Abstract
BACKGROUND AND OBJECTIVE
Periodontal disease is a highly complex chronic inflammatory disease of the oral cavity. Multiple factors influence periodontal disease including socioeconomic status, genetics, age, however, inflammation elicited by the presence of specific bacteria in the subgingival space is thought to drive the majority of soft and hard tissue destruction. Porphyromonas gingivalis is closely associated with periodontal disease. Toll-like receptors (TLRs) and their intracellular signaling pathways play roles in host responses to P. gingivalis. The focus of current study was to use microarray analysis to define the contributions that TLR adaptor molecules MyD88 and TRIF, and aging have on TLR pathway associated mRNA expression in response to P. gingivalis.
MATERIALS AND METHODS
Bone marrow derived macrophages (BMØ) from wild type (Wt), MyD88-KO and TrifLps2 mice at 2-months and 12-months of age were cultured with P. gingivalis. Expression of genes in BMØ cultured with P. gingivalis was determined in comparison to medium alone control.
RESULTS
Using a two-fold cut-off in mRNA expression criteria, differential expression of 32 genes was observed when Wt BMØ from 2-month old mice were cultured with P. gingivalis compared with medium alone control. When compared with 2-month old Wt, 21 and 12 genes were differentially expressed (P<0.05) as a result of MyD88 or TRIF mutations respectively. The expression of 5 genes was significantly (P<0.05) reduced in the 12-month group compared to the 2-month group in Wt BMØ following culture with P. gingivalis. Age also influenced expression of genes in MyD88-KO and TrifLps2 mice challenged with P. gingivalis.
CONCLUSION
Our results indicate that P. gingivalis induces differential expression of TLR pathway associated genes, and both MyD88, and TRIF play roles in the expression of these genes. Age also played a role in the expression of TLR-associated genes following stimulation of BMØ with P. gingivalis.
Keywords: MRNA expression, Innate immunity, Macrophage, MyD88, Porphyromonas gingivalis, TLRs, TRIF
INTRODUCTION
Periodontal disease is one of the most common among chronic infectious diseases in humans. This disease cause progressive loss of the hard and soft tissue supporting the teeth, and in severe cases leads to tooth loss [1]. Periodontal disease is caused by multiple factors including socioeconomic condition, genetic status, age and the presence and composition of bacteria in subgingival plaque is thought to play an important role in the destruction of hard periodontal tissue. Subgingival plaque is a complex biofilm that harbors over 400 distinct organisms [2], yet despite this complexity, only a relatively few of these organisms are highly associated with clinical periodontal parameters [3]. Porphyromonas gingivalis is a Gram-negative anaerobic bacterium implicated as one of the primary periodontal pathogens [4]. This organism possesses an array of virulence factors including lipopolysaccharide, fimbriae, gingipains, capsular polysaccharide and others that are implicated in the pathogenesis of periodontal disease [5]. Clinical investigation has identified innate immune activation during periodontal disease. A complex influx of inflammatory cells including neutrophils and monocytes occurs in the periodontium [6,7], as well as elevations in levels of cytokines, chemokines, [8,9] and receptors including toll-like receptors (TLR) [10] are reported in disease. Microarray analysis has been used to investigate mRNA expression profiles in the context of experimental gingivitis [11], periodontal disease [12] and host responses to periodontal bacteria including P. gingivalis. Yu et al. [13] reported that P. gingivalis, as well as its FimA protein, and LPS modulate the expression of a large set of genes in macrophages that are involved in cell proliferation, differentiation and innate immune response.
The TLR family of innate immune receptors is comprised of at least 10 members in human and 12 in mouse [14–19]. These molecules provide innate immune recognition of conserved pathogen-associated molecular patterns, or PAMPS, including lipopeptides (TLR2), lipopolysaccharide (TLR4), CpG DNA (TLR9), and others [14], as well as modified endogenous host ligands [20]. Following ligand binding to a TLR receptor, a specific intracellular adaptor molecule either myeloid differentiation factor (MyD) 88, TRIF (Toll/IL-1 receptor domain-containing adaptor inducing IFN-β; the protein product of the lps2 gene), Tirap/Mal (TIR domain containing-adaptor protein), and or Tram (TRIF-related adaptor molecules) are recruited to the TLR TIR domain, to initiate intracellular signaling cascades that culminate in activation of innate immune response [21]. All TLRs with the exception of TLR3 signal through MyD88 [22,23]. TLR3 signals through TRIF, while TLR4 signaling occurs through both MyD88 and TRIF [24]. Expression of TLRs is associated with the progression of periodontal disease. A recent study reported higher levels of TLR2, TLR4 and TLR9 expression in the gingival tissue of periodontal patients compared with healthy controls, with levels of TLR2 and TLR9 closely associated with the presence of P. gingivalis in periodontal patients [25]. A Tlr4 gene polymorphism has been identified in human periodontal disease [26]; furthermore, elevated TLR2 and TLR4 expression in human periodontal tissues has been reported [10]. In agreement with clinical observations, experimental findings support the contribution of TLRs to the host inflammatory response to P. gingivalis and its antigens [27–33]. Previous in vivo studies have indicated that MyD88-dependent and -independent pathways play important roles in development of inflammation and clearance of P. gingivalis [34]. Recently we reported that the cytokine response of bone marrow derived macrophages (BMØ) to P. gingivalis in the presence of low-density lipoproteins (LDL) was dependent on MyD88 and to a lesser extent on TRIF [35]. A previous study by Zhang et al. reported that P. gingivalis induced cytokine production in pre-osteoclasts primarily dependent on MyD88 although TRIF had a minor role in this response [36]. Our previous studies indicated that the functional activation of IRF3 is required for the induction of BMØ TNF-α levels in response to P. gingivalis [37]. Further, the expression of IL-6, IL-8 and CXCL12 in human gingival fibroblasts and human periodontal ligament fibroblasts in response to lipopolysaccharide (LPS) from P. gingivalis was shown to be dependent on both MyD88 and TRIF [38]. Similarly, Hemagglutinin B, a virulence factor of P. gingivalis and known TLR4 ligand, signals through both MyD88 and TRIF pathways in T-cells and dendritic cells [39]. Taken together these studies suggest that both MyD88 and TRIF play important roles in the innate immune response to P. gingivalis.
Aging is associated with increased incidence of infections [40]. It is thought that immune senescence and/or immune dysregulation contributes to the failure to effectively control infection with advancing age [41,42]. Periodontal disease commonly presents in adults in the third to fourth decades of life and progresses with age [43]. At the clinical level, why adults show onset and progression of periodontal disease with advancing age is poorly understood. At the cellular level, shifts in host immune response as a result of age trend toward immune senescence. Liang et al. showed that P. gingivalis induces differential expression of several genes encoding macrophage innate immune receptors, as well as intracellular regulators as a result of age [44]. Moreover, BMØ from 2-year old C57BL-6 (Wt) mice produced significantly lower levels of TNF-α, and IL-6 compared with BMØ from 2-month old mice following P. gingivalis stimulation [45]. Taken together these data support the concept that aging contributes to the immunological response of the host to the periodontal pathogen P. gingivalis. However, little is known regarding the effect of age on the inflammatory pathway activated by periodontal pathogens including expression of TLR-pathway associated genes. Moreover, it is not clear what changes in TLR adapter molecule usage may contribute to shifts in inflammation associated with host response to periodontal pathogens such as P. gingivalis with advancing age. In the present study, we used microarray analysis to examine the expression profiles of TLR signaling pathway-associated genes in BMØ isolated from 2- and 12-month old Wt, MyD88-KO and TrifLps2 mice in response to P. gingivalis.
MATERIALS AND METHODS
Mice and generation of bone marrow macrophages (BMØ)
C57BL-6 wild type (Wt) mice were purchased from Jackson Laboratories (Bar Harbor, ME) at 6 weeks of age. MyD88-KO mice [46] were from Dr. Shizuo Akira (Osaka University, Osaka, Japan). TrifLps2 mice [47] (a point mutation in Lps2 gene rendering TRIF protein non-functional) were provided by Dr. Bruce Beutler (University of Texas Southwestern Medical Center, Dallas, TX). All studies were performed in accordance with Boston University IACUC approved protocols and animals received normal chow diet and water ad libitum. Groups of mice were used at 2- and 12-months of age. Bone marrow cells were harvested from femurs (N=1; one mouse per group per experiment and three experiments were performed), and were differentiated in vitro into macrophages (BMØ) in RPMI-1640 + 10% FBS supplemented with 20%-conditioned medium from L929 cells (American Type Culture Collection, cat# NCTC clone 929 {L cell, L-929, derivative of Strain L} (ATCC® CCL-1™) as a source of macrophage colony-stimulating factor (M-CSF) and 1% penicillin/streptomycin [35]. After 7 days, BMØ were collected, suspended in medium containing only FBS, without M-CSF-free cell culture medium and antibiotics and were added to wells of a 6-well tissue culture plates at 5×105 cells/mL 2h prior to P. gingivalis challenge.
