorhabditis elegans, a single RGM gene also has been predicted, but the putative protein is <30% identical to mammalian RGMs, lacks several of the conserved cysteine residues found in mammalian RGM proteins, and unlike vertebrate RGM proteins, does not contain either an RGD or RGN sequence [59]. Although a single RGM has been reported in molluscs (California brown sea slug, Aplysia californica) [10], definitive genomic evidence is lacking. Clearly, further analysis of putative RGM genes and their encoded proteins in invertebrates is needed for more complete understanding of the evolution and functions of the RGM family.
STRUCTURE–FUNCTION RELATIONSHIPS AMONG RGM PROTEINS
Three-dimensional structures can provide critical insights into structure–function relationships within a protein family. Although no such information is available yet for the RGM family, emerging computational methods such as comparative modelling [60,61], fold recognition [62], and ab initio techniques [63,64] have the potential to help overcome this deficiency. Comparative modelling can approximate the three-dimensional structure of a target protein for which only the amino acid sequence is available, provided that an empirical three-dimensional ‘template’ structure is available from a protein with >30% sequence identity. Alternatively, threading methods, which search for an optimal fit of query sequences onto known three-dimensional structures of proteins in databases, can be used when a comparative modelling approach is unsuccessful. However, neither comparative modelling nor threading techniques were able to identify appropriate templates for RGM proteins. As a consequence, we constructed initial structural models for the RGM family with ab initio approaches, which use the physical properties of the primary amino acid sequence to predict structures. We employed ‘Rosetta’ ab initio modeling software, because it has been the most consistent and accurate in predicting structures of folded domains in a series of trials (CASP: critical assessment of techniques for protein structure predictions [63–70]). For the RGM family, structural segments were generated using the Rosetta fragment server with input amino acid sequence information derived from 22 RGM proteins (see legend to Figure 9). One thousand independent simulations were generated and were organized into clusters according to structural similarities, as outlined in the legend to Figure 9. All ab initio models analysed suggest that RGM proteins adopt a two-lobed structure (Figure 9).
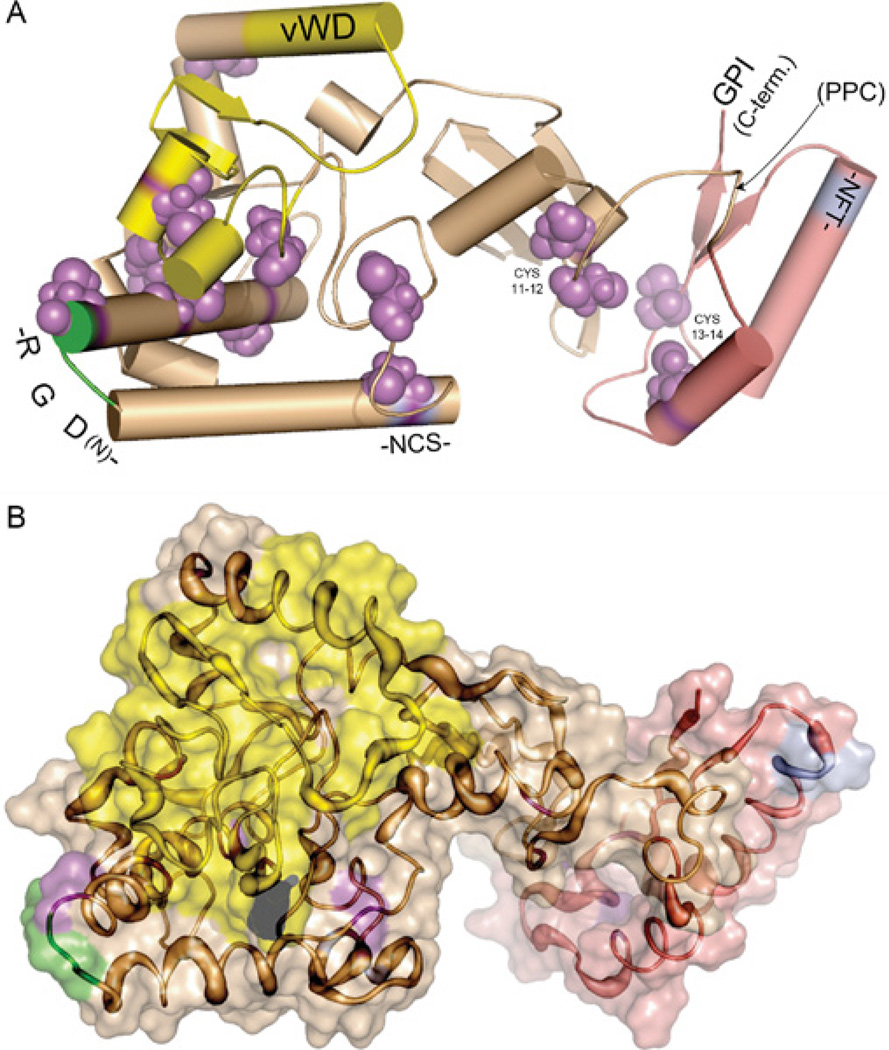
Figure 9. Ab initio model for RGM proteins.
