Abstract
Over the last decade, compelling evidence has linked the development of Alzheimer’s disease (AD) to defective intracellular trafficking of the amyloid precursor protein (APP). Faulty APP trafficking results in an overproduction of Aβ peptides, which is generally agreed to be the primary cause of AD-related pathogenesis. LR11 (SorLA), a type I transmembrane sorting receptor, has emerged as a key regulator of APP trafficking and processing. It directly interacts with APP and diverts it away from amyloidogenic processing. The 54-residue cytosolic domain of LR11 is essential for its proper intracellular localization and trafficking which, in turn, determines the fate of APP. Here, we have found a surprising membrane-proximal amphipathic helix in the cytosolic domain of LR11. Moreover, a peptide corresponding to this region folds into an α-helical structure in the presence of liposomes and transforms liposomes to small vesicles and tubule-like particles. We postulate that this amphipathic helix may contribute to the dynamic remodeling of membrane structure and facilitate LR11 intracellular transport.
Keywords: Alzheimer’s disease, LR11, SorLA, Amyloid precursor protein (APP), intracellular trafficking, amphipathic helix, membrane curvature, membrane remodeling, NMR
1. Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease that currently affects more than 36 million people worldwide. It is generally accepted that an accumulation of amyloid-β peptides (Aβ) in the brain is the primary cause of AD [1–3]. While the chemistry of Aβ generation from sequential proteolysis of the amyloid precursor protein (APP) by β- and γ-secretases is well understood, the underlying causes of Aβ peptide overproduction are much more complex [4]. In fact the vast majority of AD patients do not have genetic defects that would disrupt these proteolytic processes. Recent studies have established a link between aberrant subcellular trafficking of APP and increased Aβ production [4–7]. This is conceivable since both APP and secretases are transmembrane proteins and sorted through multiple subcellular organelles (e.g. trans-Golgi network (TGN), plasma membrane, endosomes) [8–10]. As a result, their spatial and temporal subcellular distributions are subjected to transport regulations. Indeed, altered expression of several trafficking factors results in abnormal Aβ levels [11–13]; variants in genes associated with endocytosis and retromer sorting pathways have been identified as potential AD risk factors [14].
A key regulator of APP trafficking is LR11 (also known as SorLA) [15–17]. LR11 is a 250-kDa type-1 membrane protein highly expressed in the brain and belongs to the family of vacuolar protein sorting 10 (Vps10) receptors [18–22]. Vps10 proteins interact with the retromer complex for protein intracellular trafficking. LR11 directly interacts with APP via the cluster of complement-type repeats in its ectomain domain [23] and diverts it away from amyloidogenic processing to Aβ peptides. LR11 displays tight binding to APP in vitro and co-localizes with APP in living cells, as seen in co-immunoprecipitation and fluorescence-lifetime imaging microscopy (FLIM) experiments [15, 23, 24]. The importance of LR11 in the pathophysiology of AD is highlighted by the observations of poor LR11 expression in the brain of patients suffering from sporadic AD [25–27]. A recent report indicates that subtle changes in the level of LR11 expression could significantly affect the production of Aβ peptides [28]. Furthermore, variants of the LR11 gene have been associated with potential risks for the development of AD [29].
LR11 consists of a large ectodomain, a transmembrane domain (TM), and a cytosolic domain (CT). Its proper subcellular localization to the TGN and trafficking itineraries, which rely on sorting motifs within the CT, are required for regulating the final fate of APP. The 54-residue LR11 CT is highly conserved among mammals (~95% sequence identity) and harbors multiple functionally important motifs, including an acidic-dileucine-like motif (DDVPMVIA) and an acid cluster-based motif (DDLGEDDED) (Figure 1(A). These motifs interact with adaptor proteins that mediate transports between the trans-Golgi network (TGN) and endosomes, such as GGA proteins, AP-1, AP-2, and PACS-1 [30–32]. Disruptions of LR11-GGA and LR11-AP1 interactions lead to the aberrant trafficking of LR11 and result in faulty APP trafficking and processing [33]. Furthermore, the LR11-mediated reduction of Aβ peptide is dependent on the phosphorylation of a serine residue in CT [34]. It also has been suggested that the LR11 CT may directly regulate transcription after metalloprotease TACE and γ-secretase cleavages [35].
