Abstract
Objective(s):
OCT4 is a transcription factor required for pluripotency during early embryogenesis and the maintenance of identity of embryonic stem cells and pluripotent cells. Therefore, the effective expression regulation of this gene is highly critical. UTR regions are of great significance to gene regulation. In this study, we aimed to investigate the potential regulatory role played by 5´UTR and 3´UTR of the Oct4 gene in mouse BMSC and P19 cells.
Materials and Methods:
The Oct4 5´UTR and 3´UTR sequences were cloned into pGL3 luciferase plasmid which led to the generation of pGL3 5´-UTR, pGL3 5´&3´-UTRs and pGL3 3´-UTR vectors. The vectors were transfected into BMSC and P19 cells followed by luciferase assay.
Results:
The assay of luciferase expression exhibited a direct link between the presence of Oct4 3´- UTR and the decrease of luciferase count in both cell lines; whereas 5´UTR indicated diverse behaviors in two cells. This discrepancy could be explained in view of the difference of cellular contexts in which the Oct4 UTRs act.
Conclusion:
This study sheds some light on the role of UTR regions of mouse Oct4 in regulating post-transcriptional gene expression in pluripotent cells. These data represent potential to be used for the development of novel therapeutic approaches for a variety of malignancies.
Keywords: BMSC, Luciferase, OCT4, P19, Post-transcriptional regulation, UTR
Introduction
Embryonic stem cells (ESCs) are available sources for clinical therapies because of their unlimited self-renewal ability and potential to generate differentiated cell types (pluripotency) (1). Self-renewal and pluripotency properties are regulated by an array of protein–coding genes such as those encoding transcription factors and chromatin remodeling enzymes (2). OCT4, a transcription factor belonging to the family of POU transcription factors lies at the center of this circuitry and controls a wide range of downstream genes (3).
Extensive studies have indicated that OCT4 with other pluripotency factors are required for self-renewal and pluripotency of embryonic stem cells and embryonic carcinoma cells (ECC) (4-6). In developing mouse embryo, OCT4 is expressed in early cleavage stage, inner cell mass, primitive ectoderm and primordial germ cells and downregulated in the trophectoderm (7). The suppression of OCT4 expression in developing embryo or in cultured human or mouse ES and EC cells leads to the loss of pluripotency and subsequently differentiation into trophectoderm (7, 8), whereas transgene-mediated overexpression of OCT4 triggers differentiation of embryonic cells into endodermal or mesodermal structures (4, 9). In addition to embryonic cells, germ cells and germ cell tumors, OCT4 expression has been reported in several adult somatic cells (10) and in various cancer cell lines and primary tumors (11, 12). OCT4 expression in somatic cells has been suggested to be restricted to small populations of multipotent cells with high self-renewal capacity, namely the adult stem cells in normal tissues and cancer stem cells in tumor samples (10, 13, 14). Recently, OCT4 expression has also been reported in terminally differentiated peripheral blood mononuclear cells (pBMSC) (15).
Although many features of an mRNA can contribute to its translation, most control elements are located within untranslated regions (UTRs)(16). Three-prime untranslated region (3´ UTR) is a particular section of mRNA that follows the coding region and is not translated to protein. 3´UTR has several regulatory roles. In 3´ UTR, there exist regulatory sequences such as polyA adenylation signals, binding sites for miRNAs that may cause the suppression of mRNA expression and binding sites for proteins that may affect the mRNA stability or location in the cell. Genetic alterations in 3' UTR can be used to increase or decrease the half-life of mRNA, leading to higher or lower protein levels (17-20). Overall translation rates are also affected by characteristics of 5´ UTR, including length and start site consensus sequences as well as the presence of secondary structures, upstream AUGs, upstream open reading frames (uORFs) and internal ribosome entry sites (IRES). Also, 5´ UTRs can contain sequences functioning as binding sites for regulatory proteins. These regulatory proteins may affect the mRNA stability or translation. For instance, iron responsive elements are able to regulate gene expression in response to iron (19, 21, 22).
Alterations in UTR cis-regulatory sequences could affect the expression of specific genes at the level of translation. Such modifications may turn the physiological balance from healthy to diseased state, evoking a variety of conditions such as cancer. This information tends to establish the prominent role of UTRs, perhaps as much as those of coding sequences, in health and disease (23). Findings accumulated over the substantial role of UTRs in gene regulation render the investigation of these regulatory sequences imperative. This could be overwhelmingly notable in pluripotent cells offering insights to unravel the mechanisms underlying pluripotency state.