The model was generated using Rosetta [64–67,70,83], using the following steps: First, 1000 independent structures were predicted from a fragment library prepared with the Robetta Fragment server [63,68,69]. Structures were clustered for similarity based on their root mean square deviations. The centres of the three largest clusters were chosen as the best models, defined as having the lowest standard deviation of the mean among positions of carbon atoms of all residues to all other simulations in a cluster. Selected structures were minimized using CharmM [84,85] and analysed for consistency with known experimental data as described in [86]. A single model is illustrated. (A) Cartoon version of the model. Cylinders represent α-helical regions, thick lines with arrows represent β-sheets, and thin lines represent unstructured regions. The model suggests that members of the RGM family adopt a two-lobe structure. The RGD domain is depicted in green, the partial vWD domain is in yellow, cysteines are in purple, asparagine-linked glycosylation sites conserved in all 3 mammalian RGMs are in cyan (and labeled -NCS- and -NFT-), and the GPI anchor attachment site at the C-terminus (C-term.) is noted. All of the above regions appear to be surface exposed. The PPC site (found only in mammalian RGMc) is depicted by a labelled arrow. The N-terminus is not visible as it is located behind the partial vWD domain in the left lobe of the protein. An interactive three-dimensional version of (A) can be found at http://www.BiochemJ.org/422/0393/bj4220393add.him. (B) Space-filling version of the model. The increasing thickness of the tubes represents greater divergence in primary amino acid sequences among RGM family members. The protein domains are colour-coded as in (A).
Mature RGMa, RGMb and RGMc each contain 14 similarly placed cysteine residues (Figure 3A), and all appear to be disulfide-bonded proteins [9,45,47]. However, the number or location of disulfide bonds is unknown. The majority of ab initio models show a disulfide bond between Cys9 and either Cys7 or Cys8, although one model suggests two disulfide bonds (Figure 9A, cysteine residues shown as space-filling models in purple), and this could be the linkage responsible for maintaining two-chain forms of RGMa or RGMc. Both Cys11 and Cys12, and Cys13 and Cys14, are also predicted to form disulfide bonds in all models generated, and are located within the C-terminal part of the two-lobed structure (Figure 9A). Although the connectivity varies slightly between models, the majority of the predictions suggest two disulfide bonds for the N-terminal lobe between Cys1 and Cys2, and Cys4 and Cys5, for a total of 5 or 6 disulfide linkages per RGM molecule. This would leave 2–4 free cysteines in the protein (Figure 9A). Clearly, direct experiments are needed to define the actual disulfide bonding pattern for each RGM family member.
