Abstract
The influenza A virus causes a highly contagious respiratory disease that significantly impacts our economy and health. Its replication and transcription is catalyzed by the viral RNA polymerase. This enzyme is also crucial for the virus, because it is involved in the adaptation of zoonotic strains. It is thus of major interest for the development of antiviral therapies and is being intensively studied. In this article, we will discuss recent advances that have improved our knowledge of the structure of the RNA polymerase and how mutations in the polymerase help the virus to spread effectively among new hosts.
Keywords: adaptation, host restriction, influenza A, replication, RNA polymerase, structure, transcription
The Orthomyxoviridae have been among us for at least five centuries [1]. They comprise five well-described genera (Thogotovirus, Isavirus, Influenzavirus A, Influenzavirus B and Influenzavirus C) and some unclassified species that likely form a sixth genus [2]. Although all influenza genera can infect humans and cause epidemics, the influenza A virus (IAV) subtypes have been particularly harmful for our health and economy. Vaccines are available against seasonal IAV strains, as are inhibitors against the M2 and neuraminidase (NA) proteins. However, because the dynamics of IAV are difficult to predict, current vaccines are less likely to offer protection against emerging pandemic strains. For instance, the 2009 pandemic was caused by a strain that proved to have the same hemagglutinin and NA subtypes as one of the circulating seasonal strains [3]. In addition, the appearance of strains that are resistant to current drugs is rapidly becoming a problem [4].
Besides worldwide IAV surveillance, current research focuses on the development of vaccines that offer broad-spectrum, long-term protection and new inhibitors that target IAV replication [5,6]. One of the potential targets for the development of such antivirals is the trimeric RNA-dependent RNA polymerase (RdRp), the enzyme that drives the viral replication cycle. Over the past decade, this enzyme has been intensively studied using biochemical, biophysical and crystallographic techniques. This article will focus on recent advances in our knowledge of the RdRp structure and the adaptations that have been observed in the RdRp after zoonotic transfer. Together, these two aspects are vitally important for our understanding of the viral replication cycle and the prediction and prevention of future pandemics [1,7,8].
Transcription & replication in the nucleus
The genetic material of IAV can be replicated in a wide range of vertebrates, including pigs, (wild) fowl, bats and humans. It is divided into eight segments called viral RNAs (vRNAs), which are all present in a single IAV virion (Figure 1) and encode at least 16 different viral proteins, using partially overlapping open reading frames (ORFs) and alternative splicing (see [9] and the references therein). The vRNAs are single stranded, except for the last 13–14 nucleotides of the 5′ and 3′ termini, which are partially complementary [10]. In the virion and in infected cells, the partially complementary ends are bound by a single copy of the viral RdRp and, in the presence of nucleotide triphosphates (NTPs), it can serve as promoter for vRNA synthesis (e.g., see [11]). The rest of the vRNA is covered by a helical assembly of nucleoprotein (NP) monomers, which associate with the RNA in a sequence-independent manner at 24-nucleotide intervals and a Kd of approximately 20nM [12,13]. Together, the complex of vRNA, RdRp and NP is generally referred to as the viral ribonucleoprotein (vRNP) complex.
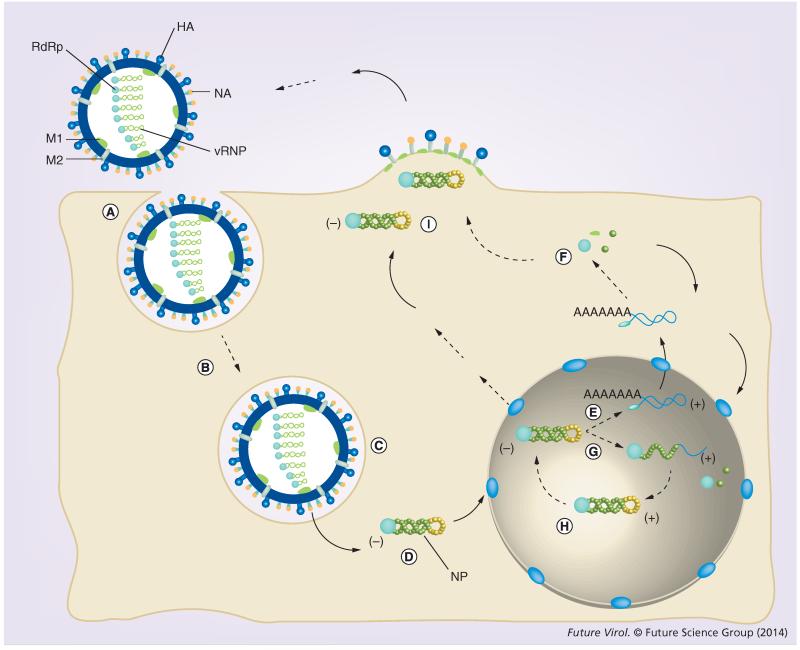
Figure 1. Influenza A virus replication cycle.
(A) A typical IAV infection starts with the binding of the HA proteins on the surface of the virus particle to the sialic acid-containing oligosaccharides on the surface of the host cell. Human influenza strains typically bind α2,6-linked sialic acid, which is present in the upper respiratory tract, while avian strains recognize α2,3-linked sialic acid, which is present in the lower respiratory tract [97]. (B) Binding to the host receptors is followed by endocytosis, (C) fusion of the endosomal membranes with the membrane of the virion and release of the vRNPs. (D) vRNPs are next transported to the nucleus of the host. NLSs on the viral proteins and host importins play a crucial role in this process. (E) In the nucleus, transcription is initiated with capped primers that are cleaved from cellular pre-mRNAs. (F) Mature viral mRNAs are exported to the cytoplasm for the synthesis of new viral proteins. (G) New viral RNP subunits are imported into the nucleus and associate with newly forming cRNAs in order to create cRNPs. (H) cRNPs next synthesize new vRNAs. (I) After several rounds of amplification, newly formed vRNPs are exported from the nucleus and transported to the cell membrane in a Rab11- and microtubule-dependent manner for packaging and budding. cRNA: Complementary RNA; cRNP: Complementary ribonucleoprotein; HA: Hemagglutinin; IAV: Influenza A virus; NLS: Nuclear localization signal; vRNA: Viral RNA; vRNP: Viral ribonucleoprotein.
Unlike the majority of RNA viruses, whose replication cycle is confined to the cytoplasm, the IAV vRNPs enter the nucleus of an infected cell in order to transcribe and replicate the viral genetic information (Figure 1) [14]. In order to facilitate transcription, the vRNPs associate with the DNA-dependent RNA polymerase II (Pol II) by binding to the C-terminal domain (CTD) of initiating Pol II complexes [15]. A hallmark of initiating Pol II is a serine-5-phosphorylated CTD, and the current evidence suggests that the RdRp recognizes these phosphopeptides specifically [15]. It is currently unknown whether replicating vRNPs form a distinct population from the transcribing vRNPs and whether the Pol II-associated vRNPs dissociate after transcription.
