SUMMARY
Mammalian oocytes can reprogram somatic cells into a totipotent state enabling animal cloning through somatic cell nuclear transfer (SCNT). However, the majority of SCNT embryos fail to develop to term due to undefined reprogramming defects. Here we identify histone H3 lysine 9 trimethylation (H3K9me3) of donor cell genome as a major epigenetic barrier for efficient reprogramming by SCNT. Comparative transcriptome analysis identified reprogramming resistant regions (RRRs) that are expressed normally at 2-cell mouse embryos generated by IVF but not SCNT. RRRs are enriched for H3K9me3 in donor somatic cells, and its removal by ectopic expression of the H3K9me3 demethylase Kdm4d not only reactivates the majority of RRRs, but also greatly improves SCNT efficiency. Furthermore, use of donor somatic nuclei depleted of H3K9 methyltransferases markedly improves SCNT efficiency. Our study thus identifies H3K9me3 as a critical epigenetic barrier in SCNT-mediated reprogramming and provides a promising approach for improving mammalian cloning efficiency.
Keywords: somatic cell nuclear transfer, reprogramming barrier, heterochromatin, H3K9me3, Suv39h
INTRODUCTION
Terminally differentiated somatic cells can be reprogrammed to the totipotent state when transplanted into enucleated oocytes by means of somatic cell nuclear transfer (SCNT) (Gurdon, 1962). Because SCNT allows the generation of a whole organism from the nucleus of single differentiated somatic cell, this technique holds great potential for agriculture, biomedical industry, and endangered species conservation (Yang et al., 2007). Indeed, since the first successful mammalian cloning was performed in sheep (Wilmut et al., 1997), more than 20 mammalian species have been cloned through SCNT (Rodriguez-Osorio et al., 2012). Moreover, because pluripotent embryonic stem cells can be established from SCNT-generated blastocysts (Wakayama et al., 2001), SCNT is a promising technique for human therapeutics (Hochedlinger and Jaenisch, 2003). Following the recent successful derivation of the first human nuclear transfer embryonic stem cells (ntESCs) (Tachibana et al., 2013) and the generation of human ntESCs from aged adult or patient cells (Chung et al., 2014; Yamada et al., 2014), this promise is now closer to reality. ntESCs can serve as a valuable cell source for in vitro disease modeling and cell/tissue-replacement therapies.
Despite its tremendous potential, several technical limitations have prevented the practical use of SCNT. One such limitation is the extremely low efficiency in producing cloned animals. For example, approximately half of mouse SCNT embryos display developmental arrest prior to implantation, and only 1–2% of embryos transferred to surrogate mothers can develop to term (Ogura et al., 2013). With the exception of bovine species, which have relatively higher rates of reproductive cloning efficiency (5 to 20%), the overall reproductive cloning efficiency in all other species is relatively low (1 to 5%) (Rodriguez-Osorio et al., 2012). Similarly, the success rate for human ntESC establishment is also low owing to poor preimplantation development (10 to 25% to the blastocyst stage; Tachibana et al., 2013; Yamada et al., 2014).
Given that developmental defects of SCNT embryos first appear at the time of zygotic genome activation (ZGA), which occurs at the 2-cell stage in mouse and at the 4- to 8-cell stage in pig, bovine and human (Schultz, 2002), it has been postulated that SCNT embryos have difficulties in ZGA due to undefined epigenetic barriers pre-existing in the genome of donor cells. Although previous studies have identified a number of dysregulated genes in mouse 2-cell SCNT embryos (Inoue et al., 2006; Suzuki et al., 2006; Vassena et al., 2007), and in the late cleavage stage human SCNT embryos (Noggle et al., 2011), the nature of the presumed “pre-existing epigenetic barriers” and their relationship with impaired ZGA in SCNT embryos remains unknown. Through comparative analysis, here we report the identification of genomic domains resistant to ZGA in SCNT embryos. These reprogramming resistant regions (RRRs) are enriched for the repressive histone modification, H3K9me3, in somatic cells and removal of this epigenetic mark either through ectopic expression of an H3K9me3-specific demethylase in oocytes or through knocking-down the H3K9 methyltransferases, Suv39h1/2, in donor cells not only attenuated the ZGA defect, but also greatly improved the reprogramming efficiency of SCNT. Our study therefore identifies Suv39h1/2-mediated H3K9me3 as one of the long sought-after “epigenetic barriers” of SCNT and provides a promising approach for improving mammalian cloning efficiency.
RESULTS
Abnormal ZGA in 2-cell SCNT embryos
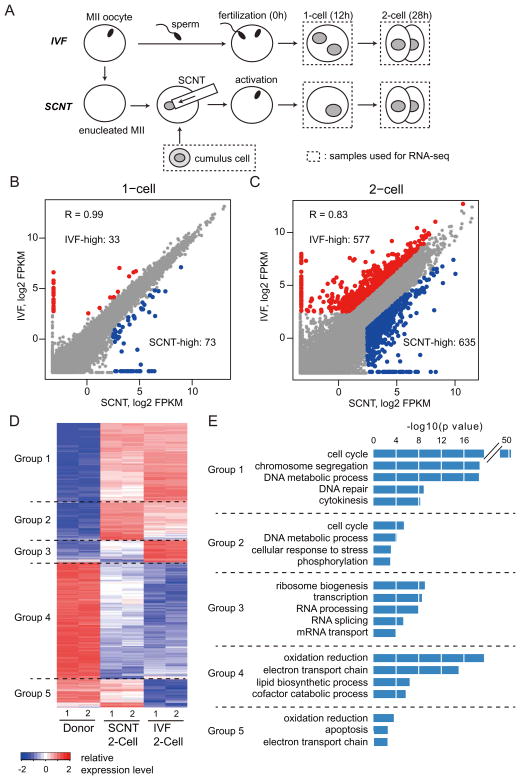
To identify the earliest transcriptional differences between mouse embryos derived through in vitro fertilization (IVF) and SCNT, we performed RNA-seq experiments using pooled embryos (25–40 embryos/sample) at 1-cell (12 hours post-activation: hpa), and late 2-cell (28 hpa) stages (Figure 1A). We obtained more than 30 million uniquely mapped reads for each sample, with the two biological replicates of each sample being highly reproducible (Figures S1A and S1B). Analysis of the 1-cell stage transcriptome revealed that SCNT and IVF embryos feature nearly identical transcriptomes (R =0.99; Figure 1B). Specifically, among the 5517 genes detected (FPKM > 5 in at least one sample), only 106 genes showed more than 3-fold difference between SCNT and IVF embryos (Figure 1B). This is consistent with the fact that ZGA largely begins after the first cleavage in mouse embryos (Schultz, 2002) and that the majority of transcripts present in 1-cell stage embryos, regardless of IVF or SCNT, are maternally stored transcripts. We therefore focused our analyses on the late 2-cell stage where the major ZGA becomes apparent in mouse embryos.
Figure 1. Abnormal gene expression of SCNT embryos at the 1- and 2-cell stage.
(A) Schematic illustration of the experimental approach. Samples used for RNA-seq are marked by dashed rectangles.
(B, C) Scatter plots comparing gene expression levels between IVF and SCNT embryos at the 1-cell stage (B) and the 2-cell stage (C). Genes expressed higher in IVF embryos (FC > 3.0, IVF-high) and higher in SCNT embryos (FC > 3.0, SCNT-high) are colored with red and blue, respectively.
(D) Heatmap illustration showing differentially expressed genes (DEGs) (FC > 5.0, FPKM > 5 in each replicates) obtained by a pairwise comparison between donor cumulus cells, IVF 2-cell and SCNT 2-cell embryos. A total of 3775 DEGs are classified into 5 groups by unsupervised hierarchical clustering.
(E) Gene ontology analysis of the 5 groups classified in (D).
See also Figure S1.