P. gingivalis cultivation
P. gingivalis strain 381 (ATCC) was grown under anaerobic conditions on blood agar plates followed by cultivation in brain heart infusion (BHI) yeast extract broth as previously reported [48]. Broth grown P. gingivalis was washed with antibiotic-free RPMI-1640 + 10% FBS, adjusted to optical density of 1 at 660 nm (approximately 1×109 bacteria/mL), and were added to BMØ cultures at a multiplicity of infection (MOI) of 100. Gram staining was used to confirm purity of P. gingivalis cultures.
BMØ stimulation assays
BMØ (5×105 cells/mL, 10 mL were used in 100 mm tissue culture dishes) from Wt, MyD88-KO, and TrifLps2 mice (N=1; one mouse per group per experiment and three experiments were performed) were stimulated with fresh antibiotic-free cell culture medium alone, or with medium containing P. gingivalis MOI 100 for 24h. The cells were then washed with PBS and lysed in RNA lysis buffer. RNA lysates were stored at −80°C. RNA was purified from lysates using RNeasy spin columns (RNeasy kit, Qiagen, Valencia, CA). RNA purity and quantity was determined using a Nanodrop 1000 (Thermo Scientific, Lafayette, CO) and was stored at −80°C until microarray assays were performed.
Microarray analysis
Microarray analysis was performed on RNA samples using mouse TLR pathway-specific microarray plates (SABiosciences, Cat# PAMM-018A, Frederick, MD) per manufacturer's instructions on the AB 7000 Q-PCR instrument (Applied Biosystems, Foster City, CA). Relative mRNA expression was determined using actin beta (Actb) as a housekeeping gene and fold change in mRNA expression in response to P. gingivalis was determined relative to expression in cells following treatment with media alone as a control using the ΔΔCt method (SABiosciences, Frederick, MD). Heat map hierarchical clustering of the expression of TLR pathway gene mRNAs were generated using fold expression data from each experiment using Qiagen RT2 Profiler PCR Array Data Analysis version 3.5 online (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Data are presented as mean value of 3 independent experiments using individual mice.
Quantitative real-time PCR validation of microarray data
To validate microarray results, quantitative real-time PCR assays were performed using Taqman assays (Life Technologies, Grand Island, NY) on a subset of genes that included Ifnβ1 (assay ID# Mm00439546_s1), Tlr8 (assay ID# Mm01157262_m1), Clec4e (assay ID# Mm00490873_m1), Ly86 (assay ID# Mm00440240_m1) and Casp8 (assay ID# Mm00802247_m1) genes. Complementary DNA (cDNA) was prepared from RNA using RNA-to-cDNA kit (Cat# 4387406, Life Technologies, Grand Island, NY) and the quantitative real-time PCR assays were performed on a StepOne real-time PCR system (Life Technologies). Fold expression was calculated using actin beta (Actb; assay ID# Mm00607939_s1), and fold change of mRNA expression in BMØ to P. gingivalis challenge compared with medium alone control was determined using the ΔΔCt method [49].
Statistical Analysis
Experiments were performed at three separate occasions with BMØ from individual mice (N=1; one mouse per group per experiment and three experiments were performed). Statistical comparisons between two groups were performed using the Student's t-test. A P<0.05 was considered significant.
RESULTS
Establishment of Actb as the housekeeping control
The microarrays consisted of 84 TLR pathway associated genes. These genes were divided into groups based on function as follows: TLRs, TLR accessory molecules, intracellular signaling, and secreted inflammatory mediators. In addition, microarrays included 7 internal assay controls, and 5 housekeeping genes; glucoronidase beta (Gusb), hypoxathine guanine phosphoribosyl transferase 1 (Hprt 1), heat shock protein 90 KDa alpha (cytosolic), class B member 1 (Hsp90ab1), glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and actin beta, cytoplasmic (Actb). Scale normalization analysis [50] of the microarray data indicated that the expression of Actb gene in BMØ was least influenced by P. gingivalis challenge than the other represented housekeeping genes. Therefore Actb was used as the housekeeping gene in this study.
Expression of TLR pathway associated genes in response to P. gingivalis
Employing a 2-fold change in mRNA expression from unchallenged controls as the cut-off, a set of 32 TLR pathway associated genes were differentially expressed in BMØ obtained from Wt mice at 2-months of age in response to P. gingivalis stimulation (Table 1 and S1). The expression of Tlr1 and Tlr2 genes was increased while that of Tlr5, Tlr8, and Tlr9 genes was reduced in response to P. gingivalis. The change in expression of Tlr3, Tlr4, Tlr6 and Tlr7 genes following P. gingivalis stimulation did not meet the 2-fold cut-off criteria (Table 1 and S1). Examining the expression of TLR accessory molecules, we observed an increase in expression of Cd14, while Ly86 (Md1) expression was reduced in Wt BMØ in response to P. gingivalis (Table 1 and Table S1). Expression of the Ly96 (Md2) gene did not reach 2-fold cut off. There were no differences in the expression of the TLR adaptor molecules, MyD88, Ticam-1 (Trif), Ticam-2 (Tram) and Tirap between the un-stimulated control and P. gingivalis stimulated groups (Table 1 and Table S1). The expression of cytokine and chemokine genes including colony stimulating factor (Csf)2, Csf3, interferon beta (Ifnb)1, interleukin (Il)1a, Il1b, Il6, Il10, Il12a, tumor necrosis factor (Tnf), and prostaglandin-endoperoxide synthase (Ptgs)2 was increased in response to P. gingivalis (Table 1). Taken collectively, these results indicate that P. gingivalis induces the expression of a large set of genes in BMØ including TLRs, TLR accessory molecules, intracellular signaling and secreted inflammatory mediator genes.
Table 1.
Microarray profile of expression of TLR pathway associated gene mRNAs in BMxØ from Wt mice of 2- and 12- months of age in response to P. gingivalis.