The RdRp initiates viral transcription by binding a cap-1 structure (m7GpppNm) of an emerging pre-mRNA and cleaving the pre-mRNA at 10–15 nucleotides from the cap [11,16]. Cleavage in vivo preferentially takes place downstream of nucleotides that closely match the 3′ vRNA terminus [17], which allows the RdRp to align the bases of the capped primer with the vRNA and extend it in a template-dependent fashion. Termination occurs when the RdRp reaches an oligo-U tract that is located just upstream of the 5′ terminus of the vRNA where it stutters, thereby adding a poly-A tail onto the viral mRNA [18] and leaving the 5′ terminus of the vRNA untranscribed.
In contrast to viral transcription, replication produces a full complementary copy of the vRNA, the complementary RNA (cRNA) (Figure 1). Akin to the vRNA, the ends of these cRNAs are partially complementary and able to base pair in order to form cRNA promoter elements. In line with these similarities, cRNAs are bound by newly produced viral NP and RdRp in order to form complementary ribonucleoprotein (cRNP) complexes, which synthesize new vRNAs. It has been shown that these cRNPs can be isolated from infected cells and used to produce new vRNA molecules in vitro [19]. Although vRNPs and cRNPs appear to be the minimal replicating and transcribing units in infected cells and in vitro [20], a recent study showed that the presence of the RdRp is sufficient to replicate and transcribe model vRNAs in the absence of NP [21,22], provided they are shorter than 149 nucleotides in length [23].
Polymerase overview
The IAV RdRp is a member of the superfamily of template-dependent nucleic acid polymerases. These enzymes have a polymerase domain that is typically <400 amino acids in length and that consists of – from N- to C-termini – the key functional motifs G, F and A–E [24]. Unlike other RNA virus RdRps, the IAV RdRp consists of three individual polypeptides called polymerase basic 2 (PB2), polymerase basic 1 (PB1) and polymerase acidic (PA). Encoded by the viral segments 1–3, respectively, these subunits make up an RdRp with a mass of approximately 250 kDa, which makes the IAV RdRp one of the largest RNA virus RNA polymerases known to date.
PB1 structure & function
Consisting of 757 amino acids, PB1 is the second largest protein of the three RdRp subunits. Sequence and mutational analyses have indicated that the conserved RdRp motifs A–F (F is sometimes referred to as premotif A) reside in the centre of PB1 (Figure 2) [25,26]. Although initial reports indicated that PB1 is able to produce short RNAs on its own, current evidence suggests that PB1 expressed in mammalian cells needs to interact with both PA and PB2 for the efficient synthesis of pppApG, the starting product of both vRNA and cRNA synthesis [27]. Moreover, and in spite of the observation that PB1 amino acids 187–216 contain a bipartite nuclear localization signal (NLS; RKRR and KRKQR), PB1 depends on an interaction with PA for its nuclear import [28].
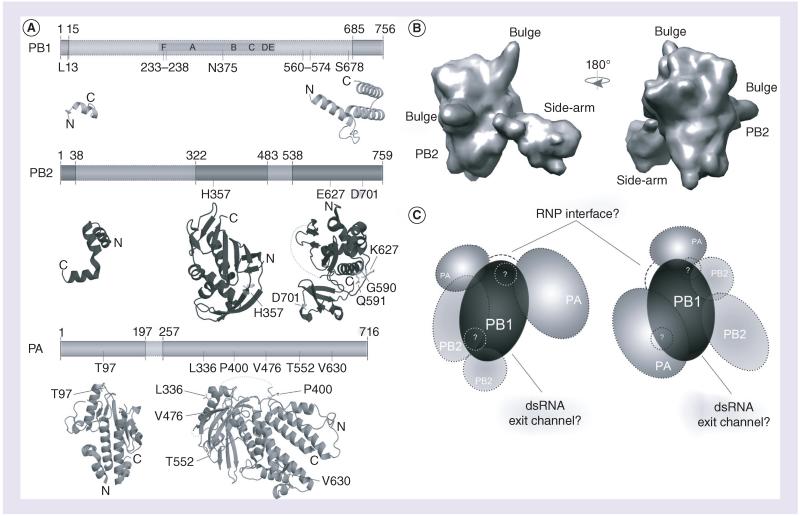
Figure 2. Structure of the influenza A virus RNA-dependent RNA polymerase.
(A) The influenza A virus (IAV) RNA-dependent RNA polymerase (RdRp) consists of the subunits PA, PB1 and PB2. Six of the seven canonical RdRp domains motifs A–F are found in PB1. Also indicated are the RNA binding sequences (gray), as are the residues involved in adaptation (black; see also Table 1). At present, significant structural information is only available for PA and PB2, which is shown below the schematics of the subunits. The visualized structural information is based on Protein Data Bank (PDB) entries 2VY6, 2W69, 2ZNL, 2ZTT, 3EBJ and 4CB4. (B) Two views of the electron microscopy model proposed by Moeller et al. [39]. Panels are based on the PDBe entry EMD-2213. (C) Two interpretations based on current electron microscopy models. On the left, the PA C-terminus presents the flexible part of the IAV RdRp, as defined by Moeller et al. [39]. It is connected to the PA endonuclease domain via a linker [37], which may wrap around PB1. An interaction through the core of PB1 would seem unlikely, as this may interfere with the right-handed fold of the polymerase active site. On the right, the PB2 C-terminus presents the flexible part of the IAV RdRp, as proposed by Torreira et al. [49], The PA domains flank PB1 on the other side, but the PA N-terminus can still interact with PB2 in accordance with recent data [47]. In both models, the flexible domain can be envisioned as blocking the dsRNA exit channel of the RdRp, which likely resides opposite – albeit at a slight angle – to the template entry grove. Both interpretations also have a similar interface consisting of PB1 and PA residues in order to interact with the RNP. Since the PA linker domain may not be part of this interaction, it is displayed in both models as a dashed line. The areas in (C) labeled with question marks refer to the bulge structures in (B), whose identity is current unknown.
PA: Polymerase acidic; PB1: Polymerase basic 1; PB2: Polymerase basic 2; RNP: Ribonucleoprotein.
At present, no structural information is available for the PB1 polymerase domain, but some information is available regarding its both its amino- and carboxyl-terminus. Cocrystallizations with PA revealed that the first 15 amino acids of the PB1 N-terminus form a small loop that interacts with PA through a number of hydrogen and hydrophobic bonds [29]. A cocrystallization with PB2 revealed that 86 amino acids of the PB1 C-terminus fold into a helix–loop–helix motif that interacts tightly with the PB2 N-terminus [30]. The two PB1 interaction interfaces are completely conserved among avian and mammalian IAV strains and are resistant to drift, probably because compensatory mutations are required in either PA and PB1 or PB1 and PB2 in order to maintain the interaction between the subunits. In turn, this also makes them interesting targets for drug design [31,32].