Transcriptome comparison between IVF and SCNT embryos at the 2-cell stage identified 1212 genes that showed more than 3-fold expression difference (Figure 1C, FPKM > 5 in at least one sample). Pairwise comparison of the transcriptome of donor cumulus cells, 2-cell IVF and 2-cell SCNT embryos identified 3775 differentially expressed genes [fold change (FC) > 5, FPKM > 5] that can be classified into 5 groups by unsupervised hierarchical cluster analysis (Figure 1D). Of these 3775 differentially expressed genes, 1549 were activated in both SCNT and IVF embryos (Groups 1 and 2). Gene ontology (GO) analysis revealed that these genes were significantly enriched in cell cycle related biological processes (Figure 1E), suggesting that SCNT embryos are as transcriptionally prepared for proper cell cycle progression as IVF embryos. Despite a portion of the highly expressed genes in donor cumulus cells still being expressed in 2-cell SCNT embryos (372 genes; Group 5), the majority of these genes were silenced following SCNT, similar to those in IVF embryos (1553 genes; Group 4). Genes in Group 4 were significantly enriched in cell metabolism related biological processes, such as oxidation/reduction and electron transport chain, suggesting that cumulus cell-specific metabolic processes are quickly terminated after SCNT. Interestingly, a group of 301 genes failed to be properly activated in SCNT embryos compared to IVF embryos (Group 3). GO analysis revealed that these genes were enriched in transcription or mRNA processing, suggesting a possible defect in activation of developmentally important regulators in SCNT embryos (Figure 1E). Given that proper activation of zygotic genes in 2-cell embryos is believed to be important for embryonic development, we focused our analysis on this group of genes and related genomic loci.
Identification of reprogramming resistant regions (RRRs) in 2-cell SCNT embryos
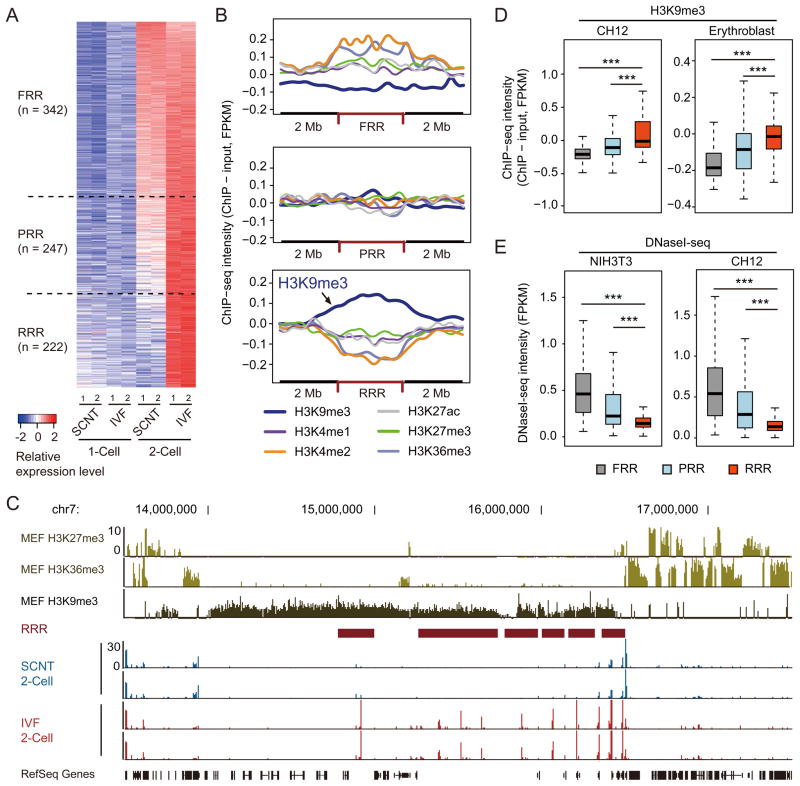
In addition to protein coding genes, previous studies have revealed that non-genic repetitive elements, such as LTR class III retrotransposons and major satellite repeats, are highly expressed in mouse preimplantation embryos, especially at the 2-cell stage (Evsikov et al., 2004; Peaston et al., 2004; Probst et al., 2010). To comprehensively characterize the transcriptome difference between IVF and SCNT 2-cell stage embryos, we applied a sliding window strategy to identify all genomic regions associated with detectable transcripts. First, we identified 811 genomic regions, ranging from 100 to 800 kb, that were significantly activated (Fisher’s exact test p value < 0.01) in 2-cell stage IVF embryos compared to 1-cell stage IVF embryos [Figure 2A, FC > 5, RPM (reads per millions of uniquely mapped reads) > 10 in IVF 2-cell embryos]. Among the 811 genomic regions, 342 regions were activated in SCNT embryos at a similar level as those in IVF embryos (FC <= 2 comparing IVF with SCNT 2-cell embryos), and these regions were termed as fully reprogrammed regions (FRRs). We also identified 247 regions, termed “partially reprogrammed regions” (PRRs), that were partially activated (FC > 2 and FC <= 5) in SCNT embryos compared to IVF embryos (Figure 2A). Interestingly, the remaining 222 regions, termed “reprogramming resistant regions” (RRRs), failed to be activated in SCNT embryos (FC > 5, Figure 2A). Notably, transcripts generated within RRRs are largely unannotated as exemplified in a representative region on chromosome 13 (Figure S2A). Indeed, RRRs are relatively gene-poor regions when compared to FRR and PRR (Figure S2B). However, RRRs are enriched for specific repeat sequences, such as LINE and LTR, but are depleted of SINE (Figure S2C). Thus, comparative transcriptome analysis allowed us to identify 222 RRRs that are refractory to transcriptional activation in 2-cell embryos generated by SCNT.
Figure 2. Reprogramming resistant regions (RRRs) are enriched for H3K9me3 in somatic cells.
(A) Heatmap illustration of the transcripts of IVF and SCNT embryos. Each tile represents an average of peaks within the region obtained by sliding-window analysis. Shown are the 811 regions that are activated from 1-cell (12 h) to 2-cell (28 h) stage IVF embryos compared to cumulus derived SCNT embryos. These regions were classified into three groups based on the fold-change (FC) in transcription levels between SCNT- and IVF 2-cell embryos. FRRs, PRRs, and RRRs indicate fully reprogrammed regions (FC <= 2), partially reprogrammed regions (2 < FC <= 5) and reprogramming resistant regions (FC > 5), respectively.
(B) The average ChIP-seq intensity of six histone modifications in MEF cells are shown within FRR, PRR and RRR compared with 2 MB flanking regions. Reads counts are normalized by input, total mapped reads and region length.
(C) Representative genome browser view of RRRs on chromosome 7.
(D, E) Box plots comparing the average intensity of H3K9me3-ChIP-seq (D) or DNaseI-seq (E) within FRR, PRR and RRR in different somatic cell types. ChIP-seq and DNaseI-seq datasets shown in (B–E) were obtained from ENCODE projects (Bernstein et al., 2012; The Encode Consortium Project, 2011).
See also Figure S2.
RRRs are enriched for H3K9me3 in somatic cells
The fact that RRRs are refractory to transcriptional activation in 2-cell SCNT embryos suggests that RRRs may possess certain epigenetic modifications that serve as a barrier for SCNT-mediated reprogramming. Given that developmental failure of SCNT embryos has been observed in different donor somatic cell types, including mouse embryonic fibroblast (MEF) cells (Ono et al., 2001), we hypothesized that such epigenetic barrier for SCNT-mediated reprogramming may be common to different somatic cell types. Because MEF cells are one of the few somatic cell types with comprehensive histone modification datasets (Bernstein et al., 2012; Chang et al., 2014; Pedersen et al., 2014), we asked whether any of the six major histone modifications are specifically enriched in the RRRs. We found that H3K9me3, but not any other modifications analyzed, was specifically enriched at RRRs, while no obvious enrichment of any histone modifications was observed in FRRs or PRRs (Figure 2B). Indeed, a careful examination of a representative region on chromosome 7 indicated that RRRs failed to activate in 2-cell SCNT embryos were clearly enriched for the H3K9me3 mark, and regions outside of H3K9me3-enriched regions were properly activated in 2-cell SCNT embryos (Figure 2C). This observation is not unique to MEF cells as similar enrichment of H3K9me3 in the RRRs was also observed in four other somatic cell- or tissue-types (CH12, Erythroblast, Megakaryocyte and whole brain) following analysis (Figures 2D and S2D) of the H3K9me3 ChIP-seq datasets from the ENCODE project (Bernstein et al., 2012). Therefore, we conclude that RRRs in somatic cells are enriched for H3K9me3.