In this work, we aimed to investigate the potential regulatory role of the Oct4 5´UTR and 3´UTR in gene expression. To this end, we cloned the Oct4 5´ UTR and 3´ UTR regions singly and in combination into pGL3 vector upstream and downstream of luciferase (as reporter gene), respectively. Subsequently, the vectors were transfected into BMSC and p19 cells and luciferase expression was evaluated.
Materials and Methods
Cloning of the Oct4 5´UTR and 3´UTR sequences in pGL3 vector
To amplify the 3´UTR sequence of murine Oct4, total RNA was extracted from CCE cell line using RNA extraction kit (Exiqon, Denmark). The extracted RNA was treated with TURBO DNAase (Ambion, Austin, TX) to remove DNA contamination, and then investigated qualitatively by electrophoresis on 1% agarose gel and quantitatively by measuring the absorbance at 260 nm. Subsequently, the amplification of 3´UTR - the fragment spanning from nucleotide 1076 to 1346 (Gene ID: 18999 pou5f1) - was conducted through RT-PCR reaction. The sequences of the primers used for RT-PCR were as follows:
Forward primer
5´TTTTTCTAGATCTGTTCCCGTCACTGCTC 3´
Reverse primer
5´TTTTCTAGAATTCAGCTATCTACTGTGTGTCCC 3´
The XbaI site was introduced into forward primer and overlapping XbaI and EcoRI sites were introduced into reverse primer. The XbaI sites are underlined and EcoRI site is boldfaced. The incorporation of restriction sites was performed for convenience of cloning procedure.
To clone the Oct4 3´UTR in pGL3 vector, the amplified 3´UTR fragment was digested with XbaI, electrophoresed on 1% agarose gel and purified from agarose gel by gel purification kit (Bioneer, South Korea). Also, pGL3 vector was digested with XbaI. The Oct4 3´UTR was inserted into the XbaI site of pGL3 vector, downstream of the LUC gene.
The second PCR reaction was performed for two purposes: to determine the orientation of 3´UTR cloned in the previous step and to amplify the Oct4 5´UTR. PCR was carried out using pFU DNA polymerase and the following forward and reverse primers on pGL3-3´UTR vector as template. It is to be mentioned that the reverse primer was the same used for amplifying the Oct4 3´UTR:
Forward primer
5´ TTTTAAGCTTAACCGTCCCTAGGTGAGCCGTCTTTCCACCAG 3´
Reverse primer
5´ TTTTCTAGAATTCAGCTATCTACTGTGTGTCCC 3´
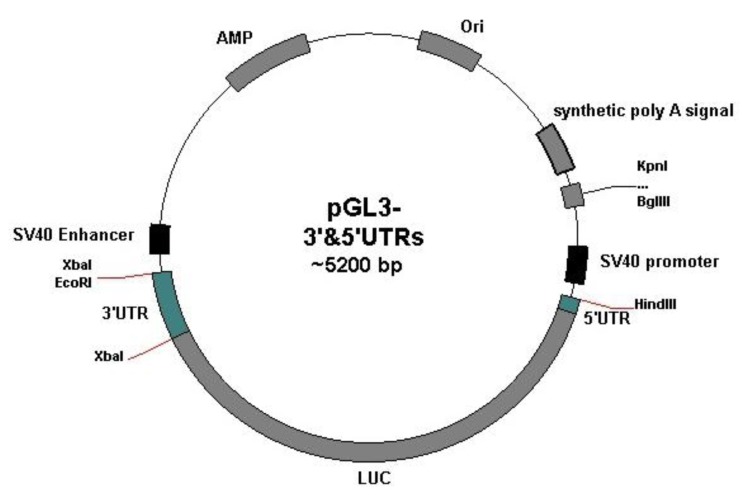
32 nucleotides of the Oct4 5´UTR were synthesized as part of forward primer. Also, AAGCTT - target site for Hind III enzyme – was introduced into forward primer. The amplified fragment contained Oct4 5´UTR, LUC gene and 3´UTR. To construct pGL3-5´&3´UTRs vector, this fragment was inserted into pGL3 vector digested with HindIII and XbaI. To obtain pGL3-5´UTR vector, pGL3-5´&3´UTR was digested with XbaI, electrophoresed on 1% agarose gel, and purified from agarose gel by gel purification kit (Bioneer Company, South Korea). The digested plasmid was then self-ligated using T4 DNA ligase. Cloning experiments were confirmed by restriction enzyme analysis and sequencing. Figure 1 indicates the schematic picture of pGL3 vector with cloned 5´UTR and 3´UTR sequences.