| Gene symbol | Gene | Fold change (Mean) | |
|---|---|---|---|
| 2M | 12M | ||
| Il6 | Interleukin 6 | 1298.64 | 2231.90 |
| Csf3 | Colony stimulating factor 3 (granulocyte) | 836.98 | 2713.51 |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 | 812.55 | 244.92 |
| Il1a | Interleukin 1 alpha | 746.41 | 2525.22 |
| Il1b | Interleukin 1 beta | 736.05 | 4344.65 |
| Clec4e | C-type lectin domain family 4, member e | 72.48 | 53.09 |
| Il12a | Interleukin 12A | 61.84 | 42.55 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 48.50 | 40.22 |
| Il10 | Interleukin 10 | 33.68 | 10.49 |
| Il1r1 | Interleukin 1 receptor, type I | 21.99 | 65.04 |
| Tnf | Tumor necrosis factor | 16.69 | 8.63 |
| Lta | Lymphotoxin A | 14.82 | 5.84 |
| Csf2 | Colony stimulating factor 2 (granulocyte-macrophage) | 10.46 | 18.90 |
| Ifnb1 | Interferon beta 1, fibroblast | 8.98 | 1.81* |
| Ifng | Interferon gamma | 8.45 | 1.20* |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 8.04 | 194.58 |
| Nfkbia | Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | 6.38 | 3.30 |
| Tnfaip3 | Tumor necrosis factor, alpha-induced protein 3 | 6.33 | 5.94 |
| Cd14 | CD14 antigen | 5.25 | 19.73 |
| Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 4.43 | 1.93* |
| Ube2v1 | Ubiquitin-conjugating enzyme E2 variant 1 | 3.92 | 0.67 |
| Tlr2 | Toll-like receptor 2 | 3.04 | 2.07 |
| Tlr1 | Toll-like receptor 1 | 2.84 | 1.78* |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 2.77 | 2.09 |
| Hspa1a | Heat shock protein 1A | 2.43 | 0.96* |
| Peli1 | Pellino 1 | 2.14 | 0.77* |
| Ticam1 | Toll-like receptor adaptor molecule 1 | 1.74* | 5.19 |
| Eif2ak2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 1.34* | 0.39 |
| Map2k4 | Mitogen activated protein kinase kinase 4 | 1.32* | 0.50 |
| Il2 | Interleukin 2 | 1.18* | 2.80 |
| Tlr3 | Toll-like receptor 3 | 1.09* | 0.46 |
| Ppara | Peroxisome proliferator activated receptor alpha | 1.04* | 0.45 |
| Map3k7 | Mitogen activated protein kinase kinase kinase 7 | 0.96* | 0.44 |
| Rel | Reticuloendotheliosis oncogene | 0.91* | 41.56 |
| Irf1 | Interferon regulatory factor 1 | 0.90* | 0.50 |
| Hmgb1 | High mobility group box 1 | 0.89* | 0.40 |
| Ube2n | Ubiquitin-conjugating enzyme E2N | 0.89* | 0.42 |
| Btk | Bruton agammaglobulinemia tyrosine kinase | 0.87* | 0.43 |
| Irak1 | Interleukin-1 receptor-associated kinase 1 | 0.86* | 0.37 |
| Fadd | Fas (TNFRSF6)-associated via death domain | 0.81* | 0.31 |
| Map3k1 | Mitogen activated protein kinase kinase kinase 1 | 0.81* | 0.28 |
| Tlr4 | Toll-like receptor 4 | 0.75* | 0.38 |
| Casp8 | Caspase 8 | 0.71* | 0.45 |
| Map2k3 | Mitogen activated protein kinase kinase 3 | 0.71* | 0.37 |
| Nr2c2 | Nuclear receptor subfamily 2, group C, member 2 | 0.71* | 0.41 |
| Tlr7 | Toll-like receptor 7 | 0.67* | 0.42 |
| Hprt1 | Hypoxanthine guanine phosphoribosyl transferase 1 | 0.63* | 0.36 |
| Gusb | Glucuronidase, beta | 0.61* | 0.48 |
| Fos | FBJ osteosarcoma oncogene | 0.57* | 0.26 |
| Il6ra | Interleukin 6 receptor, alpha | 0.45 | 57.23 |
| Pglyrp1 | Peptidoglycan recognition protein 1 | 0.38 | 0.40 |
| Tlr9 | Toll-like receptor 9 | 0.36 | 0.13 |
| Tlr8 | Toll-like receptor 8 | 0.34 | 0.23 |
| Ly86 | Lymphocyte antigen 86 | 0.22 | 0.15 |
| Tlr5 | Toll-like receptor 5 | 0.07 | 0.10 |
BMØ from Wt mice of 2-months (2M) and 12-months (12M) of age were stimulated with P. gingivalis (MOI 100) for 24h. mRNA expression was normalized to Actb expression levels. Mean fold change in the expression in response to P. gingivalis stimulation in comparison to un-stimulated control was determined by ΔΔCt method. Data presented as mean value of 3 independent experiments using individual mice.
= mRNA expression did not meet the 2-fold criteria.
The effect of age on the expression of TLR pathway associated genes in response to P. gingivalis in Wt BMØ
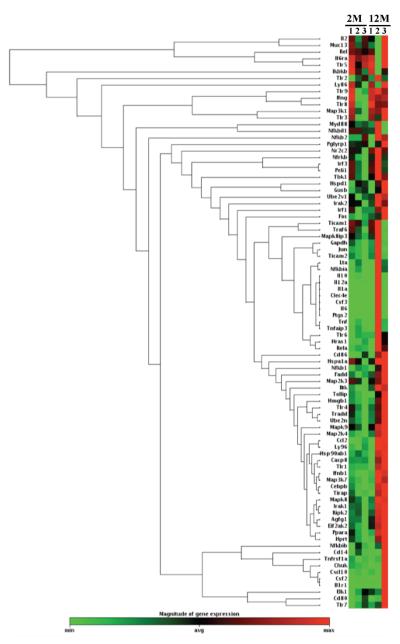
We compared mRNA expression profiles of TLR pathway associated genes in BMØ isolated from 2- and 12-month old Wt mice in response to P. gingivalis. We observed an overall trend of reduced mRNA expression in the 12-month old group; however, increased expression of a set of genes in response to P. gingivalis stimulation in 12-months group compared with 2-months group was observed (Fig 1; Table 1 and S1). These genes include Il1a, Il1b, Il6, Ccl2, Ccl3, and Cxcl10. Compared with the response of 2-month old mice, the expression of Il10, Il12a, Tnf, Ifnb, Ifng and Ptgs2 genes tended to be reduced in BMØ from 12-month old Wt but did not reach statistical significance (Table 1). Reduced expression of intra-cellular signaling genes including mitogen activated protein kinases Map2k3, Map2k4 and Map3k7 was observed with age compared to 2-months group. In addition, P. gingivalis stimulation showed a trend of reduction in the expression of 5 genes: Bruton's tyrosine kinase (Btk), eukaryotic translation inhibition factor 2-alpha kinase 2 (Eif2ak2), Map2k4, HIV rev binding protein (Hrb) gene and Fas (Tnfrsf6) associated via death domain (Fadd) genes between the two age groups, although expression levels did not meet the 2-fold cut-off criteria (Table S1).
Fig. 1. Heat map hierarchical clustering of BMØ TLR pathway associated gene mRNA expression from Wt mice with 2- and 12-months of age in response to P. gingivalis.
Heat map hierarchical clustering of mRNA expression of TLR pathway associated genes. BMØ from Wt mice of 2- and 12-months of age were stimulated with P. gingivalis (MOI 100) for 24h. Expression of genes was normalized using Actb. Mean fold change in the mRNA expression to P. gingivalis stimulation in comparison with un-stimulated control was determined by the ΔΔCt method. Heat map hierarchical clustering was generated using Qiagen RT2 Profiler PCR Array Data Analysis version 3.5 online at http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php. Data presented as mean 679 value of 3 independent experiments using individual mice.
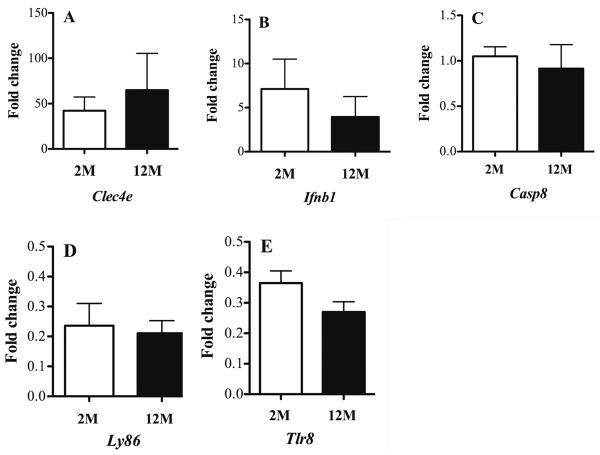
We validated the microarray findings of a subset of genes using Taqman mRNA expression assays in RNA used for microarray. Quantitative real-time PCR data was in agreement with microarray results confirming increased expression of the Clec4e, Ifnb1 genes, decreased expression of Ly86 (Md1) and Tlr8 genes, and no changes in expression of Casp8 in response to P. gingivalis (Table 1 and Fig 2). Validation data were also in agreement with microarray data finding no effect of age on the expression of these genes (Fig 2; all P>0.05).
Fig. 2. Quantitative real-time PCR verification of microarray mRNA expression of Wt BMØ to P. gingivalis.
BMØ from C57BL-6 Wt mice were cultured for 24h in medium alone, or with P. gingivalis MOI 100. Quantitative real-time PCR was performed on cDNA generated from RNA employed in microarrays using gene specific Taqman assays. Genes with increased expression A- Clec4e and B- Ifnb1, gene with no- change in the expression C- Casp8, and genes with decreased expression D- Ly86 and E- Tlr8. Expression of target gene was normalized to that of actin beta (Actb) gene. Mean fold change in expression in response to P. gingivalis stimulation over control was determined by ΔΔCt method. Open bars = 2-months of age (2M); filled bars = 12- months of age (12M). N = 1; one mouse per group per experiment and 3 experiments were performed. Data are presented as mean ± SEM of three independent experiments. Comparisons between groups were performed using Student's t-test. No significant differences between age groups were observed (P>0.05).