Previous studies have also shown that H3K9me3 is generally enriched in tightly-packaged large domains of “heterochromatin regions” (Lachner et al., 2001). One possible explanation for the failure of RRRs to be activated in 2-cell SCNT embryos is its chromatin inaccessibility. To test this possibility, we analyzed the DNaseI hypersensitivity of six different somatic cell types using the data derived from the ENCODE project. Remarkably, we found that RRRs were significantly less sensitive to DNaseI compared to FRR and PRR in all somatic cell- or tissue-types analyzed (Figures 2E and S2E). Collectively, these results suggest that RRRs possess features of heterochromatin that generally exist in somatic cell types.
Removal of H3K9me3 by Kdm4d restores transcriptional reprogramming in SCNT embryos
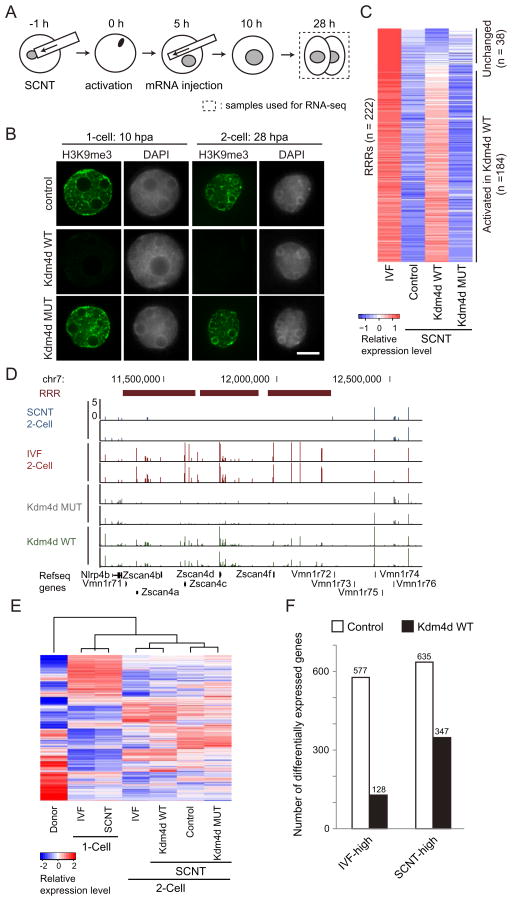
Having established a correlation between RRRs and H3K9me3 enrichment, we next attempted to address whether removal of H3K9me3 could facilitate transcriptional reprogramming of RRRs in SCNT embryos. To this end, we synthesized mRNAs encoding an H3K9me3-specific histone demethylase, Kdm4d (Krishnan and Trievel, 2013), and injected the mRNAs into SCNT embryos at 5 hpa (Figure 3A). Immunostaining revealed that injection of wild-type, but not a catalytic defective mutant, Kdm4d mRNAs greatly reduced H3K9me3 levels in SCNT embryos (Figure 3B).
Figure 3. Injection of Kdm4d mRNA removes H3K9me3 of transferred somatic cells and derepresses silenced genes in 2-cell SCNT embryos.
(A) Schematic illustration of the experimental procedure. SCNT embryos derived from cumulus cells were injected with wild-type or a catalytic defective Kdm4d mRNA at 5 hours post activation (hpa). Samples used for RNA-seq are marked by dashed rectangles.
(B) Representative nuclear images of 1-cell and 2-cell stage SCNT embryos stained with anti-H3K9me3 and DAPI. Shown in each panel is a nucleus of a single blastomere. Scale bar, 10 μm.
(C) Heatmap comparing transcription levels of the 222 RRRs at the 2-cell stage. The expression level of 184 out of the 222 RRRs are significantly (FC > 2) increased in response to wild-type, but not the catalytic mutant, Kdm4d injection.
(D) A genome browser view of an example of RRRs on chromosome 7.
(E) Hierarchical clustering of all samples used in this study. Note that 2-cell SCNT embryos injected with wild-type Kdm4d were clustered together with 2-cell IVF embryos based on their transcriptome analysis.
(F) Bar graph illustrates reduced number of differentially expressed genes (FC > 3) between IVF and SCNT 2-cell embryos after Kdm4d injection.
See also Figure S3.
To examine the effects of Kdm4d-mediated H3K9me3 removal on the transcriptional outcome of 2-cell SCNT embryos, we performed RNA-seq analysis focusing on the 222 RRRs that failed to be activated in SCNT. Compared to control SCNT embryos, 83% (184/222) of RRRs were activated by the injection of wild-type, but not a catalytic defective mutant, Kdm4d (Figure 3C, FC > 2). This result indicates that erasure of H3K9me3 facilitates transcriptional activation within RRRs. As exemplified in Figure 3D, an RRR on chromosome 7 containing the Zscan4 gene cluster was markedly activated by injection of wild-type Kdm4d, but not a catalytic defective mutant. Notably, not only protein coding genes, but also the majority of non-annotated transcripts from RRRs were also activated upon Kdm4d mRNA injection (Figures S3A and S3B). Interestingly, hierarchical clustering transcriptome analysis revealed that the transcriptome of SCNT embryos injected with wild-type Kdm4d was more similar to that of IVF embryos than that of control SCNT embryos, or of mutant Kdm4d injected embryos (Figure 3E). Indeed, the total number of differentially expressed genes (FC > 3) between SCNT and IVF 2-cell embryos decreased from 1212 to 475 by Kdm4d injection (Figures 3F and S3C), suggesting that removal of H3K9me3 from transferred somatic nuclei not only restores transcriptional activation of RRRs but also restores the global transcriptome of SCNT embryos.
Injection of Kdm4d mRNA greatly improves development of SCNT embryos
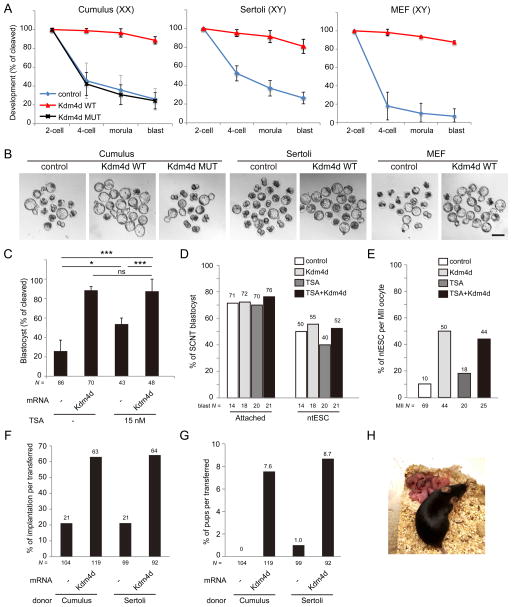
To examine the biological consequence of transcriptional restoration of SCNT embryos following Kdm4d injection, we first analyzed the developmental potential of SCNT embryos with Kdm4d mRNA injection using cumulus cells as donor cells. In control SCNT embryos, the developmental rate began to decline after the first cleavage with only 26.0 % of cleaved embryos successfully developing to the blastocyst stage after 96 h of culturing (Figures 4A and 4B, and Table S1), a finding consistent with previous studies (Kishigami et al., 2006). Strikingly, SCNT embryos injected with wild-type Kdm4d mRNA rarely arrested during 2- to 4-cell and 4-cell to morula stage transition, and developed to the blastocyst stage with high efficiency (88.6%; Figures 4A and 4B, and Table S1). In contrast, injection of a catalytic defective mutant Kdm4d mRNAs had no significant impact on the developmental rate of the SCNT embryos, indicating the improvement of Kdm4d injection on SCNT embryo development depends on its enzymatic activity. Given that H3K9me3 enrichment in RRRs appears to be a general phenomenon in different somatic cell types, we anticipated that the positive effect of Kdm4d on SCNT embryo development should be able to be extended to other somatic donor cell types. Indeed, Kdm4d mRNA injection also significantly improved the developmental efficiency of SCNT embryos when Sertoli cells or C57BL/6 background MEF cells were used as donor cells (Figures 4A and 4B, Table S1). Together, these results demonstrate that H3K9me3 removal by Kdm4d mRNA injection can significantly improve preimplantation development of SCNT embryos, regardless of the somatic donor cell type.