PB2 structure & function
The partial PB2 structures available to date suggest that the protein is made up of four domains. The most N-terminal sequences (1–35) are involved in PB1 binding, whereas mutational studies of residues 737–755 showed that the C-terminus contains a bipartite NLS (RKR and KRIR). This latter observation was confirmed with a cocrystal structure of the PB2 C-terminus with importin α5 [33]. The C-terminal part of PB2 also contains the so-called 627 domain (residues 535–684) [33], which is involved in host adaptation and virulence (see below). Interestingly, the two CTDs can be separately expressed as soluble proteins, but they pack together when fused. In this packed conformation, the 627 domain can influence the binding of the NLS to importin [34].
The middle domain of PB2, residues 318–483, was cocrystallized with cap analog m7GTP [35]. This demonstrated that PB2 is able to sandwich the m7-methylated guanine base using H357, F323 and F404 residues, which is similar to, but structurally distinct from, the binding mechanism used by other known cap binding proteins [35]. Base recognition is achieved through Q406, D361 and K376, while the phosphates of the cap are coordinated by hydrogen bonds with H432, H357, K339 and N429 residues [35]. Combined with the fact that cap binding is essential for IAV transcription and thus viral protein synthesis, PB2 presents a good target for the development of antiviral drugs. Based on the x-ray structure, docking studies have been performed in order to find guanine derivatives with an increased affinity for the PB2 ligand binding site [36]. Unfortunately, none of the identified competitive inhibitors has shown antiviral activity in infected cells, probably due to the negative charge present on the compounds, which reduced their uptake by the cell [36].
PA structure & function
Of the three influenza subunits, only the PA subunit can be expressed at sufficiently soluble levels in order to facilitate biochemical studies. Tryptic digestion of the PA subunit produces two fragments of 25 and 55 kDa, which are likely connected through an approximately 20-amino acid linker [37]. The largest of these fragments contains the PA C-terminus and is involved in the binding of the PB1 N-terminus through residues 408–412, 617–623, 666–673 and 706–710 [29]. The large fragment also contains several residues that have been linked with RdRp activity, including residues W406, E410, G502 and E524 [38]. In recent electron microscopy (EM) structures of free RdRp and isolated RNPs, the PA CTD was proposed to stick out of the globular RdRp structure [39], probably as a result of the relatively flexible nature of the linker region. Although this proposal has so far not been confirmed with antibody binding studies, it has been reported that neither the N- nor C-termini of PA can form a fully stable interaction with PB1 without the linker, which suggests that this sequence may help to stabilize the PA-PB1 interface [37].
The smallest PA cleavage fragment contains the N-terminal endonuclease domain. Crystallization studies have shown that the structure of this domain is similar to the fold of type II restriction endonucleases [40,41] and is able to coordinate Mn2+ or Mg2+ through residues H41, E80, D108, E119 and K134. For the cleavage of capped pre-mRNAs or primers, the enzyme preferentially uses substrates containing guanosine residues [42] and Mn2+ as cofactors, which matches the terminal 3′-UC of the vRNA. PA residue H510 is also involved in viral cap snatching, putatively by contributing to pre-mRNA binding [38]. So far, a number of compounds have been identified that can act as inhibitors of the PA endonuclease activity in vitro, including four diketo isolates and green tea chatechin [43], but none have shown sufficient antiviral activity in order to justify pursuing clinical development. Recent crystal structures of the PA active site with potential inhibitors revealed that the endonuclease domain contains various pockets that can be occupied by inhibitors [43]. However, some of these pockets display high plasticity, which will need to be considered for the development of optimized endonuclease inhibitors.
Overall structure & assembly of the RdRp
Various studies have demonstrated that the heterotrimeric RdRp assembles head to tail (i.e., from the amino- to carboxyl-termini) in the order PA–PB1–PB2 (e.g., [27,29,30]. Import into the nucleus of the host is crucial for this assembly and it is therefore also partially dependent on adaptation to the host [44,45]. In the current model, the PA and PB1 subunits form a dimer in the cytoplasm before being efficiently imported into the nucleus by RanBP5 [28]. By contrast, the PB2 subunit is imported separately, as assessed through correlation microscopy [46], and associates with the PA–PB1 dimer in the nucleus by binding to the PB1 C-terminus [30]. Additional interactions, such as those between PB2 and the PA N-terminus [47], probably stabilize the complex further.
No atomic reconstruction of the IAV RdRp has so far been made and, at present, only EM models are available, with the best offering a resolution of 13 Å [39]. Nevertheless, these models do give us an impression of the outline of the RdRp (Figure 2B). Overall, the IAV RdRp has a globular appearance when associated with the RNP [13,22,39,48]. In solution, two small bulges and one side-arm become visible as well, probably as a result of structural rearrangements (Figure 2B) [39,49]. Using antibody mapping, Moeller et al. showed that one of these bulges was part of PB2, whereas the CTD of PA was fitted into the side-arm [39]. Together with biochemical evidence suggesting that the two domains of PA are connected via a linker region that improves PB1 binding [37], these observations suggest that the C-terminus of PA is flexible and only conditionally associated with the body of the RdRp. By contrast, however, Torreira et al. concluded based on comparisons with a previous study that had involved anti-body binding experiments that the C-terminus of PB2, rather than a portion of PA, is part of a flexible arm of the RdRp [49,50]. In support of this, He et al. [51] managed to dock the crystal structure of the PA C-terminus into the EM model of Torreira et al. and observed that such a domain organization would be similar to the reovirus λ3 RdRp. Hence, in this model, PA would be tightly associated with PB1, whereas PB2 would be (partially) flexible. It is unclear at present whether the above two views, which we have tried to capture schematically in Figure 2C, are mutually exclusive or whether the RdRp harbors two flexible domains. At present, we assume that at least one flexible element resides in the IAV RdRp and speculate that it may be part of the RdRp’s initiation platform. Similar to other de novo-initiating RNA virus RdRps, such as the reovirus RdRp, the IAV RdRp may contain residues that support the formation of the first dinucleotide bond during replication and it may need to undergo a conformational change in order to allow the RdRp to transit from initiation to elongation.
Binding of the viral promoter
The binding of the partially complementary termini of the vRNAs or cRNAs is crucial for the stability and activity of the RdRp [52]. This is supported by a comparison of EM structures, in which template-free RdRps were found to be more prone to aggregation and had a less compact appearance than RdRps that were part of vRNPs [22,49]. These latter results also suggest that the enzyme complex undergoes some structural rearrangements upon promoter binding, which makes the residues involved in this step a potential target for antiviral strategies.
Over the years, a number of residues have been identified that directly influence vRNA or cRNA promoter binding. Only PB1 appears to be critically involved in the binding of the 5′ end of the vRNA promoter, in particular through residues 559–574 and 233–238 (e.g., see [53]). It is presently unclear which residues coordinate the 3′ end, since it cannot be efficiently bound in the absence of the 5′ end. However, when it is present, all three subunits can be cross-linked to the 3′ end (e.g., see [53]), suggesting that all three subunits contribute to the formation of the template binding channel. In contrast to the above, the 5′ strand of the cRNA promoter appears to be chiefly coordinated by the PA subunit, most notably via residues 164–176 [54].
