Stargazin enhances closure of the AMPA receptor ligand-binding domain, thereby facilitating channel activation.
Abstract
Transmembrane AMPA receptor (AMPAR) regulatory proteins (TARPs) markedly enhance AMPAR function, altering ligand efficacy and receptor gating kinetics and thereby shaping the postsynaptic response. The structural mechanism underlying TARP effects on gating, however, is unknown. Here we find that the prototypical member of the TARP family, stargazin or γ-2, rescues gating deficits in AMPARs carrying mutations that destabilize the closed-cleft states of the ligand-binding domain (LBD), suggesting that stargazin reverses the effects of these mutations and likely stabilizes closed LBD states. Furthermore, stargazin promotes a more closed conformation of the LBD, as indicated by reduced accessibility to the large antagonist NBQX. Consistent with the functional studies, luminescence resonance energy transfer experiments directly demonstrate that the AMPAR LBD is on average more closed in the presence of stargazin, in both the apo and agonist-bound states. The additional cleft closure and/or stabilization of the more closed-cleft states of the LBD is expected to translate to higher agonist efficacy and could contribute to the structural mechanism for stargazin modulation of AMPAR function.
INTRODUCTION
AMPA receptors (AMPARs) are responsible for the majority of fast excitatory synaptic signaling in the mammalian central nervous system. Given their role in rapid synaptic events, their gating properties shape the dynamics of synaptic transmission and have been studied extensively. The receptors can be composed of either homo- or heterotetramers of GluA1-GluA4 subunits, each of which exists in differentially spliced versions and in RNA-edited variants (Hollmann et al., 1989; Sommer et al., 1990, 1991; Lomeli et al., 1994). Originally, it was thought that the composition of the pore-forming subunits of the receptor is the primary contributor to their gating properties and accounts for known functional heterogeneity. However, the discovery of the family of related transmembrane AMPAR regulatory proteins (TARPs) has made it clear that TARPs add an additional layer of functional complexity, play critical roles in shaping many aspects of AMPAR operation, and make vital contributions to excitatory synaptic signaling (Tomita et al., 2005; Cho et al., 2007; Menuz et al., 2008, 2009).
TARPs were initially identified as playing important roles in receptor trafficking (Schnell et al., 2002). But more recently, the kinetic and biophysical effects of TARP modulation have been explored. Rapid kinetic measurements reveal that TARPs slow AMPAR deactivation and desensitization (Priel et al., 2005; Tomita et al., 2005; Cho et al., 2007) as well as increase the potency of glutamate (Tomita et al., 2005; Kott et al., 2007; Cokić and Stein, 2008). Additionally, at the single-channel level, they increase channel openings to large conductance states and prolong the bursting activity of single channels (Tomita et al., 2005; Shelley et al., 2012; Zhang et al., 2014). In addition to the effects on responses to the neurotransmitter glutamate, a striking feature of TARPs is their dramatic enhancement of the efficacy of the partial agonist kainate (KA) (Tomita et al., 2005; Cho et al., 2007; Kott et al., 2007; Cokić and Stein, 2008) and the conversion of the competitive antagonist CNQX into a partial agonist (Menuz et al., 2007; Cokić and Stein, 2008; Maclean and Bowie, 2011).
The most parsimonious explanation for these effects is that TARPs reduce the energy barrier for the transitions leading to gating of the channel (Howe, 2013; MacLean, 2013). Crystallographic (Armstrong et al., 2003; Jin et al., 2003), fluorescence (Ramanoudjame et al., 2006; Gonzalez et al., 2008; Ramaswamy et al., 2012) and electrophysiological (Robert et al., 2005; Zhang et al., 2008) studies have supported the conclusion that the extent to which the ligand-binding domain (LBD) is closed and the stability of the resulting closed-cleft conformations govern the efficacy of a given iGluR agonist. Therefore, one hypothesis for the mechanism of TARP action is that TARPs stabilize LBD states that are more closed, thereby lowering the energy barrier to activation. To test this idea, we used a combination of electrophysiology and luminescence resonance energy methods. Our results show that the LBD is on average more closed in the presence of stargazin (γ-2, the founding member of the TARP family), thus providing a structural mechanism for the known increase in efficacy in the presence of TARPs.
MATERIALS AND METHODS
Cell culture and transfection
HEK293 tsA201 cells were transiently transfected using Lipofectamine 2000 (Life Technologies), as described previously (Ramaswamy et al., 2012). In brief, for electrophysiology, GluA2 and all mutations were flip versions with a glutamine at the Q/R site on plasmids encoding enhanced GFP (eGFPS65T) behind an internal ribosomal entry site (provided by M. Mayer, National Institutes of Health, Bethesda, MD). GluA2/eGFP was transfected at a ratio of 5 µg cDNA/10 ml of media or with γ-2 at a ratio of 5:7.5 µg cDNA/10 ml media. Transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions and changing to fresh media (MEM plus Pen/Strep and 10% FBS) after 8–12 h to include 20 µM NBQX. For whole-cell experiments (GluA4*(Q) flip; see Results and Fig. 4), cells were subsequently replated at a lower density onto poly-lysine–coated dishes 24 h before experiments. Recordings were performed 24–48 h posttransfection. For luminescence resonance energy transfer (LRET) experiments, tsA201 cells were cultured in 10-cm dishes and transfected in 20 µM NBQX with 4 µg cDNA/10 ml for AMPAR alone, or with an added 8 µg γ-2 cDNA. LRET experiments were performed 36–48 h posttransfection.