Role of MyD88 on the expression of TLR pathway associated genes in response to P. gingivalis
Microarray analysis identified a set of 32 TLR pathway associated genes that were differentially expressed in BMØ from 2-month old MyD88-KO mice compared to Wt mice. Based on the 2-fold expression cut-off criteria, we observed reduced mRNA expression profiles in BMØ from MyD88-KO mice compared with Wt in response to P. gingivalis stimulation. Genes with reduced expression included Csf2, Csf3, Ifng, Il1a, Il1b, Il6, Ptgs2, and Tnf (Table 2). Employing statistical significance rather than the 2-fold cut-off, the expression of 21 genes was significantly different between BMØ from Wt and MyD88-KO mice following P. gingivalis stimulation (Table 3; P<0.05). In the 2- month group, expression of seven genes met the 2-fold cut-off based on Wt expression, and had significant differences between Wt and MyD88 and include Cebpb, Csf2, Csf3, Cxcl10, Il1r1, Tlr1, and Tlr5 genes. Expression of Tlr1 and Tlr6 genes were reduced in BMØ from MyD88-KO compared to Wt, although they did not meet the 2-fold cut-off (Table 3). A trend for reduced expression of Tlr2 gene was observed as a result of MyD88 oblation, but did not reach significance (Table 2). Expression of Tlr5 was reduced in BMØ from both Wt and MyD88-KO mice but the level of reduction was significantly less in BMØ from MyD88-KO mice compared to Wt (Table 3). The expression of genes encoding the transcriptional factors, interferon regulatory factor (Irf)1, and nuclear factor of kappa light chain gene enhancer in B-cells 1 (Nfkb1), were significantly reduced in the MyD88-KO group (Table 3 and S2). Expression of Csf2, and Csf3 was reduced significantly in the MyD88-KO group compared with Wt (Table 3). A trend for a similar pattern was observed with the expression of Tnf, Il6, Ptsg2, and Ifng although they did not reach significance (Table 2 and Table S3). We observed an increase in the levels of chemokine Cxcl10 and Ifnb1 expression in MyD88-KO BMØ compared with Wt (Table 2).
Table 2.
Microarray profile of expression of TLR pathway associated gene mRNAs in BMØ from Wt, MyD88-KO and TrifLps2 mice of 2-months of age in response to P. gingivalis.
| Gene symbol | Gene | Fold change (Mean) | ||
|---|---|---|---|---|
| Wt | MyD88-KO | TrifLps2 | ||
| Il6 | Interleukin 6 | 1298.64 | 3.26 | 836.68 |
| Csf3 | Colony stimulating factor 3 (granulocyte) | 836.98 | 1.08 | 724.50 |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 | 812.55 | 9.67 | 381.26 |
| Il1a | Interleukin 1 alpha | 746.41 | 6.25 | 687.91 |
| Il1b | Interleukin 1 beta | 736.05 | 0.69 | 654.97 |
| Clec4e | C-type lectin domain family 4, member e | 72.48 | 14.16 | 49.64 |
| Il12a | Interleukin 12A | 61.84 | 0.69 | 53.47 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 48.50 | 6.31 | 45.73 |
| Il10 | Interleukin 10 | 33.68 | 0.47 | 6.91 |
| Il1r1 | Interleukin 1 receptor, type I | 21.99 | 0.96 | 12.24 |
| Tnf | Tumor necrosis factor | 16.69 | 4.58 | 10.58 |
| Lta | Lymphotoxin A | 14.82 | 1.48 | 8.14 |
| Csf2 | Colony stimulating factor 2 (granulocyte-macrophage) | 10.46 | 0.69 | 3.43 |
| Ifnb1 | Interferon beta 1, fibroblast | 8.98 | 17.07 | 4.26 |
| Ifng | Interferon gamma | 8.45 | 0.69 | 0.68 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 8.04 | 19.41 | 2.45 |
| Nfkbia | Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | 6.38 | 1.91 | 3.98 |
| Tnfaip3 | Tumor necrosis factor, alpha-induced protein 3 | 6.33 | 3.60 | 4.90 |
| Cd14 | CD14 antigen | 5.25 | 1.21 | 4.98 |
| Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 4.43 | 1.20 | 2.63 |
| Ube2v1 | Ubiquitin-conjugating enzyme E2 variant 1 | 3.92 | 0.90 | 0.88 |
| Tlr2 | Toll-like receptor 2 | 3.04 | 2.04 | 3.17 |
| Tlr1 | Toll-like receptor 1 | 2.84 | 0.97 | 2.33 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 2.77 | 7.12 | 2.38 |
| Hspa1a | Heat shock protein 1A | 2.44 | 2.48 | 0.98 |
| Peli1 | Pellino 1 | 2.14 | 1.30 | 0.68 |
| Il6ra | Interleukin 6 receptor, alpha | 0.45 | 0.41 | 0.27 |
| Pglyrp1 | Peptidoglycan recognition protein 1 | 0.38 | 0.83 | 0.46 |
| Tlr9 | Toll-like receptor 9 | 0.36 | 0.48 | 0.17 |
| Tlr8 | Toll-like receptor 8 | 0.34 | 0.50 | 0.29 |
| Ly86 | Lymphocyte antigen 86 | 0.22 | 0.38 | 0.08 |
| Tlr5 | Toll-like receptor 5 | 0.07 | 0.28 | 0.12 |
BMØ from Wt, MyD88-KO, and TrifLps2 mice of 2-months of age were stimulated with P. gingivalis (MOI 100) for 24h. mRNA expression was normalized to Actb mRNA expression levels. Mean fold change in the mRNA expression in response to P. gingivalis stimulation in comparison with un-stimulated control was determined by ΔΔCt method. Data presented as mean value of 3 independent experiments using individual mice.
Table 3.
Expression of TLR pathway associated gene mRNAs in BMØ from Wt and MyD88-KO mice with 2-months of age to P. gingivalis.
| Gene symbol | Gene | Fold change (Mean) | P value# | |
|---|---|---|---|---|
| Wt | MyD88-KO | |||
| Csf3 | Colony stimulating factor 3 (granulocyte) | 837 | 1.08 | 0.01 |
| Il1r1 | Interleukin 1 receptor, type I | 21.99 | 0.96 | 0.002 |
| Csf2 | Colony stimulating factor 2 (granulocyte-macrophage) | 10.45 | 0.69 | 0.04 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 8.03 | 19.41 | 0.03 |
| Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 4.42 | 1.19 | 0.03 |
| Tlr1 | Toll-like receptor 1 | 2.83 | 0.96 | 0.0001 |
| Tlr6 | Toll-like receptor 6 | 1.95* | 0.93 | 0.01 |
| Hrb | HIV-1 Rev binding protein | 1.93* | 1 | 0.02 |
| Hras1 | Harvey rat sarcoma virus oncogene 1 | 1.9* | 1.19 | 0.02 |
| Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | 1.49* | 0.93 | 0.02 |
| Hsp90ab1 | Heat shock protein 90 alpha (cytosolic), class B member 1 | 1.46* | 1.03 | 0.04 |
| Ly96 | Lymphocyte antigen 96 | 1.41* | 0.72 | 0.008 |
| Nfkb1 | Nuclear factor of kappa light chain gene enhancer in B-cells | 1.38* | 0.77 | 0.01 |
| Map2k4 | Mitogen activated protein kinase kinase 4 | 1.32* | 0.9 | 0.01 |
| Myd88 | Myeloid differentiation primary response gene 88 | 1.31* | 0.7 | 0.01 |
| Chuk | Conserved helix-loop-helix ubiquitous kinase | 1.26* | 0.97 | 0.02 |
| Ticam2 | Toll-like receptor adaptor molecule 2 | 1.18* | 0.76 | 0.01 |
| Irf1 | Interferon regulatory factor 1 | 0.9* | 1.66 | 0.04 |
| Casp8 | Caspase 8 | 0.7* | 0.78 | 0.008 |
| Fos | FBJ osteosarcoma oncogene | 0.57* | 1.12 | 0.002 |
| Tlr5 | Toll-like receptor 5 | 0.06* | 0.27 | 0.004 |
BMØ from Wt and MyD88-KO mice of 2-months of age were stimulated with P. gingivalis (MOI 100) for 24h. Expression was normalized to Actb mRNA expression levels. Mean fold change in the mRNA expression to P. gingivalis stimulation in comparison with un-stimulated control was determined by ΔΔCt method. Comparisons between groups were performed using Student T-test. Data presented as mean value of 3 independent experiments using individual mice.
= mRNA expression did not meet the 2-fold criteria.
