Calcium signaling mediated by STIM1 and Orai1 activates Src to promote invadopodium assembly while simultaneously promoting MT1-MMP recycling to the plasma membrane to promote ECM degradation.
Abstract
Ca2+ signaling has been increasingly implicated in cancer invasion and metastasis, and yet, the underlying mechanisms remained largely unknown. In this paper, we report that STIM1- and Orai1-mediated Ca2+ oscillations promote melanoma invasion by orchestrating invadopodium assembly and extracellular matrix (ECM) degradation. Ca2+ oscillation signals facilitate invadopodial precursor assembly by activating Src. Disruption of Ca2+ oscillations inhibited invadopodium assembly. Furthermore, STIM1 and Orai1 regulate the proteolysis activity of individual invadopodia. Mechanistically, Orai1 blockade inhibited the recycling of MT1–matrix metalloproteinase (MMP) to the plasma membrane and entrapped MT1-MMP in the endocytic compartment to inhibit ECM degradation. STIM1 knockdown significantly inhibited melanoma lung metastasis in a xenograft mouse model, implicating the importance of this pathway in metastatic dissemination. Our findings provide a novel mechanism for Ca2+-mediated cancer cell invasion and shed new light on the spatiotemporal organization of store-operated Ca2+ signals during melanoma invasion and metastasis.
Introduction
Focalized proteolysis by invasive cells is essential for the remodeling of ECM in multiple physiological processes, including bone resorption, immune surveillance, and organ development (Gimona et al., 2008). This feature is exploited by malignant cells to promote invasion and metastasis during cancer progression (Sabeh et al., 2009; Murphy and Courtneidge, 2011). Invadopodia are actin-rich membrane protrusions mediating focal ECM degradation in malignant cancer cells (Linder, 2007; Wolf et al., 2007; Murphy and Courtneidge, 2011). The assembly of invadopodia is initiated in response to the focal generation of phosphatidylinositol-3,4-biphosphate and the activation of the nonreceptor tyrosine kinase Src, which recruits adaptor protein TKS5 and cortactin to initiate assembly of the actin core of invadopodium (Seals et al., 2005; Artym et al., 2006; Oikawa et al., 2008; Oser et al., 2009; Yamaguchi and Oikawa, 2010). Upon maturation, invadopodia recruit and secrete proteinases such as membrane type 1 (MT1)–matrix metalloproteinase (MMP), MMP2, and MMP9 to degrade ECM and facilitate invasion (Artym et al., 2006; Clark et al., 2007; Clark and Weaver, 2008; Oser et al., 2009). Signaling molecules downstream of the ubiquitous secondary messenger Ca2+ have been previously implicated in invadopodium regulation (Baldassarre et al., 2003; Alexander et al., 2008; Cortesio et al., 2008). However, the role of Ca2+ signaling in invadopodium modulation is not known.
Store-operated calcium entry (SOCE) is a Ca2+-entry mechanism regulated by extracellular stimuli (Putney, 1986). SOCE is induced in response to the activation of plasma membrane receptors and subsequent Ca2+ release from the endoplasmic reticulum (Hogan et al., 2010). Upon Ca2+ release, the endoplasmic reticulum Ca2+ sensor STIM1 oligomerizes and translocates to the junction between plasma membrane and endoplasmic reticulum to activate the plasma membrane pore-forming unit Orai1, which induces SOCE (Liou et al., 2005; Roos et al., 2005; Feske et al., 2006; Vig et al., 2006).
We previously reported that store-operated calcium channel proteins STIM1 and Orai1 were critical for breast cancer cell migration, invasion, and metastasis (Yang et al., 2009), and there was accumulating evidence suggesting that hyperactive SOCE promotes cancer progression (Berry et al., 2011; Chen et al., 2011, 2013a,b; Hou et al., 2011; Hu et al., 2011; Huang et al., 2011; Chang et al., 2012; Fedida-Metula et al., 2012; Wang et al., 2012, 2015; Chantôme et al., 2013). More recently, N-methyl-d-aspartate receptor (NMDA)– and TRPM7-mediated Ca2+ signals were shown to promote cancer invasion and metastasis (Middelbeek et al., 2012; Li and Hanahan, 2013; Davis et al., 2014). However, it remains unclear how Ca2+ signals are organized, spatially or temporally, to mobilize cancer invasion machinery and to promote metastasis.
Here, we examine the hypothesis that dysregulated Ca2+ signals in cancer cells promote invasion through focalized proteolysis and ECM remodeling. We unexpectedly discovered that STIM1- and Orai1-mediated SOCE in melanoma cells was organized in the form of persistent Ca2+ oscillations and regulated both the assembly and activity of invadopodium. Our findings bring insight into spatiotemporal organization of Ca2+ signals during cancer invasion and metastasis and shed new light on the role of dysregulated Ca2+ signals in cancer malignancy.
Results
Ca2+ is required for invadopodium formation and ECM degradation
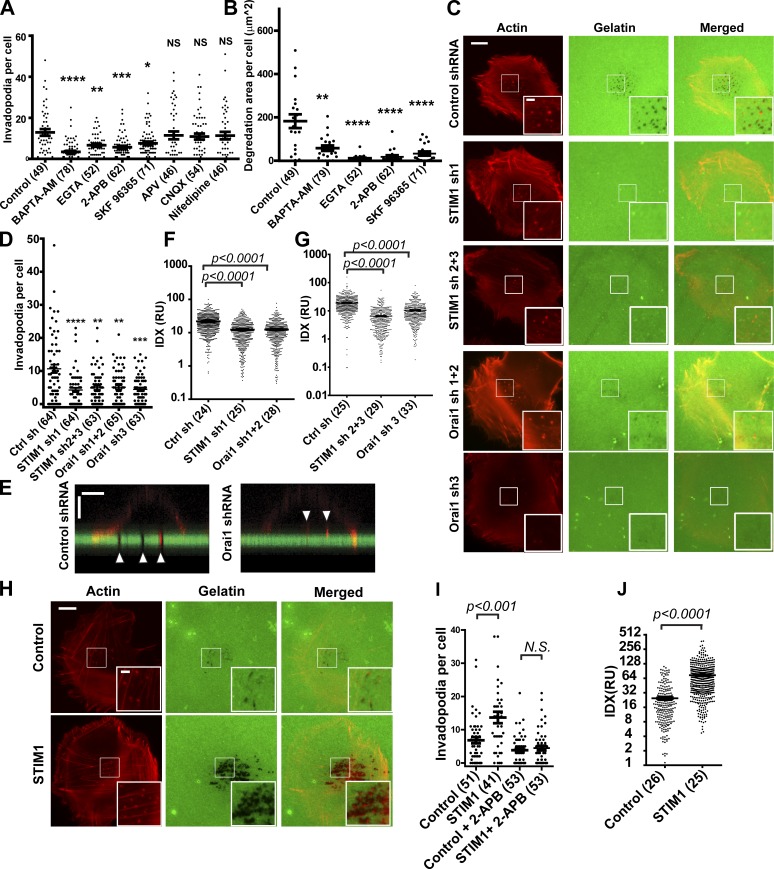
When plated on gelatin-coated coverslips, WM793 human melanoma cells assembled invadopodia within 4 h (Fig. S1, A and B). To investigate the role of Ca2+ in invadopodium regulation, WM793 cells were treated with the membrane-permeable Ca2+ chelator BAPTA-AM. Buffering of cytosolic Ca2+ remarkably reduced the number of invadopodia in WM793 cells and focalized gelatin degradation, suggesting that Ca2+ signals were a critical regulator of invadopodium formation and function (Fig. S1 C and Fig. 1, A and B). Similar inhibition of invadopodium formation and activity was observed when the extracellular Ca2+ was buffered with 0.5 mM EGTA (Fig. S1 C and Fig. 1, A and B). Treatment with nifedipine (L-type voltage-gated Ca2+ channel blocker), APV (NMDA blocker), and CNQX (AMPA receptor blocker) had no noticeable effects on invadopodium number or gelatin degradation (Fig. S1 C and Fig. 1, A and B). In contrast, the SOCE inhibitor 2-APB significantly decreased invadopodium number in WM793 cells and almost abolished focalized proteolysis by WM793 cells (Fig. S1 C and Fig. 1, A and B). The importance of SOCE in the modulation of invadopodium formation and ECM degradation was further confirmed by using a different SOCE blocker (SKF96365) in CHL-1 and WM245 melanoma cell lines (Fig. S1, D–G). Collectively, these data implied that Ca2+ influx mediated by SOCE channels is a critical regulator of invadopodium formation and ECM degradation.
Figure 1.
STIM1 and Orai1 are crucial for invadopodium formation and activity. (A and B) Scattered dot plot showing the effects of Ca2+ chelators and Ca2+ blockers on invadopodium number per cell (A) and gelatin degradation area per cell (B). Horizontal bars represent means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 as calculated by two-tailed Mann–Whitney test. NS indicates not statistically significant. (C) Representative fluorescence micrographs showing effects of STIM1 and Orai1 shRNA on invadopodium formation and focalized proteolysis in WM793 cells. (D) Scattered dot plot showing that depletion of STIM1 and Orai1 with shRNAs decreased the mean number of invadopodia per cell when compared with WM793 cells expressing control shRNA. (E) Orthogonal views of confocal z stacks showing Orai1 shRNA inhibited gelatin degradation and penetration into Alexa Fluor 488–gelatin coating by invadopodia (arrowheads) in WM793 cells, with WM793 cells expressing control shRNA as a control. Bars: (horizontal) 10 µm; (vertical) 2 µm. (F and G) Scattered dot plot showing that inhibition of SOCE with STIM1 shRNA and Orai1 shRNA inhibited the focalized proteolysis activity of individual invadopodium when compared with control shRNA, as determined by the quantification of IDX. (H) Representative fluorescence micrographs showing that ectopic STIM1 overexpression promoted invadopodium formation and focalized proteolysis of Alexa Fluor 488–gelatin. (I) Scattered dot plot showing that ectopic STIM1 increased numbers of invadopodia per cell, which was abrogated by 100 µM 2-APB; n = 51, 47, 52, and 52 for control, STIM1, control + 2-APB, and STIM1 + 2-APB, respectively. (J) Effects of STIM1 overexpression on the degradation activity of individual invadopodium in WM793 cells. Insets in C and H are magnified views of the boxed areas in the main images. RU, relative unit. Bars (main images) 10 µm; (insets) 2 µm. Two-tailed p-values were determined by Mann-Whitney test or by unpaired Student’s t test after log transformation. Horizontal bars represent means ± SEM. The numbers of cells used for quantitation are indicated in the parenthesis of respective figure labeling, and representative results from at least three similar independent experiments are presented. Ctrl sh, control shRNA.
