Abstract
Nuclear RNA interference is an important regulator of transcription and epigenetic modification, but the underlying mechanisms remain elusive. Using a genome-wide approach in the fission yeast S. pombe we have found that Dcr1, but not other components of the canonical RNAi pathway, promotes the release of Pol II from the 3’ end of highly transcribed genes, and, surprisingly, from antisense transcription of rRNA and tRNA genes, which are normally transcribed by Pol I and Pol III. These Dcr1-terminated loci correspond to sites of replication stress and DNA damage, likely resulting from transcription-replication collisions. At the rDNA loci, release of Pol II facilitates DNA replication and prevents homologous recombination, which would otherwise lead to loss of rDNA repeats especially during meiosis. Our results reveal a novel role for Dcr1-mediated transcription termination in genome maintenance and may account for widespread regulation of genome stability by nuclear RNAi in higher eukaryotes.
Introduction
Nuclear RNA interference (RNAi) has emerged as an important regulator of gene expression and epigenetic inheritance in eukaryotes, and studies of fission yeast centromeres have provided mechanistic insight into the process by which RNAi directs epigenetic modification (Bühler and Gasser, 2009; Castel and Martienssen, 2013; Goto and Nakayama, 2012; Grewal, 2010; Lejeune et al., 2011). In S. pombe, RNAi is required to direct H3 lysine 9 dimethylation (H3K9me2) and H3K4 demethylation within the heterochromatic repeats flanking each centromere (Volpe et al., 2002). Tightly regulated transcription within these repeats (Djupedal et al., 2005) leads to the production of double stranded RNA (dsRNA), in part via RNA dependent RNA polymerase (Rdp1), that is processed into small interfering RNA (sRNA) by the sole Dicer in S. pombe, Dcr1 (Colmenares et al., 2007). sRNA are loaded into Argonaute (Ago1), guiding it back to complementary nascent RNA (from transcribing RNA Polymerase II) where it directs the deposition of H3K9me2 through the histone methyltransferase Clr4 via “co-transcriptional gene silencing” (Bühler et al., 2006; Irvine et al., 2006).
Similar mechanisms of RNAi based silencing have been discovered in higher eukaryotes. In Arabidopsis the RNA-directed DNA methylation (RdDM) pathway co-transcriptionally directs de novo cytosine methylation at loci transcribed by RNA Pol V (Law and Jacobsen, 2010). In the C. elegans germline the 21U small RNA pathway directs H3K9 methylation through 22G-loaded Argonautes in the nucleus, closely resembling S. pombe (Shirayama et al., 2012). Classically, these silencing pathways have been thought to act on heterochromatic repetitive elements, such as transposons, but more recently a broader role at euchromatic genes has been discovered. Studies in Arabidopsis (Liu et al., 2012), Drosophila (Cernilogar et al., 2011), C. elegans (Guang et al., 2010), and S. pombe (Gullerova et al., 2011; Gullerova and Proudfoot, 2008; Woolcock et al., 2012) have implicated nuclear small RNA pathways in the regulation of RNA Pol II (Pol II) at individual euchromatic genes. In fission yeast, this conserved function of RNAi (Pol II regulation) is particularly important in the context of DNA replication. Centromeric repeat units in S. pombe are transcribed during S-phase, the time at which DNA replication occurs and epigenetic marks must be re-established (Chen et al., 2008; Kloc et al., 2008). This leads to a collision between Pol II and the replisome that is resolved by RNAi through the release of Pol II (Zaratiegui et al., 2011). In the absence of RNAi stalled replication forks are restarted through homologous recombination (HR), and this results in the loss of epigenetic modifications (Zaratiegui et al., 2011).