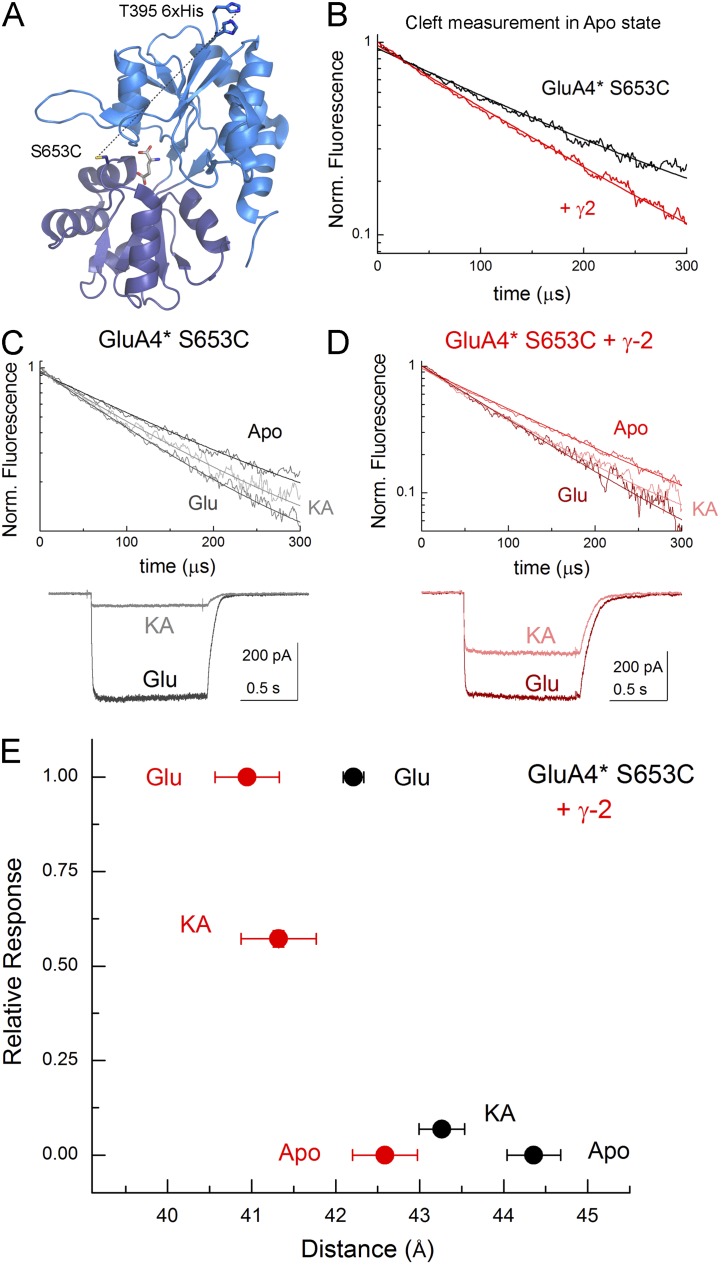
Figure 4.
TARPs induce more cleft closure in LRET experiment. (A) Structure of GluA2 LBD showing the positions of fluorophore labels for LRET (Protein Data Bank accession no. 1FTJ). (B) LRET lifetime of apo state for GluA4* S653C alone (black) or cotransfected with γ-2 (red), with each composed of at least six separate experiments. (C) LRET lifetimes (top) and electrophysiology (bottom) from GluA4* S653C alone with the indicated ligands. LRET data are aggregates of three separate experiments per ligand and six for apo. (D) LRET lifetimes (top) and electrophysiology (bottom) from GluA4* S653C plus γ-2 with the indicated ligands. (E) Plot of mean ± SEM distances as estimated from the Förster equation versus mean ± SEM electrophysiological responses. n for electrophysiology is four cells for GluA4* S653C and five cells for GluA4* S653C plus γ-2.
Electrophysiology
Both outside-out patches and whole-cell recordings were obtained from eGFP-expressing HEK293T cells with thick-walled borosilicate glass pipettes of 3–5 MΩ coated with bees wax, fire-polished, and filled with a solution that contained (mM): 135 CsF, 33 CsOH, 11 EGTA, 10 HEPES, 2 MgCl2, and 1 CaCl2, pH 7.4. External solutions were composed of (mM): 150 NaCl, 10 HEPES, 1 MgCl2, and 1 CaCl2, pH 7.4 with 5 N NaOH. The external solution for whole-cell recordings also contained 2.5 mM KCl. All recordings were performed at a holding potential of −60 mV using an amplifier (Axopatch 200B; Molecular Devices). Outside-out patch data were acquired at 30–40 kHz and filtered at 10 kHz (eight-pole Bessel). Whole-cell data were acquired at 10 kHz and filtered at 3 kHz, with both under the control of pCLAMP 10 software (Molecular Devices). Series resistances (3–12 MΩ) were routinely compensated by >95% (>65% for whole-cell experiments) where the amplitude exceeded 100 pA. Rapid application for patches was performed using home-built theta (Warner Instruments) or multi-barrel (Vitrocom) glass application pipettes, pulled to 100–150 µm, and translated using a piezoelectric microstage (Burleigh Instruments). Solution exchange times estimated from open-tip potentials were 50–300 µs (10–90% rise time). For wild-type receptors, 10 mM glutamate (Fig. 1–3) and 1 mM quisqualate (Figs. 1 and 2) were used; for T686S and T686A, 50 mM glutamate and 10 mM quisqualate were used. Solutions for whole-cell experiments were locally applied using a computer-controlled valve switcher (VC-6; Warner Instruments) and a homebuilt application pipette with a common exit port. 100 µM cyclothiazide (CTZ) was continuously present in both control and agonist solutions with 10 mM glutamate and 1 mM KA as agonists.
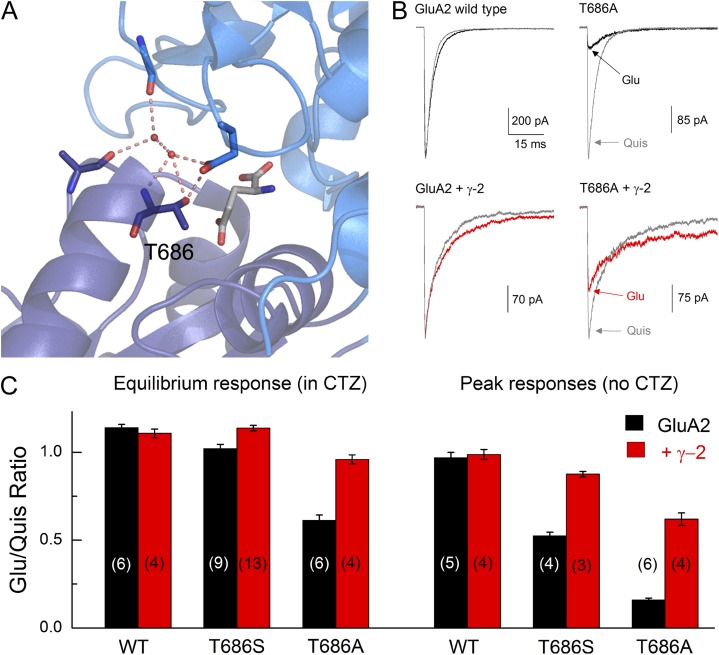
Figure 1.
TARPs rescue the gating of T686S/A. (A) Structure of GluA2 LBD with glutamate bound, indicating the Thr686 position and its interactions with neighboring residues (Protein Data Bank accession no. 1FTJ). (B) Representative responses from GluA2 wild-type (left) or T686A (right) alone or coexpressed with γ-2 (bottom) evoked by quisqualate (gray) or glutamate (black or red). (C) Mean ± SEM ratio from experiment in B performed in the continual presence (left) or absence (right) of 100 µM CTZ for wild-type, T686S, or T686A constructs. n values are given in brackets.