STIM1 and Orai1 are critical for invadopodium formation and activity
To investigate the role of SOCE in invadopodium regulation, we used shRNA to knock down the expression of STIM1 and Orai1, two key components of store-operated Ca2+ channels. The inhibition of SOCE in WM793 cells by STIM1 and Orai1 shRNA was confirmed with the use of a Fluo4-based Ca2+ assay (Fig. S1 H). We next investigated the effects of STIM1 and Orai1 depletion on invadopodium formation and ECM degradation. As shown in Fig. 1 (C and D), Orai1 shRNA and STIM1 shRNA treatment resulted in ∼40–50% reduction in the mean number of invadopodia per cell when compared with control shRNA cells. Moreover, the area of gelatin degradation per cell was inhibited by 70–80% when STIM1 and Orai1 were depleted (Fig. S1 I).
We noted that the inhibitory effects of SOCE blockade, by shRNA or by pharmacological inhibitors, were more robust on gelatin degradation than on invadopodium number. The invadopodia in Orai1 shRNA cells had shallower degradation of ECM when compared with control cells, as revealed by confocal microscopy (Fig. 1 E), implicating lower proteolysis activity for these invadopodia. To evaluate the effect of STIM1 and Orai1 on invadopodium activity, we use a new quantification method, invadopodium degradation index (IDX), to measure activity of each individual invadopodium. WM793 cells were allowed to attach to gelatin-coated coverslips in the presence of broad spectrum MMP inhibitor GM6001 for 12 h. The gelatin degradation activity of invadopodia was inhibited in the presence of GM6001. The degradation of Alexa Fluor 488–labeled gelatin was initiated by washing away GM6001. After 4-h degradation, the proteolysis activity of individual invadopodia was determined through quantifying IDX, which corresponds to total gelatin degraded by individual invadopodia in a given period of time. The mean degradation activity of individual invadopodium was inhibited by ∼50% when STIM1 or Orai1 was depleted by shRNA (Fig. 1, F and G), as a result of decreases in both degradation area and Δ intensity per invadopodium (Fig. S1, J and K).
Next, we investigated whether activation of SOCE in malignant cancer cells was sufficient to promote ECM degradation. The ectopic expression of STIM1, but not Orai1, has been previously shown to promote SOCE (Soboloff et al., 2006). As shown in Fig. 1 (H–J), ectopic STIM1 expression increased the number of invadopodia per cell and enhanced the proteolysis activity of individual invadopodium. The stimulation of invadopodium formation by STIM1 was abrogated by SOCE blocker 2-APB (Fig. 1 I). Collectively, these data indicated that STIM1 and Orai1 regulated invadopodium formation as well as the proteolysis activity of individual invadopodium.
SOCE mediates Ca2+ oscillation to regulate the assembly of invadopodial precursor
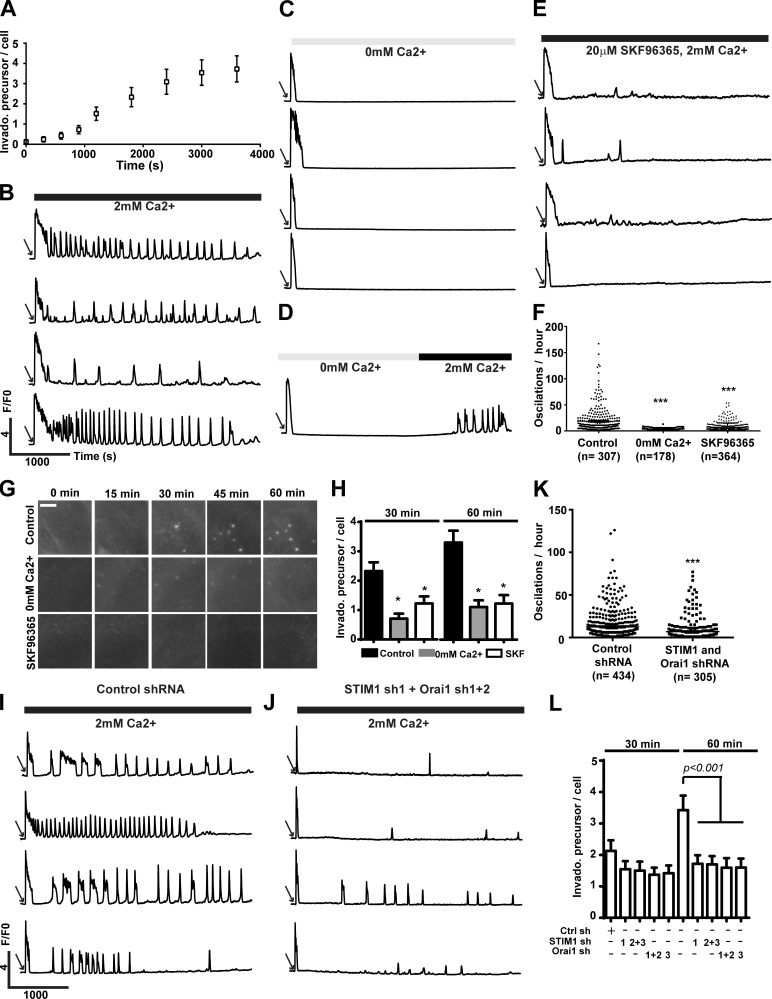
Invadopodia are assembled as actin-rich invadopodial precursors, which recruit MT1-MMP upon maturation to degrade ECM (Artym et al., 2006). When stimulated with 10% FBS, serum-starved WM793 cells started assembling invadopodial precursor 10–15-min after stimulation (Fig. 2 A). Intriguingly, serum treatment also stimulated immediate but transient Ca2+ release and sustained Ca2+ signals in the form of Ca2+ oscillation (Fig. 2 B and Video 1). Buffering extracellular Ca2+ with EGTA didn’t affect the transient Ca2+ release but almost eliminated the Ca2+ oscillation signals (Fig. 2 C and Video 2). Importantly, Ca2+ oscillation was restored after adding back Ca2+ to the extracellular medium, strongly suggesting that Ca2+ influx was crucial for Ca2+ oscillation (Fig. 2 D and Video 3). To determine whether SOCE was responsible for Ca2+ oscillation, we also examined the effects of SKF96365 on serum-stimulated Ca2+ oscillations in WM793 cells (Fig. 2 E and Video 4). EGTA buffering and SOCE blockade with SKF96365 significantly reduced the Ca2+ oscillation frequencies from 18.6 ± 1.4/h to 3.3 ± 0.1/h and 7.2 ± 0.4/h, respectively, suggesting that SOCE was a major mediator of serum-stimulated Ca2+ oscillation (Fig. 2 F). We further determine the role of Ca2+ oscillation in the assembly of invadopodial precursor through live cell imaging and fluorescent staining on fixed cells (Fig. 2, G and H; and Videos 5–7). Inhibition of Ca2+ oscillations with EGTA or SKF96365 remarkably decreased the numbers of invadopodia precursor assembly (Fig. 2, G and H). To further critically evaluate the role of SOCE in Ca2+ oscillation and invadopodia precursor assembly, we used shRNAs to knockdown STIM1 and Orai1 in WM793 cells. As expected, STIM1 and Orai1 shRNA significantly decreased the frequency of calcium oscillation when compared with control shRNA-expressing cells (P < 0.001; Fig. 2, I–K). Inhibition of Ca2+ oscillation with STIM1 or Orai1 shRNA also decreased the number of serum-stimulated invadopodial precursor (Fig. 2 L). Collectively, our data suggested that SOCE-mediated Ca2+ oscillations are critical for assembly of invadopodial precursors.
Figure 2.
SOCE-mediated Ca2+ oscillations regulate invadopodial precursor assembly. (A) WM793 cells were starved overnight, and invadopodial (Invado.) precursor assembly were initiated by stimulation with 10% FBS. Data are presented as means ± SEM (n = 33). (B, C, and E) Representative traces from single cell Ca2+ imaging experiments. WM793 cells were stimulated with 10% FBS in the presence of 2 mM Ca2+ (B, n = 307), 0 mM Ca2+ (C, n = 178), or 2 mM Ca2+ and 20 µM SKF96365 (E, n = 364). (D) Representative traces showing restoring the extracellular Ca2+ from 0 to 2 mM also restored Ca2+ oscillations (n = 150). (F) Scattered dot plot showing quantification of Ca2+ oscillation frequencies in control (2 mM Ca2+; B), 0 mM Ca2+ (C), or 20 µM SKF96365 (E) groups. Each data point represents oscillation frequency in a single cell (means ± SEM; n = 86). ***, P < 0.001 as determined by two-tailed Mann–Whitney test. (G) Live cell imaging to show serum-stimulated assembly of invadopodial precursors in control condition or in conditions that suppressed Ca2+ oscillation (0 mM Ca2+ and 20 µM SKF96365). Bar, 5 µm. (H) Cells were serum starved overnight, stimulated with 10% FBS under different conditions as indicated, fixed after 30 or 60 min, and stained with phalloidin for invadopodial precursors. *, P < 0.05 as determined by two-tailed Mann–Whitney test. Data presented are means ± SEM. (I and J) Representative trace showing Ca2+ oscillations in WM793 cells expressing control shRNA (I, n = 434) or STIM1 and Orai1 shRNA (J, n = 305). (K) Quantification of Ca2+ oscillation frequencies in control shRNA (I) or STIM1- and Orai1 shRNA–expressing (J) WM793 cells. Data presented are means ± SEM. (L) Effects of STIM1 and Orai1 shRNA on invadopodium precursor assembly (n = 86). Data presented are mean ± SEM. Cells were treated as described in H. Arrows in B–E, I, and J indicate stimulation with 10% FBS. Ctrl sh, control shRNA. F/F0 is defined as ratio between fluorescence at a given time (F) and fluorescence at time 0 (F0).
To determine whether SOCE regulate invadopodium lifetime, WM793 cells stably expressing Lifeact-mAPPLE were stimulated with 10% FBS after overnight starvation, and the assembly and disassembly of invadopodia were recorded by time-lapse live cell imaging. The effects of SOCE manipulation on invadopodium lifetime were analyzed by Kaplan–Meier survival analysis (Fig. S2). Neither SOCE activation (through STIM1 overexpression) nor inhibition (through 2-APB treatment or STIM1 and Orai1 knockdown) had a significant effect on invadopodium lifetime in WM793 cells.