von Willebrand factor is a glycoprotein that helps mediate platelet adhesion at damaged blood vessels through interactions with blood clotting Factor VIII [24,71]. It contains five distinct structural domains (vWA, B, C, D and CK) [24], and one of these motifs (type D) has been recognized in all RGM proteins [3]. Our ab initio models suggest that this partial vWD domain is highly structured, and contains surface exposed α-helices and β-strands (yellow region in Figure 9). These are consistent with the crystal structure of the entire vWD domain [RCSB (Research Collaboratory for Structural Bioinformatics) protein structural data base accession number 1ijb] [72]. The RGM partial vWD region contains the site of intramolecular proteolytic cleavage to generate two-chain forms of RGMa and RGMc (see Figures 3A and 3B), and this cleavage has been hypothesized to occur by acid-labile hydrolysis between an aspartic acid and proline residue [47]. In the model depicted in Figure 9, these two amino acids are located on the surface of the protein (surface of space-filling model in 9B). Of note, a substitution of this aspartic acid residue to glutamic acid in human RGMc (D172E) causes juvenile haemochromatosis [73], and in biochemical experiments the mutant protein does not form a two-chain molecule [9,46]. Another disease-causing amino acid substitution in human RGMc of G320V also appears to block production of the two-chain protein [9,46]. The ab initio model depicted in Figure 9 suggests that Gly320 is located on a surface that is in proximity to Asp172. On the basis of the model it thus appears possible that the G320V substitution, which increases the side-chain volume and hydrophobicity, may inhibit interactions with some unknown protein/protease to prevent proteolysis at residue Asp172. Alternatively, the substitution may induce certain conformational changes that indirectly impair proteolytic cleavage at Asp172.
RGMa and RGMc each contain a RGD motif, a tripeptide classically identified as an integrin-binding element [23], whereas RGMb does not [3,23]. Structurally, RGD motifs are found at or near the end of an α-helix [74], and our ab initio models map the RGM RGD sequence to a loop between two α-helices on the surface of the protein (Figure 9A). The exact function of this motif in RGMa or RGMc is not known, although amino acid substitutions of glycine to valine or arginine (G99V or G99R) appear to cause juvenile haemochromatosis in humans [6,73], and the analogously mutated mouse RGMc (G92V) was unable to bind BMP-2 in biochemical assays [46].
RGM proteins contain several putative asparagine-linked glycosylation sites, and have been shown to be glycoproteins [2,9,26], although the functional role of glycosylation has not been established for any RGM family member yet. In our ab initio structural models, at least two of these sites map to the surface of the molecule (Figure 9). As noted earlier, RGMc but not RGMa or RGMb contains a pro-protein convertase recognition and cleavage site near the C-terminus of the mature protein (Figure 3). As seen in Figure 9(A), this part of the protein in our ab initio model also maps to a surface loop, and thus potentially would be readily accessible to targeted proteolysis by furin or other pro-protein convertases.
SUMMARY AND CHALLENGES FOR THE FUTURE
The RGM family appears to have been composed of three genes early in vertebrate evolution, being present in a common ancestor to mammals and fish. Each gene is expressed in a distinct developmental and tissue-specific pattern, with RGMa and RGMb being produced in different parts of the central nervous system, and RGMc being synthesized in striated muscle and liver. The molecular mechanisms governing such diverse tissue-restricted gene expression have not been established, and little is known about the structure or function of RGM gene promoters, about their mechanisms of transcriptional regulation, or about control of RGM mRNA processing or stability. At the protein level, the three RGM family members share several motifs and are predicted to have similar three-dimensional structures based on our ab initio modeling, but the respective proteins appear to undergo distinct biosynthetic and processing steps, whose regulation has not been characterized. From the perspective of function, all three RGM proteins appear capable of binding selected BMPs, although binding domains have not been mapped. It appears that interactions with selected BMPs may mediate at least some of the biological effects of RGMc to control hepcidin gene expression, but to date no role for BMPs has been defined in the actions of RGMa or RGMb. To date only RGMa and RGMc have been shown to bind to neogenin, and although signalling through neogenin is critical for the biological effects of RGMa on repulsive axonal guidance and on neuronal survival, its role in the actions of RGMc remains to be elucidated. Similarly, the full spectrum of biological functions of the three RGMs has not been completely characterized yet, and will remain an active topic of ongoing investigation.