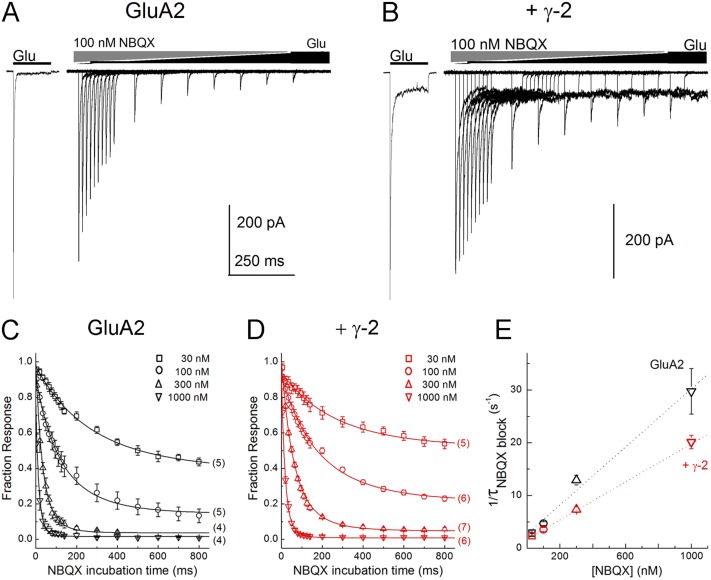
Figure 3.
TARPs slow the apparent forward binding rate of NBQX. (A and B) Responses from GluA2 wild-type alone (A) or with γ-2 (B) to progressively longer preincubations with 100 nM NBQX before a 10-mM Glu test pulse. (C and D) Summary of time course of NBQX inhibition for GluA2 alone (C) or with γ-2 (D) over a range of NBQX concentrations. Note that in every case, γ-2 slows the binding of NBQX. n for a given concentration ranges from 4 to 7 and is indicted in brackets. (E) Plot of the inverse of the time constants from fits in C and D (Eq. 1) versus NBQX concentration for GluA2 alone (black) or with γ-2 (red). Dotted lines are fits using Eq. 2. Error bars show mean data ± SEM.
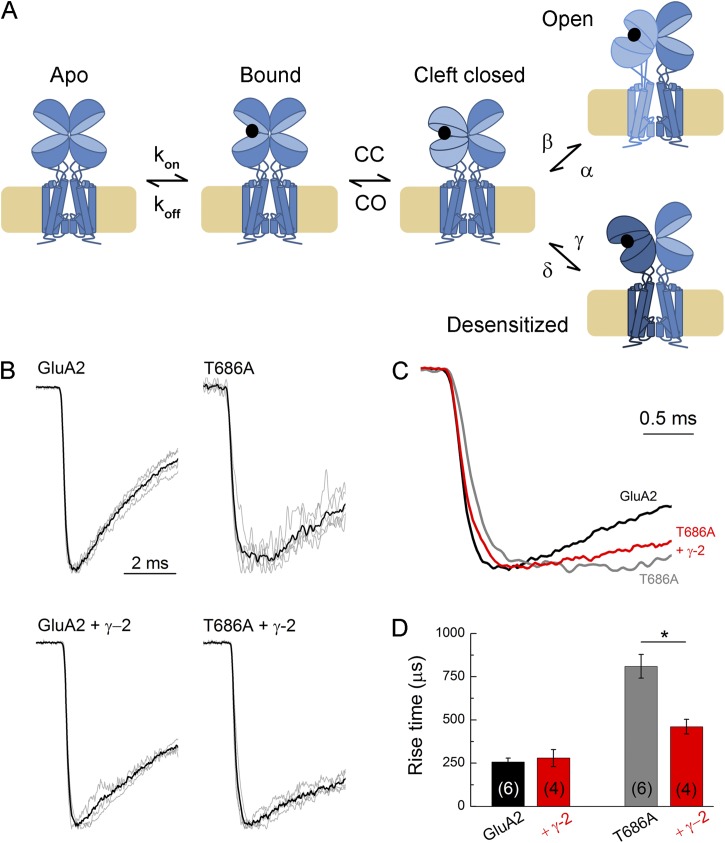
Figure 2.
TARPs restore the rise time of T686A. (A) Cartoon depicting the gating reaction of the AMPAR. “CC” stands for cleft closure, and “CO” stands for cleft open. (B) All responses from GluA2 wild-type (left) or T686A (right) alone or coexpressed with γ-2 (bottom) evoked by glutamate. Gray traces are averages from individual patches; black trace is a super-average across patches. (C) Overlay of super-averages from B for GluA2 wild-type (black), T686A (gray), and T686A plus γ-2 (red). (D) Summary of rise times (10–90%) for each condition. n values are given in brackets. Error bars show mean data ± SEM. *, P < 0.01.
LRET
The GluA4 flip protein with the N-terminal domain deleted was mutated to convert the endogenous exposed Cys residues at sites 447 and 550 to Ser. A hex-His tag was added in front of Thr395, followed by the thrombin cleavage signal (LVPRGS), which replaced the original amino acids beginning at site 394. Three to four 10-cm dishes of transfected HEK293T cells were labeled with 200 nM each of donor (terbium chelate; Invitrogen) and acceptor (Ni(NTA)2-Cy3, prepared as described previously [Sirrieh et al., 2013], using bis-reactive Cy3 purchased from GE Healthcare) at room temperature for 1 h. Cells were washed three to four times with PBS before the LRET measurements. A cuvette-based fluorescence spectrometer (Quanta Master QM3-SS; Photon Technology International) measured the lifetimes of the fluorophores. A high power, pulsed xenon lamp was used for excitation, and the emitted light was passed through a monochromator onto the detector. Experiments were performed at 15°C. Fluorescan software (Photon Technology International) was used for data acquisition. The donor-only lifetime measurements were obtained at 545 nm, whereas the sensitized acceptor lifetimes for Ni(NTA)2-Cy3 were obtained at 565 nm. For each sample, LRET lifetimes were obtained with and without agonist before digestion with thrombin, with the background LRET after digestion was subtracted from the initial signal to obtain the LRET specific to labeled AMPARs. The data in Fig. 4 are averages of at least three samples, with each sample composed of at least 297 separate scans. Distances between the donor and acceptor fluorophores were calculated using the LRET lifetime (τDA) and donor-only (τD) lifetime using the Förster equation (see Eq. 3 and Data analysis below). The largest error in the distances determined by LRET is thought to arise from the orientation factor included in the calculation of R0. For lanthanides that are isotropic, this error is reduced to being at most ±10% (dos Remedios and Moens, 1995).