Figure 4. Injection of Kdm4d mRNA improves developmental potential of SCNT embryos.
(A) Kdm4d mRNA injection greatly improves preimplantation development of SCNT embryos derived from cumulus cells, Sertoli cells and MEF cells. Shown is the percentage of embryos that reaches the indicated stages. XX and XY indicate the sex of donor mice. Error bars indicate s.d.
(B) Representative images of SCNT embryos after 120 hours of culturing in vitro. Scale bar, 100 μm.
(C) Kdm4d mRNA injection has additional effect over the treatment with Trichostatin A (TSA; 15 nM). Shown is the percentage of embryos that reached the blastocyst stage at 96 hpa. * P < 0.05, ** P < 0.01, *** P < 0.001. ns, not significant.
(D) Bar graph showing the efficiency of attachment to the feeder cells and ntESC derivation of SCNT blastocysts.
(E) Bar graph showing the efficiency of ntESC derivation. The efficiency was calculated based on the total number of MII oocytes used for the generation of SCNT embryos.
(F, G) Implantation rate (F) and birth rate (G) of SCNT embryos examined by caesarian section on E19.5.
(H) An image of an adult female mouse derived by SCNT of a cumulus cell with Kdm4d mRNA injection and its pups generated through natural mating with a wild-type male.
See also Tables S1–3.
Previous studies have demonstrated that temporal treatment with histone deacetylase (HDAC) inhibitors can also significantly improve developmental efficiency of SCNT embryos (Kishigami et al., 2006; Van Thuan et al., 2009). To explore a possible relationship between HDAC inhibitors and Kdm4d, we performed a combinatorial treatment of SCNT embryos with the HDAC inhibitor, TSA, and Kdm4d. Treatment of TSA alone improved the blastocyst rate from 26.0% to 53.8% (Figure 4C and Table S1), similar to previous report (Kishigami et al., 2006). Kdm4d mRNA injection combined with TSA further increased the blastocyst rate to 87.5%, which is statistically similar to Kdm4d injection alone (88.6%; Figure 4C and Table S1). This result indicates that TSA treatment and Kdm4d mRNA injection does not have synergistic effect on SCNT reprogramming, at least in the preimplantation stage, and that TSA treatment may exert its effects through a similar pathway as that of Kdm4d.
We also examined the efficiency of ntESC derivation from the blastocysts. When control SCNT blastocysts were cultured on feeder MEF cells with ES cell derivation medium, 71% of the blastocysts attached to the feeder cells and 50% of the blastocysts eventually gave rise to the established ntESC lines (Figure 4D and Table S2). Blastocysts generated through Kdm4d mRNA injection, TSA treatment, or a Kdm4d/TSA combination did not show any significant difference in efficiency of attachment or ntESC derivation when compared to control (Figure 4D and Table S2). Importantly, efficiency was greatly improved by Kdm4d injection when calculations were based on the total number of MII oocytes used for SCNT (Figure 4E and Table S2).
To examine whether the positive effect of Kdm4d on preimplantation development could be maintained through postimplantation development, we transferred 2-cell stage SCNT embryos generated from cumulus cells into the oviducts of pseudopregnant female mice. Caesarian section at E19.5 (the day of term) revealed that the rate of implantation, evidenced by implantation sites, was 3-fold higher in Kdm4d-injected SCNT embryos (63.0%) than in control SCNT embryos (21.2%, Figure 4F). Importantly, 7.6% (9/119) of transferred Kdm4d-injected 2-cell SCNT embryos developed to term, while none of the 104 transferred control embryos developed to term under the same conditions (Figure 4G and Table S3). Similar experiments using Sertoli cell-derived SCNT embryos also demonstrated the positive effect of Kdm4d on the implantation rate (21% vs 64%) and the development to term rate (1% vs 8.7%) (Figures 4F, G and Table S3). Furthermore, SCNT pups generated through Kdm4d-injection grew normally to adulthood and generated offspring by natural mating (Figure 4H). These results demonstrate that H3K9me3 in somatic cells is a barrier for oocyte-mediated genomic reprogramming and removal of H3K9me3 by Kdm4d injection at very early stages of SCNT embryo development can significantly improve the overall efficiency of mouse reproductive cloning.
Candidate genes responsible for the poor developmental phenotype of SCNT embryos
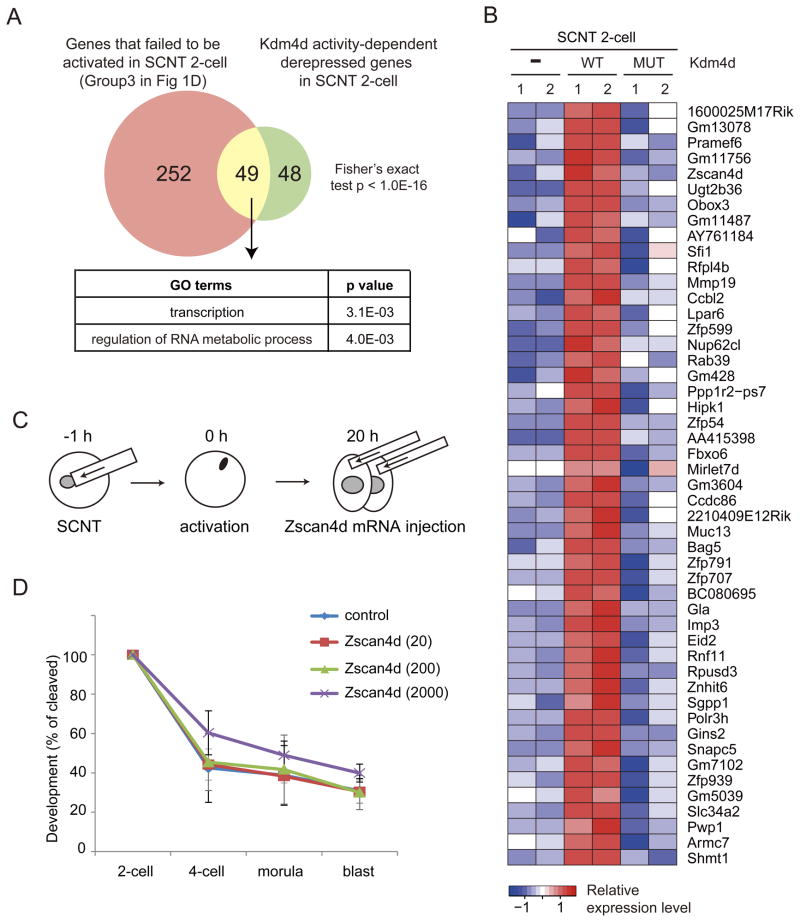
We next asked which of the genes repressed by H3K9me3 could be responsible for the poor developmental phenotype of SCNT embryos. Given that Kdm4d overexpression greatly increases the rate of SCNT embryos reaching the blastocyst stage, the responsible genes must be derepressed in the wild-type Kdm4d-injected SCNT embryos. Analysis of the genes that failed to be activated in the 2-cell SCNT embryos (Group 3 genes in Figure 1D), and the genes derepressed in wild-type Kdm4d-injected, but not the mutant Kdm4d-injected 2-cell SCNT embryos, allowed us to identify 49 common genes (Figures 5A and 5B, FC > 5). GO analysis indicated that this group of genes are enriched for genes involved in transcription and RNA metabolic processes (Figure 5A). While the function of most of the 49 genes in preimplantation development is unknown, the 2-cell specific Zscan4 family member, Zscan4d, has been shown to be important for preimplantation development (Falco et al., 2007). Therefore, we examined whether supplement of exogenous Zscan4d mRNA could enhance the developmental efficiency of SCNT embryos.