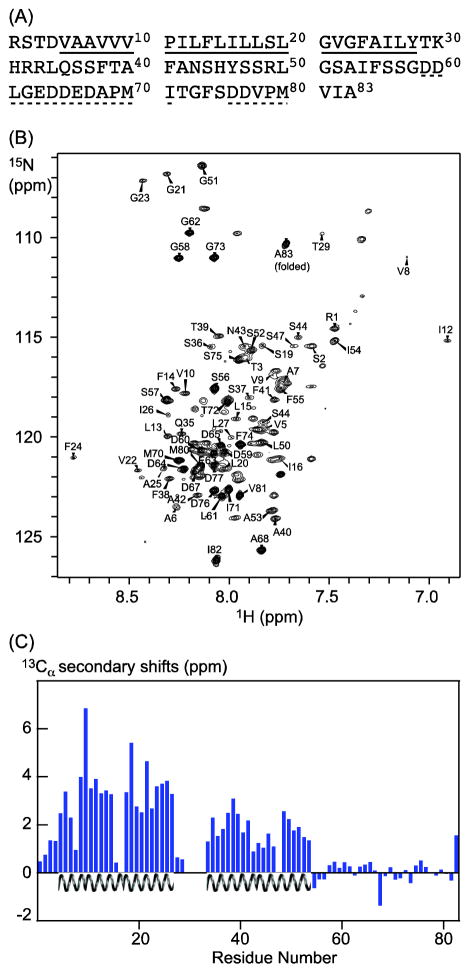
Figure 1.
Secondary structures of LR11 TMCT from NMR chemical shift analysis. (A) Primary sequence of LR11 TMCT, corresponding to residues 2132 to 2214 of the full-length protein. Amino acids underlined by a solid line are from the TM domain, and those underlined by dashed lines belong to two functionally important motifs: an acid cluster region and a GGA-binding site (dileucine-like motif), respectively. These motifs interact with adaptor proteins that mediate Golgi to endosome transport. (B) 15N-1H TROSY spectrum of 2H,13C,15N labeled LR11 TMCT in deuterated DPC micelles showing resonance assignments (resonances from tags are not labeled). (C) 13Cα secondary chemical shifts of LR11 TMCT (Cα, LR11TMCT – Cα,random coil, 2H isotope shifts were corrected using the program TALOS+) indicate two α-helical regions: a TM helix includes residues Val5 to Tyr28 and a membrane proximal helix includes residues Leu34 to Ile54.
In this study, we characterize the secondary structure of LR11 CT together with its native membrane anchor, the TM, in a membrane mimic environment using high resolution NMR spectroscopy. We identify a surprising membrane proximal amphipathic α-helix in LR11 CT. This helix interacts with liposomes mimicking the Golgi apparatus lipid composition and, moreover, deforms liposomes. We suggest that this helix may play an active role in remodeling membrane structures during vesicular trafficking and facilitating the transport of LR11 between subcellular compartments.
2. Materials and methods
Overexpression and purification of LR11 TMCT
LR11 TMCT protein was expressed and purified as previously described [36].
LR11 CT30-60 peptide
LR11 CT30-60 peptide was synthesized and purified to >90% purity by GenScript. The peptide was dissolved in either 2.5% acetic acid for circular dichroism (CD) and dynamic light scattering (DLS) experiments or 8 M urea for electron microscopy (EM) experiments at 1 mg/mL. These stock solutions were diluted to 0.1 mg/mL (33μM) with a buffer of 75 mM Hepes at pH 7.5 prior to use.
NMR spectroscopy
All NMR spectra were recorded at 37 °C on a Bruker 600 MHz spectrometer equipped with a cryoprobe unless otherwise specified. TROSY based 3D HNCA, HNCA-intra [31], HNCACB and CBCA(CO)NH data were acquired on a sample of 1 mM, U- 2H,15N,13C labeled protein in 20 mM phosphate, 100 mM NaCl, pH 7.0, and 4.5% DPC solution for backbone resonance assignments. NMR data were processed with NMRPipe and analyzed using Sparky and NMRView softwares.
Liposome preparation
Lipids in chloroform solution were purchased from Avanti Polar Lipids. Small unilamellar liposomes were prepared by either sonication or extrusion methods. First, the lipid solution was evaporated to a dried film. The dried film was then hydrated in an aqueous buffer at 40 °C for one hour to form a lipid suspension. For CD measurements, the lipid suspension, after five cycles of freezing and thawing, was sonicated in a 40 °C water bath until transparent. For DLS and EM experiments, the lipid suspension was prepared by successive extrusion through 1 μm, 0.1 μm, and 0.05 μm polycarbonate filter 11 times using a hand extruder (Avanti). Debris was removed by spinning the liposome solution at 14,000g on a desktop centrifuge for 10 minutes. Liposomes of two different compositions were prepared with either DMPC lipids or a mixture of egg PC (50%, molar ratio), DOPE (30%), Brain PS (10%), and 1,2 -DOG (10%) to mimic lipid compositions of the Golgi membrane [37, 38]. Liposomes were stored at room temperature and used within 3 days of preparation.