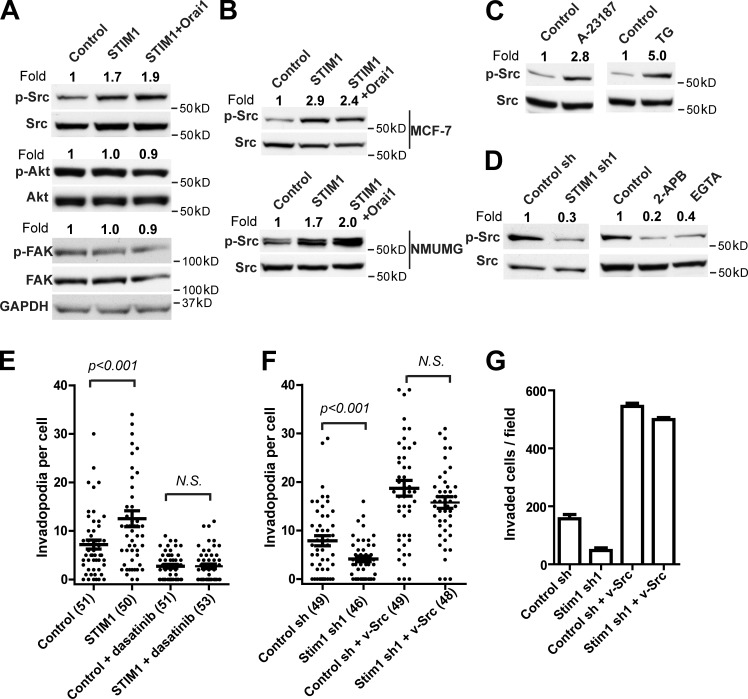
SOCE promotes invadopodium formation through Src activation
To understand the molecular mechanisms by which STIM1 and Orai1 regulate invadopodium formation, we investigated the effects of SOCE on a panel of protein kinases. As shown in Fig. 3 A, ectopic expression of STIM1 or STIM1 together with Orai1 increased the levels of phosphotyrosine 416 Src (pY416 Src) in WM793 cells by about twofold without affecting total Src levels, suggesting activation of Src by SOCE. In contrast, the levels of phospho-FAK and phospho-Akt were not affected by ectopic STIM1 and Orai1 (Fig. 3 A). The increase in pY416 Src levels after ectopic expression of STIM1 and Orai1 was also observed in MCF-7 (a human breast cancer cell line) and NMuMG (a normal mouse mammary epithelial cell line) cells (Fig. 3 B). Induction of Ca2+ influx using thapsigargin or ionophore A23187 rapidly increased pY416 Src levels within 30 min, suggesting that Ca2+ signals were sufficient to activate Src (Fig. 3 C). Moreover, inhibition of SOCE through STIM1 shRNA, Ca2+ chelator EGTA, or pharmacological inhibitor 2-APB reduced pY416 Src levels in WM793 cells, indicating that SOCE-mediated Ca2+ oscillation was critical for maintaining basal Src activity (Fig. 3 D).
Figure 3.
STIM1 and Orai1 regulate invadopodia through Src. (A) Effects of ectopic STIM1 and Orai1 on the levels of pY416 Src, pT308 Akt, and pY397 FAK in WM793 cells. Adherent WM793 cells expressing control vector, STIM1, or STIM1 plus Orai1 were lysed on plates, and 50 µg total cell lysate was loaded onto each lane. Western blotting was performed with respective antibodies as indicated, with GAPDH as a loading control. (B) Effects of ectopic STIM1 and Orai1 on pY416 Src levels in MCF-7 and NMuMG cells; 50 µg total cell lysate was loaded onto each lane. (C) Induction of Ca2+ influx with A23187 and thapsigargin (TG) increased Src activity. Adherent WM793 cells were treated with 5 µM A23187 and 2 µM thapsigargin for 30 min before being lysed. (D) Inhibition of SOCE with STIM1 shRNA, 100 µM 2-APB, or 0.5 mM EGTA inhibited Src activity. (E) Scattered dot plot showing that STIM1-mediated invadopodia formation was abrogated by treatment with 2.5 nM dasatinib; n = 51, 50, 51, and 53 for control, STIM1, control + dasatinib, and STIM1 + dasatinib, respectively. (F) Scattered dot plot showing ectopic expression of v-Src in WM793 cells rescued invadopodium formation in STIM1 knockdown cells; n = 49, 46, 49, and 48 for control shRNA, STIM1 shRNA, control shRNA + v-Src, and STIM1 shRNA + v-Src, respectively. (G) Ectopic expression of v-Src rescued the inhibition WM793 cell invasion by STIM1 knockdown. The relative levels of p-Src, p-Akt, and p-FAK in A–D were determined through densitometry using ImageJ software. Horizontal bars in E–G represent means ± SEM. Two-tailed p-values are determined by Mann–Whitney test. The numbers of cells used for quantitation were indicated in the parenthesis of respective figure labeling, and representative results from at least three similar independent experiments were presented. sh, shRNA.
To investigate the hypothesis that SOCE modulates invadopodium through Src, we used constitutively active v-Src and the Src-specific kinase inhibitor dasatinib to manipulate Src activity in WM793 cells. Inhibition of Src activity with dasatinib abrogated STIM1-promoted invadopodium formation (Fig. 3 E and Fig. S3 A). Furthermore, ectopic expression of v-Src was able to rescue invadopodium formation and Matrigel invasion after STIM1 knockdown (Fig. 3. F and G; and Fig. S3 B). Collectively, these data indicated that Src was required for the regulation of invadopodia by SOCE.
SOCE blockade inhibits the plasma membrane localization of MT1-MMP
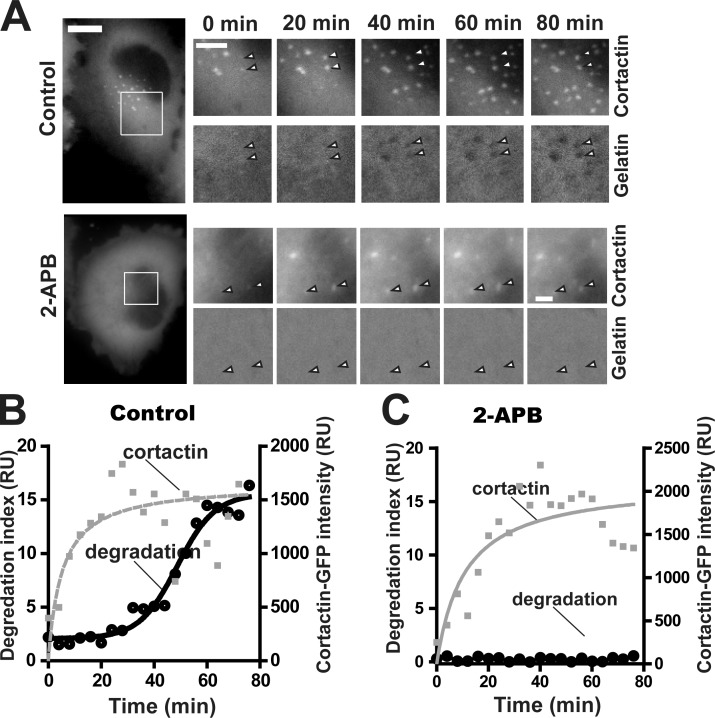
We used WM793 cells expressing cortactin-GFP to investigate the effects of SOCE blockade on invadopodium assembly and ECM degradation kinetics. WM793 cells started assembling invadopodia ∼60–90 min after being plated onto gelatin-coated glass coverslips. The cortactin-GFP signals in invadopodial precursor increased steadily after initiation, reaching a plateau at around 20 min (Fig. 4, A and B). An accelerated ECM degradation phase was detected when the cortactin-GFP signal reached the plateau, suggesting the recruitment of proteinase and maturation around this time. Intriguingly, although SOCE blockade with 2-APB dramatically reduced the number of invadopodium precursors, 2-APB treatment had no obvious effect on the kinetics of the invadopodium assembly (Fig. 4, A and C). Importantly, no degradation of gelatin was detected in the 2-APB–treated cells even 60 min after the cortactin-GFP signal reached plateau, suggesting that SOCE blockade also impaired invadopodium maturation (Fig. 4, A and C).
Figure 4.
SOCE blockade inhibits invadopodium maturation. (A) Live cell imaging showing invadopodium precursor assembly and gelatin degradation by mature invadopodia in control (top) and 100 µM 2-APB–treated (bottom) cells. Arrowheads indicate nascent invadopodia. Bars: (main images) 10 µm; (magnified views) 5 µm. (B and C) Quantification of nascent invadopodium assembly (cortactin-GFP fluorescence intensity) and ECM degradation by nascent invadopodia in control (B) or 2-APB–treated cells (C). Lines in B and C are curve fittings of cortactin-GFP intensity and gelatin degradation index data. The data shown are representative of three experimental repeats. RU, relative unit.
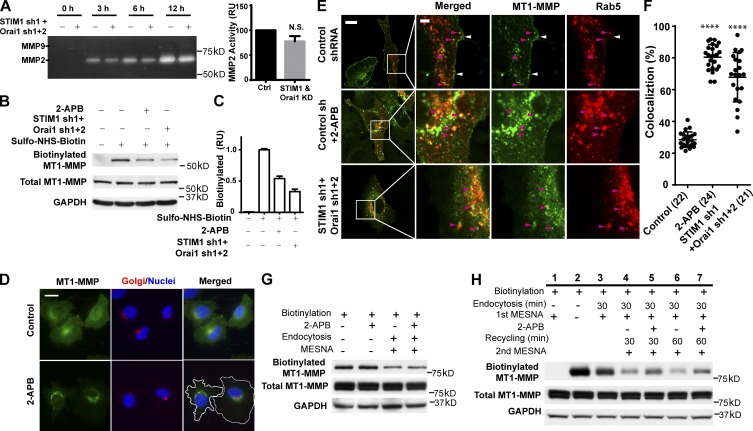
To understand how SOCE regulated the proteolysis activity of invadopodia, we investigated the effects of SOCE blockade on levels of secreted soluble MMPs and membrane-bound MT1-MMP (Fig. 5). There were very little MMP9 present in the conditioned medium, suggesting that WM793 cells secrete mostly MMP2 instead of MMP9. SOCE blockade with STIM1 and Orai1 knockdown had only very modest effect on the levels of secreted MMP2, indicating SOCE regulates invadopodium activity through mechanisms other than soluble MMP (Fig. 5 A).
Figure 5.