When vRNA is bound, the IAV RdRp is able to synthesize both mRNA and cRNA in infected cells [55] and in vitro [20], suggesting that no clear molecular switch between the two activities exists. It is possible that the pathway of mRNA production simply depends on the – putatively stochastic – association of vRNPs with a serine-5-phosphorylated CTD of an initiating Pol II complex inside the nucleus of the host cell. Any non-CTD-associated vRNPs would then be dedicated to cRNA synthesis. However, other mechanisms appear to play a role as well, including NTP concentrations in in vitro assays [56], the presence of short 5′ end-derived small vRNAs [57] and the ‘hinge’ residue on the vRNA promoter, which can be targeted in order to uncouple replication from transcription [58].
In the absence of the viral RdRp, the viral promoter RNA element forms a partially complementary structure that consists of the conserved 5′ and 3′ terminal sequences present in each genome segment [10]. A similar sequence element can be found in the genomic ends of the genomes of the Arenaviridae and Bunyaviridae. Binding to the RdRp creates structural rearrangements in the promoter, which force it to adopt a corkscrew-like structure. This view is now supported by both mutagenesis and single-molecule experiments. As mentioned above, structural rearrangements are also observed in the RdRp upon promoter binding [49]. So far, the exact location of the promoter has not been conclusively determined, owing in part to the low resolution of current EM structures and the relatively small mass of the promoter (~10 kDa) relative to the RdRp (~250 kDa).
Although naturally occurring mutations in the noncoding regions of the vRNA have so far not been observed, probably because they play roles in transcription, replication and packaging, man-made mutations can affect the activity of the viral RdRp. Most notably, promoter mutations can allow an avian RdRp to function efficiently in mammalian cells without the need for protein adaptations [59]. The most significant improvements in RdRp activity are seen for mutations found at positions 3, 5 and 8 of the vRNA promoter, the so-called ‘promoter up’ mutations. In a viral context, these mutations generally result in attenuated recombinant viruses, explaining the lack of these mutations in natural isolates.
NP structure & function
IAV NP is essential for the replication of viral genome segments and the transcription of their encoded genes. However, as mentioned above, it is not crucial for the replication and transcription of model vRNAs [22] or the activity on model promoters [21]. In crystal structures, NP forms a trimeric complex that contains a body (residues 1–148, 277–396 and 464–498), head (residues 150–263 and 437–451) and tail domain (397–436; reviewed in [60]). Interactions between NP monomers are mediated through the insertion of a tail domain loop of one NP into a specific body domain groove of a second NP. Residues in the NP head domain, most crucially R204, W207 and R208, were found to be required for the interaction between NP and the viral polymerase [61], which is associated with the RNP at the end of the partially double-stranded promoter (Figure 1).
Current evidence suggests that phosphorylation could play a role in the oligomerization dynamics of NP, since NP monomers are typically loaded onto the growing NP chain in new RNPs [23]. In the vRNP [13,39] and cRNP [19], the NP molecules assemble into a double-helical arrangement (six pairs of NP per turn) that shows major and minor grooves not dissimilar to dsDNA and dsRNA. At the side of the minor groove, antiparallel-running NP molecules interact with each other via the N-terminus, while intrastrand NP neighbors bind via the tail loop–body groove interaction [13,39].
Adaptation of viral RdRp subunits
Avian IAV strains can infect and replicate in a multitude of hosts. However, efficient replication depends on the interaction of viral proteins with various host proteins, such as importins and the cellular transcription complex Pol II [15,44], but also with host-specific enhancing factors [62]. Moreover, the virus may also encounter host-specific antiviral responses. Together, these factors impose a strong selective pressure in order to improve the efficacy of viral replication after zoonotic transfer [63].
To date, adaptive mutations have been found in avian strains isolated from human, swine, equine or canine hosts (Table 1). In all cases, point mutations in the hemagglutinin and NA proteins were identified as important factors in determining host restriction (e.g., see [7,64]). However, it has also been observed that mutations in the viral RdRp can contribute significantly to the pathogenicity of avian strains in mammalian hosts, in part because avian RdRps have limited activity in mammalian cells (discussed below). In turn, such impaired polymerase activities can result in reduced viral transcription and replication rates and concomitantly low viral titers. In addition, lower rates of replication also limit the number of new variant vRNAs that the RdRp can produce in order to allow the virus to explore beneficial mutations and overcome any host-associated bottlenecks.
Table 1. Key adaptive mutations in the influenza A virus viral polymerase subunits that enhance the replication, spread or pathogenicity of avian strains in mammalian cells, where known affects on replication or transcription are specified.
| Subunit | Mutation | Affects | Ref. |
|---|---|---|---|
| PB1 | L13P | cRNA synthesis | [66] |
| S678N | RNA synthesis | [66] | |
| N375S | Pathogenicity, RNA synthesis | [65] | |
|
| |||
| PB2 | E627K | RNA synthesis, nuclear import | [67] |
| H357N | Pathogenicity, RNA synthesis | [85] | |
| D701N | Pathogenicity cRNA, mRNA synthesis | [66] | |
| E677G | Pathogenicity, RNA synthesis | [81] | |
| T271A | Pathogenicity, RNA synthesis | [81] | |
| S714R | Pathogenicity, cRNA and mRNA synthesis | [66] | |
| G590S | Pathogenicity, RNA synthesis | [81] | |
| Q591R | Pathogenicity, RNA synthesis | [81] | |
|
| |||
| PA | P400L | RNA synthesis | [84] |
| T552S | Pathogenicity, RNA synthesis | [84] | |
| T85I | RNA synthesis | [82] | |
| G186S | RNA synthesis | [82] | |
| L336M | Pathogenicity, RNA synthesis | [82] | |
| A36T | RNA synthesis | [85] | |
| T97I | Pathogenicity, RNA synthesis | [83] | |
| K615N | Pathogenicity, vRNA synthesis | [66] | |
cRNA: Complementary RNA; PA: Polymerase acidic; PB1: Polymerase basic 1; PB2: Polymerase basic 2; vRNA: Viral RNA.
PB1 adaptations
For PB1, only a few adaptive mutations have been identified. The most widely observed residue concerns position 375, which resides just upstream of the polymerase domain. In avian strains, an asparagine is typically found at this position, whereas strains adapted to human cells have a serine (N375S) [65]. Interestingly, the separation between avian and human strains is not strict and avian strains with a serine at position 375 have been found. Although the mechanism by which position 375 influences host restriction has not yet been identified, it seems that additional mutations may play a role. These possibly include adaptive mutations such as L13P and S678N [66]. L13P was observed to cause an increase in cRNA synthesis in mammalian cells, whereas S678N dramatically increased RNA synthesis in general [66], lending credence to the concept that an increase in polymerase activity is correlated with virulence.