Table 4. Abbreviations in genomic loci.
| Abbreviation | Definition |
|---|---|
| NR2F2 | Nuclear receptor subfamily 2, group F, member 2 |
| MCTP | Multiple C2 domains, transmembrane |
| RGM | Repulsive guidance molecule |
| CHD | Chromodomain helicase DNA binding protein 2 |
| ST8SIA2 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase |
| SLCO3A1 | Solute carrier organic anion transporter family, member 3A1 |
| GNRH-R4HS | Gonadotropin-releasing hormone receptor GnRH-R4SHS |
| LNPEP | Leucyl/cystinyl aminopeptidase |
| LIX1 | Protein limb expression 1 |
| RIOK2 | Right open reading frame kinase 2 |
| PRDM9 | PR domain-containing 9 |
| TXNIP | Thioredoxin-interacting protein |
| POLR3GL | Polymerase (RNA) III (DNA directed) polypeptide G-like |
| ANKRD34 | Ankyrin repeat domain 34 |
| MTX1 | Metaxin 1 |
| THBS3 | Thrombospondin 3 |
| RBM8A | RNA-binding motif protein 8A |
| PEX11B | Peroxisomal biogenesis factor 11β |
| ZNF364 (Zfp364) | Zinc finger protein 364 |
| FCGR1A | Fc fragment of IgG, high affinity Ia, receptor (CD64) |
ACKNOWLEDGEMENT
We thank Kevin Kendall at MacVector for advice and guidance.
FUNDING
This work was supported by the National Institutes of Health [grant numbers R01 DK42748 (to P.R.), T32 HL007781 and F30 HL095327 (to C. J.S.)] and the National Science Foundation [grant number NSF-0746589 (to U.S.)].
Abbreviations used
- BMP
bone morphogenetic protein
- Chd
chromodomain helicase DNA-binding protein
- DRG
dorsal root ganglion
- E
embryonic day
- GPI
glycosylphosphatidylinositol
- Lix1
Limb expression 1
- Mctp2
multiple C2 domains, transmembrane 2
- PI-PLC
phosphoinositide-specific phospholipase C
- Polr3gl
polymerase (RNA) III (DNA directed) polypeptide G-like
- PPC
pro-protein convertase
- RGD motif
arginine-glycine-aspartic acid
- RGM
repulsive guidance molecule
- Slco/SLCO
solute carrier organic anion transporter family
- St8sia/ST8SIA
ST8 α-N-acetyl-neuraminide α-2, 8-sialyltransferase
- TGF
transforming growth factor
- Txnip
thioredoxin interacting protein
- Unc
unco-ordinated
- UTR
untranslated region
- vWD
von Willebrand type D
REFERENCES
- 1.Niederkofler V, Salie R, Sigrist M, Arber S. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J. Neurosci. 2004;24:808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, Mueller BK. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419:392–395. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- 3.Schmidtmer J, Engelkamp D. Isolation and expression pattern of three mouse homologues of chick Rgm. Gene Expr. Patterns. 2004;4:105–110. doi: 10.1016/s1567-133x(03)00144-3. [DOI] [PubMed] [Google Scholar]
- 4.Samad TA, Srinivasan A, Karchewski LA, Jeong SJ, Campagna JA, Ji RR, Fabrizio DA, Zhang Y, Lin HY, Bell E, Woolf CJ. DRAGON: a member of the repulsive guidance molecule-related family of neuronal- and muscle-expressed membrane proteins is regulated by DRG11 and has neuronal adhesive properties. J. Neurosci. 2004;24:2027–2036. doi: 10.1523/JNEUROSCI.4115-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuninger D, Kuzmickas R, Peng B, Pintar JE, Rotwein P. Gene discovery by microarray: identification of novel genes induced during growth factor-mediated muscle cell survival and differentiation. Genomics. 2004;84:876–889. doi: 10.1016/j.ygeno.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat. Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 7.Matsunaga E, Tauszig-Delamasure S, Monnier PP, Mueller BK, Strittmatter SM, Mehlen P, Chedotal A. RGM and its receptor neogenin regulate neuronal survival. Nat. Cell Biol. 2004;6:749–755. doi: 10.1038/ncb1157. [DOI] [PubMed] [Google Scholar]
- 8.Oldekamp J, Kramer N, Alvarez-Bolado G, Skutella T. Expression pattern of the repulsive guidance molecules RGM A, B and C during mouse development. Gene Expr. Patterns. 2004;4:283–288. doi: 10.1016/j.modgep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Kuninger D, Kuns-Hashimoto R, Kuzmickas R, Rotwein P. Complex biosynthesis of the muscle-enriched iron regulator RGMc. J. Cell Sci. 2006;119:3273–3283. doi: 10.1242/jcs.03074. [DOI] [PubMed] [Google Scholar]