Figure 5. Candidate genes responsible for the poor developmental phenotype of SCNT embryos.
(A) Venn diagram showing the overlap between the genes that failed to be activated in SCNT 2-cell embryos (Group3 in Figure 1D) and Kdm4d enzyme activity-dependently derepressed genes in SCNT 2-cell embryos. GO enrichment analysis was performed on the 49 overlap genes.
(B) Heatmap showing the expression pattern of 49 overlap genes in (A).
(C) Schematic illustration of the experimental procedure. Zscan4d mRNA was injected into both of 2-cell blastomeres of SCNT embryos at 20 hpa (early 2-cell stage).
(D) Preimplantation development rate of SCNT embryos injected with Zscan4d mRNA at 0, 20, 200 or 2000 ng/μl. Error bars indicate s.d. of three biological replicates.
We microinjected mRNA encoding full-length Zscan4d into SCNT embryos at the early 2-cell stage (20 hpa: Figure 5C) following the expression pattern of endogenous Zscan4d. However, injection of Zscan4d mRNA failed to rescue the poor developmental phenotype of SCNT embryos, regardless of concentration (Figure 5D and Table S4). Therefore, a defect in Zscan4d activation in 2-cell SCNT embryos is unlikely to be solely responsible for the poor preimplantation development of SCNT embryos. Rather, complicated gene networks including transcripts derived from non-genic repetitive elements (Figure S4) may underlie the defects of SCNT embryonic development.
Suv39h1 and Suv39h2 establish the H3K9me3 barrier in somatic cells
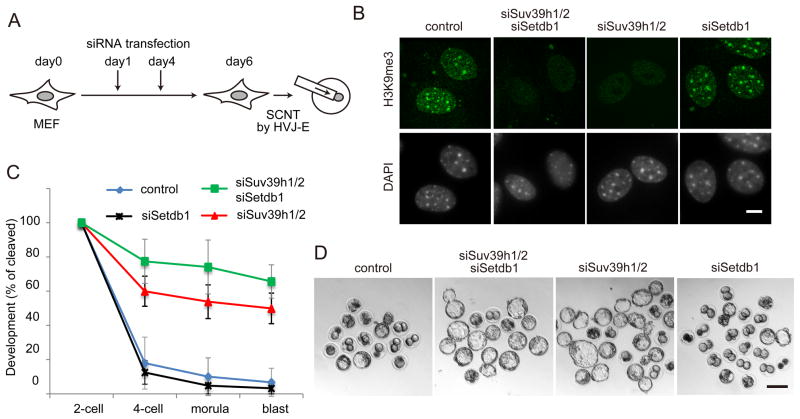
Having demonstrated that H3K9me3 is an epigenetic barrier of SCNT-mediated reprogramming, we next attempted to identify the histone methyltransferase(s) responsible for the deposition of H3K9me3 within RRRs in somatic genomes. Previous studies have revealed that at least three histone lysine methyltransferases (KMTs), Suv39h1, Suv39h2 and Setdb1, can catalyze the generation of H3K9me3 in mammalian cells (Matsui et al., 2010; Peters et al., 2001). First, we depleted the three H3K9me3 methyltransferases in MEF cells by transfecting a mixture of short interfering RNAs (siRNAs) targeting Suv39h1, Suv39h2 and Setdb1 (Figure 6A). RT-qPCR analysis confirmed a knockdown efficiency of 80–60% was achieved 48 hours after transfection (Figure S5). Immunostaining showed that transfection of these siRNAs (twice within 6-days of cell culture) could greatly reduce H3K9me3 levels in MEF cells (Figures 6A and 6B), demonstrating the three H3K9me3 methyltransferases, and not other unidentified enzymes, as the responsible factors for the H3K9me3 deposition in the somatic cells. Using triple KD MEF cells as donors, we generated SCNT embryos and examined their preimplantation development. We found that while only 6.7% of control SCNT embryos developed to the blastocyst stage after 96 h culture (Figures 6C, 6D, and Table S1), 65.6% of the triple knockdown MEF-derived embryos developed to blastocyst stage (Figures 6C, 6D and Table S1). This result not only confirms that somatic H3K9me3 is an epigenetic barrier of SCNT-mediated reprogramming, but also demonstrates that these three enzymes are responsible for generating this epigenetic barrier.
Figure 6. Suv39h1/2 is responsible for the establishment of the H3K9me3 barrier.
(A) Schematic illustration of SCNT using siRNA transfected MEF cells (see Extended Experimental Procedures for details).
(B) Representative images of MEF cells stained with anti-H3K9me3 antibody and DAPI at day 6 of transfection. Scale bar, 10 μm.
(C) Preimplantation development rate of SCNT embryos derived from different knockdown MEF cells. Error bars indicate s.d. of three biological replicates.
(D) Representative images of SCNT embryos after 120 hours of culturing in vitro. Scale bar, 100 μm.
We next examined which of the three histone methyltransferases could be responsible for establishing the H3K9me3 reprogramming barrier by individually depleting them in MEF cells. Since Suv39h1 and Suv39h2 have redundant functions in H3K9 trimethylation (Peters et al., 2001), we knocked down both genes at the same time. Immunostaining at day 6 of knockdown demonstrated that Suv39 KD reduced global H3K9me3 levels, especially at pericentric regions, while Setdb1 knockdown did not cause a global change in H3K9me3 levels (Figure 6B), a finding consistent with a previous report (Matsui et al., 2010). When these knockdown MEF cells were used as donors for SCNT analysis, the developmental rate to the blastocyst stage was greatly improved from 6.7 % in control to 49.9% in the Suv39h KD MEF group, very close to that of the triple knockdown MEF group (Figures 6C, 6D, and Table S1). In contrast, knockdown of Setdb1 did not significantly alter the developmental rate (Figures 6C, 6D, and Table S1). Collectively, these results suggest that Suv39h1/2 are primarily responsible for establishing H3K9me3 in somatic cells, which functions as a barrier in genomic reprogramming of SCNT embryos.