Circular dichroism spectroscopy
CD spectra in the far UV (200–250 nm) were recorded at 25 °C using a quartz cell with an optical path length of 0.02 cm on a Jasco 710 J-spectropolarimeter. The instrument was set at 2 nm bandwidth and 2 sec response time. LR11 CT30-60 peptide at 33 μm was incubated with liposomes for 5 minutes in a buffer of 11.25 mM HEPES, pH 7.5 before CD measurement. For each CD spectrum, 6 scans were collected and averaged with a scan rate of 50 nm/min and 1 nm interval. All spectra were corrected for background signals from an aqueous liposome solution.
Dynamic light scattering
DLS data were collected at 25 °C on a Viscotek 802 DLS instrument (Malvern). LR11 CT30-60 peptide at 8 μM was incubated with 27.2, 109.6, and 438.4 μM lipids, respectively, for 5 minutes before measurement. Data was processed and analyzed using OmniSize software.
Electron microscopy
Micrographs were taken at an initial magnification of 28,500 with a Jeol JEM 1400 transmission electron microscope (Phillips) operating at 60 kV. Before staining, 21.5 μM LR11 CT30-60 peptide was incubated with 500 μM liposomes for 10 minutes at room temperature. Negatively stained samples were prepared by applying 5 μL drops of sample to Formvar-coated 400 mesh copper grids for 2 minutes, then blotting excess sample, followed by staining, 20 seconds each, in 3 successive drops of 1% (w/v) PTA pH 7.0, and a final blotting of excess stain.
3. Results
3.1 NMR secondary structure analysis of LR11 TMCT in DPC micelles
Using an MBP-fusion construct (Figure S1) we have succeeded in preparing recombinant human LR11 TMCT protein as described previously [36]. The fusion protein, mainly expressed in E. coli membranes, was extracted from membrane with detergents and first purified using a Ni-NTA column. LR11 TMCT was then cleaved from the fusion partner and further purified and reconstituted into a DPC micelle solution. A 2D 1H-15N TROSY spectrum of 2H,13C,15N-labeled LBT-LR11 TMCT preparation is shown in Figure 1(B). The spectrum displays good quality with typical chemical shift dispersion for a helical protein. 8 out of 9 expected glycines are observed. Furthermore, the spectrum resembles the data collected from LR11 TMCT in bilayer-like bicelle solution (Figure S2a), where the protein displays expected interactions with the VHS domain of GGA (Figure S2b) [30, 39]. Thus, LR11 TMCT likely maintains its native state in DPC micelles.
We have assigned ~90% of backbone residues using several TROSY-based triple resonance experiments. Most of the unassigned residues are in regions between TM and CT domains. Analysis of the secondary shifts of assigned 13Cα indicates two helical segments: a transmembrane helix spanning residues Val5 to Tyr28 as predicted, and an unanticipated membrane proximal helix at the N-terminal region of CT extending from residues Leu34 to Ile54 (Figure 1(C). The rest of the LR11 CT (from residues Ser56 to Ala83) appears to lack stable regular secondary structure. These predictions are further supported by the backbone torsion angles derived from the TALOS+ program (listed in supplemental Table S1) and the chemical shift index (CSI) analysis of assigned chemical shifts of Cα, Cβ, and C′ (Figure S3). In addition, resonances from unstructured regions at the C-terminal half of LR11 CT consistently show strong intensities.