We have found that Dcr1 coordinates transcription and replication outside of pericentromeres, identifying a novel role for Dcr1 in transcription termination and maintaining genome stability. Pol II accumulation is a hallmark of polymerase collision, and we found that Pol II accumulates in dcr1Δ cells at previously uncharacterized loci including protein coding genes, tDNA, and rDNA. Dcr1-dependent sRNA were detected at these loci, but transcriptional termination was not dependent on sRNA biogenesis or other RNAi pathway components, demonstrating for the first time a Dcr1-specific role in Pol II release. These loci are strongly correlated with sites of replication pausing, and thus likely represent collisions between transcription and replication (Bermejo et al., 2012). We focused on one particularly striking and unexpected site of Pol II regulation, the subtelomeric rDNA repeats, where we found that Dcr1 is required for rDNA copy number maintenance. Our findings suggest that in S. pombe Dcr1 has a genome wide role in terminating transcription by releasing Pol II at sites of collision between transcription and replication, and thus maintains genome stability.
Results
Dcr1 has a genome wide role in Pol II regulation
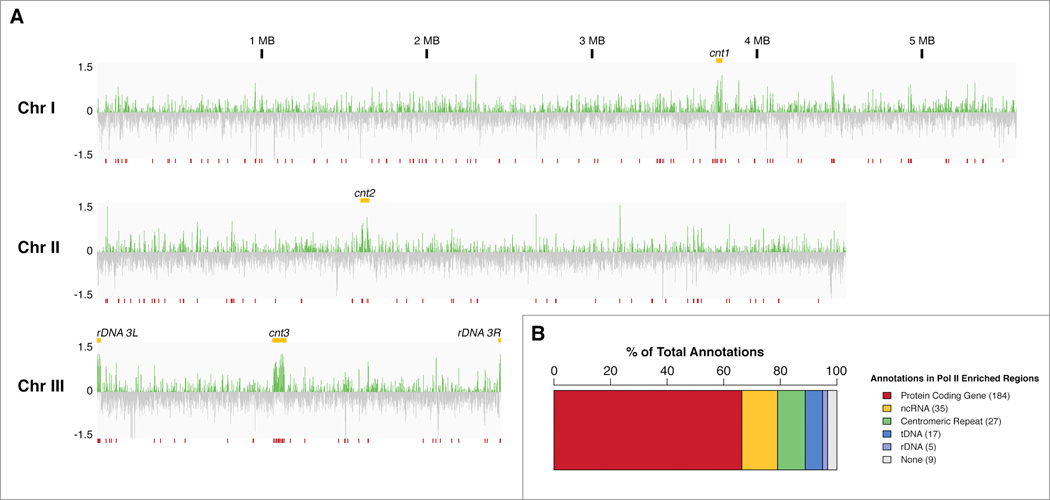
To identify sites transcriptionally regulated by Dcr1 we profiled Pol II enrichment in WT and dcr1Δ mitotic cells by ChIP-seq using antibodies raised against both “poised” (S5 phosphorylated, pS5) and “elongating” (S2 phosphorylated, pS2) forms of Pol II. We observed genome wide effects on Pol II enrichment in the absence of Dcr1 that were not limited to centromeric repeats (Figure 1a). Indeed, striking regions of accumulation were visible not only at the centromeres, but also within the subtelomeric rDNA repeats on chromosome III, which are normally transcribed by Pol I. A total of 224 high-confidence (FDR <= 0.01) regions of Pol II accumulation in dcr1Δ as compared to WT were identified using both antibodies and replicates of each (Table S1). Features found within these regions largely contained protein coding genes, non-coding RNA (ncRNA), centromeric repeats, tDNA, and rDNA (Figure 1b).
Figure 1. Pol II transcriptional profile by ChIP-seq reveals novel Dcr1-regulated loci.
A) Chromosome-wide view of log2(ratio) between dcr1Δ and WT Pol II enrichment. Repeat features including centromeres and rDNA clusters are indicated (yellow). Regions of significant (FDR <= 0.01) enrichment across combined initiating and elongating Pol II replicates are indicated (red), and listed in Table S1. B) Count of annotated features contained within regions of increased Pol II enrichment. See Figure S2 for the effect of other canonical silencing factors on Pol II transcription at some of these features.