SOCE blockade inhibits the membrane expression of MT1-MMP. (A) WM793 cells expressing control shRNA or STIM1 and Orai1 shRNA were plated onto a 6-well plate overnight. The cells were washed and incubated with starvation RPMI 1640 medium. The conditioned medium was collected at various time points as indicated. (left) MMP2 and MMP9 activities in the conditioned medium determined through gelatin zymography. (right) Quantitation of MMP2 zymography activity in control (ctrl) or STIM1- and Orai1 shRNA–expressing cells (STIM1 & Orai1 knockdown [KD]) at 6 h (mean ± SD from three independent repeats). Ctrl, control; RU, relative unit. (B) Effects of SOCE blockade with 100 µM 2-APB or STIM1 and Orai1 shRNA on plasma membrane expression of endogenous MT1-MMP. The plasma membrane MT1-MMP in control WM793 cells, 2-APB–treated cells, or STIM1 and Orai1 knockdown cells was labeled with sulfo-NHS-SS-biotin, affinity purified with NeutrAvidin, and detected by Western blotting. (C) Quantification of the results in B with densitometry (means ± SD from three measurements). (D) WM793 cells stably expressing MT1-MMP–EGFP were treated with or without 100 µM 2-APB for 2.5 h and stained with anti-p230 trans-Golgi antibody and DAPI to visualize trans-Golgi network and nuclei. White lines indicate cell boundaries. Bar, 20 µm. (E) Confocal micrographs showing that SOCE blockade with 2-APB or STIM1 and Orai1 knockdown resulted in entrapment of MT1-MMP in the Rab5-positive endosomes. WM793 cells stably expressing MT1-MMP–EGFP were transiently transfected with mRFP-Rab5 to visualize endosomal compartments. White and magenta arrowheads indicate membrane and endosomes, respectively. Bars: (main images) 10 µm; (magnified views) 2 µm. (F) Quantitation of the colocalization between MT1-MMP–EGFP and mRFP-Rab5 in WM793 cells using LAS AF lite V2.0 (means ± SD). Each cell was manually segmented using the region of interest selection tool. The background threshold was identical for all the images. The numbers of cells used for quantitation are indicated in the parenthesis. Representative results from three independent experiments are presented. ****, P < 0.0001. (G) Biotinylated MT1-MMP–EGFP was allowed to be endocytosed in the presence or absence of 100 µM 2-APB. The plasma membrane biotinylation was removed by MESNA treatment, and the endocytosed MT1-MMP–EGFP were purified with NeutrAvidin beads and detected by Western blotting. (H) MT1-MMP–EGFP recycling experiment showing the effects of SOCE blockade. Plasma membrane MT1-MMP–EGFP was labeled through biotinylation. Cells were lysed immediately (lane 2), lysed after an immediate MESNA treatment without endocytosis (lane 1), or allowed to undergo endocytosis for 30 min and then treated with MESNA (lane 3–7). Cells were then either lysed immediately (lane 3) or allowed to undergo recycling for 30 or 60 min in the presence or absence of 100 µM 2-APB as indicated and then treated again with MENSA (lanes 4–7). Biotinylated MT1-MMP–EGFP were purified with NeutrAvidin beads and detected by Western blotting.
Next, we investigated the effect of SOCE blockade on MT1-MMP total protein levels and subcellular localization. SOCE blockade with 2-APB or STIM1 and Orai1 double knockdown had no noticeable effect on total protein levels of MT1-MMP (Fig. 5 B). However, surprisingly, SOCE blockade reduced the plasma membrane fraction of MT1-MMP (biotinylated MT1-MMP) by 50–70% (Fig. 5, B and C), suggesting that SOCE regulates invadopodium activity mainly through the subcellular localization of MT1-MMP. Indeed, when stably expressed in WM793 cells, MT1-MMP–EGFP localized to the plasma membrane and perinuclear compartments in the vicinity of trans-Golgi network (Fig. 5 D). Strikingly, after 2-APB treatment for 2.5 h, MT1-MMP–EGFP signals were drastically reduced in the plasma membrane and increased in the perinuclear region, suggesting the translocation of plasma membrane MT1-MMP to intracellular compartments that are reminiscent of endosomes (Fig. 5 D and Video 8).
To investigate whether MT1-MMP was trapped in the endocytic compartment after SOCE blockade, we used mRFP-Rab5 and confocal microscopy to visualize endosomes in WM793 cells. As shown in Fig. 5 E, strong MT1-MMP–EGFP signals in the control cells were detected mostly on the plasma membrane (Fig. 5 E, white arrowheads), although MT1-MMP signals were also detectable in some Rab5-positive endosomes (Fig. 5 E, magenta arrowheads). In sharp contrast, most of the MT1-MMP signals were present at the Rab5 endosomes instead of plasma membrane in 2-APB–treated cells or STIM1 Orai1 double knockdown cells (Fig. 5, E and F).
We reasoned that the entrapment of MT1-MMP in the endocytic compartments could be caused by accelerated endocytosis, depressed recycling, or both. To understand how SOCE regulates the subcellular localization of MT1-MMP, we next examined the effect of SOCE blockade on MT1-MMP endocytosis. The biotinylated MT1-MMP was endocytosed at essentially the same rate in SOCE inhibitor-treated cells as in control cells (Fig. 5 G), suggesting that the accumulation of MT1-MMP in the endocytic compartment might be caused by defective recycling back to the plasma membrane. To examine this possibility, biotinylated MT1-MMP were allowed to be endocytosed, and the remaining biotinylation on the plasma membrane MT1-MMP was removed with 2-mercaptoethane sulfonate (MESNA; Fig. 5 H, lanes 1–3). MESNA treatment before endocytosis completely removed biotinylation on MT1-MMP, confirming the efficacy of this approach (Fig. 5 H, lane 1). After 30-min endocytosis at 37°C, MESNA was able to only partially remove biotin, suggesting endocytosis of plasma membrane MT1-MMP (Fig. 5 H, lanes 2 and 3). The biotinylated MT1-MMP in the endocytic compartment were then allowed to be recycled back to the plasma membrane in the presence of SOCE blocker or vehicle control (Fig. 5 H, lanes 4–7). After 60-min (or 30 min) recycling, there were ∼20% (or 30%) of biotinylated MT1-MMP retained in the intracellular compartments of the control cells (Fig. 5 H, lanes 4 and 6). In contrast, when SOCE was blocked with 2-APB, >40% (or 50%) of biotinylated MT1-MMP remained intracellular after a 60-min (or 30 min) recycling, indicating inhibition of MT1-MMP recycling by SOCE blockade. Collectively, our data suggest that SOCE blockade entraps MT1-MMP in the endocytic compartment through interfering with its recycling to the plasma membrane, which inhibits the ECM degradation activity of invadopodia.
SOCE is critical for melanoma metastasis
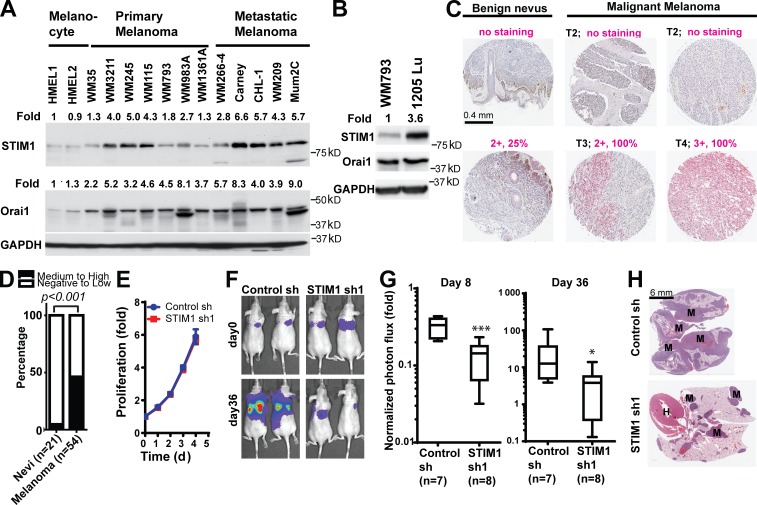
Gaining invasiveness is one of the first and most critical steps of metastasis (Fidler, 2003; Nürnberg et al., 2011). Prognosis for melanoma patients significantly worsens with deeper levels of dermal invasion and when melanoma progresses from a radial growth phase to vertical growth phase (Clark, 1991). Our data suggested that dysregulated SOCE may promote melanoma metastasis and progression. To examine this hypothesis, we first determined the expression of STIM1 and Orai1 in a panel of melanoma cell lines. As shown in Fig. 6 A, STIM1 and Orai1 were overexpressed in most melanoma cells when compared with normal epidermal human melanocytes. The expression levels of STIM1 in melanoma cells derived from metastatic melanoma were higher than cells from primary melanoma (Fig. 6 A). We further compared the STIM1 and Orai1 expression levels between WM793 cells and its highly metastatic 1205Lu subline, which was selected based on its ability to consistently metastasize to the lung in a spontaneous metastasis mouse model (Juhasz et al., 1993). Orai1 levels were similar between the two cell lines, but STIM1 protein expression was further increased by three- to fourfold in 1205Lu cells, implicating a role for STIM1 in melanoma metastasis (Fig. 6 B).
Figure 6.
SOCE is critical for melanoma metastasis. (A) Expression levels of STIM1 and Orai1 in human melanocytes and melanoma cell lines derived from primary melanoma or distant metastases. The relative levels of STIM1 and Orai1 were determined through densitometry using ImageJ software. (B) STIM1 expression was up-regulated in the highly metastatic 1205Lu subline of WM793 cells. (C) Immunohistochemistry staining showing STIM1 overexpression in malignant melanoma when compared with benign nevus. Staining intensity and percentage of positively stained lesion (in magenta) and melanoma T stages (T1–T4, in black) are indicated. (D) Proportions of benign nevi (n = 21) and malignant melanoma (n = 53) with medium to high expression of STIM1; P < 0.001 as determined by two-tailed Fisher’s exact test. (E) STIM1 knockdown had no effect on the proliferation of 1205Lu cells in vitro. Data are shown as means ± SD from triplicates. The data shown are from a single experiment out of three repeats. (F–H) Inhibition of 1205Lu cell lung metastasis by STIM1 knockdown. (F) Representative bioluminescence images showing the lung metastasis of 1205Lu cells stably expressing control shRNA or STIM1 shRNA. (G) Quantification of 1205Lu cell lung metastasis using bioluminescence imaging. Normalized photon flux showed the fold increase in bioluminescence signals (on day 8 and day 36, respectively) over day 0. Box and whiskers are 25–75th and 10–90th percentiles, respectively. *, P < 0.05; ***, P < 0.001. (H) Representative hematoxylin and eosin staining showing the presence of 1205Lu metastases in the lung of nude mice. M, melanoma metastases in the lung; H, heart; sh, shRNA.