Data analysis
Electrophysiology data were analyzed using a combination of Clampfit 10.2 (Molecular Devices), Excel (Microsoft), and Origin 9.0 (OriginLab Corp.). The onset of NBQX block was fit with Eq. 1 based on previous work (Jones et al., 1998):
| (1) |
where It is the fraction of peak current for a given NBQX equilibration time t, Iss is the fraction of peak current remaining at steady-state inhibition, I0 is the initial peak current in the absence of NBQX (repeatedly measured throughout the experiment to correct for rundown), and τ is the time constant for the onset of NBQX inhibition. Raising the right-hand terms to the fourth power corresponds to the four binding sites needed for maximal conductance. Attempts to fit the onset of NBQX block with single- or double-binding site equations resulted in visibly poor fits. The rate of onset of block (i.e., the inverse of the time constant) at various NBQX concentrations was fit with Eq. 2:
| (2) |
where τ is the time constant for inhibition from Eq. 1 for a given concentration of NBQX, kon is the forward rate constant for binding in M−1s−1, and koff is the rate constant for unbinding in s−1.
LRET data were analyzed using Origin 4.0 or 9.0, and the distances were calculated using the Förster equation (Eq. 3):
| (3) |
where R is the distance between donor and acceptor fluorophore, R0 is the distance yielding half-maximal energy transfer for a given fluorophore pair (in this instance, 65 Å), τD is the measured lifetime of the donor when bound to the protein, and τDA is the measured lifetime of the sensitized emission of the acceptor in the presence of donor.
Statistical analysis
For all electrophysiological experiments, n was a single patch or cell. Between 5 and 10 individual traces from a single patch were averaged for representative traces. For LRET experiments, n was a single experiment in which three to four 10-cm dishes of transfected cells were pooled, labeled, and scanned at least 297 times (see LRET section above). For statistical significance, an independent sample two-tailed t test was used for all experiments, with P < 0.05 considered significant. An exception to this is certain measurements of LRET lifetime within a receptor population (i.e., apo vs. KA in Fig. 4, C and D), which were evaluated using a repeated measure t test. Estimates of kon and koff (Fig. 3 and Eq. 2) were obtained by fitting Eq. 1 to results from individual patches with varying concentrations of NBQX, averaging the resulting fit values (plotted in Fig. 3 E), and then using Eq. 2 to estimate kon and koff from the averaged fits. Because curves at varying concentrations were composed of between four and seven separate patches, statistical differences in kon and koff estimates used the conservative lower value of n = 4. All summary plots show mean data ± SEM.
RESULTS
Effect of γ-2 on gating of T686 mutations
To investigate if γ-2 (stargazin) increases responses from AMPARs by stabilizing closed-bound states of the LBD, we asked whether γ-2 could rescue gating in mutant AMPARs known to have destabilized closed-cleft conformations. To do this, we took advantage of mutations at the Thr686 position of GluA2 receptors. As seen in Fig. 1 A, the hydroxyl moiety of the Thr residue at 686 makes cross-cleft interactions, thus stabilizing the closed-cleft state of the LBD. Mutation of this Thr to a Ser residue or Ala disrupts these cross-cleft interactions and dramatically accelerates the deactivation of GluA2 responses as well as right shifts the peak dose–response curve, as shown previously and as expected from mutations that destabilize the closed-cleft conformations of the LBD (Robert et al., 2005; Zhang et al., 2008). Recent single molecule Förster resonance energy transfer (FRET) data on the soluble LBD find that the glutamate-bound form of the T686S mutation exhibits a wide range of cleft closures, thus reducing the fraction of time that individual receptors spend in the conformation(s) from which pore opening can proceed (Landes et al., 2011). Therefore, if γ-2 is able to rescue gating in this mutation, it would be consistent with TARPs stabilizing LBD conformations from which channel opening is likely.
To test if TARPs can rescue the deficits of the mutants at the Thr686 position, we compared the efficacy of glutamate (Glu) and quisqualate (Quis) in outside-out patches. Previous work has found that the effect of Thr686 mutations on gating is strongly agonist specific, with responses to glutamate being reduced substantially more than those to quisqualate (Zhang et al., 2008). Therefore, the ratio of the responses to these two agonists reflects the degree of rescue of the T686S or T686A phenotype. As expected, wild-type GluA2 receptors yield identical peak responses to both saturating quisqualate and glutamate (Glu/Quis ratio: 0.97 ± 0.03, n = 6; Fig. 1 B), and this ratio was not altered by the presence of γ-2 (Glu/Quis ratio: 0.99 ± 0.03, n = 4; Fig. 1, B and C). Consistent with previous work, both GluA2 T686S and T686A substantially reduced the Glu/Quis peak response ratio (T686S: 0.52 ± 0.02, n = 9; T686A: 0.16 ± 0.01, n = 6; Fig. 1 C) (Zhang et al., 2008). Interestingly, coexpression of γ-2 partially rescues this deficit in glutamate responses, restoring the Glu/Quis peak response ratio to ∼0.90 for T686S and 0.60 for T686A (T686S plus γ-2: 0.87 ± 0.01, n = 13, P < 0.001 vs. T686S alone; T686A plus γ-2: 0.62 ± 0.03, n = 4, P < 0.001 vs. T686A alone; Fig. 1 C). This partial rescue of the T686A phenotype could suggest that TARPs stabilize or promote closed-cleft conformations of the LBD of AMPARs. Moreover, rescue of glutamate efficacy by γ-2 was also present when desensitization was suppressed using CTZ. Under these conditions, wild-type GluA2 alone or with γ-2 gave identical responses to both glutamate and quisqualate (GluA2 alone: steady-state ratio Glu/Quis: 1.14 ± 0.02, n = 5; plus γ-2: 1.11 ± 0.02, n = 4; Fig. 1 C). As expected, T686S showed a slight reduction in the Glu/Quis ratio (1.02 ± 0.02, n = 4; Fig. 1 C), which was rescued by coexpression with γ-2 (1.14 ± 0.02, n = 3, P < 0.01 vs. T686S alone in CTZ). As in the absence of CTZ, the T686A mutation showed a greater reduction in the Glu/Quis ratio (0.61 ± 0.03, n = 6; Fig. 1 C), which was also largely rescued by γ-2 (0.96 ± 0.02, n = 4, P < 0.001 vs. T686A alone in CTZ). As with experiments in the absence of CTZ, this illustrates that γ-2 is able to ameliorate deficits in gating caused by destabilizing closed-cleft conformations of the LBD.