3.2 Membrane induced α-helical folding of the N-terminal region of LR11 CT
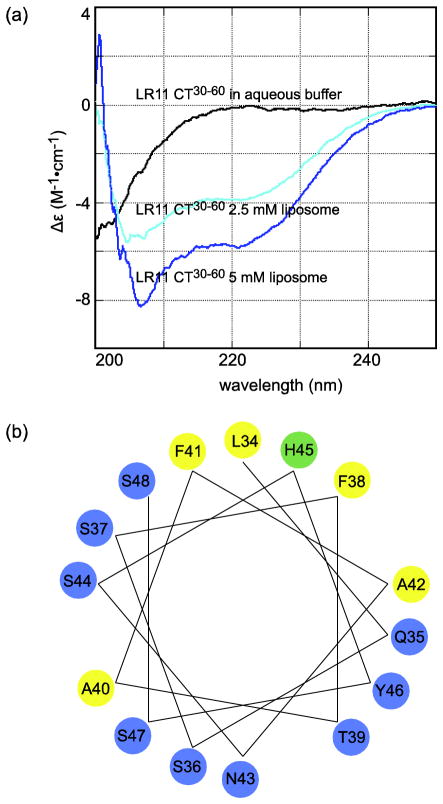
While previous studies have identified two functionally important motifs at the C-terminal half of LR11 CT [31], little is known about the significance of the N-terminal region of LR11 CT except that the sequence of FANSHY (residues 41 to 46) may be a recognition motif for the VPS26 subunit of the retromer complex [40]. To further characterize the putative N-terminal membrane proximal helix of LR11 CT, a peptide that corresponds to residues K30 to D60, LR11 CT30-60, was synthesized. CD spectra were collected in aqueous buffer and in liposome solution in order to determine if this peptide can form an α-helical structure in the absence of LR11 TM. As shown in Figure 2(A), the CD spectrum of LR11 CT30-60 peptide in aqueous solution at a concentration of 33.3 μM displays typical features of a random coil structure. In contrast, in the presence of liposomes, this peptide produces negative ellipticity at 208 and 222 nm, clearly indicating that the peptide folds to α-helical structures. Thus the membrane proximal region of LR11 CT has an intrinsic propensity to adopt helical structures in lipid environments, independent of its transmembrane domain. In addition, this folding process appears to depend on the lipid composition of membrane vesicles. While liposomes with lipid compositions that resemble those of Golgi membrane induce α-helical folding of LR11 CT30-60, the peptide remains unstructured in the presence of DMPC-liposomes (data not shown). This is likely due to loose lipid packing of liposomes with a Golgi-like composition as opposed to the presence of negative charges lipids [38], since this peptide also folds to a helical structure in neutral DPC micelles (Figure S4).
Figure 2.
(A) CD spectra of LR11 CT30-60 peptide in aqueous solution and liposomes mimicking the Golgi apparatus lipid composition. The peptide concentration is 33.3 μM. (B) Helical wheel plot of LR11 CT34-48.
A helical wheel analysis of this membrane proximal region provided some molecular insights into its interactions with the lipid bilayer. As shown in Figure 2(B), the majority of this region (residues L34 to S48) adopts an amphipathic α-helix structure. The hydrophilic face of this helix mainly consists of polar residues, while the hydrophobic face includes nonpolar amino acids and a histidine. When it is uncharged, the histidine residue prefers a hydrophobic environment. Amphipathic helices are particularly suited for effectively interacting with the membrane. As they bind to the membrane, the hydrophilic side of amphipathic helices can interact with the lipid head groups while the hydrophobic surface interacts with lipid acetyl chains. Together, our data suggest that the N-terminal membrane proximal region of LR11 CT forms an amphipathic α-helix and interacts with the lipid bilayer.
3.3 Membrane remodeling by the N-terminal region of LR11 CT
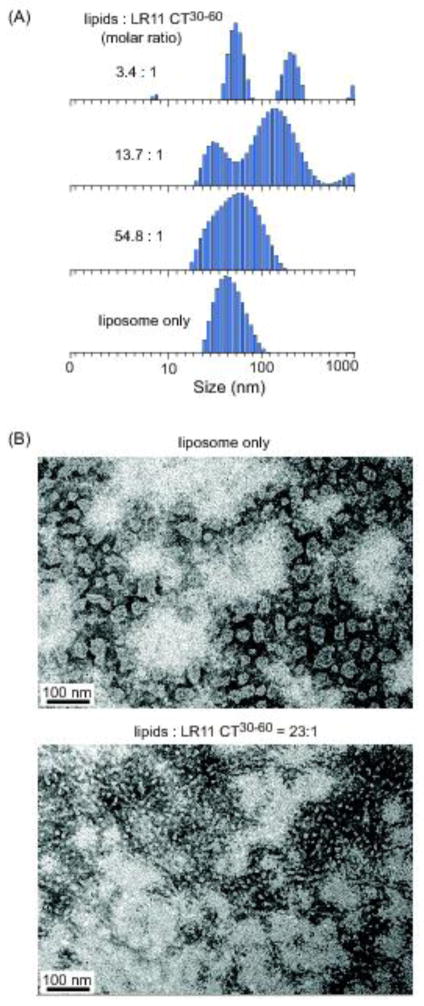
The amphipathic helix is a common motif that mediates protein interactions with membrane. Some of its classic roles include acting as a membrane anchor and membrane-destabilizing agent. Recently, amphipathic helices have been implicated in sensing and modifying membrane geometry [41–44]. To investigate the possible consequences of LR11 CT30-60 interactions with the membrane, we measured the sizes and size-distributions of liposomes incubated with and without LR11 CT30-60 peptides using DLS. Changes in these parameters provide insights into the extent of membrane perturbations upon peptide binding. For DLS experiments, relative homogenous liposomes were prepared by extrusion method. As shown in Figure 3(A), DLS measurement confirms the homogeneity of our preparations with an average mean diameter of 70 ± 20 nm. These liposomes are stable and few changes are detected for several days. In contrast, after incubating with the LR11 CT30-60 peptide for 10 minutes, dramatic changes in liposome sizes and size-distributions are observed. At a high lipid-to-peptide molar ratio of 54.8:1, the mean size of liposomes changes slightly yet the size distributions almost double. When lipid-to-peptide molar ratios decrease to below 14:1, DLS data indicate that the solution becomes polydisperse. In a control experiment, solution with LR11 CT30-60 peptides alone at the same concentration remains monodisperse (data not shown). Thus DLS data suggest that the binding of LR11 CT30-60 to liposomes may alter their shapes and sizes.