= P<0.05; fold change in the mRNA expression compared between Wt and MyD88-KO groups.
We examined the effect of age on the MyD88-dependent expression of TLR pathway genes in response to P. gingivalis stimulation. Overall, the trend observed in the 12-month group was a reduction in the expression of inflammatory mediator genes including cytokines and chemokines while expression of intracellular signaling molecules including Map2k3, Map2k4, Map3k1 and Map3k7 was increased in MyD88-KO mice compared to Wt (Table 4 and S3). We observed significant differences in the expression of 12 genes in BMØ from 12-month old MyD88-KO mice compared to Wt to P. gingivalis stimulation (Table S3). Expression of Tlr3, 4, 8 and 9 genes were significantly increased (P<0.05) in BMØ from MyD88-KO compared with Wt, although the expression levels did not meet 2-fold cut-off criteria (Table S3). The expression of Map2k3 and Map3k1 genes were significantly (P<0.05) increased in BMØ from MyD88-KO mice compared with Wt (Table S3). In addition expression of the Ifng gene was significantly reduced in MyD88-KO mice compared with Wt, however age did not affect the expression of Ifng gene as similar levels were observed between 2- and 12-months of age (Table S3). In contrast, the expression of Ifnb1 was increased in BMØ from MyD88-KO mice compared with Wt in response to P. gingivalis and its expression was independent of age (Table 2 and S3). Comparing mRNA expression profiles of BMØ from 2-month and 12-month old MyD88-KO mice identified 18 genes that were differentially expressed as a result of age (Table S4). Among the TLR genes, expression of Tlr1, Tlr4 and Tlr6 were significantly increased with age compared with 2-month group. In contrast Tlr5 mRNA expression was significantly reduced in the 12-months group compared with the 2-months group, although different, changes in expression levels were below 2-fold cut-off criteria (Table S4). In the case of inflammatory mediator genes, the expression of Ifng, Il10 were increased while Ptgs2 mRNA expression was reduced as a result of age (Table S4).
Table 4.
Microarray profiling of expression of TLR pathway associated gene mRNAs in BMØ from Wt, MyD88-O and TrifLps2 mice of 12- months of age in response to P. gingivalis.
| Gene symbol | Gene | Fold change (Mean) | ||
|---|---|---|---|---|
| Wt | MyD88-KO | TrifLPS2 | ||
| Il1b | Interleukin 1 beta | 4344.65 | 1.01 | 3299.31 |
| Csf3 | Colony stimulating factor 3 (granulocyte) | 2713.51 | 0.47 | 2951.98 |
| Il1a | Interleukin 1 alpha | 2525.22 | 8.57 | 4134.11 |
| Il6 | Interleukin 6 | 2231.90 | 5.45 | 2888.33 |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 | 244.92 | 1.63 | 128.69 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 194.58 | 48.95 | 1.64 |
| Il1r1 | Interleukin 1 receptor, type I | 65.04 | 0.55 | 5.48 |
| Il6ra | Interleukin 6 receptor, alpha | 57.23 | 0.49 | 0.27 |
| Clec4e | C-type lectin domain family 4, member e | 53.09 | 23.63 | 113.89 |
| Il12a | Interleukin 12A | 42.55 | 0.85 | 79.49 |
| Rel | Reticuloendotheliosis oncogene | 41.56 | 1.12 | 0.88 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 40.22 | 5.27 | 58.74 |
| Cd14 | CD14 antigen | 19.73 | 0.79 | 4.41 |
| Csf2 | Colony stimulating factor 2 (granulocyte-macrophage) | 18.90 | 1.18 | 36.77 |
| Il10 | Interleukin 10 | 10.49 | 0.81 | 17.24 |
| Tnf | Tumor necrosis factor | 8.63 | 3.77 | 26.04 |
| Tnfaip3 | Tumor necrosis factor, alpha-induced protein 3 | 5.94 | 3.68 | 8.41 |
| Lta | Lymphotoxin A | 5.84 | 1.67 | 11.00 |
| Ticam1 | Toll-like receptor adaptor molecule 1 | 5.19 | 1.03 | 1.15 |
| Nfkbia | Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | 3.30 | 2.03 | 6.31 |
| Il2 | Interleukin 2 | 2.80 | 0.85 | 0.89 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 2.09 | 1.48 | 1.93 |
| Tlr2 | Toll-like receptor 2 | 2.07 | 1.92 | 2.58 |
| Irf1 | Interferon regulatory factor 1 | 0.50 | 1.72 | 0.80 |
| Map2k4 | Mitogen activated protein kinase kinase 4 | 0.50 | 0.94 | 1.05 |
| Gusb | Glucuronidase, beta | 0.48 | 1.18 | 0.90 |
| Tlr3 | Toll-like receptor 3 | 0.46 | 1.92 | 0.38 |
| Casp8 | Caspase 8 | 0.45 | 0.95 | 0.82 |
| Ppara | Peroxisome proliferator activated receptor alpha | 0.45 | 0.85 | 0.89 |
| Map3k7 | Mitogen activated protein kinase kinase kinase 7 | 0.44 | 0.93 | 1.00 |
| Btk | Bruton agammaglobulinemia tyrosine kinase | 0.43 | 0.84 | 0.71 |
| Tlr7 | Toll-like receptor 7 | 0.42 | 0.70 | 0.60 |
| Ube2n | Ubiquitin-conjugating enzyme E2N | 0.42 | 0.66 | 0.73 |
| Nr2c2 | Nuclear receptor subfamily 2, group C, member 2 | 0.41 | 0.45 | 0.51 |
| Pglyrp1 | Peptidoglycan recognition protein 1 | 0.40 | 0.76 | 0.73 |
| Hmgb1 | High mobility group box 1 | 0.40 | 0.68 | 0.77 |
| Eif2ak2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 0.39 | 1.63 | 0.60 |
| Tlr4 | Toll-like receptor 4 | 0.38 | 1.24 | 0.76 |
| Irak1 | Interleukin-1 receptor-associated kinase 1 | 0.37 | 1.01 | 0.79 |
| Map2k3 | Mitogen activated protein kinase kinase 3 | 0.37 | 0.73 | 0.49 |
| Hprt1 | Hypoxanthine guanine phosphoribosyl transferase 1 | 0.36 | 0.66 | 0.70 |
| Fadd | Fas (TNFRSF6)-associated via death domain | 0.31 | 1.16 | 0.75 |
| Map3k1 | Mitogen activated protein kinase kinase kinase 1 | 0.28 | 0.73 | 0.40 |
| Fos | FBJ osteosarcoma oncogene | 0.26 | 0.81 | 0.24 |
| Tlr8 | Toll-like receptor 8 | 0.23 | 0.50 | 0.20 |
| Ly86 | Lymphocyte antigen 86 | 0.15 | 0.35 | 0.05 |
| Tlr9 | Toll-like receptor 9 | 0.13 | 0.51 | 0.10 |
| Tlr5 | Toll-like receptor 5 | 0.10 | 0.11 | 0.02 |
BMØ from Wt, MyD88-KO and TrifLps2 mice of 12-months of age were stimulated with P. gingivalis (MOI 100) for 24h. Expression was normalized to Actb mRNA expression levels. Mean fold change in the mRNA expression in response to P. gingivalis stimulation in comparison with un-stimulated control was determined by ΔΔCt method. Data presented as mean value of 3 independent experiments using individual mice.
Role of TRIF on the expression of TLR pathway associated genes in response to P. gingivalis stimulation
Using the 2-fold expression cut-off, microarray indicated a trend for reduction in the expression of 32 TLR pathway associated genes in BMØ from 2-month old TrifLps2 mice compared with Wt in response to P. gingivalis stimulation (Table 2). In addition, a total of 12 genes were differentially expressed in BMØ from TrifLps2 mice compared with Wt in response to P. gingivalis although expression levels were less than 2-fold (Table 5). The expression of Il1r, and Tlr1 genes met the 2-fold cut-off based on Wt expression, and the expression was significantly reduced in BMØ from TrifLps2 mice compared with Wt (Table 2 and 5). Among these genes, only Tlr1 and Tlr6 were significantly reduced as a result of Trif mutation (Table 5). No difference in the expression of Tlr2 and Tlr4 genes was observed between Wt and TrifLps2 BMØ (Table S5). In the case of TLR adaptor protein genes, expression of MyD88 was slightly increased, and expression levels of Tirap and Ticam1 (Trif) were reduced in response to P. gingivalis in the absence of functional TRIF (Table S5). No difference in expression of Ticam2 (Tram) was observed in BMØ between Wt and TrifLps2 mice (Table S5). Expression levels of Cxcl10 gene were reduced as a result of the Trif mutation compared to Wt (Table 2). Expression of Map2k4, as well as Il1r1 and Tnfarsf1a were decreased significantly as a result of Trif mutation (Table 5).