DISCUSSION
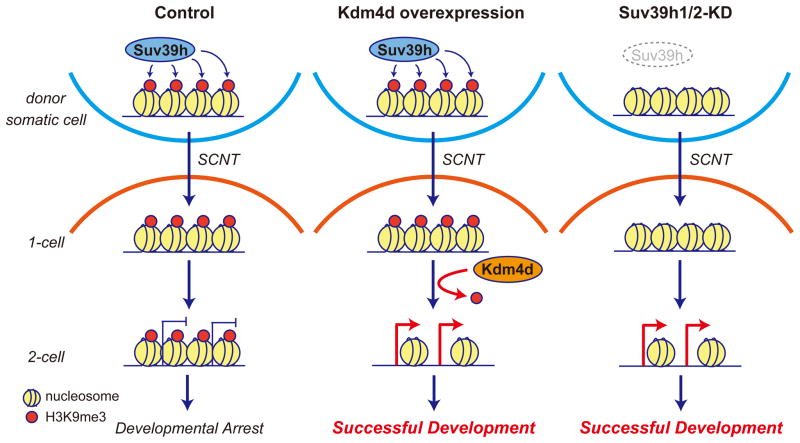
More than 50 years have passed since the first demonstration of animal cloning through somatic cell nuclear transfer in Xenopus eggs (Gurdon, 1962). Despite tremendous efforts, cloning efficiency has remained relatively low in most of the species, and the mechanism underlying epigenetic reprogramming following SCNT has remained poorly understood. In this study, through comparative transcriptome and integrated epigenomic analysis, we have identified that Suv39h1/2-deposited H3K9me3 in donor somatic cells functions as an epigenetic barrier for somatic cell nuclear reprogramming in mouse oocytes. By comparing to the transcriptome of IVF embryos, we identified 222 genomic regions, termed as RRRs (reprogramming resistant regions), resistant to transcriptional reprogramming in SCNT embryos. RRRs are characterized by significant enrichment of Suv39h1/2-deposited H3K9me3 and low DNase I accessibility, both of which are general features of heterochromatin, in several somatic cell types analyzed. Efficient activation of transcripts within RRRs appears to be instrumental for the development of SCNT embryos, as removal of H3K9me3 either by Suv39h1/2 knockdown or by expression of exogenous Kdm4d results in activation of RRRs and significant improvement in the development of SCNT embryos. Thus, our data support a model where Suv39h1/2-deposited H3K9me3 in somatic cells serves as a barrier for the activation of developmentally important genes in oocytes, leading to developmental arrest of SCNT embryos (Figure 7). Removal of this epigenetic barrier either by exogenous Kdm4d following SCNT, or by depletion of Suv39h1/2 in donor cells allows the expression of developmental genes and thus improves the development of SCNT embryos (Figure 7).
Figure 7. A model illustrating how the H3K9me3 reprogramming barrier can be overcome.
Suv39h-deposited H3K9me3 in somatic cells serves as a transcriptional barrier for SCNT-mediated reprogramming which affects normal embryonic development (Left). Removal of this barrier either by the expression of exogenous Kdm4d (Middle) or by prevention of H3K9me3 establishment by Suv39h knockdown (Right) can lead to activation of developmental regulators in SCNT embryos, resulting in successful embryonic development.
How does H3K9me3 impede reprogramming? Previous studies have demonstrated that H3K9me3 can be recognized and bound by the heterochromatin protein, HP1 (Bannister et al., 2001; Lachner et al., 2001), which can nucleate the formation of heterochromatin (Canzio et al., 2013). Interaction between heterochromatin and nuclear lamina can tether heterochromatin to the nuclear periphery leading to epigenetic silencing (Poleshko and Katz, 2014). Therefore, H3K9me3-initiated heterochromatin assembly can prevent access to reprogramming and transcriptional factors, and thereby prevent the activation of developmentally important genes in RRRs. In addition to preventing access to reprogramming and transcription factors, H3K9me3 may inhibit subsequent deposition of activation marks, such as H3K9 acetylation and H3K4 methylation, as we have demonstrated previously (Wang et al., 2001).
It is likely that H3K9me3-mediated heterochromatin formation may function as a general reprogramming barrier, which is supported by the recent demonstration that both H3K9me3 (Chen et al., 2013; Soufi et al., 2012) and HP1 (Sridharan et al., 2013) in MEF cells inhibit iPS cell generation. Nonetheless, several differences exist regarding the barrier between SCNT and iPS reprogramming. First, the H3K9me3-barrier in mouse iPS reprogramming is established primarily by Setdb1(Chen et al., 2013; Sridharan et al., 2013). In contrast, our results clearly point to Suv39h1/2, but not Setdb1, as critical enzymes that establish the H3K9me3 barrier for SCNT reprogramming. Second, the downstream gene networks necessary for successful reprogramming, repressed by the H3K9me3 barrier, are likely different. In iPS reprogramming, the key downstream factors within the H3K9me3 barrier are the core pluripotency network genes, such as Nanog and Sox2, which operate during relatively late stages of reprogramming (Chen et al., 2013; Sridharan et al., 2013). In contrast, in SCNT reprogramming, transcripts that play a critical function at the 2-cell stage are the key factors repressed by H3K9me3 (discussed below). This distinction most likely stems from the differences in the set of transcription factors required for successful reprogramming in each context. Indeed, core transcription factors of iPS reprogramming, Oct4/Pou5f1, have recently been demonstrated to be dispensable in SCNT reprogramming (Wu et al., 2013). Therefore, although H3K9m3 is a common reprogramming barrier, its deposition, and how it affects the reprogramming process are likely different between iPS and SCNT.
We have listed 49 candidate genes that are potentially responsible for the poor developmental phenotype of SCNT embryos. Overexpression of one of the candidate genes, Zscan4d, did not improve the blastocyst rate. It is thus likely that other factors activated at 2-cell stage IVF embryos are necessary for SCNT embryos to successfully develop to the blastocyst stage. In addition to the protein coding genes we listed, the deregulated RRRs also harbor many un-annotated transcripts and repeat sequences whose repression in 2-cell SCNT embryos exhibits H3K9me3 dependency. For example, expression of the major satellite repeats were greatly suppressed in 2-cell SCNT embryos, and this repression was completely restored by the wild-type, but not the catalytic mutant, Kdm4d injection (Figure S4). Similarly, Kdm4d injection also partially relieved SCNT-induced repression of class III retrotransposon elements, such as MERVL, in 2-cell SCNT embryos (Figure S4). Given that activation of both repeat sequences is important for preimplantation development (Kigami, 2003; Probst et al., 2010), transcriptional deregulation of these repeat sequences in 2-cell SCNT embryos may also contribute to the developmental defects observed in these embryos. Therefore, it is likely that defective activation of protein coding genes and repeat sequences that harbor the H3K9me3 mark inherited from somatic cells is collectively responsible for the developmental failure of SCNT embryos.
In addition to better understanding the detailed mechanism of how H3K9me3 removal contributes to increased SCNT efficiency, whether or not our observation can be generally applied to other animal species warrants future investigation. Similar to mice, developmental defects of SCNT embryos appear concurrently with ZGA in other mammalian species such as rabbit (Li et al., 2006), pig (Zhao et al., 2009), bovine (Akagi et al., 2011) and human (Noggle et al., 2011), with abnormal heterochromatin or H3K9me3 status observed in SCNT embryos of these species (Pichugin et al., 2010; Santos et al., 2003; Yang et al., 2009). Therefore, the H3K9me3 reprogramming barrier might be conserved among different species. If so, Kdm4d mRNA injection has the potential to enhance cloning efficiency in a broad range of mammalian species, including humans. Importantly, injection of Kdm4d mRNA greatly enhanced ntESC derivation efficiency in mice. If this intervention can be successfully translated to human ntESC derivation, our method could hold great promise for human therapeutic cloning (Hochedlinger and Jaenisch, 2003; Yang et al., 2007). The simplicity of Kdm4d mRNA injection during SCNT makes the testing of our approach worthwhile. Furthermore, when a Suv39h-specific inhibitor becomes available, a simple incubation of the donor cells with such inhibitor prior to transferring to enucleated oocytes could drastically improve cloning efficiency.