Figure 3.
LR11 CT30-60 peptide remodels liposomes mimicking the Golgi apparatus lipid composition. (A) DLS data of extruded liposome solution in the absence and presence of the peptide. The peptide concentration is at 8 μM. (B) Electron micrographs of negatively stained vesicle in the absence and presence of the peptide. The liposome concentration is 500 μM.
We directly visualized the effect of LR11 CT30-60 binding on liposomes using negative staining and EM. Extruded liposomes show vesicles of relatively similar sizes with an average size of about 60 nm (Figure 3(A). By contrast, at a lipid-to-peptide molar ratio of 23:1 these liposomes are clearly deformed into significantly smaller vesicles coexisting with some tubule-like particles as seen in Figure 3(B). These tubule-like particles are likely responsible for the observed increase in vesicle sizes seen in Figure 3(A) since DLS measurements are more sensitive to larger particles. Together, our results indicate that the LR11 CT30-60 peptide may efficiently reshape membrane structures in vitro.
4. Discussion
Intracellular transport requires the constant budding and fusion of membrane-bound trafficking vesicles. During this process, membrane geometry is concomitantly remodeled. The generation, recognition, and regulation of membrane structure depend on a complex interplay between proteins and lipids, proteins and proteins, and lipids and lipids. Despite its structural simplicity, the amphipathic helix has emerged as one of the most common motifs invoked to sense and modulate curved membranes [45–50]. Once folded, amphipathic helices usually reside within the interfacial zone with its hydrophilic face interacting with lipid polar groups and its hydrophobic side penetrating into the hydrocarbon core of the bilayer. Insertion of a hydrophobic region into one leaflet of the bilayer changes the bilayer symmetry and perturbs lipid packing, thereby presenting the potential for sensing and inducing membrane curvature. Increasing numbers of membrane-bending amphipathic helices have been found in proteins involved in endocytosis and vesicular trafficking, in viral proteins, and in several peripheral membrane proteins. Examples include N-BAR domain containing proteins such as amphiphysin and endophilin [51], small GTP-binding proteins such as Arf and Sar1 [52], M2 protein from Influenza virus [53], Tip protein from Herpesvirus saimiri [54], Pex11 protein involved in peroxisome proliferation [55], and CTP: phosphocholine cytidylyltransferanse which regulates phosphatidylcholine synthesis [56]. Here we report that a putative membrane-proximal amphiphathic helix in LR11 CT deforms liposomes in vitro. This helix is short and has a small hydrophobic face (Figure 2(B), similar to the membrane-bending H0 helix in Epsin [45, 51], a protein that contributes to the formation of clathrin-coated vesicles. However, unlike the H0 helix, the hydrophilic face of the LR11 CT helix does not contain charged residue, a feature that was previously ascribed to a class of amphipathic helices that senses but does not induce membrane curvature [42, 57]. It remains to be seen if the LR11 CT helix can also function as a curvature sensor. On the other hand, our current knowledge about membrane structure rearrangement remains qualitative; a detailed understanding of the underlying molecular mechanism requires further study [58–60].