A prominent feature of the T686A mutation is that the destabilizing effect of the alanine mutation on closed-cleft conformations delays channel opening (Zhang et al., 2008). In single-channel experiments, this has been observed as a substantial increase in latency to first opening. This has been explicitly modeled previously using the scheme in Fig. 2 A, where T686A increases CO, the rate of cleft reopening, which causes the closed-pore receptor to oscillate several times between open-cleft and closed-cleft conformations, thus delaying transition to the open state (Robert et al., 2005; Zhang et al., 2008). Consistent with these findings, we observed changes in the macroscopic proxy of latency to first opening, namely the rise time of the onset of the peak current response. As seen in Fig. 2, wild-type GluA2 receptors, either alone or with γ-2, activated rapidly with rise times (10–90%) on the order of 250 µsec (GluA2: 260 ± 20 µsec, n = 6; GluA2 plus γ-2: 280 ± 50 µsec, n = 4). In contrast, GluA2 T686A receptors showed markedly slower rise times (810 ± 70 µsec, n = 6). This is best seen in Fig. 2, where averages from the results from all the patches for GluA2 wild-type and GluA2 T686A are illustrated. If γ-2 rescues the gating of the T686A mutation by acting on the cleft-closure step in the reaction, this slowed rising phase should also be rescued. This is in fact the case as seen in Fig. 2, where GluA2 T686A coexpressed with γ-2 shows rise times that are faster than GluA2 T686A alone and closer to wild-type levels (460 ± 40 µsec, n = 4, P = 0.0055 vs. T686A alone). These results are consistent with TARP stabilization of receptor conformations in which the LBD is closed. This stabilization could reflect changes in rate constants (decreased CO, increased β; Fig. 2 B) for transitions between states that exist in the absence of TARPs (Howe, 2013; MacLean, 2013), or alternatively result from an effect of TARPs to promote LBD conformations that are inherently more closed and rarely visited in the absence of TARPs. To test this further, we investigated the effect of TARPs on the behavior of antagonists, which do not cause pore opening but retain a ligand-bound pre-open step where the LBD is closed.
Accessibility of the LBD to antagonists in the presence of γ-2
Previous work found that coexpression of γ-2 slightly reduces the apparent affinity of AMPAR for both CNQX and NBQX (Maclean and Bowie, 2011). One proposed explanation is that TARPs slightly close the LBD, or more precisely shift the ensemble distribution of LBD states toward more closed conformations, reducing the accessibility of large, rigid, competitive antagonists like NBQX to their binding pocket (Maclean and Bowie, 2011). Such a mechanism is not expected to have a large effect on the rates of small agonists such as glutamate. Therefore, to determine the ensemble accessibility or openness of the LBD in GluA2 wild-type receptors alone or with γ-2, we measured the forward-binding rate of the large antagonist NBQX using a “triple-jump” protocol. Receptors were first exposed to NBQX for variable times before jumping into glutamate to measure the fractional inhibition of the peak current (Fig. 3, A and B). As expected, 100 nM NBQX inhibited both GluA2 alone and GluA2 with γ-2, reaching equilibrium in ∼750 ms. The extent of inhibition by 100 nM was greater for GluA2 alone than when γ-2 was present (GluA2 alone: 14 ± 4% Glu peak, n = 5; GluA2 plus γ-2: 24 ± 1% Glu peak, n = 7, P < 0.02), consistent with results with GluA1 and γ-2 (Maclean and Bowie, 2011). Importantly, the time constant for inhibition was slower when γ-2 was present than for GluA2 alone (τ from Eq. 1 for GluA2 alone: 220 ± 20 ms; τ for GluA2 plus γ-2: 300 ± 20 ms, P < 0.02). Measuring the time constants of inhibition over a range of NBQX concentrations (30–1,000 nM; n = 4–7 patches per concentration) showed that the presence of γ-2 resulted in a slower rate of onset and reduced inhibition at all NBQX concentrations tested (Fig. 3, C and D).
To estimate the forward and reverse rate constants for NBQX binding, we plotted the time constants of inhibition as a function of NBQX concentration, where the slope of this plot yields the apparent kon and the intercept corresponds to the apparent koff (Jones et al., 1998; Zheng et al., 2001). As clearly seen in Fig. 3 E, the slope of this plot when γ-2 is present is markedly shallower than the corresponding plot for GluA2 alone, demonstrating that the apparent forward rate of binding of NBQX is slowed ∼30% (GluA2 kon: 2.8 ± 0.2 × 107 M−1 s−1; GluA2 + γ-2 kon: 1.86 ± 0.07 × 107 M−1 s−1; P = 0.007). The apparent koff values extracted from this analysis were not significantly different, although NBQX was likewise slower in the presence of γ-2 (GluA2 + γ-2 koff = 1.78 ± 0.03 s−1 vs 3 ± 1 s−1 for GluA2 alone, P = 0.26). The mean kon and koff values obtained in this way give equilibrium dissociation constants from NBQX binding to GluA2 and GluA2 with γ-2 of 110 and 95 nM, respectively, which are excellent agreement with the values obtained for steady-state NBQX inhibition of peak currents for GluA1 and GluA1 coexpressed with γ-2 (Maclean and Bowie, 2011). Because NBQX binding occurred before exposure of the receptor to agonist, the reduced kon values in the coexpression experiments directly supports an effect of TARPs on the apo conformation of the LBD.