- 10.Camus LM, Lambert LA. Molecular evolution of hemojuvelin and the repulsive guidance molecule family. J. Mol. Evol. 2007;65:68–81. doi: 10.1007/s00239-006-0241-5. [DOI] [PubMed] [Google Scholar]
- 11.Catchen JM, Conery JS, Postlethwait JH. Inferring ancestral gene order. Methods Mol. Biol. 2008;452:365–383. doi: 10.1007/978-1-60327-159-2_17. [DOI] [PubMed] [Google Scholar]
- 12.Wheelan SJ, Church DM, Ostell JM. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 16.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga E, Nakamura H, Chedotal A. Repulsive guidance molecule plays multiple roles in neuronal differentiation and axon guidance. J. Neurosci. 2006;26:6082–6088. doi: 10.1523/JNEUROSCI.4556-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J. Biol. Chem. 2005;280:29820–29827. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- 20.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinks H, Conrad S, Vogt J, Oldekamp J, Sierra A, Deitinghoff L, Bechmann I, Alvarez-Bolado G, Heimrich B, Monnier PP, et al. The repulsive guidance molecule RGMa is involved in the formation of afferent connections in the dentate gyrus. J. Neurosci. 2004;24:3862–3869. doi: 10.1523/JNEUROSCI.5296-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doering TL, Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- 23.Ruoslahti E. RGD and other recognition sequences for integrins. Annu. Rev. Cell. Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 24.Sadler JE. Biochemistry and genetics of von Willebrand Factor. Annu. Rev. Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 25.Matsunaga E, Chédotal A. Repulsive guidance molecule/neogenin: a novel ligand-receptor system playing multiple roles in neural development. Dev. Growth Differ. 2004;46:481–486. doi: 10.1111/j.1440-169x.2004.00768.x. [DOI] [PubMed] [Google Scholar]
- 26.Stahl B, Muller B, von Boxberg Y, Cox EC, Bonhoeffer F. Biochemical characterization of a putative axonal guidance molecule of the chick visual system. Neuron. 1990;5:735–743. doi: 10.1016/0896-6273(90)90227-7. [DOI] [PubMed] [Google Scholar]
- 27.Hata K, Fujitani M, Yasuda Y, Doya H, Saito T, Yamagishi S, Mueller BK, Yamashita T. RGMa inhibition promotes axonal growth and recovery after spinal cord injury. J. Cell Biol. 2006;173:47–58. doi: 10.1083/jcb.200508143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajagopalan S, Deitinghoff L, Davis D, Conrad S, Skutella T, Chedotal A, Mueller BK, Strittmatter SM. Neogenin mediates the action of repulsive guidance molecule. Nat. Cell Biol. 2004;6:756–762. doi: 10.1038/ncb1156. [DOI] [PubMed] [Google Scholar]
- 29.Cirulli V, Yebra M. Netrins: beyond the brain. Nat. Rev. Mol. Cell. Biol. 2007;8:296–306. doi: 10.1038/nrm2142. [DOI] [PubMed] [Google Scholar]
- 30.Conrad S, Genth H, Hofmann F, Just I, Skutella T. Neogenin-RGMa signaling at the growth cone is bone morphogenetic protein-independent and involves RhoA, ROCK, and PKC. J. Biol. Chem. 2007;282:16423–16433. doi: 10.1074/jbc.M610901200. [DOI] [PubMed] [Google Scholar]
- 31.Hata K, Kaibuchi K, Inagaki S, Yamashita T. Unc5B associates with LARG to mediate the action of repulsive guidance molecule. J. Cell Biol. 2009;184:737–750. doi: 10.1083/jcb.200807029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endo M, Yamashita T. Inactivation of Ras by p120GAP via focal adhesion kinase dephosphorylation mediates RGMa-induced growth cone collapse. J. Neurosci. 2009;29:6649–6662. doi: 10.1523/JNEUROSCI.0927-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaffar G, Taniguchi J, Brodbeck T, Meyer AH, Schmidt M, Yamashita T, Mueller BK. LIM-only-protein 4 (LMO4) interacts directly with the RGM A receptor Neogenin. J. Neurochem. 2008;107:418–431. doi: 10.1111/j.1471-4159.2008.05621.x. [DOI] [PubMed] [Google Scholar]