The cytosolic domain of LR11 contains multiple motifs that are responsible for its intracellular trafficking and subcellular localization [30]. Functional interactions of these motifs with adaptor proteins that mediate Golgi body-endosome transports determine the proper subcellular location and intracellular trafficking of LR11, which in turn are critical for protecting APP from amyloidogenic processing. As shown previously, faulty trafficking of LR11 leads to aberrant APP processing and enhanced Aβ peptide production. LR11 is recruited to carrier vesicles primarily by adaptor proteins such as GGA via the acidic-dileucine-like motif in its cytosolic domain [30, 61]. While the basics of vesicular biogenesis have been described, the precise molecular mechanisms responsible for membrane deformation and vesicle budding have not been clearly elucidated. It is generally believed that Arf and coat proteins mediate these processes [62–64]. In the present study, we found that the membrane proximal helix of LR11 CT deforms liposomes, suggesting that this sorting receptor may also actively participate in altering membrane structure for vesicle formation. This hypothesis needs to be tested both in vivo and in vitro using the full length LR11, but it is worth noting that an amphipathic helix within the cytosolic tail of the M2 protein alone is sufficient for Influenza virus scission [53].
A recent study reported that mutations in the F41ANSHY46 motif, in particular the F41A mutant, within the putative helix of LR11 CT disrupted its interaction with the retromer complex and resulted in altered LR11 subcellular localization [40]. This membrane proximal region may perform dual functional roles. Previously, an amphipathic helix preceding the transmembrane domain of the Herpersviral protein Tip has been shown to mediate both lipid raft localization and membrane deformation [54]. On the other hand, aromatic residues are known to be critical for protein-lipid interactions. The F41A mutation will also likely affect the binding of the LR11 CT helix to the membrane bilayer besides abolishing its interaction with the retromer complex.
In summary, we have identified a membrane proximal helix at the N-terminal region of LR11 CT from an analysis of NMR chemical shifts. We have shown that the folding of this helix at the membrane surface is independent of the LR11 TM helix. This helix has characteristic features of an amphipathic helix and interacts with liposomes, transforming them to small vesicles and tubule-like particles. Since change in membrane geometry is an inherent part of vesicular transport, we speculate that LR11 may play an active role in remodeling membrane structure and facilitating the intracellular trafficking process.
Supplementary Material
Highlights.
A membrane proximal amphipathic helix in the LR11 cytosolic domain is identified.
This region folds to an α-helix without the LR11 transmembrane domain.
This helix transforms liposomes to small vesicles and tubule-like particles.
Acknowledgments
We are grateful for financial support from the National Institutes of Health (R01GM105963-01A1) and the Penn State University College of Medicine. We thank Dr. J. M. Flanagan at the Penn State University College of Medicine for helpful discussions, Drs. A. Benesi and E. Hatzakis at the NMR facility of the Penn State University, University Park for the assistance of the use of 850 MHz instrument, Dr. T. Fox at the Penn State University College of Medicine for help in liposome preparation, Mr. R. Myers at the Penn State College of Medicine Core Electron Microscopy Imaging Facility for technical assistance of EM experiment. We also would like to thank the Penn State Automated Biological Calorimetry Facility - University Park, PA for use of DLS instrument.
Abbreviations
- TM
transmembrane domain
- CT
cytoplasmic domain
- APP
amyloid precursor protein
- AD
Alzheimer’s disease
- MBP
Maltose binding protein
- DMPC
1,2-dimyristoyl-sn-glycerophosphocholine
- DOPE
1,2-dioleoyl-sn-Glycero-3-Phosphoethanolamine
- 1,2-DOG
1,2-dioleoyl-sn-glycerol
- PTA
phosphotungstic acid
- PS
phosphatidylserine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hardy JA, Higgins GA. Alzheimer’s disease: The amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathway to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajendran L, Annaert W. Membrane trafficking pathways in Alzheimer’s disease. Traffic. 2012;13:759–770. doi: 10.1111/j.1600-0854.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 6.Mayeux R, Hyslop PSG. Alzeimer’s disease: advances in trafficking. Neurology. 2008;7:2–3. doi: 10.1016/S1474-4422(07)70298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sannerud R, Annaert W. Trafficking, a key player in regulated intramembrane proteolysis. Semin Cell Dev Biol. 2009;20:183–190. doi: 10.1016/j.semcdb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Chia PZC, Toh WH, Sharples R, Gasnereau I, Hill AF, Gleeson PA. Intracellular itinerary of internalised β-secretase, BACE1, and its potential impact on β-amyloid peptide biogenesis. Traffic. 2013;14:997–1013. doi: 10.1111/tra.12088. [DOI] [PubMed] [Google Scholar]