Figure 1.
The schematic picture of pGL3 vector containing the Oct4 5´UTR and 3´UTR (developed by Plasmidomics 0.2 software)
Cell culture and vector transfection
Bone marrow stem cells (BMSC) were isolated from mouse femur and tibia by flushing the bone shafts with special buffer (phosphate-buffered saline supplemented with 0.5 % bovine serum albumin, pH=7.2) as described previously (24). Mouse BMSCs were maintained in α-MEM medium (Gibco, USA) supplemented with 20% FBS in a 5% CO2 incubator. P19, mouse embryonic carcinoma cell line, is a teratocarcinoma cell line derived from transplant epiblast mouse embryonic cells. P19 cells were grown in α-MEM medium (Gibco, USA) supplemented with 10% FBS, 100 unit/ml of penicillin, and 100 µg/ml of streptomycin. Cells were plated 24 hr before transfection in 24-well plates and transfected with lipofectamin 2000 (Invitrogen, USA) following the manufacturer’s protocol. After 48 hr, cells were harvested and lysed in 1X passive lysis buffer.
Luciferase assay
48 hrs after transfection, cells were washed with PBS twice and lysis buffer was added to each well. After 15 min of incubation at 4°C temperature, the cells were harvested and transferred to tubes. Following vortexing for 15 sec, luciferase activity was measured in terms of relative light unit (RLU). The activity of luciferase was measured using Sirius tube luminometer (Berthold detection system). Each transfection experiment was performed in duplicate and repeated at least twice.
Statistical analysis
All experiments were replicated three times. The results of luciferase expression were analyzed by ANOVA and t-test in order to determine the difference of luciferase count (indicated as RLU) among different vectors transfected into cells (SPSS 13 software and GraphPad Prism 6). Statistical significance was considered as P<0.05.
Results
Construction of UTR-containing vectors
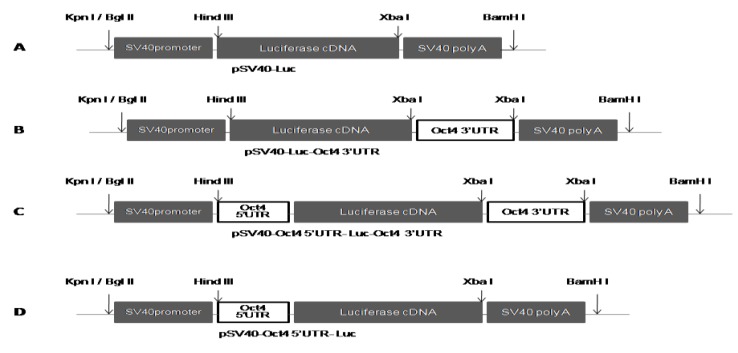
Construction of three vectors pGL3- 5´UTR, pGL3- 5´& 3´UTRs and pGL3-3´UTR was conducted to analyze the regulatory roles of UTR sequences of mouse Oct4 gene. The amplified Oct4 3´UTR was cloned into XbaI site downstream of LUC gene in pGL3 vector (Figure 2A and 2B) to generate pGL3-3´UTR. 5´UTR-LUC-3´UTR fragment - amplified from pGL3-3´UTR by using forward primer containing 32 nucleotides of 5´UTR as an overhang tail and reverse primer complementary to a part of 3´ UTR - was used for ligation into pGL3 vector. Both PCR product and vector were digested with XbaI and Hind III enzymes. This resulted in construction of the vector pGL3-5´& 3´UTRs (Figure 2C). For generation of pGL3 harboring solely Oct4 5´UTR, pGL3- 5´& 3´UTRs vector was digested with XbaI and then self-ligated which led to the production of pGL3-5´UTRvector (Figure 2D). The cloning of UTR sequences and construction of plasmids were confirmed by restriction enzyme analysis and sequencing.
Figure 2.