The average extent of cleft closure in the LBD in the presence of γ-2
The experiments with NBQX support the hypothesis that TARPs close the LBD of the AMPAR in the apo state. To examine the average LBD conformation of ligand-bound active states, we used LRET methodology as described previously (Du et al., 2005; Gonzalez et al., 2008; Rambhadran et al., 2011). For these measurements, we used a similar construct to those described previously (Du et al., 2005; Gonzalez et al., 2008), where the accessible non-disulphide–bonded cysteines were mutated to serines and a cysteine was introduced at site 653 for donor labeling with a 6xHistag before site Thr395 for acceptor labeling (Fig. 4 A). This residue, like Thr686, participates in cross-cleft LBD interactions in agonist-bound states (Armstrong and Gouaux, 2000), and the FRET signal can be used as a readout of the average degree of the extent of LBD closure (Du et al., 2005; Gonzalez et al., 2008; Rambhadran et al., 2011). However, unlike Thr686, it is the backbone of Ser653 that is involved in cross-cleft interactions, and hence mutation or fluorophore addition does not measurably alter AMPAR function (Gonzalez et al., 2008; Ramaswamy et al., 2012). Additionally, a thrombin recognition sequence was introduced after the acceptor site to allow for background subtraction (see Materials and methods) (Du et al., 2005; Gonzalez et al., 2008; Rambhadran et al., 2011). Importantly, none of the amino acid substitutions noticeably impacted the function of the AMPAR (Du et al., 2005; Gonzalez et al., 2008; Lau et al., 2013), and the presence of fluorophores at position S653C also does not noticeably impact receptor activation (Ramaswamy et al., 2012). However, in GluA2, these manipulations did result in very poor surface expression, making differences in LRET lifetimes from this construct difficult to confidently discern. We therefore performed similar modifications on GluA4, which we have used previously in LRET experiments (Du et al., 2005; Gonzalez et al., 2008) and which is similarly modulated by TARPs (Körber et al., 2007; Kott et al., 2007), including at the level of single-channel currents (Zhang et al., 2014). Moreover, in additional experiments with the resulting construct (GluA4* S653C), we demonstrated that KA remained a partial agonist (7 ± 2% of steady-state glutamate response in CTZ, n = 5 cells; Fig. 4 C), and the known TARP enhancement of KA efficacy was intact as coexpression of γ-2 markedly increased the relative efficacy of KA (57 ± 2% of steady-state glutamate response in CTZ, n = 6 cells, P < 0.001; Fig. 4 C).
With both function and TARP modulation intact, we proceeded to measure cross-cleft distances using LRET with terbium chelate as a donor binding to the 6xHis tag and Ni(NTA)2-Cy3 as an acceptor binding to S653C. As seen in Fig. 4 B, in the absence of added ligand, LRET lifetime signals from GluA4* S653C were well fit with a single-exponential function, consistent with LRET originating predominantly from one subunit and not from cross-talk between subunits. Specifically, the apo state of GluA4* S653C yielded a distance of 44.4 ± 0.3 Å (n = 6 separate experiments; Fig. 4 D), in excellent agreement with previous reports and with the distances measured from crystal structures when fluorophore length is considered. Adding KA or glutamate produced a significant acceleration of the LRET decay, demonstrating that these agonists bring T395 6xHis and S653C closer together (Fig. 4 C). As expected, glutamate closed the LBD more than KA, with glutamate producing 2.2 Å of domain closure (42.2 ± 0.1 Å with glutamate, n = 3, P < 0.02 vs. apo) and KA yielding 1.1 Å of closure (43.3 ± 0.3 Å with KA, n = 3, P < 0.05 vs. apo; Fig. 4 D).
We next repeated the LRET measurements on cells cotransfected with γ-2. As seen in Fig. 4 B, the LRET signals from GluA4* S653C cotransfected with γ-2 were also well fit by a single-exponential function, demonstrating that the presence of γ-2 does not introduce any contamination of the signal by additional Cys labeling or Trp quenching. Estimates of cleft conformation from the apo state with γ-2 coexpression yielded a distance of 42.6 ± 0.4 Å (n = 6, P < 0.001 vs. GluA4* S653C), which is 1.8 Å more closed than the same condition in the absence of γ-2 (Fig. 4, B and D). This independent result provides direct support for our conclusion that NBQX binds more slowly to TARP-associated AMPARs because γ-2 promotes apo states of the LBD in which the binding cleft is on average more closed (Fig. 3). We also found for both KA- and glutamate-bound GluA4* S653C plus γ-2 receptors, the LBD was consistently 1–2 Å more closed for each agonist than the corresponding condition in the absence of γ-2. Specifically, the glutamate-bound receptors were 1.2 Å more closed in the presence of γ-2 than in its absence (41.0 ± 0.4 Å in glutamate, n = 3, P < 0.05 vs. GluA4* S653C glutamate), and the KA-bound TARP-associated receptor displayed an additional 2.0 Å of LBD closure (41.3 ± 0.4 Å in KA, n = 3, P < 0.05 vs. GluA4* S653C KA; Fig. 4 D). Collectively, these data clearly demonstrate that the presence of the prototypical TARP, γ-2, induces additional LBD cleft closure both in the absence and presence of ligands.
DISCUSSION
The results reported here provide the first direct evidence that TARPs alter the conformation of the LBD of pore-forming GluA subunits. The partial rescue of the effects of the Thr686 mutations on glutamate efficacy and activation kinetics, as well as the shorter LRET lifetimes we observed in the presence of both glutamate and KA, are consistent with an effect of TARPs to promote LBD closure in agonist-bound states. Although these results could arise from stabilization of either closed or open states, as suggested from earlier work (Howe, 2013; MacLean, 2013), the slower rate of NBQX binding and the shorter LRET lifetimes we measured in the apo state of full-length receptors demonstrate that TARPs promote LBD cleft closure even in the absence of agonists. Given the established relationship between the extent/stability of LBD closure and agonist efficacy (Jin et al., 2003; Gonzalez et al., 2008; Zhang et al., 2008; Ramaswamy et al., 2012), the additional LBD closure demonstrated here is likely to contribute importantly to TARP modulation of receptor function.
The shorter LRET lifetimes we observe in the absence of agonist could arise either because TARPs promote apo states that are physically distinct and more closed than those in the absence of TARPs, or because the LBD normally visits multiple conformations with different extents of cleft closure and TARP association favors conformations that are more closed. We have found evidence for this latter possibility in previous single-molecule FRET studies on the isolated LBD (Landes et al., 2011). Recently, analysis of unitary kinetics in fast application studies has shown that TARPs promote distinct gating behavior where individual receptors switch between a high Popen mode (characterized by more openings per activation and a greater frequency of large-conductance openings) and normal gating on a time scale of tens of seconds (Zhang et al., 2014). The slow switching between these gating modes in the presence of TARPs suggests that TARPs may promote access of the receptor to a set of states that are physically distinct from those visited in the absence of TARPs. The two mechanisms of TARP modulation suggested by the single-molecule data may act in concert. For example, by shifting the equilibrium of apo LBD states toward those that are initially more cleft closed, TARPs might promote access to agonist-bound states that favor activation over desensitization. Further resolution of these issues awaits single-molecule FRET data on intact receptors and AMPAR structures that include both pore-forming subunits and TARPs. At present, the data presented here are consistent with an emerging view that TARP modulation of AMPAR function may be associated with alterations of the entire conformational landscape (Landes et al., 2011; Lau and Roux, 2011; Yao et al., 2013) rather than acting only on discrete steps in the reaction pathway that leads to receptor activation.