Dcr1 releases Pol II at the 3’ end of highly transcribed genes
Many sites of significant Pol II accumulation fell within protein coding genes, and within these genes enrichment was most often found within the open reading frame (ORF) and at the 3’ end, rather than in the promoter region (Figure 2a). We calculated Pol II enrichment specifically at all protein coding genes and found that 235 genes showed significant (FDR <= 0.01) accumulation in dcr1Δ as compared to WT (Table S2). Importantly, the few protein coding genes where Dicer regulation has been experimentally confirmed (hsp16, hsp104, hsp9) (Woolcock et al., 2012) were present in our list, validating the approach.
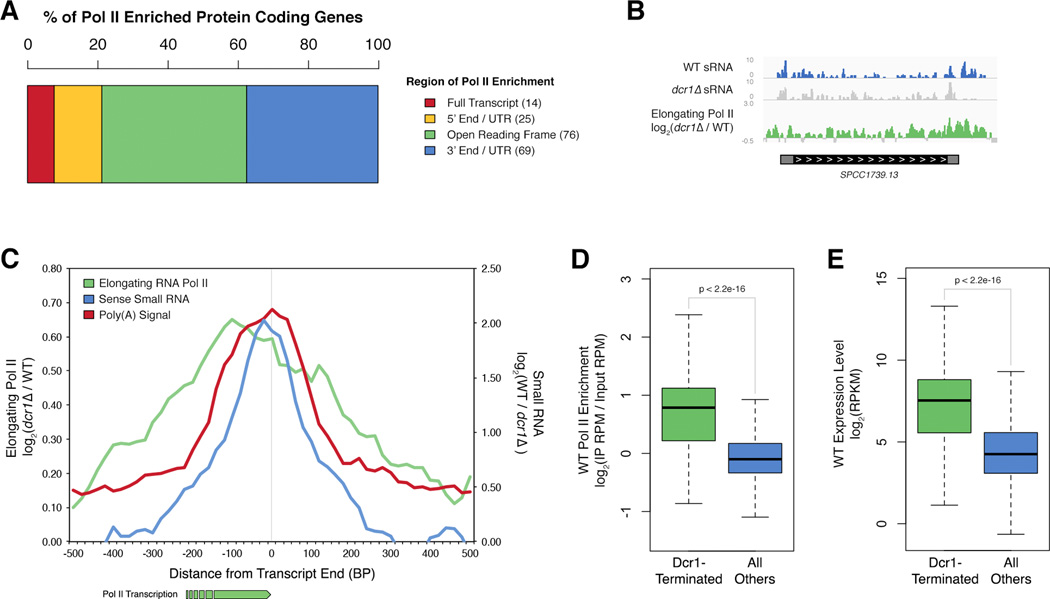
Figure 2. Dcr1 releases Pol II at the 3’ end of highly transcribed genes.
A) Region of Pol II accumulation within protein coding genes identified in Figure 1b. B) Example of a Dcr1-terminated gene (SPCC1739.13, ssa2). Normalized (RPM) sense sRNA reads in WT (blue) and dcr1Δ (grey), elongating Pol II accumulation in dcr1Δ vs WT (green). C) Average elongating Pol II accumulation in dcr1Δ vs WT (green), Dcr1-dependent sRNA (blue), and Poly(A) signal frequency (red) (Schlackow et al., 2013), at the 3’ end of all Dcr1-regulated protein coding genes (listed in Table S2). Values averaged over a 100 bp sliding window for ease of viewing. D–E) Boxplot representation of WT Pol II enrichment (average of initiating (pS5) and elongating (pS2)) at (D), and expression level of (E) Dcr1-terminated (green) vs all other protein coding genes (blue). See Table S3 for GO Term enrichment of Dcr1-terminated genes. See also Figure S1.