PB2 adaptations
The majority of adaptive mutations found in the IAV RdRp occur within PB2, where they localize to an N-terminal and a C-terminal cluster. The most common of these mutations occurs at residue 627 [67]. In normal avian strains, albeit with some exceptions [68], this residue is typically a glutamate. However, in avian H5N1 strains isolated from humans, this residue has been mutated to a lysine (i.e., E627K) in slightly over 30% of the isolates [69]. K627 is also the signature of the 1918 pandemic H1N1 strain and the recently isolated H7N9 virus [8,70]. More importantly, the introduction of this mutation is sufficient to rescue avian RdRp activity in mammalian cells, although it is not yet fully understood through which mechanism this occurs. Current evidence suggests that the mutation may confer multiple advantages.
Residue 627 resides in the surface-exposed ‘627 domain’ (Figure 2A), which is part of the C-terminus of PB2 and located near the PB2 NLS and the α-importin binding region [33]. The overall charge of this region is negative in avian PB2 variants and the introduction of a lysine to this surface can disrupt this charge [33,34]. Although the change in charge does not appear to affect viral transcription rates in mammalian cells per se [71], E627K does have clear effects on RdRp activity. One of the directly measurable effects of the E627K PB2 mutation is an increased RdRp activity in vivo at temperatures of approximately 33°C [72]. This is in line with the difference between the temperature of the human upper respiratory tract (33–35°C) and the avian intestinal tract (38–40°C), the site where avian influenza strains thrive. Similar results were obtained in vitro [73]. The E627K mutation has also been proposed to improve interactions with host factors. One such example concerns the DEAD-box RNA helicase DDX17/p72, which was identified in a large-scale siRNA study [74]. In particular, it was found that the presence of the human homolog of this protein stimulated IAV RdRp containing a PB2-E627 variant, whereas the chicken homolog only stimulated the transcription and replication activity of the PB2-K627 variant.
An improved RNP assembly in the human host may also explain the effect of the E627K mutation, since some evidence has been interpreted as an enhanced interaction between NP and PB2. This view is mostly based on the observation that recombinant avian PB2-E627 viruses only acquire the E627K mutation in the presence of a NP-R65 variant [68]. However, there is also evidence that enhanced coprecipitation levels of PB2 and NP can simply be achieved by increasing the amount of vRNA present in cells [75], suggesting that E627K chiefly affects replication. Interestingly, using short model vRNAs that can be replicated independently of NP [23], recombinant RdRps containing either PB2-E627 or -K627 were demonstrated to have equal activity in cell culture assays at 37°C [59]. Similar results were obtained by simply introducing mutations in the vRNA promoter that stimulated viral replication and transcription [59], suggesting in turn that the catalytic activity of the RdRp is unaffected by the mutation and that a host factor is limiting the initiation or elongation, or both.
Although considerable research has focused on the effects of PB2-E627K, other PB2 mutations can also assist the virus in overcoming host restriction. Moreover, introduction of an E627K mutation in the 2009 H1N1 pandemic strain did not enhance viral replication or pathogenicity at all [76]. Instead of E627K, it was found that a D701N mutation was able to enhance the nuclear localization of PB2 and improve the pathogenicity of the virus and its transmission in ferrets [44,77,78]. Based on the partial PB2 structure that is currently available, D701 appears to be part of the importin-α5 binding domain of PB2 [34]. Since it is presently assumed that human strains depend on importin-α7 for their import, whereas avian strains utilize importin-α3 [79], avian RdRp assembly may be strongly impaired in mammalian cells due to limited PB2-D701 import. This import may be improved after the mutation of aspartate 701 to asparagine (D701N).
Aside from D701N, numerous other mutations have been found that are involved in host restriction (Table 1). On the same domain as E627, residues 590 (G590S) and 591 (Q591R) (Figure 2A) introduce a similar change in charge as E627K [80], and together with T271A, they improve viral replication and virulence [81]. Further PB2 mutations target residues 158, 199, 253 and 256, but no mechanistic details are available for these mutations at present.
PA adaptations
The role of PA in host adaptation is less well established than the role of PB2, but similar to PB2, some PA adaptations enhance the ability of the polymerase to replicate at higher temperatures (Table 1) [82]. Various other PA mutations have also been reported to improve the polymerase activity of avian strains in mammalian cells, such as T97I and T552S [83-85]. Furthermore, some of these mutations have been observed in isolates of H5 and H7 strains [83]. In the PA subunit of the H7N7 strain, the adaptive mutation K615N results in improved replicative kinetics of avian IAV in mammalian cells, but not in avian cells. In addition, a T515A mutation has been implicated in pathogenicity. Interestingly, this change does not affect pathogenicity in ferrets or mice, but it has been shown to reduce virulence in fowl without affecting replication or transmission [86]. Unfortunately, little is known about the mechanisms behind these adaptations, but they do challenge the concept that mutations that increase polymerase activity directly enhance pathogenicity.
Adaptation of other segment 1–3-encoded proteins
Alongside the RdRp proteins, various other viral proteins influence IAV host restriction and viral adaptation. Some of these are encoded by minor ORFs in the polymerase-encoding segments [87-89]. On the PA segment (segment 3), for instance, three additional protein-coding ORFs have been found. One of these proteins is produced as a result of a +1 frameshift at codon 191 of the PA-coding sequence [89]. Since the ORFs of PA and PA-X share the region that encodes the endonuclease domain of PA, it has been proposed that PA-X is involved in host shut-off. Supporting evidence for this role comes from metabolic labeling studies showing reduced host mRNA and protein synthesis in the presence of PA-X. Moreover, mouse studies with PA-X-deletion mutants demonstrated a wide upregulation of host genes, including those involved in antiviral response in particular [89].
The C-terminal region of PA-X that is encoded downstream of the frameshift differs substantially in sequence and length from PA [89]. Sequence analysis has shown that, in particular, the variabilities in sequence and length of this region are different in pathogenic and nonpathogenic strains, which hints at the involvement of this region in host shut-off. One example of this is the 2009 pandemic H1N1 strain, in which the PA-X C-terminus is 20 amino acids shorter (41 vs 61) than many nonpandemic human strains [90].
In addition to PA-X, segment 3 encodes two other proteins: PA-N155 and PA-N182 [88]. These proteins can be produced by leaky ribosomal scanning and initiation from the 11th and 13th start codons in the PA reading frame, respectively [88]. Sequence analysis showed that both codons are highly conserved among IAV strains. Furthermore, mutation of these initiation sites to CUA (methionine to leucine) resulted in a drop in pathogenicity and cell culture replication [88], suggesting that these minor viral proteins are involved in host restriction as well. However, these results are difficult to interpret, since such codon changes (M155L and M182L) also alter the PA and PA-X sequences.