Although we have shown that the changes in LBD closure and/or stability play an important role in TARP modulation of AMPARs, it is likely that there are additional structural modulations that may be at play as well. For example, a recent study has found an important role of the C-tail of γ-2 in control of polyamine block. Specifically, removal of the γ-2 C-tail reduces the attenuation of polyamine block by TARPs while simultaneously increasing the effect on weighted conductance (Soto et al., 2014). Moreover, the structural basis for TARP’s ability to accelerate recovery from desensitization has yet to be addressed. Cryoelectron microscopy studies have argued that the desensitized states of AMPARs exhibit a transition from twofold to fourfold symmetry at the LBD layer (Meyerson et al., 2014). It seems likely that TARPs must exert some additional structural effects beyond LBD cleft closure to more rapidly reverse this symmetry transition. Here, we have provided the first direct evidence that TARPs alter the conformations of the AMPAR LBD, but it is presently unknown if this mechanism also underlies modulation by the other auxiliary proteins such as cornichons (Schwenk et al., 2009; Coombs et al., 2012), CMPK44 (von Engelhardt et al., 2010), and GSG-1L (Shanks et al., 2012), as well as the Neto class of proteins, which regulate KARs (Zhang et al., 2009; Copits et al., 2011). Future studies may uncover other mechanisms in these cases, which may ultimately help in gaining a more complete understanding of the structural mechanism of iGluR modulation by auxiliary proteins.
Acknowledgments
This work is dedicated to the memory of Prof. John F. MacDonald.
This work was supported in part by the National Institutes of Health (grant GM094246), American Heart Association (grant 11GRNT7890004), Harry S. and Isabel C. Cameron Foundation Fellowship (to S.S. Ramaswamy), and American Heart Association Postdoctoral Fellowship (to D.M. MacLean).
The authors declare no competing financial interests.
Author contributions: D.M. MacLean, S.S. Ramaswamy, and M. Du performed the experiments. D.M. MacLean, J.R. Howe, and V. Jayaraman designed the experiments, interpreted the results, and wrote the manuscript.
Sharona E. Gordon served as editor.
Note added in proof. While this work was in press, the following publication provided evidence for direct interactions between stargazin and the LBD (and NTD) of GluA2 subunits: Cais et al. 2014. Cell Reports. http://dx.doi.org/10.1016/j.celrep.2014.09.029
Footnotes
Abbreviations used in this paper:
- AMPAR
- AMPA receptor
- CTZ
- cyclothiazide
- FRET
- Förster resonance energy transfer
- KA
- kainate
- LBD
- ligand-binding domain
- LRET
- luminescence resonance energy transfer
- TARP
- transmembrane AMPAR regulatory protein
References
- Armstrong N., and Gouaux E.. 2000. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 28:165–181 10.1016/S0896-6273(00)00094-5 [DOI] [PubMed] [Google Scholar]
- Armstrong N., Mayer M., and Gouaux E.. 2003. Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc. Natl. Acad. Sci. USA. 100:5736–5741 10.1073/pnas.1037393100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C.H., St-Gelais F., Zhang W., Tomita S., and Howe J.R.. 2007. Two families of TARP isoforms that have distinct effects on the kinetic properties of AMPA receptors and synaptic currents. Neuron. 55:890–904 10.1016/j.neuron.2007.08.024 [DOI] [PubMed] [Google Scholar]
- Cokić B., and Stein V.. 2008. Stargazin modulates AMPA receptor antagonism. Neuropharmacology. 54:1062–1070 10.1016/j.neuropharm.2008.02.012 [DOI] [PubMed] [Google Scholar]
- Coombs I.D., Soto D., Zonouzi M., Renzi M., Shelley C., Farrant M., and Cull-Candy S.G.. 2012. Cornichons modify channel properties of recombinant and glial AMPA receptors. J. Neurosci. 32:9796–9804 10.1523/JNEUROSCI.0345-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copits B.A., Robbins J.S., Frausto S., and Swanson G.T.. 2011. Synaptic targeting and functional modulation of GluK1 kainate receptors by the auxiliary neuropilin and tolloid-like (NETO) proteins. J. Neurosci. 31:7334–7340 10.1523/JNEUROSCI.0100-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Remedios C.G., and Moens P.D.. 1995. Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. J. Struct. Biol. 115:175–185 10.1006/jsbi.1995.1042 [DOI] [PubMed] [Google Scholar]
- Du M., Reid S.A., and Jayaraman V.. 2005. Conformational changes in the ligand-binding domain of a functional ionotropic glutamate receptor. J. Biol. Chem. 280:8633–8636 10.1074/jbc.C400590200 [DOI] [PubMed] [Google Scholar]
- Gonzalez J., Rambhadran A., Du M., and Jayaraman V.. 2008. LRET investigations of conformational changes in the ligand binding domain of a functional AMPA receptor. Biochemistry. 47:10027–10032 10.1021/bi800690b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M., O’Shea-Greenfield A., Rogers S.W., and Heinemann S.. 1989. Cloning by functional expression of a member of the glutamate receptor family. Nature. 342:643–648 10.1038/342643a0 [DOI] [PubMed] [Google Scholar]
- Howe J.R.2013. CrossTalk proposal: TARPs modulate AMPA receptor gating transitions. J. Physiol. 591:1581–1583 10.1113/jphysiol.2012.247486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R., Banke T.G., Mayer M.L., Traynelis S.F., and Gouaux E.. 2003. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 6:803–810 10.1038/nn1091 [DOI] [PubMed] [Google Scholar]
- Jones M.V., Sahara Y., Dzubay J.A., and Westbrook G.L.. 1998. Defining affinity with the GABAA receptor. J. Neurosci. 18:8590–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körber C., Werner M., Kott S., Ma Z.L., and Hollmann M.. 2007. The transmembrane AMPA receptor regulatory protein gamma 4 is a more effective modulator of AMPA receptor function than stargazin (gamma 2). J. Neurosci. 27:8442–8447 10.1523/JNEUROSCI.0424-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott S., Werner M., Körber C., and Hollmann M.. 2007. Electrophysiological properties of AMPA receptors are differentially modulated depending on the associated member of the TARP family. J. Neurosci. 27:3780–3789 10.1523/JNEUROSCI.4185-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landes C.F., Rambhadran A., Taylor J.N., Salatan F., and Jayaraman V.. 2011. Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nat. Chem. Biol. 7:168–173 10.1038/nchembio.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.Y., and Roux B.. 2011. The hidden energetics of ligand binding and activation in a glutamate receptor. Nat. Struct. Mol. Biol. 18:283–287 10.1038/nsmb.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.Y., Salazar H., Blachowicz L., Ghisi V., Plested A.J., and Roux B.. 2013. A conformational intermediate in glutamate receptor activation. Neuron. 79:492–503 10.1016/j.neuron.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H., Mosbacher J., Melcher T., Höger T., Geiger J.R., Kuner T., Monyer H., Higuchi M., Bach A., and Seeburg P.H.. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 266:1709–1713 10.1126/science.7992055 [DOI] [PubMed] [Google Scholar]