We noticed a striking pattern of Pol II accumulation at the 3’ end of Dcr1-regulated genes accompanied by sense Dcr1-dependent small RNA (sRNA), exemplified in Figure 2b. Meta-analysis of Dcr1-regulated protein coding genes showed an increase in elongating Pol II peaking sharply at the 3’ end, and strongly correlated with Poly(A) signal frequency (Schlackow et al., 2013) (Figure 2c), suggesting a defect in transcriptional termination. This peak was accompanied by a peak in Dcr1-dependent sRNA sense to protein coding transcripts indicative of direct Dcr1 activity. Dcr1-dependent sense sRNA at the 3’ end of Dcr1-regulated genes was significant when compared to expression-matched control genes (Figure S1a) and matched the expected size distribution for Dcr1 products (Figure S1b). The presence of only sense sRNA suggested that Dcr1 might be acting on hairpins at the 3’ end of the transcripts. Supporting this, the entire sRNA producing region of the example gene shown in Figure 2b was predicted to be highly double stranded (Figure S1c).
We performed strand specific RNA-seq to distinguish termination defects from transcriptional increase because both can cause an increase in Pol II enrichment, but failure to release Pol II during termination results in no change, or else a decrease in RNA transcript levels (Padmanabhan et al., 2012; West and Proudfoot, 2009). In dcr1Δ cells we saw a decrease of at least 20% in transcript level at 176 of the 235 Dcr1-regulated genes, supporting a defect in termination, while an increase of an equivalent level was only seen at 31 of the 235 (Figure S1d). The expression of Dcr1-regulated genes was significantly reduced when compared to expression-matched controls (Figure S1e). Consistently, analysis of dcr1Δ expression data from Hansen et al. showed an increase at only 32 of our Dcr1-regulated genes. Genes with increased transcript level were enriched for stress response genes as might be expected (p = 0.00961). We saw no evidence of read-through transcription at Dcr1-regulated genes when compared to expression-matched controls (Figure S1f). The general lack of both transcript accumulation and read-through transcription at Dicer targets suggests a termination defect during the release phase, rather than the pausing phase in the absence of Dcr1.
A GO annotation analysis of all Dcr1-terminated genes revealed enrichment in many core cellular processes, most substantially in translation (Table S3). These categories contain many highly transcribed genes, and suggested that this may be a common feature. Indeed Dcr1-terminated genes were highly transcribed in WT cells when compared to global gene transcription levels as measured both by Pol II enrichment (Figure 2d) and expression level (Figure 2e, Figure S1d).
Dcr1 releases Pol II from antisense tDNA and rDNA transcription