Segment 2 encodes two additional proteincoding ORFs that are important for the regulation of the RdRp and viral replication [87,91]. The first of these encoded proteins is PB1-F2, which is transcribed from a +1 frameshift [91]. PB1-F2 is not found in all IAV isolates. However, in strains that express it, the full-length protein can increase pathogenesis [92]. Moreover, although deletion of PB1-F2 does not appear to have a strong effect on virus growth [91], mutation of position 66 (S66N) can attenuate pathogenetic viruses, such as the 1918 strain, in mice [92]. Current research indicates that PB1-F2 localizes to both the outer mitochondrial membrane (where it can interact with proapoptotic factors) and the nucleus (where it interacts with PB1 and is thought to modulate polymerase activity) [92].
The second protein encoded by segment 2 is PB1-N40, which is expressed from the fifth AUG in the PB1 frame as a result of leaky ribosome scanning. The level of PB1-N40 expression is approximately 5–10% of PB1 and is thought to interact with PB2, PB1, NP and PB1-F2 in the nucleus. It is not yet known exactly what the impacts of the segment 2 proteins are on adaptation and host interaction, since their expressions are strongly intertwined [91]. However, primer extension analysis has shown that their relative expression is very important for virus growth and host adaptation, which suggests that adaptive mutations might occur in this protein after zoonotic transfer [91].
NP adaptations
The adaptation N319K of NP enhances vRNA synthesis in mammalian cells [66]. Current evidence suggests that it alters the interaction of NP with host α-importin isoforms [44]. NP is also involved in the interferon-stimulated activities of MxA, a member of the Dynamin-like large GTPase family. This host protein has multiple antiviral effects against IAV, including inhibition of nuclear localization of vRNPs through the formation of an MxA oligomeric ring around the vRNPs that prevents them from entering the nucleus through – putatively – size restriction [93]. The sensitivity of particular IAV strains to MxA was recently found to determine, in part, their pathogenicity and to be directly related to the origin of the encoded NP [94,95]. NP mutations associated with MxA sensitivity, including G16D, L283P, F313Y and Q357K, have been studied in silico, but have not been validated in cell culture assays at present.
Conclusion
The IAV RdRp is a target for the development of antivirals and an important viral complex that needs to acquire adaptive mutations in order to permit avian IAV strains to replicate and spread efficiently in mammalian cells. The general trend, however, is that, with the exception of the PB1-L13P mutation [66], mRNA and cRNA syntheses are typically affected at similar levels. Given the number of mutations and adaptations found, the virus thus has the ability to gradually adapt its RdRp and modulate vRNA levels in order to buffer the loss of fitness that results from mutations in other viral proteins. Furthermore, it gives the virus the ability to explore adaptations to the new host without a loss of fitness in the old host.
Recent advancements in our understanding of the viral RdRp have shown that the PA endonuclease site, the PB2 cap binding site and the PA–PB1 and PB1–PB2 interactions are excellent targets for the structure-based design of new anti-IAV drugs and they thus deserve significant attention in the near future. However, the process of IAV replication and transcription not only depends on the viral replicative machinery; its activity is also heavily intertwined with various host and viral factors, which leaves us with a wealth of alternative targets that can be explored in the future (e.g., see [4]).
Future perspective
The first studies with isolated IAV RdRps were performed over four decades ago [96]. Since then, our technologies have improved significantly, most notably through the purification of recombinant IAV RdRp, and our knowledge of IAV replication and transcription has followed suit. The most significant advances in the last 5 years were the x-ray structures of fragments of the PA and PB2 subunits. In the next 5 years, we will likely see the appearance of atomic resolution structures for the complete IAV RdRp. This will shine new light on the structure of the PB1 subunit, the transfer of the ‘snatched’ primer from PA to the active site of PB1 and how the RdRp interacts with the Pol II CTD. We may also expect to see the first single-molecule studies of IAV transcription and replication, which may help us to answer outstanding questions and create more detailed models for these two activities.
EXECUTivE SUMMARY.
RNA-dependent RNA polymerase structure
Electron microscopy structures of the influenza A virus (IAV) RNA-dependent RNA polymerase are available at 13 Å, but atomic resolution is only available for the domains of polymerase basic 2 and polymerase acidic proteins.
The partial crystal structures currently available have stimulated the design of new, potential anti-IAV inhibitors.
RNA-dependent RNA polymerase adaptation
Key residues involved in the adaptation of avian IAV strains to mammalian cells have been discovered, allowing us to understand this adaptation and monitor the appearance of such mutations in potentially pandemic strains.
Acknowledgements
We apologize to colleagues whose work could not be cited due to length restrictions.
Financial & competing interests disclosure
The authors’ research is supported by grant 825.11.029 from The Netherlands Organization for Scientific Research (NWO), grant 098721/Z/12/Z from the Wellcome Trust, a Kemp postdoctoral fellowship from Lincoln College Oxford and a BBSRC DTP studentship. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Taubenberger JK, Morens DM. Pandemic influenza – including a risk assessment of H5N1. Rev. Sci. Tech. 2009;28(1):187–202. doi: 10.20506/rst.28.1.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Presti RM, Zhao G, Beatty WL, et al. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. 2009;83(22):11599–11606. doi: 10.1128/JVI.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girard MP, Tam JS, Assossou OM, Kieny MP. The 2009 A (H1N1) influenza virus pandemic: a review. Vaccine. 2010;28(31):4895–4902. doi: 10.1016/j.vaccine.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Das K. Antivirals targeting influenza A virus. J. Med. Chem. 2012;55(14):6263–6277. doi: 10.1021/jm300455c. [DOI] [PubMed] [Google Scholar]
- 5.Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr. Opin. Virol. 2012;2(2):134–141. doi: 10.1016/j.coviro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krammer F, Palese P. Universal influenza virus vaccines: need for clinical trials. Nat. Immunol. 2014;15(1):3–5. doi: 10.1038/ni.2761. [DOI] [PubMed] [Google Scholar]
- 7.Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi L, Davis AS, Jagger BW, et al. Analysis by single-gene reassortment demonstrates that the 1918 influenza virus is functionally compatible with a low-pathogenicity avian influenza virus in mice. J. Virol. 2012;86(17):9211–9220. doi: 10.1128/JVI.00887-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois J, Terrier O, Rosa-Calatrava M. Influenza viruses and mRNA splicing: doing more with less. MBio. 2014;5(3):e00070–14. doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson JS. 5 and 3 terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Res. 1979;6(12):3745–3758. doi: 10.1093/nar/6.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994;68(6):4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baudin F, Bach C, Cusack S, Ruigrok RW. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13(13):3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arranz R, Coloma R, Chichón FJ, et al. The structure of native influenza virion ribonucleoproteins. Science. 2012;338(6114):1634–1637. doi: 10.1126/science.1228172. [•• Stunning study reporting electron microscopy (EM) structures of the helical nature of the influenza A virus ribonucleoproteins (RNPs).] [DOI] [PubMed] [Google Scholar]
- 14.Jackson DA, Caton AJ, McCready SJ, Cook PR. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature. 1982;296(5855):366–368. doi: 10.1038/296366a0. [DOI] [PubMed] [Google Scholar]