To evaluate the clinical significance of SOCE in melanoma progression, we examined a melanoma tissue microarray for levels of STIM1 expression (Fig. 6 C). There was 10-fold higher frequency of medium to high levels of STIM1 expression in malignant melanoma (46%, 25 out of 54 cases) compared with benign nevi (5%, 1 out of 21 cases; Fig. 6 D), suggesting dysregulation of SOCE in melanomas. To define the role of SOCE in melanoma metastasis, we used a luciferase reporter to label 1205Lu cells expressing control shRNA or STIM1 shRNA. STIM1 knockdown did not affect the proliferation of 1205Lu cells in vitro (Fig. 6 E). Luciferase-labeled cells were injected via tail vein into nude mice, and lung metastasis of 1205Lu melanoma cells was monitored using bioluminescence imaging. As shown in Fig. 6 (F and G), we detected significant lung metastasis of 1205Lu control cells by bioluminescence imaging 36 d after mouse xenografting, which was consistent with the highly metastatic nature of this WM793 subline. The lung metastasis of 1205Lu cells were reduced by >80% after STIM1 knockdown, as determined by measuring normalized photon flux from luciferase-labeled melanoma cells on day 8 and day 36 (Fig. 6, F and G). The inhibition of melanoma lung metastasis by STIM1 shRNA was further confirmed by hematoxylin and eosin staining of the lungs harvested from nude mice (Fig. 6 H). Collectively, our data suggested that STIM1 overexpression in melanoma was critical for melanoma metastasis.
Constitutive Ca2+ influx is not able to promote invadopodium formation and melanoma invasion
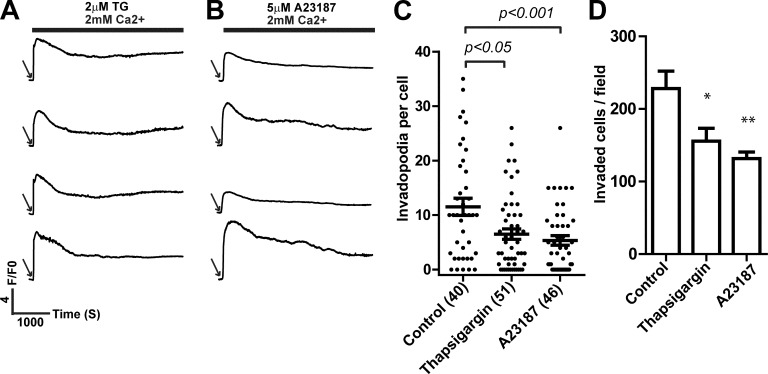
Our data demonstrated that SOCE signals in melanoma cells were organized in oscillatory patterns to regulate invadopodia. It was not clear, however, whether oscillatory organization per se was needed or whether oscillatory frequency was merely indicative of the requirement of net increase in cytosolic Ca2+ during a certain time period. Because constitutive Ca2+ influx, induced by thapsigargin and A23187, was sufficient to activate Src activity (Fig. 3 C), we set out to determine the effects of constitutive Ca2+ influx in melanoma cells. We confirmed that treatment with thapsigargin and A23187 induced constitutive instead of oscillatory increase in cytosolic Ca2+ (Fig. 7, A and B). Intriguingly, treatment with thapsigargin and A23187 inhibited, instead of promoted, invadopodium formation and melanoma invasion (Fig. 7, C and D). These data indicated that the oscillatory organization is required for SOCE to coordinate invadopodium assembly and ECM remodeling.
Figure 7.
Inhibition of invadopodium formation and melanoma cell invasion by constitutive increase in cytosolic Ca2+. (A and B) Representative traces showing the effects of 2 µM thapsigargin (A) and 5 µM A23187 (B) on cytosolic Ca2+ (n > 100, from three independent experiments). Arrows indicate addition of 10% FBS and thapsigargin or A23187. (C and D) Effects of thapsigargin and A23187 induced constitutive Ca2+ increases on invadopodium formation (C) and Matrigel invasion (D) in WM793 cells. The numbers of cells used for quantitation in C are indicated in the parenthesis. Data presented are means ± SE. Representative results from at least three similar independent experiments are presented. *, P < 0.05; **, P < 0.01. F/F0 is defined as ratio between fluorescence at a given time (F) and fluorescence at time 0 (F0). TG, thapsigargin.
Discussion
In this study, we present evidence showing that STIM1- and Orai1-mediated SOCE promotes melanoma invasion and ECM degradation by increasing invadopodium formation and activity. Using a novel image quantification method, we were able to demonstrate that SOCE regulates the proteolysis activity of individual invadopodium. Surprisingly, SOCE regulates invadopodium activity not through the total protein levels of MT1-MMP or secreted soluble MMPs but through the subcellular localization of MT1-MMP to the plasma membrane. The plasma membrane MT1-MMP is believed to be recruited to invadopodia through incorporation of MT1-MMP–containing vesicles at the ECM degradation sites (Bravo-Cordero et al., 2007; Steffen et al., 2008). Although SOCE blockade did not affect MT1-MMP endocytosis, it did dramatically reduce the recycling of endocytosed MT1-MMP back to the plasma membrane, which results in the entrapment of MT1-MMP in Rab5-positive endosomes. SOCE was previously reported to be required for the exocytosis of cytotoxic granules in mast cells and natural killer cells (Ma et al., 2008; Vig et al., 2008; Maul-Pavicic et al., 2011). It is possible that SOCE blockade decreased the membrane expression of MT1-MMP by inhibiting the plasma membrane insertion of MT1-MMP vesicles. It is also worth noting the SOCE regulation of exocytosis appeared to be selective, as the exocytosis of the soluble MMP2 was only very modestly inhibited by SOCE blockade.
Unexpectedly, SOCE signals in melanoma cells were temporally organized in the form of persistent Ca2+ oscillations. It was proposed that Ca2+ oscillations might serve as a mode of digital signaling that dictates the specificity and robustness of downstream signaling cascades through oscillation frequency and amplitude (Bird et al., 2009; Dupont et al., 2011). For any supralinear calcium-dependent process, the time-averaged effect of an oscillatory calcium signal is always greater than the effect of a steady calcium signal of the same net strength. For example, certain threshold-dependent phenomena can be selectively activated in an oscillation frequency-dependent manner. Moreover, because constitutive increase in cytosolic Ca2+ is deleterious and may lead to cell death (Orrenius et al., 2003), Ca2+ oscillations provide the Ca2+ signals necessary for invadopodia formation and activity without causing cytotoxicity. It is also conceivable that invadopodia formation and function require coordinated cycles of high and low calcium signals provided by oscillation, as recently demonstrated in the case of mast cell exocytosis (Wollman and Meyer, 2012). Although constitutive Ca2+ increase was effective in the activation of Src, it inhibited invadopodia formation and melanoma invasion. This is consistent with the notion that temporal oscillation is critical for Ca2+ signals to coordinate invasion and ECM degradation. The SOCE activated by repetitive discharge of store Ca2+ during oscillation also creates subplasmalemmal Ca2+ microdomains to regulate enzyme activity and gene transcription (Chang et al., 2008; Di Capite et al., 2009). Future effort to determine spatial and temporal organization of subplasmalemmal Ca2+ microdomains during melanoma invasion and ECM remodeling is warranted.
Invasion is a critical step during melanoma progression. The prognosis of melanoma patients significantly worsens with deeper levels of invasion, after melanoma breaches the basement membrane separating epidermis from dermis and progresses from radial growth phase to vertical growth phase (Clark, 1991). Our data suggested that SOCE was hyperactivated during melanoma progression, with STIM1 expression increased in malignant melanomas when compared with benign nevi. The overexpression of STIM1 and hyperactivation of SOCE in melanoma likely promoted melanoma invasion and metastasis. Indeed, when STIM1 was knocked down in 1205Lu cells, the lung metastasis of this highly metastatic melanoma cell was dramatically reduced. Our data, together with recently reported findings in breast and cervical cancer (Yang et al., 2009; Chen et al., 2011; Chantôme et al., 2013), suggested that hyperactive SOCE pathways in various cancer might be targeted to inhibit metastasis and progression.
Materials and methods
Antibodies
We used the following antibodies in this study: STIM1 antibody for immunohistochemistry (MA1-19451; Thermo Fisher Scientific), STIM1 antibody for Western blotting (mouse monoclonal; 610954; BD), Orai1 (rabbit polyclonal; O8264; Sigma-Aldrich), GAPDH (mouse monoclonal; G8795; Sigma-Aldrich), Src (mouse monoclonal; clone GD11 05-184; EMD Millipore), p-Src (Tyr416) (rabbit polyclonal; 2101; Cell Signaling Technology), Akt (mouse monoclonal; 2966; Cell Signaling Technology), p-AKT (Ser473) (rabbit monoclonal; 193H12; Cell Signaling Technology), FAK (rabbit polyclonal; 3285; Cell Signaling Technology), p-FAK (Tyr397) (rabbit polyclonal; 3283; Cell Signaling Technology), Tubulin (mouse monoclonal; T6199; Sigma-Aldrich), cortactin (mouse monoclonal; clone 4F11 05-180; EMD Millipore), and p230 trans-Golgi (mouse monoclonal; 611281; BD).
Cell culture
The cell culture media used were RPMI 1640 (for all melanoma cells) and DMEM (for MCF-7 and NMuMG). All cell culture media were supplemented with 10% FBS and penicillin/streptomycin.