Table 5.
Expression of TLR pathway associated gene mRNAs in BMØ from Wt and TrifLps2 mice of 2-months of age to P. gingivalis
| Gene symbol | Gene | Fold change (Mean) | P value# | |
|---|---|---|---|---|
| Wt | TrifLps2 | |||
| Il1r1 | Interleukin 1 receptor, type I | 21.99 | 12.23 | 0.049 |
| Tlr1 | Toll-like receptor 1 | 2.84 | 2.33 | 0.014 |
| Tlr6 | Toll-like receptor 6 | 1.96* | 1.27 | 0.044 |
| Hras1 | Harvey rat sarcoma virus oncogene 1 | 1.9* | 1.34 | 0.033 |
| Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | 1.49* | 0.9 | 0.029 |
| Ly96 | Lymphocyte antigen 96 | 1.41* | 0.86 | 0.03 |
| Nfkb1 | Nuclear factor of kappa light chain gene enhancer in B-cells 1, p105 | 1.39* | 0.88 | 0.035 |
| Map2k4 | Mitogen activated protein kinase kinase 4 | 1.32* | 0.99 | 0.033 |
| Chuk | Conserved helix-loop-helix ubiquitous kinase | 1.27* | 0.91 | 0.034 |
| Rela | V-rel reticuloendotheliosis viral oncogene homolog A (avian) | 0.98* | 0.69 | 0.007 |
| Tirap | Toll-interleukin 1 receptor (TIR) domain-containing adaptor protein | 0.98* | 0.55 | 0.034 |
| Hmgb1 | High mobility group box 1 | 0.89* | 0.64 | 0.042 |
BMØ from Wt and TrifLps2 mice of 2-months of age were stimulated with P. gingivalis (MOI 100) for 24h. Expression of genes was normalized using Actb. Mean fold change in the mRNA expression to P. gingivalis stimulation in comparison with un-stimulated control was determined by ΔΔCt method. Comparisons between groups were performed using Student t-test. Data presented as mean value of 3 independent experiments using individual mice.
= mRNA expression did not meet the 2-fold criteria.
= P<0.05; fold change in the mRNA expression compared between Wt and TrifLps2 groups.
Focusing on the influence of age on the expression of TLR pathway associated genes as a result of the Trif mutation, we did not observe major changes in expression in BMØ between Wt and TrifLps2 mice to P. gingivalis stimulation (Table 4 and S6). However, there was a change in mRNA expression due to aging between macrophages isolated from 2- and 12-month old TrifLps2 mice. A total of 29 genes had altered expression in response to P. gingivalis. Among these genes, the expression of Tlr1, 4 and 6 was increased, while the expression of Tlr3 decreased in BMØ from the 12-months group compared with 2-months group (Table S7). Expression of interleukin-1 receptor-associated kinase (Irak)1, Irak2, Nfkb1, Nfkb2, Nfkb inhibitor (Nfkbi)a, Nfkbib were all significantly increased in the 12-months group. Similarly the expression of Il1a, Il1b, Il6 and Il10 genes increased with age as a result of Trif mutation (Table S7).
DISCUSSION
In this study, we used murine BMØ and microarray analysis to define the roles of two major TLR adaptor molecules, MyD88 and TRIF, and the effect of age on the expression of TLR pathway associated genes in response to P. gingivalis stimulation. In this study we performed P. gingivalis stimulation for 24h. A total of 32 genes were differentially expressed at least 2-fold in BMØ from Wt mice following P. gingivalis stimulation compared with unchallenged cells. Previous studies have begun to define the mRNA expression profiles of BMØ from either young, or young and old Wt mouse strains to P. gingivalis or its antigens [13,44]. In the context of BMØ from Wt mice of 2-months of age, we observed increased expression of Tlr1 and Tlr2, while expression of Tlr5, Tlr8 and Tlr9 were decreased in response to P. gingivalis. Our results with regard to the expression of Tlr 1, 2 and 5 are in broad agreement with a recent study by Liang et al. [44]. No difference in the expression of Tlr4 in response to P. gingivalis was observed and it is in agreement with a recent study reporting no differences in the expression of Tlr4 gene in response to P. gingivalis exposure [13]. However, our findings are in contrast to those reported by Liang et al. in which reduced expression of the Tlr4 gene was observed in BMØ following stimulation with P. gingivalis [44]. Differences in experimental design between these studies may account for the discrepancy. We employed BMØ from C57BL/6 mice, whereas Liang et al. used BMØ from BALB/c mice. In addition, the strain of P. gingivalis employed was different. Interestingly, similar findings of a strong reduction in Tlr5 mRNA expression were observed in our study and the study by Liang et al. [44] despite differences in mouse and P. gingivalis strains. TLR5 recognizes flagella from both Gram-positive and -negative bacteria and stimulates the production of pro-inflammatory cytokines including TNFα, through signaling pathways mediated by the TLR adaptor protein, MyD88 [51,52]. Recently, Feng et al. [53] reported reduced expression of Tlr5 in mouse bone marrow derived dendritic cells in response to a set of TLR ligands including enteric lipopolysaccharide (TLR4 ligand), CpG ODN (TLR9 ligand) and flagellin (TLR5 ligand). These findings are consistent with our results of reduced Tlr5 expression in response to P. gingivalis. The significance of TLR5 signaling in the context of innate immune response to P. gingivalis is not clear as this organism does not possess flagella. Interestingly microarray analysis indicated that the expression of TLR4 accessory protein genes, Cd14 and Ly96 were increased in response to P. gingivalis, despite no change in the expression of Tlr4. We observed a general trend of an increase in the expression of both cytokines and chemokine genes in response to P. gingivalis. These results are in broad agreement with previous studies from our group as well as others that P. gingivalis induces robust levels of secreted inflammatory mediators in murine macrophages [28,44,45,54].
Clinical studies indicate that patients first present with periodontal clinical parameters between the ages of 30–49 years [43]. Using P. gingivalis, recent studies from our group and others employing macrophage models suggest that age may lead to immune senescence in response to periodontal pathogens [44,45]. We were interested in understanding the contribution of age on the expression of TLR pathway genes in response P. gingivalis. A large set of TLR pathway associated genes were differentially expressed by more than two fold between 2- and 12-month age groups and a trend for a reduction in the expression of TLR pathway genes was also observed with increase in age. However expression of a small subset consisting of 5 genes involved in intracellular signaling showed a trend for reduced mRNA expression with an increase in age, indicating that in BMØ, aging contributes to alterations in intracellular signaling events in response to P. gingivalis. Our results support the findings reported by Liang et al. of age-related changes in the expression of TLR pathway genes in macrophages [44]. Similarly, our findings broadly agree with a previous study reporting that both peritoneal and splenic macrophages from aged (18–24 months of age) mice exhibited reduced TLR, as well as IL6 and TNFα expression compared to macrophages from young (2–3 months of age) [55] following stimulation with several TLR ligands [55]. We noted a trend for decreased Tnf mRNA expression accompanied by increased Il6 mRNA expression in BMØ from 12-month old mice challenged with P. gingivalis. In the case of TLR4 costimulatory molecules Cd14 and Ly96 (Md2), our results are in a broad agreement with a study showing macrophages from aged mice had reduce levels of these genes compared with macrophages from young mice in response to enteric LPS stimulation [56]. Collectively, our results and those from other investigators indicate that age influences the elicited immune response to P. gingivalis and bacterial antigens.