- 15.Engelhardt OG, Smith M, Fodor E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 2005;79(9):5812–5818. doi: 10.1128/JVI.79.9.5812-5818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krug RM, Broni BA, Bouloy M. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell. 1979;18(2):329–334. doi: 10.1016/0092-8674(79)90052-7. [•• Classic paper describing the cap-snatching activity of the influenza A virus polymerase.] [DOI] [PubMed] [Google Scholar]
- 17.Geerts-Dimitriadou C, Goldbach R, Kormelink R. Preferential use of RNA leader sequences during influenza A transcription initiation in vivo. Virology. 2011;409(1):27–32. doi: 10.1016/j.virol.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Poon LL, Pritlove DC, Fodor E, Brownlee GG. Direct evidence that the poly (A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 1999;73(4):3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.York A, Hengrung N, Vreede FT, Huiskonen JT, Fodor E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc. Natl. Acad. Sci. USA. 2013;110(45):E4238–E4245. doi: 10.1073/pnas.1315068110. [• Study showing that complementary RNPs can be isolated from infected cells and studied in vitro.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vreede FT, Brownlee GG. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 2007;81:2196–2204. doi: 10.1128/JVI.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee MT, Bishop K, Medcalf L, Elton D, Digard P, Tiley L. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acid. Res. 2002;30:429–438. doi: 10.1093/nar/30.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Resa-Infante P, Recuero-Checa MA, Zamarreño N, Llorca O, Ortín J. Structural and functional characterization of an influenza virus RNA polymerase–genomic RNA complex. J. Virol. 2010;84(20):10477–10487. doi: 10.1128/JVI.01115-10. [• Elegant study of the preparation of influenza A virus RNA-dependent RNA polymerase–RNA complexes for EM studies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turrell L, Lyall JW, Tiley LS, Fodor E, Vreede FT. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat. Commun. 2013;4:1591. doi: 10.1038/ncomms2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31(7):1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu C, Fan S, Li C, et al. Functional analysis of conserved motifs in influenza virus PB1 protein. PLoS ONE. 2012;7:e36113. doi: 10.1371/journal.pone.0036113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas SK, Nayak DP. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 1994;68(3):1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng T, Sharps J, Fodor E, Brownlee GG. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 2005;79(13):8669–8674. doi: 10.1128/JVI.79.13.8669-8674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 2004;78(17):9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obayashi E, Yoshida H, Kawai F, et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature. 2008;454(7208):1127–1131. doi: 10.1038/nature07225. [DOI] [PubMed] [Google Scholar]
- 30.Sugiyama K, Obayashi E, Kawaguchi A, et al. Structural insight into the essential PB1–PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 2009;28(12):1803–1811. doi: 10.1038/emboj.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mänz B, Götz V, Wunderlich K, et al. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza A virus. J. Biol. Chem. 2011;286(10):8414–8424. doi: 10.1074/jbc.M110.205534. [•• One of the first studies to show that the interaction between the influenza A virus RNA-dependent RNA polymerase subunits can be targeted for antiviral treatment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muratore G, Goracci L, Mercorelli B, et al. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc. Natl. Acad. Sci. USA. 2012;109(16):6247–6252. doi: 10.1073/pnas.1119817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarendeau F, Crepin T, Guilligay D, Ruigrok RW, Cusack S, Hart DJ. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 2008;4(8):e1000136. doi: 10.1371/journal.ppat.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boivin S, Hart DJ. Interaction of the influenza A virus polymerase PB2 C-terminal region with importin alpha isoforms provides insights into host adaptation and polymerase assembly. J. Biol. Chem. 2011;286(12):10439–10448. doi: 10.1074/jbc.M110.182964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilligay D, Tarendeau F, Resa-Infante P, et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008;15(5):500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 36.Pautus S, Sehr P, Lewis J, et al. New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J. Med. Chem. 2013;56(21):8915–8930. doi: 10.1021/jm401369y. [DOI] [PubMed] [Google Scholar]
- 37.Guu TS, Dong L, Wittung-Stafshede P, Tao YJ. Mapping the domain structure of the influenza A virus polymerase acidic protein (PA) and its interaction with the basic protein 1 (PB1) subunit. Virology. 2008;379(1):135–142. doi: 10.1016/j.virol.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 38.Fodor E, Crow M, Mingay LJ, et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 2002;76(18):8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [•• Stunning study reporting EM structures of the helical nature of the influenza A virus RNPs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dias A, Bouvier D, Crépin T, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458(7240):914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 41.Yuan P, Bartlam M, Lou Z, et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458(7240):909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 42.Datta K, Wolkerstorfer A, Szolar OH, Cusack S, Klumpp K. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res. 2013;41(17):8289–8299. doi: 10.1093/nar/gkt603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar OH, Ruigrok RW, Cusack S. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 2012;8(8):e1002831. doi: 10.1371/journal.ppat.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4(2):e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resa-Infante P, Jorba N, Zamarreño N, Fernández Y, Juárez S, Ortín J. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS ONE. 2008;3(12):e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huet S, Avilov SV, Ferbitz L, Daigle N, Cusack S, Ellenberg J. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 2010;84(3):1254–1264. doi: 10.1128/JVI.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hemerka JN, Wang D, Weng Y, et al. Detection and characterization of influenza A virus PA–PB2 interaction through a bimolecular fluorescence complementation assay. J. Virol. 2009;83(8):3944–3955. doi: 10.1128/JVI.02300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortín J, Martín-Benito J. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5(6):e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torreira E, Schoehn G, Fernández Y, et al. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res. 2007;35(11):3774–3783. doi: 10.1093/nar/gkm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Area E, Martín-Benito J, Gastaminza P, et al. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl Acad. Sci. USA. 2004;101(1):308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He X, Zhou J, Bartlam M, et al. Crystal structure of the polymerase PA(C)–PB1(N) complex from an avian influenza H5N1 virus. Nature. 2008;454(7208):1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 52.Brownlee GG, Sharps JL. The RNA polymerase of influenza a virus is stabilized by interaction with its viral RNA promoter. J. Virol. 2002;76(14):7103–7113. doi: 10.1128/JVI.76.14.7103-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung TE, Brownlee GG. A new promoter-binding site in the PB1 subunit of the influenza A virus polymerase. J. Gen. Virol. 2006;87(Pt3):679–688. doi: 10.1099/vir.0.81453-0. [DOI] [PubMed] [Google Scholar]