Schematic representation of the section of pGL3 vector used for cloning of 5´UTR and 3´UTR sequences: A) pGL3 vector without UTR sequences B) pGL3-3´UTR vector with pSV40-LUC- Oct4 3´UTR construct C) pGL3- 5´& 3´UTRs vector with pSV40- Oct4 5´UTR-LUC-Oct4 3´UTR construct D) pGL3- 5´UTR vector with pSV-Oct4 5´UTR-LUC
Cell transfection and luciferase assay
Transfection of cells with constructed vectors was used to evaluate the effects of Oct4 5´UTR and 3´UTR on gene regulation. Luciferase expression (in terms of relative light unit; RLU) was compared among pGL3-5´UTR, pGL3-5´& 3´UTRs and pGL3-3´ UTR vectors with pGL3 as control which was lacking any UTR sequences. This property leads to the resistance of control vector to regulation factors (such as microRNAs) interacting with UTR.
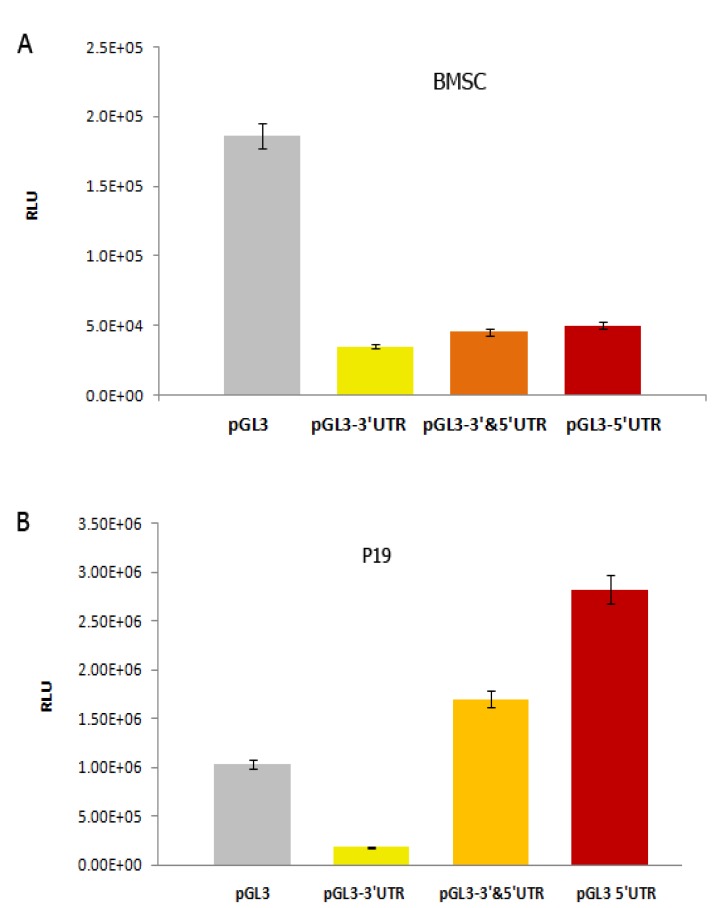
Evaluation of reporter gene expression was conducted 48 hr post-transfection. One-way ANOVA test indicated a significant decrease (P<0.05) in luciferase expression in BMSC cells following transfection with pGL3-5´UTR, pGL3-5´& 3´UTRs and pGL3-3´ UTR (Figure 3A). This analysis also exhibited that transfection of P19 cells with pGL3-3´UTR leads to a significant reduction (P<0.05) in luciferase expression. However, transfection with pGL3-5´ UTR and pGL3-5´& 3´UTRs resulted in the enhancement of luciferase expression (Figure 3B).
Figure 3.
Graphs indicating the effects of transfected vectors on luciferase expression. A) In upper graph, transfection of BMSCs with pGL3-3´UTR, pGL3- 5´& 3´ UTRs and pGL3- 5´UTR vectors resulted in a significant decrease of luciferace expression (P<0.05). B) In lower graph, trasfection of P19 with pGL3 3´UTR vector led to a significant reduction (P<0.05), while transfection with pGL3-5´&3´UTRs and pGL3-5´UTR induced an increase of luciferase expression. The vertical axis shows luciferase expression in terms of RLU (relative light unit)
Discussion
OCT4, as a POU transcription factor, resides at the top of a transcriptional hierarchy that is responsible for the initiation and/or maintenance of the expression of a group of genes positively regulating the pluripotent phenotype and repressing those genes associated with differentiation (25). Thus conferring a prominent position in the ranking of transcriptional regulators of mammalian cell fate to OCT4 (7). Expression of OCT4 is regulated at the transcriptional level by cis-acting elements located upstream of the Oct4 gene and methylation of chromatin structure. Although transcriptional networks controlled by OCT4 have been delineated and several proteins that interact with OCT4 have been identified, the mechanisms through which OCT4 is post-transcriptionally regulated have largely remained unexplored (2, 26) and this area has continued to be a conspicuous black box in our understanding of the circuitry which controls pluripotency. Therefore, comprehending the post-transcriptional regulatory mechanisms of OCT4 in stem and cancerous cells which assists in understanding the molecular basis of the pluripotent phenotype is fundamental to realizing their therapeutic potential and unraveling the molecular and cellular mechanisms of pluripotency. This information ultimately leads to novel treatments for certain malignancies.