RNA interference
RNA interference of STIM1 and Orai1 was performed using pSUPER.Retro.puro vector (Oligoengine) encoding shRNA. The target sequences were as follows: 5′-AGAAGGAGCTAGAATCTCAC-3′ (STIM1sh1), 5′-TCGGCCTGATCTTTATCGT-3′ (Orai1sh1), and 5′-CCAGCATTGAGTGTGTACA-3′ (Orai1sh2). To efficiently knockdown Orai1, two shRNAs targeting two different regions of the same gene were used simultaneously. In some experiments, two previously described STIM1 shRNA (sh2 and sh3, targeting 5′-GGCTCTGGATACAGTGCTC-3′ and 5′-GGATGCTGTCATTTTTTGA-3′; Yang et al., 2009) and a new Orai1 sh3 (targeting 5′-GGCTCTGGATACAGTGCTC-3′) were used to confirm STIM1 and Orai1 knockdown effects.
cDNA constructs
The retrovirus vectors encoding STIM1 and Orai1 were constructed by subcloning human STIM1 and Orai1 fragments into pLNCX2 vector (cytomegalovirus promoter; Takara Bio Inc.) between Bgl II and Stu I, as described previously (Yang et al., 2009). Lifeact-mAPPLE was generated by first subcloning mAPPLE into pLNCX2 between HindIII and StuI and then subcloning Lifeact between Xho I and Hind III. MT1-MMP–EGFP in the pEGFP-N1 vector was a gift from M. Montoya (Spanish Nacional Cancer Research Center, Madrid, Spain; Bravo-Cordero et al., 2007). pEGFP–MT1-MMP was digested with Hind III and Not I, and the MT1-MMP–EGFP fragment was inserted between Hind III and Not I sites in pLPCX vector (cytomegalovirus promoter; Takara Bio Inc.). mRFP-Rab5 in pmRFP-C3 backbone was obtained from Addgene (14437).
Fluorescent gelatin labeling
Fluorescent gelatin was synthesized by incubating bovine skin gelatin with amine reactive fluorescent dyes. Alexa Fluor 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester (Invitrogen), or Texas red-X succinimidyl ester (Invitrogen) was dissolved in DMSO at 10 mg/ml. Dye solution (50 µl) was added slowly to 1 ml of bovine skin gelatin solution (Sigma-Aldrich; 40 mg/ml; dissolved in 0.1 M sodium bicarbonate buffer, at pH 8.3 for succinimidyl ester or pH 9.0 for tetrafluorophenyl ester) while vortexing. After incubation at room temperature for 1 h, the free dyes in the mixture were removed with a HiTrap desalting column (GE Healthcare). The desalted fluorescent gelatin solution can be stored at 4°C for short-term storage or at −80°C with 10% glycerol as a cryoprotectant for long-term storage.
Invadopodia activity assay
The invadopodia activity assay protocol was adapted from a previous protocol (Artym et al., 2006) with modifications (Sun et al., 2013) by plating cancer cells onto glass coverslips coated with a thin film of fluorescent gelatin. To coat the coverslips with fluorescent gelatin, acid-washed glass coverslips were incubated with 50 µg/ml poly-d-lysine for 20 min, washed with PBS, and cross-linked with 0.5% glutaraldehyde for 20 min. After extensive wash with PBS, the coverslips were incubated with diluted fluorescent gelatin (mixing 0.2% unlabeled bovine skin gelatin solution with fluorescent gelatin at 8:1 ratio) for 10 min. The coverslips were then washed with PBS, and the residual glutaraldehyde was quenched with freshly made NaBH4 solution (5 mg/ml) for 15 min.
To evaluate invadopodium formation and focalized proteolysis activity, 9 × 104 melanoma cells in RPMI 1640 growth medium were plated onto fluorescent gelatin-coated glass coverslips and allowed to attach for 2 h in a 37°C CO2 incubator. Ca2+ chelators and channel blockers and other inhibitors or control vehicle were added at indicated concentrations to the growth medium and incubated for another 2 h. Cells were fixed 4 h after plating with 4% paraformaldehyde. Alternatively, melanoma cells in growth medium were allowed to attach to fluorescent gelatin coverslips in the presence of broad spectrum inhibitor GM6001 (10 µM) for 12 h. In the presence of GM6001, no gelatin degradation was detected. To initiate focalized proteolysis of gelatin, melanoma cells were washed three times with RPMI 1640 medium and incubated with RPMI 1640 growth medium containing control vector or appropriate inhibitors, blockers, or Ca2+ chelators in a 37°C CO2 incubator for 4 h before being fixed with 4% paraformaldehyde. The fixed melanoma cells were then permeabilized in antibody diluting buffer (2% BSA and 0.1% Triton X-100 in PBS) and followed by incubation with Alexa Fluor 594– or Alexa Fluor 488–labeled phalloidin (1:100 dilution from a 20-U/ml stock solution) for 30 min. Extensive washes with PBS were performed after each step. The coverslips were then mounted onto slides in mounting medium (150 mM Tris, pH 8.0, and 90% glycerol). Fluorescent micrographs were obtained with an upright fluorescence microscope (Axio Observer.Z1; Carl Zeiss) equipped with 63× oil immersion objective.
Gelatin degradation was quantified using ImageJ software (National Institutes of Health) by setting signal threshold for gelatin fluorescence in each individual cell. A region of interest was selected along the outline of each individual cell based on phalloidin staining, and the degraded area with gelatin fluorescence signal below the set threshold was measured by ImageJ. We quantified 20–30 cells from five random 63× fields. Data are shown as scattered dots, with each dot representing area (in square micrometers) degraded in a single melanoma cell.
To evaluate the degradation activity of individual invadopodium, we quantified the IDX. A region of interest was selected around the actin core of an invadopodium to include all the degraded area, and the mean gelatin fluorescence intensity (Fi) and total area (Ai) of the region (in pixels) were quantified using ImageJ. Mean fluorescence intensity in a no-degradation reference area (Fr) in the vicinity was also measured. IDX of each invadopodium was calculated according to the following equation: IDX = (1 − Fi/Fr) × Ai. IDX is proportional to the total amount of gelatin degraded by individual invadopodium and is comparable between different coverslips assuming similar gelatin coating thickness. Data are presented as scatter dots, with each dot representing the IDX of a single invadopodium.
Immunofluorescence staining
Melanoma cells (9 × 104) in RPMI 1640 growth medium were plated onto gelatin-coated glass coverslips for 12 h and fixed with 4% paraformaldehyde. The cells were then permeabilized in antibody diluting buffer (2% BSA and 0.1% Triton X-100 in PBS) and incubated with mouse anti-cortactin (clone 4F11 05-180; EMD Millipore) at 1:1,000 dilution, for 1 h. The cells were incubated with Alexa Fluor 488–conjugated anti–mouse IgG (1:300) or anti–rabbit IgG (1:300) and Alexa Fluor 594–labeled phalloidin (1:100) for 30 min. An extensive wash was performed between each step. The coverslips were then mounted onto slides and imaged using an upright fluorescence microscope (Axio Observer.Z1) or laser scanning confocal microscope (TCS SP5 Acousto-Optical Beam Splitter; Leica).
To quantify the number of invadopodia per cell, melanoma cells stained with cortactin antibody (clone 4F11 05-180) and phalloidin were visualized under an upright fluorescence microscope (Axio Observer.Z1) with a 63× oil immersion objective. Invadopodia are defined as round dots positive for both cortactin staining and actin staining on the ventral side of cells (Yamaguchi et al., 2005; Artym et al., 2006). Typically, invadopodium numbers from around 50 cells from 10 random 63× fields were counted for each experimental group. Because essentially all of the ventral actin dots were positively for cortactin, in some experiments, only the actin channel was used for quantification. Data are presented as scattered dots, with each dot representing the number of invadopodia in a single cell.
Live cell time-lapse recording
WM793 cells stably expressing cortactin-EGFP were plated on Texas red gelatin-coated glass-bottomed 35-mm tissue culture dishes (MatTek Corporation) 90 min before imaging. Cells were maintained at 37°C in Ringer’s solution supplemented with 100 mM Hepes buffer, pH 7.4. Oxyfluor (1:100 dilution; Oxyrase, Inc.) and 10 mM sodium lactate were added throughout the observation period to reduce phototoxicity.
Invadopodial precursor quantitation
To quantitate invadopodial precursor formation, WM793 cells were plated onto Alexa Fluor 488–gelatin-coated coverslips in growth medium (10% FBS) and allowed to attached for 1 h. The medium were then changed to starvation medium (1% FBS), and the cells were starved overnight. To induce assembly of invadopodial precursor, FBS were added directly to the well to the final concentration of 10%. Cells were fixed with 4% PFA 30 or 60 min after induction and stained with phalloidin. Invadopodial precursors were defined as invadopodium-like actin dots at the ventral side of the cell without gelatin degradation activity.
For live cell imaging of invadopodial precursors, WM793 cells expressing Lifeact-mAPPLE were plated on gelatin-coated glass-bottomed 35-mm tissue culture dishes for 4 h. Cells were then washed by RPMI 1640 containing 1% FBS and incubated overnight. Invadopodia was induced by adding 10% FBS, and cells were subjected to live cell imaging in a live cell incubator (XL S1 full enclosure incubator; Carl Zeiss) immediately. The invadopodium formation process was recorded for 60 min at 30-s intervals. The time-lapse recordings were examined frame by frame to identify newly assembled invadopodial precursors.
Membrane protein biotinylation assay
WM793 cells grown to 90% confluence were washed twice with ice-cold PBS, and freshly prepared 1 mM EZ-link sulfo-NHS-SS-biotin (Thermo Fisher Scientific) in ice-cold PBS was added. After incubation on ice for 30 min, cells were washed twice with ice-cold PBS. The unreacted biotin was quenched for 10 min by incubation with ice-cold PBS containing 50 mM glycine. Cells were then washed three times with ice-cold PBS and lysed in 0.5 ml immunoprecipitation (IP) buffer (PBS, pH 7.4, 5 mM EDTA, 5 mM EGTA, 10 mM sodium pyrophosphate, 50 mM NaF, 1 mM NaVO3, and 1% Triton X-100). Cell lysate was briefly sonicated (5 s) and centrifuged at 14,000 rpm on a 4°C bench top centrifuge. The supernatant was incubated with NeutrAvidin beads (Thermo Fisher Scientific) at 4°C for 3 h. The beads were extensively washed with IP buffer, eluded with SDS-PAGE loading buffer, and subjected to Western blotting.
To examine MT1-MMP endocytosis, quenched cells were incubated at 37°C in cell cultural incubator for 30 min in the presence of 100 µM 2-APB or vehicle control to allow the internalization of biotinylated MT1-MMP. The internalization was stopped by washing the cells with ice-cold PBS for three times. Biotin present on the cell surface was cleaved off by incubating the cells with PBS, pH 8.2, containing 100 mM MESNA for 15 min. Cells were washed three times with ice-cold PBS and then lysed for affinity purification.
For the recycling assay, MESNA-treated cells were incubated in a cell culture incubator at 37°C for 30 or 60 min to allow recycling of the endocytosed biotinylated MT1-MMP. Recycling was stopped by washing the cells with ice-cold PBS for three times. Biotin reexposed to the cell surface was cleaved off by a second MESNA treatment. Finally, cells were washed with ice-cold PBS for three times and lysed in IP buffer for affinity purification.