- 9.Willnow TE, Andersen OM. Sorting receptor SorLA - a trafficking path to avoid Alzheimer disease. J Cell Sci. 2013;126:2751–2760. doi: 10.1242/jcs.125393. [DOI] [PubMed] [Google Scholar]
- 10.Marks N, Berg MJ. BACE and γ-secretase characterization and their sorting as therapeutic targets to reduce amyloidogenesis. Neurochem Res. 2010;35:181–210. doi: 10.1007/s11064-009-0054-1. [DOI] [PubMed] [Google Scholar]
- 11.Burgos PV, Mardones GA, Rojas AL, daSilva LLP, Prabhu Y, Hurley JH, Bonifacino JS. Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev Cell. 2010;18:425–436. doi: 10.1016/j.devcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choy RWY, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid beta (A beta) production in the trans-Golgi network. Proc Natl Acad Sci U S A. 2012;109:E2077–E2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 14.Vardarajan BN, Bruesegem SY, Harbour ME, St George-Hyslop P, Seaman MNJ, Farrer LA. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. 2012;33 doi: 10.1016/j.neurobiolaging.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, Arnim CAF, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor SorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid β production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S, Yu G. Sorting APP prevents harmful processing. Mol Interventions. 2006;6:74–76. doi: 10.1124/mi.6.2.4. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki H, Bujo H, Kusunoki J, Seimiya K, Kanaki T, Morisaki N, Schneider WJ, Saito Y. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem. 1996;271:24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
- 19.Jacobsen L, Madsen P, Moestrup SK, Lund AH, Tommerup N, Nykjaer A, Sottrup-Jensen L, Gliemann J, Petersen CM. Molecular characterization of a novel human hybrid-type receptor that binds the a2-macroglobulin receptor-associated protein. J Biol Chem. 1996;271:31379–31383. doi: 10.1074/jbc.271.49.31379. [DOI] [PubMed] [Google Scholar]
- 20.Jacobsen LJ, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem. 2001;276:222788–222796. doi: 10.1074/jbc.M100857200. [DOI] [PubMed] [Google Scholar]
- 21.Hermey G. The Vps10p-domain receptor family. Cell Mol Life Sci. 2009;66:2677–2689. doi: 10.1007/s00018-009-0043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors - regulators of neuronal viability and function. Nat Rev Neurosci. 2008;9:899–909. doi: 10.1038/nrn2516. [DOI] [PubMed] [Google Scholar]
- 23.Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- 24.Spoelgen R, Von Arnim CAF, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Anderson DE, Willnow TE, Hyman BT. Interaction of the cytosolic domains of SorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scherzer CR, Offe K, Gearing M, Rees HD, Fang GF, Heilman C, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 27.Sager KL, Wuu J, Herskowitz JH, Mufson EJ, Levey AI, Lah JJ. Neuronal LR11 Expression Does Not Differentiate between Clinically-Defined Alzheimer’s Disease and Control Brains. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt V, Baum K, Lao A, Rateitschak K, Schmitz Y, Teichmann A, Wiesner B, Petersen CM, Nykjaer A, Wolf J, Wolkenhauer O, Willnow TE. Quantitative modelling of amyloidogenic processing and its influence by SorLA in Alzheimer’s disease. EMBO J. 2012;31:187–200. doi: 10.1038/emboj.2011.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogaeva E, Meng Y, Lee JH, Gu YJ, Kawarai T, Zou FG, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen FS, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, George-Hyslop PS. The neuronal sortilin-related receptor SorL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;2:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen L, Madsen P, Nielsen MS, Geraerts WPM, Gliemann J, Smit AB, Petersen CM. The SorLA cytoplasmic domain interacts with GGA1 and -2 and defines minimum requirements for GGA binding. FEBS Lett. 2002;511:155–158. doi: 10.1016/s0014-5793(01)03299-9. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Sorting by the cytoplasmic domain of the amyloid precursoe protein binding receptor SorLA. Mol Cell Biol. 2007;27:6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgert T, Schmidt V, Caglayan S, Lin F, Fuchtbauer A, Fuchtbauer E, Nykjaer A, Carlo A, Willnow TE. SorLA-dependent and -independent functions for PACS1 in control of amyloidogenic processes. Mol Cell Biol. 2013;33:4308–4320. doi: 10.1128/MCB.00628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herskowitz JH, Offe K, Deshpande A, Kahn RA, Levey AI, Lah JJ. GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol Biol Cell. 2012;23:2645–2657. doi: 10.1091/mbc.E12-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herskowitz JH, Seyfried NT, Gearing M, Kahn RA, Peng J, Levey AI, Lah JJ. Rho kinase II phosphorylation of the lipoprotein receptor LR11/SorLA alters amyloid-β production. J Biol Chem. 2011;286:6117–6127. doi: 10.1074/jbc.M110.167239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohm C, Seibel NM, Henkel B, Steiner H, Haass C, Hampe W. SorLA signaling by regulated intramembrane proteolysis. J Biol Chem. 2006;281:14547–14533. doi: 10.1074/jbc.M601660200. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, RLG, Zhu Q, Tian F. Bacterial expression, purification, and model membrane reconstitution of the transmembrane and cytoplasmic domains of the human APP binding protein LR11/SorLA for NMR studies. Protein Expres Purif. 2011;77:224–230. doi: 10.1016/j.pep.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Meer G, de Kroon AIPM. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 38.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiba T, Takatsu H, Nogi T, Matsugaki N, Kawasaki M, Igarashi N, Suzuki M, Kato R, Earnest T, Nakayama K, Wakatsuki S. Structural basis for recognition of acid-cluster dileucine sequence by GGA1. Nature. 2002;415:937–941. doi: 10.1038/415937a. [DOI] [PubMed] [Google Scholar]
- 40.Fjorback AW, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, Willnow TE, Christensen EI, Mobley WB, Nykjaer A, Andersen OM. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci. 2012;32:1467–1480. doi: 10.1523/JNEUROSCI.2272-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madsen KL, Bhatia VK, Gether U, Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 42.Antonny B. Mechanisms of membrane curvature sensing. In: Kornberg RD, Raetz CRH, Rothman JE, Thorner JW, editors. Annu Rev Biochem. 2011. pp. 101–123. [DOI] [PubMed] [Google Scholar]
- 43.Derganc J, Antonny B, Copic A. Membrane bending: the power of protein imbalance. Trends Biochem Sci. 2013;38:576–584. doi: 10.1016/j.tibs.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Baumgart T, Capraro BR, Zhu C, Das SL. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu Rev Phys Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt NW, Wong GCL. Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr Opin Solid ST M. 2013;17:151–163. doi: 10.1016/j.cossms.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sc. 2006;7:539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 48.McMahon HT, Kozlov MM, Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 49.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Bio. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 51.Kweon DH, Shin YK, Shin JY, Lee JH, Lee JB, Seo JH, Kim YS. Membrane topology of helix 0 of the epsin N-terminal homology domain. Mol Cells. 2006;21:428–435. [PubMed] [Google Scholar]
- 52.Prinz WA, Hinshaw JE. Membrane-bending proteins. Crit Rev Biochem Mol Biol. 2009;44:278–291. doi: 10.1080/10409230903183472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossman JS, Jing XH, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Min C, Bang S, Cho B, Choi Y, Yang J, Lee S, Seong S, Kim K, Kim S, Jung JU, Choi M, Kim I, Cho N. Role of amphipathic helix of a Herpesviral protein in membrane deformation and T cell receptor downregulation. Plos Pathog. 2008;4 doi: 10.1371/journal.ppat.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Opalinski L, Kiel JAKW, Williams C, Veenhuis M, van der Klei IJ. Membrane curvature during peroxisome fission requires Pex11. EMBO J. 2011;30:5–16. doi: 10.1038/emboj.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taneva SG, Lee JMC, Cornell RB. The amphipathic helix of an enzyme that regulates phosphatidylcholine synthesis remodels membranes into highly curved nanotubules. Biochim Biophys Acta-Biomembr. 2012;1818:1173–1186. doi: 10.1016/j.bbamem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 58.Frolov VA, Zimmerberg J. Cooperative elastic stresses, the hydrophobic effect, and lipid tilt in membrane remodeling. FEBS Lett. 2010;584:1824–1829. doi: 10.1016/j.febslet.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itoh T, Takenawa T. Mechanisms of membrane deformation by lipid-binding domains. Prog Lipid Res. 2009;48:298–305. doi: 10.1016/j.plipres.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka M, Takamura Y, Kawakami T, Aimoto S, Saito H, Mukai T. Effect of amino acid distribution of amphipathic helical peptide derived from human apolipoprotein A-I on membrane curvature sensing. FEBS Lett. 2013;587:510–515. doi: 10.1016/j.febslet.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 62.Lundmark R, Doherty GJ, Vallis Y, Peter BJ, McMahon HT. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J. 2008;414:189–194. doi: 10.1042/BJ20081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krauss M, Jia JY, Roux A, Beck R, Wieland FT, De Camilli P, Haucke V. Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J Biol Chem. 2008;283:27717–27723. doi: 10.1074/jbc.M804528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beck R, Sun Z, Adolf F, Rutz C, Bassler J, Wild K, Sinning I, Hurt E, Brugger B, Bethune J, Wieland F. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc Natl Acad Sci U S A. 2008;105:11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.