- 34.Xia Y, Yu PB, Sidis Y, Beppu H, Bloch KD, Schneyer AL, Lin HY. Repulsive guidance molecule RGMa alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J. Biol. Chem. 2007;282:18129–18140. doi: 10.1074/jbc.M701679200. [DOI] [PubMed] [Google Scholar]
- 35.Massague J. TGF-b signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 36.Ding YQ, Yin J, Xu HM, Jacquin MF, Chen ZF. Formation of whisker-related principal sensory nucleus-based lemniscal pathway requires a paired homeodomain transcription factor, Drg11. J. Neurosci. 2003;23:7246–7254. doi: 10.1523/JNEUROSCI.23-19-07246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito T, Greenwood A, Sun Q, Anderson DJ. Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets molecular and cellular neuroscience. Mol. Cell. Neurosci. 1995;6:280–292. doi: 10.1006/mcne.1995.1022. [DOI] [PubMed] [Google Scholar]
- 38.Schnichels S, Conrad S, Warstat K, Henke-Fahle S, Skutella T, Schraermeyer U, Julien S. Gene expression of the repulsive guidance molecules/neogenin in the developing and mature mouse visual system: C57BL/6J vs. the glaucoma model DBA/2J. Gene Expr. Patterns. 2007;8:1–11. doi: 10.1016/j.modgep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Xia Y, Sidis Y, Mukherjee A, Samad TA, Brenner G, Woolf CJ, Lin HY, Schneyer A. Localization and action of Dragon (repulsive guidance molecule b), a novel bone morphogenetic protein coreceptor, throughout the reproductive axis. Endocrinology. 2005;146:3614–3621. doi: 10.1210/en.2004-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong S-J, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, et al. DRAGON, a bone morphogenetic protein co-receptor. J. Biol. Chem. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- 41.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, Babitt JL. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J. Clin. Invest. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuninger D, Kuns-Hashimoto R, Nili M, Rotwein P. Pro-protein convertases control the maturation and processing of the iron-regulatory protein, RGMc/hemojuvelin. BMC Biochem. 2008;9:9. doi: 10.1186/1471-2091-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111:924–931. doi: 10.1182/blood-2007-07-100677. [DOI] [PubMed] [Google Scholar]
- 45.Lin L, Nemeth E, Goodnough JB, Thapa DR, Gabayan V, Ganz T. Soluble hemojuvelin is released by proprotein convertase-mediated cleavage at a conserved polybasic RNRR site. Blood Cells Mol. Dis. 2008;40:122–131. doi: 10.1016/j.bcmd.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuns-Hashimoto R, Kuninger D, Nili M, Rotwein P. Selective binding of RGMc/hemojuvelin, a key protein in systemic iron metabolism, to BMP-2 and neogenin. Am. J. Physiol. Cell Physiol. 2008;294:C994–C1003. doi: 10.1152/ajpcell.00563.2007. [DOI] [PubMed] [Google Scholar]
- 47.Zhang AS, West AP, Jr, Wyman AE, Bjorkman PJ, Enns CA. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J. Biol. Chem. 2005;280:33885–33894. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]
- 48.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 49.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J. Clin. Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 51.Papanikolaou G, Tzilianos M, Christakis JI, Bogdanos D, Tsimirika K, MacFarlane J, Goldberg YP, Sakellaropoulos N, Ganz T, Nemeth E. Hepcidin in iron overload disorders. Blood. 2005;105:4103–4105. doi: 10.1182/blood-2004-12-4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803–1806. doi: 10.1182/blood-2004-08-3042. [DOI] [PubMed] [Google Scholar]