- MacLean D.M.2013. CrossTalk opposing view: TARPs modulate AMPA receptor conformations before the gating transitions. J. Physiol. 591:1585–1586 10.1113/jphysiol.2012.247726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean D.M., and Bowie D.. 2011. Transmembrane AMPA receptor regulatory protein regulation of competitive antagonism: a problem of interpretation. J. Physiol. 589:5383–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K., Stroud R.M., Nicoll R.A., and Hays F.A.. 2007. TARP auxiliary subunits switch AMPA receptor antagonists into partial agonists. Science. 318:815–817 10.1126/science.1146317 [DOI] [PubMed] [Google Scholar]
- Menuz K., O’Brien J.L., Karmizadegan S., Bredt D.S., and Nicoll R.A.. 2008. TARP redundancy is critical for maintaining AMPA receptor function. J. Neurosci. 28:8740–8746 10.1523/JNEUROSCI.1319-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K., Kerchner G.A., O’Brien J.L., and Nicoll R.A.. 2009. Critical role for TARPs in early development despite broad functional redundancy. Neuropharmacology. 56:22–29 10.1016/j.neuropharm.2008.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson J.R., Kumar J., Chittori S., Rao P., Pierson J., Bartesaghi A., Mayer M.L., and Subramaniam S.. 2014. Structural mechanism of glutamate receptor activation and desensitization. Nature. 514:328–334 10.1038/nature13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A., Kolleker A., Ayalon G., Gillor M., Osten P., and Stern-Bach Y.. 2005. Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J. Neurosci. 25:2682–2686 10.1523/JNEUROSCI.4834-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanoudjame G., Du M., Mankiewicz K.A., and Jayaraman V.. 2006. Allosteric mechanism in AMPA receptors: A FRET-based investigation of conformational changes. Proc. Natl. Acad. Sci. USA. 103:10473–10478 10.1073/pnas.0603225103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Cooper D., Poddar N., MacLean D.M., Rambhadran A., Taylor J.N., Uhm H., Landes C.F., and Jayaraman V.. 2012. Role of conformational dynamics in α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. J. Biol. Chem. 287:43557–43564 10.1074/jbc.M112.371815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A., Gonzalez J., and Jayaraman V.. 2011. Conformational changes at the agonist binding domain of the N-methyl-d-aspartic acid receptor. J. Biol. Chem. 286:16953–16957 10.1074/jbc.M111.224576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A., Armstrong N., Gouaux J.E., and Howe J.R.. 2005. AMPA receptor binding cleft mutations that alter affinity, efficacy, and recovery from desensitization. J. Neurosci. 25:3752–3762 10.1523/JNEUROSCI.0188-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E., Sizemore M., Karimzadegan S., Chen L., Bredt D.S., and Nicoll R.A.. 2002. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc. Natl. Acad. Sci. USA. 99:13902–13907 10.1073/pnas.172511199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J., Harmel N., Zolles G., Bildl W., Kulik A., Heimrich B., Chisaka O., Jonas P., Schulte U., Fakler B., and Klöcker N.. 2009. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 323:1313–1319 10.1126/science.1167852 [DOI] [PubMed] [Google Scholar]
- Shanks N.F., Savas J.N., Maruo T., Cais O., Hirao A., Oe S., Ghosh A., Noda Y., Greger I.H., Yates J.R. III, and Nakagawa T.. 2012. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Reports. 1:590–598 10.1016/j.celrep.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley C., Farrant M., and Cull-Candy S.G.. 2012. TARP-associated AMPA receptors display an increased maximum channel conductance and multiple kinetically distinct open states. J. Physiol. 590:5723–5738 10.1113/jphysiol.2012.238006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh R.E., MacLean D.M., and Jayaraman V.. 2013. Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-d-aspartate (NMDA) receptor. J. Biol. Chem. 288:22555–22564 10.1074/jbc.M113.482356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Keinänen K., Verdoorn T.A., Wisden W., Burnashev N., Herb A., Köhler M., Takagi T., Sakmann B., and Seeburg P.H.. 1990. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 249:1580–1585 10.1126/science.1699275 [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., and Seeburg P.H.. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 67:11–19 10.1016/0092-8674(91)90568-J [DOI] [PubMed] [Google Scholar]
- Soto D., Coombs I.D., Gratacòs-Batlle E., Farrant M., and Cull-Candy S.G.. 2014. Molecular mechanisms contributing to TARP regulation of channel conductance and polyamine block of calcium-permeable AMPA receptors. J. Neurosci. 34:11673–11683 10.1523/JNEUROSCI.0383-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S., Adesnik H., Sekiguchi M., Zhang W., Wada K., Howe J.R., Nicoll R.A., and Bredt D.S.. 2005. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 435:1052–1058 10.1038/nature03624 [DOI] [PubMed] [Google Scholar]
- von Engelhardt J., Mack V., Sprengel R., Kavenstock N., Li K.W., Stern-Bach Y., Smit A.B., Seeburg P.H., and Monyer H.. 2010. CKAMP44: A brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 327:1518–1522 10.1126/science.1184178 [DOI] [PubMed] [Google Scholar]
- Yao Y., Belcher J., Berger A.J., Mayer M.L., and Lau A.Y.. 2013. Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure. 21:1788–1799 10.1016/j.str.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Cho Y., Lolis E., and Howe J.R.. 2008. Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J. Neurosci. 28:932–943 10.1523/JNEUROSCI.3309-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., St-Gelais F., Grabner C.P., Trinidad J.C., Sumioka A., Morimoto-Tomita M., Kim K.S., Straub C., Burlingame A.L., Howe J.R., and Tomita S.. 2009. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 61:385–396 10.1016/j.neuron.2008.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Devi S.P., Tomita S., and Howe J.R.. 2014. Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors. Eur. J. Neurosci. 39:1138–1147 10.1111/ejn.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Erreger K., Low C.M., Banke T., Lee C.J., Conn P.J., and Traynelis S.F.. 2001. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat. Neurosci. 4:894–901 10.1038/nn0901-894 [DOI] [PubMed] [Google Scholar]