Clinical and epidemiological studies have identified elevated levels of TLRs, particularly TLR2 and 4, in the gingival tissues obtained from periodontal patients indicating that TLRs may play important roles in the pathogenesis of periodontal disease [10]. Moreover, involvement of TLR2 [28,32,57,58], TLR4 [59], MyD88 [34] and TRIF [35,60] in host immune responses to P. gingivalis has been reported. However very little is known regarding the MyD88-dependent and -independent pathways on the expression of TLR pathway associated genes in response to P. gingivalis. A recent study using the P. gingivalis subcutaneous chamber model reported that secreted levels of TNFα, IL1β in response to P. gingivalis required TLR2 but was not dependent on MyD88 [34]. However, levels of secreted IL10 were dependent on both TLR2 and MyD88 [34]. In addition, functional TRIF appears to be involved in the full expression of TNFα, IL1β and IL10 in response to P. gingivalis [34]. The clearance of P. gingivalis in vivo appears to involve a MyD88-dependent and TLR4-independent mechanism [34]. Our microarray data identified a trend of increased expression of the MyD88 mRNA in response to P. gingivalis and the levels reduced with age. These results are in a broad agreement with a previous study by Chelvarajan et al. [61], reporting reduced expression of MyD88 in macrophages following LPS exposure as a result of aging. Our data also identified 21 genes significantly reduced as a result of MyD88 deletion compared to Wt in response to P. gingivalis. The expression of secreted inflammatory mediators including Tnf, Il6, Ptsg2, Csf2 and Csf3 was reduced in BMØ from MyD88-KO mice compared with Wt, indicating that the MyD88 pathway is required for full expression of inflammatory mediators including cytokine and chemokine genes in response to P. gingivalis. These results support our previous studies that MyD88 is required for full production of cytokines and chemokines in response to P. gingivalis [35]. Previously it has been shown that the expression of cytokine and chemokine genes in BMØ from MyD88-KO were reduced compared with Wt in response to enteric lipopolysaccharide [62]. Interestingly the expression of Cxcl10 (Ip10) and Ifnb1 genes in BMØ from 2-month old MyD88-KO mice were increased compared with Wt in response to P. gingivalis. Expression of Cxcl10, and Ifnb1 are regulated through a MyD88-independent and TRIF/IRF3 -dependent signaling pathway [14,24]. This increase in expression of Ifnb1 and Cxcl10 could be the result of a compensatory mechanism in which the TRIF pathway is activated in the absence of MyD88. We also examined the influence of age on the expression of these genes as a result of MyD88 mutation. Expression profiles of these genes were similar between Wt and MyD88 except for a subset of genes including Map2k3, Map3k1, Tlr3 and Tlr4. In this subset, expression was modestly increased in BMØ from 12-month old MyD88-KO mice compared to Wt as a result of age. The expression of Cxcl10 was reduced and Ifnb1gene was increased as a result of MyD88 ablation compared to Wt as a consequence of age in response to P. gingivalis. Collectively our data indicate that MyD88 plays an important role in the expression of the immune response to P. gingivalis.
Less is known regarding the contribution of the MyD88-independent adaptor molecule, TRIF and its signaling on the expression of TLR pathway genes in response to P. gingivalis. Our data identified a general trend of a reduction in the expression of these genes in BMØ from 2-months old TrifLps2 mice based on 2-fold expression criteria in BMØ from Wt mice in response to P. gingivalis stimulation, while the expression of 12 genes were statistically significant as a result of the Trif mutation. These genes include intracellular signaling molecules, Map2k4 and Nfkb. In addition, a large set of genes encoding secreted inflammatory mediators including Tnf, Il6, Csf2, Csf3, Ptsg2 were reduced as a result of the Trif mutation compared with Wt, but MyD88 deletion had the strongest effect on the expression of these genes compared to TrifLps2 at the 2-months of age group. Although not reaching significance, in the 12-month age groups, BMØ from TrifLps2 mice displayed a trend for increased Tnfa mRNA expression compared with Wt. We do not understand the reason for this shift; however, this may reflect a compensatory process in BMØ from these mice. Expression levels of Cxcl10 gene were reduced as a result of the TrifLps2 mutation compared to Wt, and this finding is in agreement with previous studies that the expression of Cxcl10 is dependent on functional TRIF [63,64]. Further, these results point to our previous studies suggesting that TRIF play roles in P. gingivalis induced inflammatory response, although MyD88 exhibited a stronger effect [35].
Our microarray study investigating expression of BMØ TLR pathway associated genes in response to P. gingivalis identify important functional contributions of MyD88, and TRIF in the development of host response to this organism. Our findings also support that host immune responses to P. gingivalis are influenced by age. These studies provide a broad assessment of mRNA expression and serve as a starting point for detailed functional studies to understand the significance of MyD88 and TRIF on macrophage immune response to P. gingivalis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors report no conflicts of interest related to this work. These studies were supported by PHS grants from NIAID (P01AI078894; Project 3) and NIDCR (R01DE018318 and R21DE021497) to FCG. Additional support was provided by a Pilot Program Grant, Department of Medicine, Boston University to FCG, and Boston University CTSI Bioinformatics Core for microarray data analysis. Authors thank Dr. Robin Ingalls, Boston University Medical Center, Boston, MA for L929 cells.
REFERENCES
- 1.Oliver RC, Brown LJ, Loe H. Periodontal diseases in the United States population. J Periodontol. 1998;69:269–278. doi: 10.1902/jop.1998.69.2.269. [DOI] [PubMed] [Google Scholar]
- 2.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 5.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 6.Attstrom R. Presence of leukocytes in crevices of healthy and chronically inflamed gingivae. J Periodontal Res. 1970;5:42–47. doi: 10.1111/j.1600-0765.1970.tb01836.x. [DOI] [PubMed] [Google Scholar]
- 7.Sinden PR, Walker DM. Inflammatory cells extracted from chronically inflamed gingiva. J Periodontal Res. 1979;14:467–474. doi: 10.1111/j.1600-0765.1979.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 8.Page RC. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodontal Res. 1991;26:230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 9.Seymour GJ, Gemmell E. Cytokines in periodontal disease: where to from here? Acta Odontol Scand. 2001;59:167–173. doi: 10.1080/000163501750266765. [DOI] [PubMed] [Google Scholar]
- 10.Mori Y, Yoshimura A, Ukai T, Lien E, Espevik T, et al. Immunohistochemical localization of Toll-like receptors 2 and 4 in gingival tissue from patients with periodontitis. Oral Microbiol Immunol. 2003;18:54–58. doi: 10.1034/j.1399-302x.2003.180109.x. [DOI] [PubMed] [Google Scholar]
- 11.Jonsson D, Ramberg P, Demmer RT, Kebschull M, Dahlen G, et al. Gingival tissue transcriptomes in experimental gingivitis. J Clin Periodontol. 2011;38:599–611. doi: 10.1111/j.1600-051X.2011.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe D, Kubota T, Morozumi T, Shimizu T, Nakasone N, et al. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. J Periodontal Res. 2011;46:345–353. doi: 10.1111/j.1600-0765.2011.01349.x. [DOI] [PubMed] [Google Scholar]
- 13.Yu WH, Hu H, Zhou Q, Xia Y, Amar S. Bioinformatics analysis of macrophages exposed to Porphyromonas gingivalis: implications in acute vs. chronic infections. PLoS One. 2010;5:e15613. doi: 10.1371/journal.pone.0015613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 15.Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 16.Mishra BB, Gundra UM, Teale JM. Expression and distribution of Toll-like receptors 11–13 in the brain during murine neurocysticercosis. J Neuroinflammation. 2008;5:53. doi: 10.1186/1742-2094-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11:372–378. [PubMed] [Google Scholar]
- 18.Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11:362–371. [PubMed] [Google Scholar]
- 19.Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 20.Wagner H. Endogenous TLR ligands and autoimmunity. Adv Immunol. 2006;91:159–173. doi: 10.1016/S0065-2776(06)91004-9. [DOI] [PubMed] [Google Scholar]