- 53.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat. Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 54.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J. Clin. Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008;111:5195–5204. doi: 10.1182/blood-2007-09-111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang F, West AP, Jr, Allendorph GP, Choe S, Bjorkman PJ. Neogenin interacts with hemojuvelin through its two membrane-proximal fibronectin type III domains. Biochemistry. 2008;47:4237–4245. doi: 10.1021/bi800036h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 58.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 60.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 61.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 62.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 63.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 65.Bonneau R, Strauss CE, Rohl CA, Chivian D, Bradley P, Malmstrom L, Robertson T, Baker D. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 2002;322:65–78. doi: 10.1016/s0022-2836(02)00698-8. [DOI] [PubMed] [Google Scholar]
- 66.Bonneau R, Tsai J, Ruczinski I, Chivian D, Rohl C, Strauss CE, Baker D. Rosetta in CASP4: progress in ab initio protein structure prediction. Proteins. 2001;45(Suppl. 5):119–126. doi: 10.1002/prot.1170. [DOI] [PubMed] [Google Scholar]
- 67.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 68.Chivian D, Kim DE, Malmstrom L, Schonbrun J, Rohl CA, Baker D. Prediction of CASP6 structures using automated Robetta protocols. Proteins. 2005;61(Suppl 7):157–166. doi: 10.1002/prot.20733. [DOI] [PubMed] [Google Scholar]
- 69.Chivian D, Kim DE, Malmstrom L, Bradley P, Robertson T, Murphy P, Strauss CE, Bonneau R, Rohl CA, Baker D. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53(Suppl 6):524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- 70.Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, Khare S, Tyka MD, Bhat D, Chivian D, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69:118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 71.Jorieux S, Fressinaud E, Goudemand J, Gaucher C, Meyer D, Mazurier C. Conformational changes in the D’ domain of von Willebrand factor induced by CYS 25 and CYS 95 mutations lead to factor VIII binding defect and multimeric impairment (INSERM (Inserm Network on Molecular Abnormalities in von Willebrand Disease) group) Blood. 2000;95:3139–3145. [PubMed] [Google Scholar]
- 72.Fukuda K, Doggett TA, Bankston LA, Cruz MA, Diacovo TG, Liddington RC. Structural basis of von Willebrand Factor activation by the snake toxin botrocetin. Structure. 2002;10:943–950. doi: 10.1016/s0969-2126(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 73.Lanzara C, Roetto A, Daraio F, Rivard S, Ficarella R, Simard H, Cox TM, Cazzola M, Piperno A, Gimenez-Roqueplo AP, et al. Spectrum of hemojuvelin gene mutations in 1q-linked juvenile hemochromatosis. Blood. 2004;103:4317–4321. doi: 10.1182/blood-2004-01-0192. [DOI] [PubMed] [Google Scholar]
- 74.Villard V, Kalyuzhniy O, Riccio O, Potekhin S, Melnik TN, Kajava AV, Ruegg C, Corradin G. Synthetic RGD-containing alpha-helical coiled coil peptides promote integrin-dependent cell adhesion. J. Pept. Sci. 2006;12:206–212. doi: 10.1002/psc.707. [DOI] [PubMed] [Google Scholar]
- 75.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate larges phylogenies by maximum likelihood. Systematic Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 78.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 80.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 81.Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:W506–W511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chevenet F, Brun C, Banuls A-L, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinf. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 84.MacKerel AD, Jr, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: the energy function and its parameterization with an overview of the program. In: von Ragué Schleyer P, editor. Encyclopedia of Computational Chemistry. hichester: John Wiley & Sons; 1998. pp. 271–277. [Google Scholar]
- 85.Brooks BR, Bruccoleri RE, Olafson DJ, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 86.Subbian E, Yabuta Y, Shinde U. Positive selection dictates the choice between kinetic and thermodynamic protein folding and stability in subtilases. Biochemistry. 2004;43:14348–14360. doi: 10.1021/bi048397x. [DOI] [PubMed] [Google Scholar]