In this study, we investigated the effects of mouse Oct4 UTRs as post-transcriptional modulators on the expression of luciferase reporter gene in mouse P19 embryonal carcinoma (EC) cells and BMSCs. Transfection experiments conducted by pGL3-5´UTR, pGL3-5´&3´UTRs and pGL3- 3´UTR demonstrated that in BMSC cells all three vectors provoke the reduction of luciferase expression, whereas in p19 cells transfection with pGL3-3´UTR leads to the decline of luciferase expression and transfection with pGL3-5´UTR and pGL3-5´&3´UTRs elicits the augmentation of reporter gene expression.
The main reasons for the variation of effects of Oct4 UTR sequences on luciferase gene expression observed in our study could be explained in view of the cell context in which these UTRs act as well as the dissimilarities of 5´ UTR and 3´ UTR sequences. The combination of these parameters could determine the ultimate function of these UTRs thereby specifying whether these regulatory regions have negative or positive role in the regulation of gene expression. Overwhelming evidence supports the fundamental concept that no common regulatory network exists among various cell lineages (27). Therefore, elements within the same mRNA are likely to undergo differential regulation dependent upon disparate spatial and/or temporal contexts (16) such as various cell types utilized in our study. It has been suggested that, in spite of continuous expression of OCT4 in a variety of pluripotent cell lineages from ICM through the primitive ectoderm to PGC, its expression level is controlled by distinct mechanisms depending on the developmental stage or cell type. For instance, the OCT4 expression in ICM and PGC - ES cells is specifically activated by the distal enhancer (DE), whereas its expression in the primitive ectoderm or P19 EC cells is regulated by the proximal enhancer (PE) (28). This discrepancy in the control of gene expression also holds true at the post-transcriptional level and different results obtained from BMSCs and P19 cells is due to the fact that the environment of P19 embryonal carcinoma cell line is principally of a different kind in comparison with BMSCs. This leads to the diverse mechanisms of post-transcriptional regulation of OCT4 functioning in these different cellular contexts.
The expression of OCT4 has been reported in embryonic stem cells (29) and adult somatic tissues. Bone marrow cells of both human and mouse, particularly hematopoietic and mesenchymal stem cells (HSCs and MSCs), have been the most frequently cited source of OCT4 expression in somatic tissues. Hence, it could be inferred that OCT4 may not only be crucial for the maintenance of pluripotency in embryonic cells but may also play an important role in the self-renewal of somatic stem cells and in maintaining tissue homeostasis (30, 31).
Murine P19 embryonal carcinoma cells exhibit greater amount of Oct4 expression relative to BMSCs (somatic stem cells). This discrepancy in the expression level of Oct4 is closely correlated with difference in self-renewal and pluripotency capacity of these cells which conforms to their markedly distinguishable characteristics as embryonic and somatic stem cells. We could hypothesize that in BMSCs there exist negative regulatory elements including microRNAs required to keep the expression of Oct4 in lower levels compared to P19 cells. These microRNAs are able to bind to the Oct4 UTR and impede the expression of luciferase reporter gene.