EXPERIMENTAL PROCEDURES
SCNT and mRNA injection
Somatic cell nuclear transfer was carried out as described previously (Matoba et al., 2011). Briefly, recipient MII oocytes were collected from superovulated adult BDF1 females by a brief treatment with 300 U/ml bovine testicular hyaluronidase (Calbiochem). Isolated MII oocytes were enucleated in Hepes-buffered KSOM medium containing 7.5 μg/ml of cytochalasin B (Calbiochem # 250233). The nuclei of donor cumulus cells or Sertoli cells were injected into the enucleated oocytes using a Piezo-driven micromanipulator (Primetech # PMM-150FU). MEF cells were fused with enucleated oocytes by inactivated Sendai virus envelope (HVJ-E; Ishihara Sangyo, Japan). After 1h incubation in KSOM, reconstructed SCNT oocytes were activated by incubation in Ca-free KSOM containing 5 μg/ml cytochalasin B for 1 h and further cultured in KSOM with cytochalasin B for 4 h. Activated SCNT embryos were washed 5 h after the onset of SrCl2 treatment (hours post activation, hpa) and cultured in KSOM in a humidified atmosphere of 5% CO2 at 37.8°C. In some experiments, SCNT embryos were injected with ~10 pl of water (control), 1800 ng/μl wild-type or mutant (H189A) Kdm4d mRNA at 5–6 hpa by using a Piezo-driven micromanipulator (Primetech). Microinjection of Zscan4d mRNA was performed at 20 hpa to correspond to the early 2-cell stage. In some experiments, trichostatin A (TSA) was added to the culture medium at 15 nM from the beginning of the activation for a total of 8 hours. Preimplantation developmental rates were statistically analyzed by Student’s T-test. Further details on donor cell preparation, embryo transfer, mRNA preparation and other procedures are included in the Extended Experimental Procedures.
RNA-sequencing analysis
The embryos were directly lysed and used for cDNA synthesis using the SMARTer Ultra Low Input RNA cDNA preparation kit (Clontech # 634936). After amplification, the cDNA samples were fragmented using Covaris sonicator (Covaris # M220). Sequencing libraries were made with the fragmented DNA using NEBNext Ultra DNA Library Prep Kit for Illumina according to manufacturer’s instruction (New England Biolabs # E7370). Single end 50 bp sequencing was performed on a HiSeq 2500 sequencer (Illumina). Sequencing reads were mapped to the mouse genome (mm9) with NovoalignV3.02.00. All programs were performed with default settings (unless otherwise specified). Uniquely mapped reads (about 70% of total reads) were subsequently assembled into transcripts guided by the reference annotation (UCSC gene models) with Cufflinks v2.0.2. Expression level of each gene was quantified with normalized FPKM (fragments per kilobase of exon per million mapped fragments). Functional annotation of significantly different transcripts and enrichment analysis was performed with DAVID. Statistical analyses were implemented with R (http://www.r-project.org/). Independent 2-group Wilcoxon rank sum test were used to compare distributions using the wilcox.test function in R. Pearson’s r coefficient was calculated using the cor function with default parameters. The hierarchical clustering analysis of the global gene expression pattern in different samples was carried out using heatmap.2 function (gplots package) in R.
Identification of reprogramming resistant regions
A sliding window (size 100kb, step size 20kb) was used to assess the genome-wide expression level of 1-cell and 2-cell embryos. For each window, the expression level was quantified with normalized RPM (reads per millions of uniquely mapped reads). The significantly activated regions in 2-cell relative to 1-cell IVF embryos were identified with stringent criteria (FC > 5, Fisher’s exact test p value < 0.01, RPM > 10 in 2-cell IVF embryos), and the overlapping regions were merged. These activated regions were classed into three groups based on their expression differences in SCNT and IVF 2-cell embryos.
Supplementary Material
(A) Summary of total and uniquely mapped reads for each sample of the seven types of samples with two biological replicates used in this study.
(B) Scatter plot evaluation of the reproducibility of different biological replicates.
(A) Genome browser view of an example of RRRs on chromosome 13 showing ChIP-seq data of histone modifications in MEF cells and RNA-seq data of 2-cell embryos. Note that this RRR overlaps with a large block of H3K9me3 peaks and is located within a gene-poor region.
(B) Box plot comparing the average percentage of exonic sequences, which represents the density of protein coding genes, in FRR, PRR and RRR. *** P < 0.001.
(C) Box plot comparing the average percentage of repetitive sequence within FRR, PRR and RRR. *** P < 0.001.
(D) Box plot comparing the average values of ChIP-seq for H3K9me3 in megakaryocyte and whole brain. ChIP-seq data were obtained from ENCODE projects (Bernstein et al., 2012). H3K9me3 is significantly enriched in RRRs compared to FRRs and PRRs.
(E) Box plot comparing the average values of sequence intensity after DNaseI treatment in whole brain, T-regulatory cells, Cell_416b and Mel cells. DNaseI-seq data were obtained from ENCODE projects (The Encode Consortium Project, 2011). RRR is significantly less sensitive to DNaseI than FRR or PRR in all four types of cells/tissues. ** P < 0.01, *** P < 0.001.
(A) Genome browser view of representative RRRs on chromosome 7.
(B) Genome browser view of an example of RRRs on chromosome 13.
(C) Scatter plot comparing gene expression of Kdm4d WT injected SCNT 2-cell embryos with that of IVF 2-cell embryos. Genes express higher (FC > 3) in IVF (IVF-high) or SCNT (SCNT-high) embryos were colored as red and blue, respectively.
Bar graphs indicate the expression level (uniquely mapped read numbers) of the major satellite DNA and the mouse endogenous retrotransposon MERVL in IVF and SCNT embryos.
(A–C) RT-qPCR analysis of Suv39h1 (A), Suv39h2 (B) and Setdb1 (C) mRNA levels in MEF cells at 48 hours after transfection of each siRNA. Data shown are mean expression values relative to Gapdh. The value in control was set as 1.0. Error bars represents s.d. with three biological replicates. *** P<0.001 by Student’s T-test.
Table S1. Preimplantation development of SCNT embryos injected with Kdm4d mRNA, Related to Figures 4 and 6
Table S2.Establishment of ntESCs from SCNT embryos, Related to Figure 4
Table S3. In vivo development of SCNT embryos injected with Kdm4d mRNA, Related to Figure 4
Table S4. Preimplantation development of SCNT embryos injected with Zscan4d mRNA, Related to Figure 5
Acknowledgments
We thank Drs. Shinpei Yamaguchi, Luis M. Tuesta, and Hao Wu for critical reading of the manuscript. This project is supported by NIH U01DK089565 (to Y.Z.). S.M. and K.A.I. are supported by postdoctoral fellowship from the Japan Society for the Promotion of Science (JSPS). Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The RNA-seq datasets have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE59073.