Calcium assay
The effects of STIM1 shRNA, Orai1 shRNA, and ectopic STIM1 overexpression on store-operated Ca2+ influx were measured using Fluo4-AM as the Ca2+ indicator using a fluorescence microplate reader as described previously (Yang and Huang, 2005; Yang et al., 2009). In brief, Fluo4-AM–loaded cells in a 96-well plate were stimulated with 2 µM thapsigargin (in the presence of 0 mM extracellular Ca2+), and the change in fluorescence was monitored using a fluorescent microplate reader for 300 s. Extracellular Ca2+ was then added to a final concentration of 2 mM, and the change in Fluo4 fluorescence was monitored for another 300 s.
Calcium imaging
Calcium imaging was performed with a confocal microscope (LSM710; Carl Zeiss) with a 40×, 1.3 NA oil immersion objective and sampling speed of 1.97 s/frame (512 × 512 pixels). For chemical Ca2+ indicator loading, WM793 cells were incubated with 5 µmol/liter Fluo4-AM (Invitrogen) for 10 min at room temperature. For measurement of Fluo4, excitation was at 488 nm, and fluorescence emission was collected at 490–540 nm.
Cell invasion assay
Cell invasion assay were performed using Matrigel-coated cell invasion insert (6.5-mm diameter and 8-µm pore size; BD) as previously described (Yang et al., 2009; Sun et al., 2011). In brief, melanoma cells (105) suspended in starvation medium were added to the upper chamber of the invasion insert, and the insert was placed in a 24-well dish containing medium with or without 10% serum. Invasion assays were performed for 10 h, and cells were fixed with 3.7% formaldehyde. Cells were stained with crystal violet staining solution, and cells on the upper side of the insert were removed with a cotton swab. Three randomly selected fields (10× objectives) on the lower side of the insert were photographed, and the cells on the lower surface of the insert were counted.
Immunohistochemistry staining
Melanoma tissue microarray was purchased from Biomax. Tissue microarray slides were baked in a 60°C oven for 60 min, deparafinized, and hydrated to deionized H2O. Epitope was retrieved by incubating with 0.21% citrate buffer, pH 6, in a 99°C water bath for 30 min. After epitope retrieval, slides were washed in running water for 2–5 min, incubated with 3% H2O2 for 5 min, and washed with deionized H2O and PBS. After slides were incubated with normal serum (1:20, diluted in 2% BSA-PBS) for 10 min at room temperature in a humidity chamber, slides were then incubated overnight with 150 µl monoclonal anti-STIM1 antibody (1:300; Thermo Fisher Scientific) at 4°C in a humidity chamber. Slides were then washed with PBST (PBS with Tween 20; three times, 5 min each) and incubated with biotinylated anti–mouse secondary antibody (1:500) at room temperature for 60 min. The slides were then incubated with avidin biotin complex (alkaline phosphatase conjugated) for 45 min. After three washes with PBST (5 min each), slides were incubated with Vulcan Fast red for 10–15 min, washed in deionized H2O, and then counterstained using Harris modified hematoxylin (Thermo Fisher Scientific). Slides were then dehydrated and mounted. The tissue microarray was examined by a board-certified dermatopathologist (R. Mathew) for STIM1 staining intensity (on a 0–3 scale) and percentage of STIM1-positive cells (0–100%). A final STIM1 staining score for each sample was calculated by multiplying intensity score by percentage score. Melanoma samples with STIM1 staining score <1 were stratified to the “STIM1 negative or low” group, whereas samples with a score equal to or >1 were stratified to the “STIM1 medium or high” group.
1205Lu melanoma metastasis experiment
All animal work was performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Moffitt Cancer Center. Nu/Nu immunodeficient mice were used for experimental lung metastasis experiments. 1205Lu human melanoma cells expressing the luciferase reporter were trypsinized and washed with PBS. Subsequently, 106 cells in 0.2 ml PBS were injected into the lateral tail vein. Luciferase-based noninvasive bioluminescent imaging and analysis were performed as described previously with an imaging system (IVIS 200; Xenogen) by injecting 100 µl d-luciferin (15 mg/ml) via i.p. into each mouse (Yang et al., 2009, 2012). The mice were imaged on day 0 and then on a weekly basis thereafter. At the end of the metastasis experiment, lungs were harvested from euthanized mice, fixed in paraformaldehyde, and embedded in paraffin. Paraffin sections were stained with hematoxylin and eosin.
Data analyses
Data are expressed as means ± SEM or means ± SD as indicated. Two-tailed p-values were determined by Student’s t test, Mann–Whitney test, or Fisher’s exact test as indicated, with P < 0.05 defined as statistically significant.
Microscopy
For wide-field fluorescence microscopy, samples were examined with an automated upright microscope (Axiovert.Z1; Carl Zeiss) through a 63×/1.4 NA Plan Apochromat oil immersion objective. Images were captured using the charge-coupled device camera (AxioCam MRm3; Carl Zeiss) and AxioVision version 4.7 software suite (Carl Zeiss). Real-time live cell microscopy was performed with an automated inverted microscope (Observer Z.1; Carl Zeiss) outfitted with full enclosure incubator (37°C, 5% CO2; XL S1) in RPMI 1640 medium. Images were captured using the charge-coupled camera (AxioCam MRm3) through a 40×, 1.4 NA Plan Apochromat oil objective. AxioVision version 4.7 software suite (Carl Zeiss) was used to manage the acquired images. Definite focus was used to ensure the stability of the focal plane over time.
For confocal microscopy, samples were viewed with an inverted microscope (DMI6000; Leica), confocal scanner (TCS SP5; Leica), and a 63×, 1.4 NA Plan Apochromat oil immersion objective (Leica). Argon 488 and HeNe 594 laser lines were applied to excite the samples, and a tuneable acousto-optical beam splitter was used to minimize cross talk between fluorochromes. Gain, offset, pinhole, and lookup table settings were identical for all samples. Images were captured with photomultiplier detectors and prepared with the LAS AF software version 1.6.0 build 1016 (Leica).
Online supplemental material
Fig. S1 presents effects of Ca2+ influx blockade on invadopodium formation and ECM degradation. Fig. S2 shows the lack of effect of STIM1 overexpression, 2-APB treatment, and STIM1 and Orai1 knockdown on invadopodium lifetime. Fig. S3 demonstrates the role of Src in SOCE-mediated invadopodium formation. Videos 1, 2, and 4 show Ca2+ oscillation in WM793 cells stimulated by 10% FBS (Video 1), and the inhibition of which by EGTA (Video 2) and SKF96365 (Video 4); in Video 3, the inhibition of Ca2+ oscillation was restored by adding back Ca2+ to the medium. Videos 5–7 show invadopodium precursor assembly in WM793 cells (Video 5) and the inhibition of which by EGTA (Video 6) and SKF96365 (Video 7). Video 8 shows the translocation of MT1-MMP–EGFP from plasma membrane to intracellular vesicles after 2-APB treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407082/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Chellappan and Weber for critical reading of the manuscript, Joseph Johnson and Mark Lloyd for assistance with image acquisition, and Rasa Hamilton for editorial assistance.
This work was supported by the National Cancer Institute (R01CA175741 and P50CA168536). The research in H. Cheng’s laboratory was supported by the Major State Basic Research Program of China (2013CB531200). The Analytic Microscopy Core at Moffitt Cancer Center is supported in part by the National Cancer Institute (P30-CA76292-14).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CI
- confidence interval
- IDX
- invadopodium degradation index
- IP
- immunoprecipitation
- MESNA
- 2-mercaptoethane sulfonate
- MMP
- matrix metalloproteinase
- NMDA
- N-methyl-d-aspartate receptor
- SOCE
- store-operated calcium entry
References
- Alexander N.R., Branch K.M., Parekh A., Clark E.S., Iwueke I.C., Guelcher S.A., and Weaver A.M.. 2008. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 18:1295–1299 10.1016/j.cub.2008.07.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., and Mueller S.C.. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Baldassarre M., Pompeo A., Beznoussenko G., Castaldi C., Cortellino S., McNiven M.A., Luini A., and Buccione R.. 2003. Dynamin participates in focal extracellular matrix degradation by invasive cells. Mol. Biol. Cell. 14:1074–1084 10.1091/mbc.E02-05-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry P.A., Birnie R., Droop A.P., Maitland N.J., and Collins A.T.. 2011. The calcium sensor STIM1 is regulated by androgens in prostate stromal cells. Prostate. 71:1646–1655 10.1002/pros.21384 [DOI] [PubMed] [Google Scholar]
- Bird G.S., Hwang S.Y., Smyth J.T., Fukushima M., Boyles R.R., and Putney J.W. Jr. 2009. STIM1 is a calcium sensor specialized for digital signaling. Curr. Biol. 19:1724–1729 10.1016/j.cub.2009.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J.J., Marrero-Diaz R., Megías D., Genís L., García-Grande A., García M.A., Arroyo A.G., and Montoya M.C.. 2007. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 26:1499–1510 10.1038/sj.emboj.7601606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.C., Di Capite J., Singaravelu K., Nelson C., Halse V., and Parekh A.B.. 2008. Local Ca2+ influx through Ca2+ release-activated Ca2+ (CRAC) channels stimulates production of an intracellular messenger and an intercellular pro-inflammatory signal. J. Biol. Chem. 283:4622–4631 10.1074/jbc.M705002200 [DOI] [PubMed] [Google Scholar]
- Chang W.C., Woon P.Y., Hsu Y.W., Yang S., Chiu Y.C., and Hou M.F.. 2012. The association between single-nucleotide polymorphisms of ORAI1 gene and breast cancer in a Taiwanese population. ScientificWorldJournal. 2012:1–6 10.1100/2012/916587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantôme A., Potier-Cartereau M., Clarysse L., Fromont G., Marionneau-Lambot S., Guéguinou M., Pagès J.C., Collin C., Oullier T., Girault A., et al. 2013. Pivotal role of the lipid Raft SK3-Orai1 complex in human cancer cell migration and bone metastases. Cancer Res. 73:4852–4861 10.1158/0008-5472.CAN-12-4572 [DOI] [PubMed] [Google Scholar]
- Chen Y.F., Chiu W.T., Chen Y.T., Lin P.Y., Huang H.J., Chou C.Y., Chang H.C., Tang M.J., and Shen M.R.. 2011. Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc. Natl. Acad. Sci. USA. 108:15225–15230 10.1073/pnas.1103315108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.T., Chen Y.F., Chiu W.T., Liu K.Y., Liu Y.L., Chang J.Y., Chang H.C., and Shen M.R.. 2013a. Microtubule-associated histone deacetylase 6 supports the calcium store sensor STIM1 in mediating malignant cell behaviors. Cancer Res. 73:4500–4509 10.1158/0008-5472.CAN-12-4127 [DOI] [PubMed] [Google Scholar]