Different behaviors of Oct4 UTRs in P19 and BMSCs could also be attributed to the dissimilarity between 5´UTR and 3´UTR in some evolutionarily crucial aspects. Studies based upon quantitative analysis of UTR length have yielded some insight into the roles played by 5´UTR and 3´UTR in the control of gene expression. A computational multispecies analysis of a large UTR database suggests that the mean 3´UTR length in human transcripts is several times longer than the mean human 5´UTR length (32). It has also become evident that 5´UTR average length is constant, roughly 200 nucleotides, while more striking differences are observed for mRNAs 3'UTRs in which the length of mRNA 3´UTR is significantly greater in vertebrates than invertebrates, plants and fungi (33). In addition, the average length of 3´UTR sequences has increased during evolution, suggesting their possible contribution to organism complexity. By contrast, the 5´UTR length is remarkably consistent in organisms ranging from fungi and plants to invertebrates and vertebrates, including human. This evolutionary expansion of the 3´UTR reveals that there is substantial potential for 3´UTR-based translational regulation during evolution (34), and that such control mechanisms might be significant in determining differences among various cell types. The greater length of 3´UTR sequences in mRNAs might be connected to the fact that they have acquired new functions in evolution (33). Taken together, the more extended length of 3´UTRs during evolution among organisms and relative to 5´UTR in the same organism is indicative of their greater functional conservation in gene regulation which could explain the functional constancy of the Oct4 3´UTR in two different cellular contexts.
Taking into account recent reports which show differentiation of somatic stem cells (35) and the fact that mouse ESCs (36, 37) can be modulated by miRNAs through post-transcriptional attenuation of key pluripotent factors, the identification of miRNA target sites on the UTRs of Oct4 could be beneficial in detailing mechanisms underlying the function of these UTRs. Expression analyses and computational predictions have demonstrated miR-145 can target 3´UTRs of Oct4, Sox2, and Klf4 in luciferase assays in both HeLa cells and hESCs (14). Other miRNAs may also be involved in the negative regulation of these 3´UTRs, considering the prediction of many miRNA target sites in human Oct4, Sox2, and Klf4 3´UTRs(1). Targets of miRNA regulation are inherently difficult to identify due to the partial complementarity between the miRNAs and the target mRNA. For this reason, focus has largely been on computational prediction of targets (38). Thus, we conducted bioinformatic analysis to predict microRNAs associated with UTR regions of Oct4 mRNA. To identify the miRNA targets on the Oct4 5´UTR and 3´UTR, we took advantage of miRWalk database (39). Based on the prediction, 29 miRNAs can recognize their targets by binding to motives in the 3´UTR sequence of Oct4 and 5 miRNAs can recognize their targets by binding to motifs in the 5´UTR sequence of Oct4 (data not shown). Comparing miRNA target sites on 5´UTR and 3´UTR, we observed the larger number of target sites on the Oct4 3´UTR indicating the remarkably greater significance of miRNA regulation on the 3´UTR of Oct4 compared to the 5´UTR of Oct4. One of the microRNAs binding to the Oct4 5´UTR recognized through our analysis which was miR21 previously indicated to repress stem cell factors such as OCT4, NANOG, SOX2, and C-MYC (23). It is worthy of note that the number of microRNA target sites known through bioinformatic tools usually is lower than those identified by experimental approaches. The challenge of establishing miRNA functions and understanding the biological processes they regulate has emphasized the need for experimental approaches to identify miRNA targets (38). It is self- explanatory that the functional analysis of microRNAs underlying the regulation of Oct4 UTRs will identify novel mechanisms that determine the maintenance of P19 and BMSC pluripotecy.
Conclusion
Examination of OCT4 regulation with respect to its diverse biological functions is of great significance. This study sheds some light on the part played by the UTR regions of mouse Oct4 in regulating gene expression in BMSCs and P19 cells and is only the starting point in appreciating the processes underlying post-transcriptional regulation of OCT4 in various pluripotent cells. This information may elucidate the mode of post-transcriptional regulation of OCT4 as one of the most potent factors mediating pluripotency. Certainly, more detailed work will be required to understand the key regulatory elements affecting the UTR regions of Oct4 and also particularize the effects of these interacting factors on the activity of Oct4 mRNA. This knowledge could aid us in conceiving the fact that how OCT4 exerts its pleiotropic affects in various cell lines as well as drawing general principles that define the commonalities and differences of OCT4 regulation among different stem cells. Being of special importance, microRNAs specifically associated with Oct4 UTRs will be at the focus of our future study.
Acknowledgment
We are deeply grateful to Dr Saman Hosseinkhani for his kind support and encouragement. This work was supported by a research grant from Iranian Council of Stem Cell Technology and Tarbiat Modares University. The results described in this paper were part of student thesis.