- 54.Maier HJ, Kashiwagi T, Hara K, Brownlee GG. Differential role of the influenza A virus polymerase PA subunit for vRNA and cRNA promoter binding. Virology. 2008;370(1):194–204. doi: 10.1016/j.virol.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 55.Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 2004;78(17):9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vreede FT, Gifford H, Brownlee GG. Role of initiating nucleoside triphosphate concentrations in the regulation of influenza virus replication and transcription. J. Virol. 2008;82(14):6902–6910. doi: 10.1128/JVI.00627-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez JT, Varble A, Sachidanandam R, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl Acad. Sci. USA. 2010;107(25):11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.te Velthuis AJ, Turrell L, Vreede FT, Fodor E. Uncoupling of influenza A virus transcription and replication through mutation of the unpaired adenosine in the viral RNA promoter. J. Virol. 2013;87(18):10381–10384. doi: 10.1128/JVI.00636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paterson D, te Velthuis AJ, Vreede FT, Fodor E. Host restriction of influenza virus polymerase activity by PB2 627E is diminished on short viral templates in a nucleoprotein-independent manner. J. Virol. 2014;88(1):339–344. doi: 10.1128/JVI.02022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ng AK, Wang JH, Shaw PC. Structure and sequence analysis of influenza A virus nucleoprotein. Sci. China C Life Sci. 2009;52(5):439–449. doi: 10.1007/s11427-009-0064-x. [DOI] [PubMed] [Google Scholar]
- 61.Marklund JK, Ye Q, Dong J, Tao YJ, Krug RM. Sequence in the influenza A virus nucleoprotein required for viral polymerase binding and RNA synthesis. J. Virol. 2012;86:7292–7297. doi: 10.1128/JVI.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moncorgé O, Mura M, Barclay WS. Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J. Virol. 2010;84(19):9978–9986. doi: 10.1128/JVI.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mehle A, Doudna JA. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe. 2008;4(2):111–122. doi: 10.1016/j.chom.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437(7060):889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 66.Gabriel G, Abram M, Keiner B, Wagner R, Klenk HD, Stech J. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J. Virol. 2007;81(17):9601–9604. doi: 10.1128/JVI.00666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993;67(4):1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [•• Classic paper that describes the importance of PB2 residue 627 in host restriction.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bogs J, Kalthoff D, Veits J, et al. Reversion of PB2-627E to -627K during replication of an H5N1 clade 2.2 virus in mammalian hosts depends on the origin of the nucleoprotein. J. Virol. 2011;85(20):10691–10698. doi: 10.1128/JVI.00786-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Long JS, Howard WA, Núñez A, et al. The effect of the PB2 mutation 627K on highly pathogenic H5N1 avian influenza virus is dependent on the virus lineage. J. Virol. 2013;87(18):9983–9996. doi: 10.1128/JVI.01399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mok CK, Lee HH, Lestra M, et al. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J. Virol. 2014;88(6):3568–3576. doi: 10.1128/JVI.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Foeglein Á , Loucaides EM, Mura M, Wise HM, Barclay WS, Digard P. Influence of PB2 host-range determinants on the intranuclear mobility of the influenza A virus polymerase. J. Gen. Virol. 2011;92(Pt 7):1650–1661. doi: 10.1099/vir.0.031492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scull MA, Gillim-Ross L, Santos C, et al. Avian influenza virus glycoproteins restrict virus replication and spread through human airway epithelium at temperatures of the proximal airways. PLoS Pathog. 2009;5(5):e1000424. doi: 10.1371/journal.ppat.1000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aggarwal S, Dewhurst S, Takimoto T, Kim B. Biochemical impact of the host adaptation-associated PB2 E627K mutation on the temperature-dependent RNA synthesis kinetics of influenza A virus polymerase complex. J. Biol. Chem. 2011;286(40):34504–34513. doi: 10.1074/jbc.M111.262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bortz E, Westera L, Maamary J, et al. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. MBio. 2011;2(4):e00151–11. doi: 10.1128/mBio.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cauldwell AV, Moncorgé O, Barclay WS. Unstable polymerase–nucleoprotein interaction is not responsible for avian influenza virus polymerase restriction in human cells. J. Virol. 2013;87(2):1278–1284. doi: 10.1128/JVI.02597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herfst S, Chutinimitkul S, Ye J, et al. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J. Virol. 2010;84(8):3752–3758. doi: 10.1128/JVI.02634-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5(1):e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou B, Pearce MB, Li Y, et al. Asparagine substitution at PB2 residue 701 enhances the replication, pathogenicity, and transmission of the 2009 pandemic H1N1 influenza A virus. PLoS ONE. 2013;8(6):e67616. doi: 10.1371/journal.pone.0067616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hudjetz B, Gabriel G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-α1 and -α7. PLoS Pathog. 2012;8(1):e1002488. doi: 10.1371/journal.ppat.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamada S, Hatta M, Staker BL, et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6(8):e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Q, Qiao C, Marjuki H, et al. Combination of PB2 271A and SR polymorphism at positions 590/591 is critical for viral replication and virulence of swine influenza virus in cultured cells and in vivo. J. Virol. 2012;86(2):1233–1237. doi: 10.1128/JVI.05699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bussey KA, Desmet EA, Mattiacio JL, et al. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J. Virol. 2011;85(14):7020–7028. doi: 10.1128/JVI.00522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song M-S, Pascua PNQ, Lee JH, et al. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J. Virol. 2009;83(23):12325–12335. doi: 10.1128/JVI.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mehle A, Dugan VG, Taubenberger JK, Doudna JA. Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J. Virol. 2012;86(3):1750–1757. doi: 10.1128/JVI.06203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu W, Zhu Y, Qin K, et al. Mutations in polymerase genes enhanced the virulence of 2009 pandemic H1N1 influenza virus in mice. PLoS ONE. 2012;7(3):e33383. doi: 10.1371/journal.pone.0033383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hulse-Post DJ, Franks J, Boyd K, et al. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 2007;81(16):8515–8524. doi: 10.1128/JVI.00435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wise HM, Foeglein A, Sun J, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 2009;83(16):8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 2013;87(5):2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jagger BW, Wise HM, Kash JC, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337(6091):199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J. Virol. 2012;86(22):12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tauber S, Ligertwood Y, Quigg-Nicol M, Dutia BM, Elliott RM. Behaviour of influenza A viruses differentially expressing segment 2 gene products in vitro and in vivo. J. Gen. Virol. 2012;93(Pt 4):840–849. doi: 10.1099/vir.0.039966-0. [DOI] [PubMed] [Google Scholar]
- 92.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao S, von der Malsburg A, Dick A, et al. Structure of myxovirus resistance protein a reveals intra-and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35(4):514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 94.Zimmermann P, Mänz B, Haller O, Schwemmle M, Kochs G. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J. Virol. 2011;85(16):8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mänz B, Dornfeld D, Götz V, et al. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 2013;9(3):e1003279. doi: 10.1371/journal.ppat.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chow NL, Simpson RW. RNA-dependent RNA polymerase activity associated with virions and subviral particles of myxoviruses. Proc. Natl Acad. Sci. USA. 1971;68(4):752–756. doi: 10.1073/pnas.68.4.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matrosovich MN, Gambaryan AS, Teneberg S, et al. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233(1):224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]