- 21.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Hasan U, Chaffois C, Gaillard C, Saulnier V, Merck E, et al. Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J Immunol. 2005;174:2942–2950. doi: 10.4049/jimmunol.174.5.2942. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 25.Wara-Aswapati N, Chayasadom A, Surarit R, Pitiphat W, Boch JA, et al. Induction of Toll-Like Receptor Expression by Porphyromonas Gingivalis. J Periodontol. 2013;84:1010–1018. doi: 10.1902/jop.2012.120362. [DOI] [PubMed] [Google Scholar]
- 26.Schroder NW, Meister D, Wolff V, Christan C, Kaner D, et al. Chronic periodontal disease is associated with single-nucleotide polymorphisms of the human TLR-4 gene. Genes Immun. 2005;6:448–451. doi: 10.1038/sj.gene.6364221. [DOI] [PubMed] [Google Scholar]
- 27.Tabeta K, Yamazaki K, Akashi S, Miyake K, Kumada H, et al. Toll-like receptors confer responsiveness to lipopolysaccharide from Porphyromonas gingivalis in human gingival fibroblasts. Infect Immun. 2000;68:3731–3735. doi: 10.1128/iai.68.6.3731-3735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ukai T, Yumoto H, Gibson FC, 3rd, Genco CA. Macrophage-elicited osteoclastogenesis in response to bacterial stimulation requires Toll-like receptor 2-dependent tumor necrosis factor-alpha production. Infect Immun. 2008;76:812–819. doi: 10.1128/IAI.01241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bainbridge BW, Darveau RP. Porphyromonas gingivalis lipopolysaccharide: an unusual pattern recognition receptor ligand for the innate host defense system. Acta Odontol Scand. 2001;59:131–138. doi: 10.1080/000163501750266710. [DOI] [PubMed] [Google Scholar]
- 30.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–5051. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.d'Empaire G, Baer MT, Gibson FC., 3rd The K1 serotype capsular polysaccharide of Porphyromonas gingivalis elicits chemokine production from murine macrophages that facilitates cell migration. Infect Immun. 2006;74:6236–6243. doi: 10.1128/IAI.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S, Ratti P, et al. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell Microbiol. 2006;8:1557–1570. doi: 10.1111/j.1462-5822.2006.00730.x. [DOI] [PubMed] [Google Scholar]
- 33.Davey M, Liu X, Ukai T, Jain V, Gudino C, et al. Bacterial fimbriae stimulate proinflammatory activation in the endothelium through distinct TLRs. J Immunol. 2008;180:2187–2195. doi: 10.4049/jimmunol.180.4.2187. [DOI] [PubMed] [Google Scholar]
- 34.Burns E, Eliyahu T, Uematsu S, Akira S, Nussbaum G. TLR2-dependent inflammatory response to Porphyromonas gingivalis is MyD88 independent, whereas MyD88 is required to clear infection. J Immunol. 2010;184:1455–1462. doi: 10.4049/jimmunol.0900378. [DOI] [PubMed] [Google Scholar]
- 35.Shaik-Dasthagirisaheb YB, Huang N, Baer MT, Gibson FC., 3rd Role of MyD88-dependent and MyD88-independent signaling in Porphyromonas gingivalis-elicited macrophage foam cell formation. Mol Oral Microbiol. 2013;28:28–39. doi: 10.1111/omi.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, Liu J, Xu Q, Harber G, Feng X, et al. TLR2-dependent modulation of osteoclastogenesis by Porphyromonas gingivalis through differential induction of NFATc1 and NF-kappaB. J Biol Chem. 2011;286:24159–24169. doi: 10.1074/jbc.M110.198085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaik-Dasthagirisaheb YB, Huang N, Gibson FC., 3rd Inflammatory response to Porphyromonas gingivalis partially requires interferon regulatory factor (IRF) 3. Innate Immun. 2013 Jun 26; doi: 10.1177/1753425913492180. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morandini AC, Chaves Souza PP, Ramos-Junior ES, Souza Costa CA, Santos CF. MyD88 or TRAM knockdown regulates interleukin (IL)-6, IL-8, and CXCL12 mRNA expression in human gingival and periodontal ligament fibroblasts. J Periodontol. 2013;84:1353–1360. doi: 10.1902/jop.2012.120496. [DOI] [PubMed] [Google Scholar]
- 39.Gaddis DE, Michalek SM, Katz J. Requirement of TLR4 and CD14 in dendritic cell activation by Hemagglutinin B from Porphyromonas gingivalis. Mol Immunol. 2009;46:2493–2504. doi: 10.1016/j.molimm.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez CR, Nomellini V, Faunce DE, Kovacs EJ. Innate immunity and aging. Exp Gerontol. 2008;43:718–728. doi: 10.1016/j.exger.2008.05.0168.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castle SC, Uyemura K, Fulop T, Makinodan T. Host resistance and immune responses in advanced age. Clin Geriatr Med. 2007;23:463–479. v. doi: 10.1016/j.cger.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasto S, Candore G, Balistreri CR, Caruso M, Colonna-Romano G, et al. Inflammatory networks in ageing, age-related diseases and longevity. Mech Ageing Dev. 2007;128:83–91. doi: 10.1016/j.mad.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka S, Murakami Y, Ogiwara T, Shoji M, Seto K, et al. Frequency of reactivity for Porphyromonas gingivalis and Prevotella spp. in supra- and subgingival plaques, and periodontal clinical parameters according to subject age. J Periodontol. 2002;73:877–885. doi: 10.1902/jop.2002.73.8.877. [DOI] [PubMed] [Google Scholar]
- 44.Liang S, Domon H, Hosur KB, Wang M, Hajishengallis G. Age-related alterations in innate immune receptor expression and ability of macrophages to respond to pathogen challenge in vitro. Mech Ageing Dev. 2009;130:538–546. doi: 10.1016/j.mad.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaik-Dasthagirisaheb YB, Kantarci A, Gibson FC., 3rd Immune response of macrophages from young and aged mice to the oral pathogenic bacterium Porphyromonas gingivalis. Immun Ageing. 2010;7:15. doi: 10.1186/1742-4933-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 47.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 48.Baer MT, Huang N, Gibson FC., 3rd Scavenger receptor A is expressed by macrophages in response to Porphyromonas gingivalis, and participates in TNF-alpha expression. Oral Microbiol Immunol. 2009;24:456–463. doi: 10.1111/j.1399-302X.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen TD. Real-time quantitative PCR. Methods. 2001;25:383–385. doi: 10.1006/meth.2001.1260. [DOI] [PubMed] [Google Scholar]
- 50.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 51.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 53.Feng T, Cong Y, Alexander K, Elson CO. Regulation of Toll-like receptor 5 gene expression and function on mucosal dendritic cells. PLoS One. 2012;7:e35918. doi: 10.1371/journal.pone.0035918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Q, Desta T, Fenton M, Graves DT, Amar S. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect Immun. 2005;73:935–943. doi: 10.1128/IAI.73.2.935-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renshaw M, Rockwell J, Engleman C, Gewirtz A, Katz J, et al. Cutting edge: impaired Toll-like receptor expression and function in aging. J Immunol. 2002;169:4697–4701. doi: 10.4049/jimmunol.169.9.4697. [DOI] [PubMed] [Google Scholar]
- 56.Dunston CR, Griffiths HR. The effect of ageing on macrophage Toll-like receptor-mediated responses in the fight against pathogens. Clin Exp Immunol. 2010;161:407–416. doi: 10.1111/j.1365-2249.2010.04213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibson FC, 3rd, Genco CA. Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs. Curr Pharm Des. 2007;13:3665–3675. doi: 10.2174/138161207783018554. [DOI] [PubMed] [Google Scholar]
- 58.Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- 59.Hou L, Sasaki H, Stashenko P. Toll-like receptor 4-deficient mice have reduced bone destruction following mixed anaerobic infection. Infect Immun. 2000;68:4681–4687. doi: 10.1128/iai.68.8.4681-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaddis DE, Michalek SM, Katz J. TLR4 signaling via MyD88 and TRIF differentially shape the CD4+ T cell response to Porphyromonas gingivalis hemagglutinin B. J Immunol. 2011;186:5772–5783. doi: 10.4049/jimmunol.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chelvarajan RL, Liu Y, Popa D, Getchell ML, Getchell TV, et al. Molecular basis of age-associated cytokine dysregulation in LPS-stimulated macrophages. J Leukoc Biol. 2006;79:1314–1327. doi: 10.1189/jlb.0106024. [DOI] [PubMed] [Google Scholar]
- 62.Bjorkbacka H, Fitzgerald KA, Huet F, Li X, Gregory JA, et al. The induction of macrophage gene expression by LPS predominantly utilizes Myd88-independent signaling cascades. Physiol Genomics. 2004;19:319–330. doi: 10.1152/physiolgenomics.00128.2004. [DOI] [PubMed] [Google Scholar]
- 63.Shalova IN, Kajiji T, Lim JY, Gomez-Pina V, Fernandez-Ruiz I, et al. CD16 regulates TRIF-dependent TLR4 response in human monocytes and their subsets. J Immunol. 2012;188:3584–3593. doi: 10.4049/jimmunol.1100244. [DOI] [PubMed] [Google Scholar]
- 64.Weighardt H, Jusek G, Mages J, Lang R, Hoebe K, et al. Identification of a TLR4- and TRIF-dependent activation program of dendritic cells. Eur J Immunol. 2004;34:558–564. doi: 10.1002/eji.200324714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.