References
- 1.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 2.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;22:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 5.Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ, et al. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 6.Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 8.Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, et al. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–668. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- 9.Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, Grey C, et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell. 2006;11:535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 11.Webster JD, Yuzbasiyan-Gurkan V, Trosko JE, Chang CC, Kiupel M. Expression of the embryonic transcription factor Oct4 in canine neoplasms: a potential marker for stem cell subpopulations in neoplasia. Vet Pathol. 2007;44:893–900. doi: 10.1354/vp.44-6-893. [DOI] [PubMed] [Google Scholar]
- 12.Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trosko JE, Tai MH. Adult stem cell theory of the multi-stage, multi-mechanism theory of carcinogenesis: role of inflammation on the promotion of initiated stem cells. Contrib Microbiol. 2006;13:45–65. doi: 10.1159/000092965. [DOI] [PubMed] [Google Scholar]
- 14.Hatefi N, Nouraee N, Parvin M, Ziaee S-AM, Mowla SJ. Evaluating the expression of Oct4 as a prognostic tumor marker in bladder cancer. Iran J Basic Med Sci. 2012;15:1154. [PMC free article] [PubMed] [Google Scholar]
- 15.Zangrossi S, Marabese M, Broggini M, Giordano R, D'Erasmo M, Montelatici E, et al. Oct-4 expression in adult human differentiated cells challenges its role as a pure stem cell marker. Stem Cells. 2007;25:1675–1680. doi: 10.1634/stemcells.2006-0611. [DOI] [PubMed] [Google Scholar]
- 16.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 17.Makhanova N, Hagaman J, Kim HS, Smithies O. Salt-sensitive blood pressure in mice with increased expression of aldosterone synthase. Hypertension. 2008;51:134–140. doi: 10.1161/HYPERTENSIONAHA.107.098897. [DOI] [PubMed] [Google Scholar]
- 18.Ryan K, Bauer DL. Finishing touches: post-translational modification of protein factors involved in mammalian pre-mRNA 3' end formation. Int J Biochem Cell Biol. 2008;40:2384–2396. doi: 10.1016/j.biocel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: messages from the 3' end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 20.Hu Z, Bruno AE. The Influence of 3'UTRs on MicroRNA function inferred from human SNP Data. Comp Funct Genomics. 2011;2011:910769. doi: 10.1155/2011/910769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 22.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sangeeta C, Jayanta KP. Role of 5 - and 3 -untranslated regions of mRNAs in human diseases. Biol Cell. 2009. pp. 251–262. [DOI] [PubMed]
- 24.Krebsbach PH, Kuznetsov SA, Satomura K, Emmons RV, Rowe DW, Robey PG. Bone formation in vivo: comparison of osteogenesis by transplanted mouse and human marrow stromal fibroblasts. Transplantation. 1997;63:1059–1069. doi: 10.1097/00007890-199704270-00003. [DOI] [PubMed] [Google Scholar]
- 25.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005. pp. 947–956. [DOI] [PMC free article] [PubMed]
- 26.Matoba R, Niwa H, Masui S, Ohtsuka S, Carter MG, Sharov AA, et al. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengner CJ, Welstead GG, Jaenisch R. The pluripotency regulator Oct4: a role in somatic stem cells? Cell Cycle. 2008;7:725–728. doi: 10.4161/cc.7.6.5573. [DOI] [PubMed] [Google Scholar]
- 28.Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 29.Solter D. Mammalian cloning: advances and limitations. Nat Rev Genet. 2000;1:199–207. doi: 10.1038/35042066. [DOI] [PubMed] [Google Scholar]
- 30.Izadpanah R, Trygg C, Patel B, Kriedt C, Dufour J, Gimble JM, et al. Biologic properties of mesenchymal stem cells derived from bone marrow and adipose tissue. J Cell Biochem. 2006;99:1285–1297. doi: 10.1002/jcb.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 32.Pesole G, Liuni S, Grillo G, Licciulli F, Mignone F, Gissi C, et al. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5' and 3' untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pesole G, Grillo G, Larizza A, Liuni S. The untranslated regions of eukaryotic mRNAs: structure, function, evolution and bioinformatic tools for their analysis. Brief Bioinform. 2000;1:236–249. doi: 10.1093/bib/1.3.236. [DOI] [PubMed] [Google Scholar]
- 34.Mazumder B, Seshadri V, Fox PL. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 35.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 37.Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- 38.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 39.http://www.umm.uniheidelberg.de/apps/zmf/mirwalk/predictedmirnagene.php .