Supplemental Information including five figures, four tables and Extended Experimental Procedures can be found online at xxxx
AUTHOR CONTRIBUTIONS
SM and YZ conceived the project and designed the experiments. SM performed most of the experiments. YL analyzed RNA-seq and ChIP-seq datasets. FL prepared RNA-seq libraries. LS performed RNA-seq. KAI prepared donor MEF cells. AI assisted in some of the experiments. SM and YZ wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akagi S, Matsukawa K, Mizutani E, Fukunari K, Kaneda M, Watanabe S, Takahashi S. Treatment with a histone deacetylase inhibitor after nuclear transfer improves the preimplantation development of cloned bovine embryos. J Reprod Dev. 2011;57:120–126. doi: 10.1262/jrd.10-058a. [DOI] [PubMed] [Google Scholar]
- Bannister aJ, Zegerman P, Partridge JF, Miska Ea, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Liao M, Naber N, Pate E, Larson A, Wu S, Marina DB, Garcia JF, Madhani HD, Cooke R, et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496:377–381. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G, Gao S, Hou X, Xu Z, Liu Y, Kang L, Tao Y, Liu W, Huang B, Kou X, et al. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014;24:293–306. doi: 10.1038/cr.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Chung YG, Eum JH, Lee JE, Shim SH, Sepilian V, Hong SW, Lee Y, Treff NR, Choi YH, Kimbrel Ea, et al. Human Somatic Cell Nuclear Transfer Using Adult Cells. Cell Stem Cell. 2014;14:777–780. doi: 10.1016/j.stem.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Evsikov aV, de Vries WN, Peaston aE, Radford EE, Fancher KS, Chen FH, Blake Ja, Bult CJ, Latham KE, Solter D, et al. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res. 2004;105:240–250. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MSH. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N Engl J Med. 2003;349:275–286. doi: 10.1056/NEJMra035397. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ogonuki N, Miki H, Hirose M, Noda S, Kim JM, Aoki F, Miyoshi H, Ogura A. Inefficient reprogramming of the hematopoietic stem cell genome following nuclear transfer. J Cell Sci. 2006;119:1985–1991. doi: 10.1242/jcs.02913. [DOI] [PubMed] [Google Scholar]
- Kigami D. MuERV-L Is One of the Earliest Transcribed Genes in Mouse One-Cell Embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Mizutani E, Ohta H, Hikichi T, VanThuan N, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Trievel RC. Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure. 2013;21:98–108. doi: 10.1016/j.str.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li S, Chen X, Fang Z, Shi J, Sheng HZ. Rabbits generated from fibroblasts through nuclear transfer. Reproduction. 2006;131:1085–1090. doi: 10.1530/rep.1.01065. [DOI] [PubMed] [Google Scholar]
- Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci U S A. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova Ia, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Noggle S, Fung HL, Gore A, Martinez H, Satriani KC, Prosser R, Oum K, Paull D, Druckenmiller S, Freeby M, et al. Human oocytes reprogram somatic cells to a pluripotent state. Nature. 2011;478:70–75. doi: 10.1038/nature10397. [DOI] [PubMed] [Google Scholar]
- Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear Recent advancements in cloning by somatic cell nuclear transfer. Phil Trans R Soc B. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Shimozawa N, Ito M, Kono T. Cloned Mice from Fetal Fibroblast Cells Arrested at Metaphase by a Serial Nuclear Transfer. Biol Reprod. 2001;64:44–50. doi: 10.1095/biolreprod64.1.44. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Pedersen MT, Agger K, Laugesen A, Johansen JV, Cloos PaC, Christensen J, Helin K. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol Cell Biol. 2014;34:1031–1045. doi: 10.1128/MCB.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pichugin A, Le Bourhis D, Adenot P, Lehmann G, Audouard C, Renard JP, Vignon X, Beaujean N. Dynamics of constitutive heterochromatin: two contrasted kinetics of genome restructuring in early cloned bovine embryos. Reproduction. 2010;139:129–137. doi: 10.1530/REP-08-0435. [DOI] [PubMed] [Google Scholar]
- Poleshko A, Katz RA. Specifying peripheral heterochromatin during nuclear lamina reassembly. Nucleus. 2014;5:32–39. doi: 10.4161/nucl.28167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Osorio N, Urrego R, Cibelli JB, Eilertsen K, Memili E. Reprogramming mammalian somatic cells. Theriogenology. 2012;78:1869–1886. doi: 10.1016/j.theriogenology.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic Marking Correlates with Developmental Potential in Cloned Bovine Preimplantation Embryos. Curr Biol. 2003;13:1116–1121. doi: 10.1016/s0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Minami N, Kono T, Imai H. Zygotically activated genes are suppressed in mouse nuclear transferred embryos. Cloning Stem Cells. 2006;8:295–304. doi: 10.1089/clo.2006.8.295. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Encode Consortium Project. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Thuan N, Bui HT, Kim JH, Hikichi T, Wakayama S, Kishigami S, Mizutani E, Wakayama T. The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction. 2009;138:309–317. doi: 10.1530/REP-08-0299. [DOI] [PubMed] [Google Scholar]
- Vassena R, Han Z, Gao S, Baldwin Da, Schultz RM, Latham KE. Tough beginnings: alterations in the transcriptome of cloned embryos during the first two cell cycles. Dev Biol. 2007;304:75–89. doi: 10.1016/j.ydbio.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Tabar V, Rodriguez I, Perry aC, Studer L, Mombaerts P. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science. 2001;292:740–743. doi: 10.1126/science.1059399. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-bromage H, Borchers C, Tempst P, Zhang Y. Purification and Functional Characterization of a Histone H3-Lysine 4-Specific Methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke A, McWhir J, Kind A, Campbell K. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wu G, Han D, Gong Y, Sebastiano V, Gentile L, Singhal N, Adachi K, Fischedick G, Ortmeier C, Sinn M, et al. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, et al. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature. 2014;510:533–536. doi: 10.1038/nature13287. [DOI] [PubMed] [Google Scholar]
- Yang C, Liu Z, Fleurot R, Adenot P, Vignon X, Zhou Q, Renard J, Beaujean N. Heterochromatin reprogramming in rabbit embryos after fertilization, intra-, and inter-species SCNT correlates with preimplantation development. Reproduction. 2009;145:149–159. doi: 10.1530/REP-11-0421. [DOI] [PubMed] [Google Scholar]
- Yang X, Smith SL, Tian XC, Lewin HA, Renard J, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ross JW, Hao Y, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS. Significant Improvement in Cloning Efficiency of an Inbred Miniature Pig by Histone Deacetylase Inhibitor Treatment after Somatic Cell Nuclear Transfer. Biol Reprod. 2009;81:525–530. doi: 10.1095/biolreprod.109.077016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Summary of total and uniquely mapped reads for each sample of the seven types of samples with two biological replicates used in this study.
(B) Scatter plot evaluation of the reproducibility of different biological replicates.
(A) Genome browser view of an example of RRRs on chromosome 13 showing ChIP-seq data of histone modifications in MEF cells and RNA-seq data of 2-cell embryos. Note that this RRR overlaps with a large block of H3K9me3 peaks and is located within a gene-poor region.
(B) Box plot comparing the average percentage of exonic sequences, which represents the density of protein coding genes, in FRR, PRR and RRR. *** P < 0.001.
(C) Box plot comparing the average percentage of repetitive sequence within FRR, PRR and RRR. *** P < 0.001.
(D) Box plot comparing the average values of ChIP-seq for H3K9me3 in megakaryocyte and whole brain. ChIP-seq data were obtained from ENCODE projects (Bernstein et al., 2012). H3K9me3 is significantly enriched in RRRs compared to FRRs and PRRs.
(E) Box plot comparing the average values of sequence intensity after DNaseI treatment in whole brain, T-regulatory cells, Cell_416b and Mel cells. DNaseI-seq data were obtained from ENCODE projects (The Encode Consortium Project, 2011). RRR is significantly less sensitive to DNaseI than FRR or PRR in all four types of cells/tissues. ** P < 0.01, *** P < 0.001.
(A) Genome browser view of representative RRRs on chromosome 7.
(B) Genome browser view of an example of RRRs on chromosome 13.
(C) Scatter plot comparing gene expression of Kdm4d WT injected SCNT 2-cell embryos with that of IVF 2-cell embryos. Genes express higher (FC > 3) in IVF (IVF-high) or SCNT (SCNT-high) embryos were colored as red and blue, respectively.
Bar graphs indicate the expression level (uniquely mapped read numbers) of the major satellite DNA and the mouse endogenous retrotransposon MERVL in IVF and SCNT embryos.
(A–C) RT-qPCR analysis of Suv39h1 (A), Suv39h2 (B) and Setdb1 (C) mRNA levels in MEF cells at 48 hours after transfection of each siRNA. Data shown are mean expression values relative to Gapdh. The value in control was set as 1.0. Error bars represents s.d. with three biological replicates. *** P<0.001 by Student’s T-test.
Table S1. Preimplantation development of SCNT embryos injected with Kdm4d mRNA, Related to Figures 4 and 6
Table S2.Establishment of ntESCs from SCNT embryos, Related to Figure 4
Table S3. In vivo development of SCNT embryos injected with Kdm4d mRNA, Related to Figure 4
Table S4. Preimplantation development of SCNT embryos injected with Zscan4d mRNA, Related to Figure 5