- Chen Y.T., Chen Y.F., Chiu W.T., Wang Y.K., Chang H.C., and Shen M.R.. 2013b. The ER Ca²+ sensor STIM1 regulates actomyosin contractility of migratory cells. J. Cell Sci. 126:1260–1267 10.1242/jcs.121129 [DOI] [PubMed] [Google Scholar]
- Clark E.S., and Weaver A.M.. 2008. A new role for cortactin in invadopodia: regulation of protease secretion. Eur. J. Cell Biol. 87:581–590 10.1016/j.ejcb.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., and Weaver A.M.. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Clark W.H., Jr 1991. Human cutaneous malignant melanoma as a model for cancer. Cancer Metastasis Rev. 10:83–88 10.1007/BF00049406 [DOI] [PubMed] [Google Scholar]
- Cortesio C.L., Chan K.T., Perrin B.J., Burton N.O., Zhang S., Zhang Z.Y., and Huttenlocher A.. 2008. Calpain 2 and PTP1B function in a novel pathway with Src to regulate invadopodia dynamics and breast cancer cell invasion. J. Cell Biol. 180:957–971 10.1083/jcb.200708048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F.M., Azimi I., Faville R.A., Peters A.A., Jalink K., Putney J.W. Jr, Goodhill G.J., Thompson E.W., Roberts-Thomson S.J., and Monteith G.R.. 2014. Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene. 33:2307–2316 10.1038/onc.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Capite J., Ng S.W., and Parekh A.B.. 2009. Decoding of cytoplasmic Ca2+ oscillations through the spatial signature drives gene expression. Curr. Biol. 19:853–858 10.1016/j.cub.2009.03.063 [DOI] [PubMed] [Google Scholar]
- Dupont G., Combettes L., Bird G.S., and Putney J.W.. 2011. Calcium oscillations. Cold Spring Harb. Perspect. Biol. 3:a004226 10.1101/cshperspect.a004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedida-Metula S., Feldman B., Koshelev V., Levin-Gromiko U., Voronov E., and Fishman D.. 2012. Lipid rafts couple store-operated Ca2+ entry to constitutive activation of PKB/Akt in a Ca2+/calmodulin-, Src- and PP2A-mediated pathway and promote melanoma tumor growth. Carcinogenesis. 33:740–750 10.1093/carcin/bgs021 [DOI] [PubMed] [Google Scholar]
- Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.H., Tanasa B., Hogan P.G., Lewis R.S., Daly M., and Rao A.. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 441:179–185 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Fidler I.J.2003. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 3:453–458 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., and Linder S.. 2008. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20:235–241 10.1016/j.ceb.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Hogan P.G., Lewis R.S., and Rao A.. 2010. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu. Rev. Immunol. 28:491–533 10.1146/annurev.immunol.021908.132550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M.F., Kuo H.C., Li J.H., Wang Y.S., Chang C.C., Chen K.C., Chen W.C., Chiu C.C., Yang S., and Chang W.C.. 2011. Orai1/CRACM1 overexpression suppresses cell proliferation via attenuation of the store-operated calcium influx-mediated signalling pathway in A549 lung cancer cells. Biochim. Biophys. Acta. 1810:1278–1284 10.1016/j.bbagen.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Hu J., Qin K., Zhang Y., Gong J., Li N., Lv D., Xiang R., and Tan X.. 2011. Downregulation of transcription factor Oct4 induces an epithelial-to-mesenchymal transition via enhancement of Ca2+ influx in breast cancer cells. Biochem. Biophys. Res. Commun. 411:786–791 10.1016/j.bbrc.2011.07.025 [DOI] [PubMed] [Google Scholar]
- Huang W.C., Chai C.Y., Chen W.C., Hou M.F., Wang Y.S., Chiu Y.C., Lu S.R., Chang W.C., Juo S.H., Wang J.Y., and Chang W.C.. 2011. Histamine regulates cyclooxygenase 2 gene activation through Orai1-mediated NFκB activation in lung cancer cells. Cell Calcium. 50:27–35 10.1016/j.ceca.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Juhasz I., Albelda S.M., Elder D.E., Murphy G.F., Adachi K., Herlyn D., Valyi-Nagy I.T., and Herlyn M.. 1993. Growth and invasion of human melanomas in human skin grafted to immunodeficient mice. Am. J. Pathol. 143:528–537. [PMC free article] [PubMed] [Google Scholar]
- Li L., and Hanahan D.. 2013. Hijacking the neuronal NMDAR signaling circuit to promote tumor growth and invasion. Cell. 153:86–100 10.1016/j.cell.2013.02.051 [DOI] [PubMed] [Google Scholar]
- Linder S.2007. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 17:107–117 10.1016/j.tcb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E. Jr, and Meyer T.. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H.T., Peng Z., Hiragun T., Iwaki S., Gilfillan A.M., and Beaven M.A.. 2008. Canonical transient receptor potential 5 channel in conjunction with Orai1 and STIM1 allows Sr2+ entry, optimal influx of Ca2+, and degranulation in a rat mast cell line. J. Immunol. 180:2233–2239 10.4049/jimmunol.180.4.2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul-Pavicic A., Chiang S.C., Rensing-Ehl A., Jessen B., Fauriat C., Wood S.M., Sjöqvist S., Hufnagel M., Schulze I., Bass T., et al. 2011. ORAI1-mediated calcium influx is required for human cytotoxic lymphocyte degranulation and target cell lysis. Proc. Natl. Acad. Sci. USA. 108:3324–3329 10.1073/pnas.1013285108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelbeek J., Kuipers A.J., Henneman L., Visser D., Eidhof I., van Horssen R., Wieringa B., Canisius S.V., Zwart W., Wessels L.F., et al. 2012. TRPM7 is required for breast tumor cell metastasis. Cancer Res. 72:4250–4261 10.1158/0008-5472.CAN-11-3863 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberg A., Kitzing T., and Grosse R.. 2011. Nucleating actin for invasion. Nat. Rev. Cancer. 11:177–187 10.1038/nrc3003 [DOI] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., and Takenawa T.. 2008. Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 182:157–169 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S., Zhivotovsky B., and Nicotera P.. 2003. Regulation of cell death: the calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 4:552–565 10.1038/nrm1150 [DOI] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C.C., Bravo-Cordero J.J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A.J., and Condeelis J.. 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186:571–587 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney J.W., Jr 1986. A model for receptor-regulated calcium entry. Cell Calcium. 7:1–12 10.1016/0143-4160(86)90026-6 [DOI] [PubMed] [Google Scholar]
- Roos J., DiGregorio P.J., Yeromin A.V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J.A., Wagner S.L., Cahalan M.D., et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169:435–445 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F., Shimizu-Hirota R., and Weiss S.J.. 2009. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185:11–19 10.1083/jcb.200807195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F. Jr, Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., and Courtneidge S.A.. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 7:155–165 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Soboloff J., Spassova M.A., Tang X.D., Hewavitharana T., Xu W., and Gill D.L.. 2006. Orai1 and STIM reconstitute store-operated calcium channel function. J. Biol. Chem. 281:20661–20665 10.1074/jbc.C600126200 [DOI] [PubMed] [Google Scholar]
- Steffen A., Le Dez G., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T., and Chavrier P.. 2008. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 18:926–931 10.1016/j.cub.2008.05.044 [DOI] [PubMed] [Google Scholar]
- Sun J., He H., Xiong Y., Lu S., Shen J., Cheng A., Chang W.C., Hou M.F., Lancaster J.M., Kim M., and Yang S.. 2011. Fascin protein is critical for transforming growth factor β protein-induced invasion and filopodia formation in spindle-shaped tumor cells. J. Biol. Chem. 286:38865–38875 10.1074/jbc.M111.270413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., He H., Pillai S., Xiong Y., Challa S., Xu L., Chellappan S., and Yang S.. 2013. GATA3 transcription factor abrogates Smad4 transcription factor-mediated fascin overexpression, invadopodium formation, and breast cancer cell invasion. J. Biol. Chem. 288:36971–36982 10.1074/jbc.M113.506535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M., Peinelt C., Beck A., Koomoa D.L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., and Kinet J.P.. 2006. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 312:1220–1223 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M., DeHaven W.I., Bird G.S., Billingsley J.M., Wang H., Rao P.E., Hutchings A.B., Jouvin M.H., Putney J.W., and Kinet J.P.. 2008. Defective mast cell effector functions in mice lacking the CRACM1 pore subunit of store-operated calcium release-activated calcium channels. Nat. Immunol. 9:89–96 10.1038/ni1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Y., Chen B.K., Wang Y.S., Tsai Y.T., Chen W.C., Chang W.C., Hou M.F., Wu Y.C., and Chang W.C.. 2012. Involvement of store-operated calcium signaling in EGF-mediated COX-2 gene activation in cancer cells. Cell. Signal. 24:162–169 10.1016/j.cellsig.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Wang J.Y., Sun J., Huang M.Y., Wang Y.S., Hou M.F., Sun Y., He H., Krishna N., Chiu S.J., Lin S., et al. 2015. STIM1 overexpression promotes colorectal cancer progression, cell motility and COX-2 expression. Oncogene. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Wu Y.I., Liu Y., Geiger J., Tam E., Overall C., Stack M.S., and Friedl P.. 2007. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9:893–904 10.1038/ncb1616 [DOI] [PubMed] [Google Scholar]
- Wollman R., and Meyer T.. 2012. Coordinated oscillations in cortical actin and Ca2+ correlate with cycles of vesicle secretion. Nat. Cell Biol. 14:1261–1269 10.1038/ncb2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., and Oikawa T.. 2010. Membrane lipids in invadopodia and podosomes: key structures for cancer invasion and metastasis. Oncotarget. 1:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., and Condeelis J.. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP–Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., and Huang X.Y.. 2005. Ca2+ influx through L-type Ca2+ channels controls the trailing tail contraction in growth factor-induced fibroblast cell migration. J. Biol. Chem. 280:27130–27137 10.1074/jbc.M501625200 [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang J.J., and Huang X.Y.. 2009. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell. 15:124–134 10.1016/j.ccr.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang J.J., and Huang X.Y.. 2012. Mouse models for tumor metastasis. Methods Mol. Biol. 928:221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.