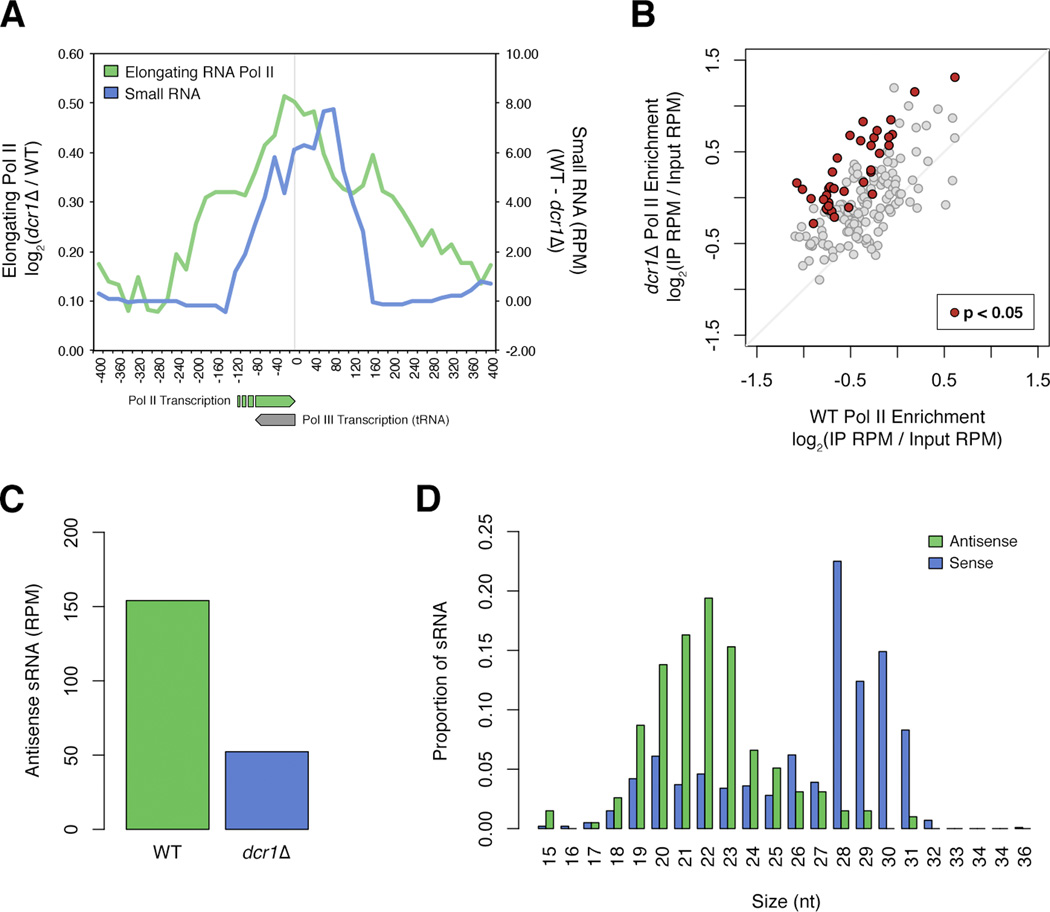
Surprisingly, many tRNA genes (tDNA), which are normally transcribed by RNA Pol III, were found within regions of Pol II accumulation. Because single tDNA are very short (<100 bp) we assessed Pol II accumulation in dcr1Δ vs WT at all chromosomal tDNA (Figure 3a). Elongating Pol II peaked at the 5’ end of tRNA genes, and there was an accompanying peak of antisense Dcr1-dependent sRNA, suggesting antisense Pol II transcription. When quantified individually, 108 of 171 tDNA showed an increase in Pol II enrichment of at least 20%, whereas only 4 showed a decrease (Figure 3b, Table S4). The increase was statistically significant across replicates for 37 of the 108, and there was no bias towards pericentromeric tDNA. sRNA that peaked at the site of Pol II accumulation was antisense to tRNA, Dcr1-dependent (Figure 3c), and fell within the expected size range for Dcr1 products when compared to sense tRNA processing fragments (Figure 3d).
Figure 3. Dcr1 releases Pol II from antisense Pol II transcription at tDNA.
A) Average elongating Pol II accumulation in dcr1Δ vs WT (green), and Dcr1-dependent sRNA level (blue) at nuclear tRNA genes. Values averaged over a 100bp sliding window for ease of viewing. Direction of Pol III tRNA transcription and antisense Pol II transcription are indicated. B) Average of pS5 and pS2 Pol II enrichment at each of the S. pombe nuclear tDNA, for both dcr1Δ and WT. tDNA with significant (p < 0.05) Pol II enrichment are indicated in red (listed in Table S4). C) Normalized read counts of antisense sRNA mapping to tDNA in WT (green) and dcr1Δ (blue). D) Size distribution of antisense (green) and sense (purple) sRNA mapping to tDNA in WT cells.
Similar to tDNA, but more striking at the genome-wide scale was Pol II accumulation within the subtelomeric rDNA repeats on chromosome 3 (Figure 1a and Figure 4a). Enrichment of both poised and elongating Pol II at rDNA repeats was significantly increased in dcr1Δ vs WT when quantified across replicates (Figure 4b). Poised Pol II peaked at the 3’ end of Pol I transcripts while elongating Pol II peaked at the 5’ end, again suggesting antisense transcription. We also identified a population of Dcr1-dependent sRNA antisense to 35S rRNA that peaked at the 3’ end of Pol II transcription (Figure 4a and c). These sRNA fell within the expected size range for Dcr1 products, unlike sense rRNA fragments, which were more evenly distributed and most likely degradation products (Figure 4d). Consistently, overexpression of dcr1 even in the absence of the RNA-dependent RNA polymerase (Rdp1) results in a dramatic increase in sRNA levels antisense to both rDNA (~38.5 fold) and tDNA (~4.5 fold) while a comparable increase in sense sRNA is not seen (Yu et al., 2013).
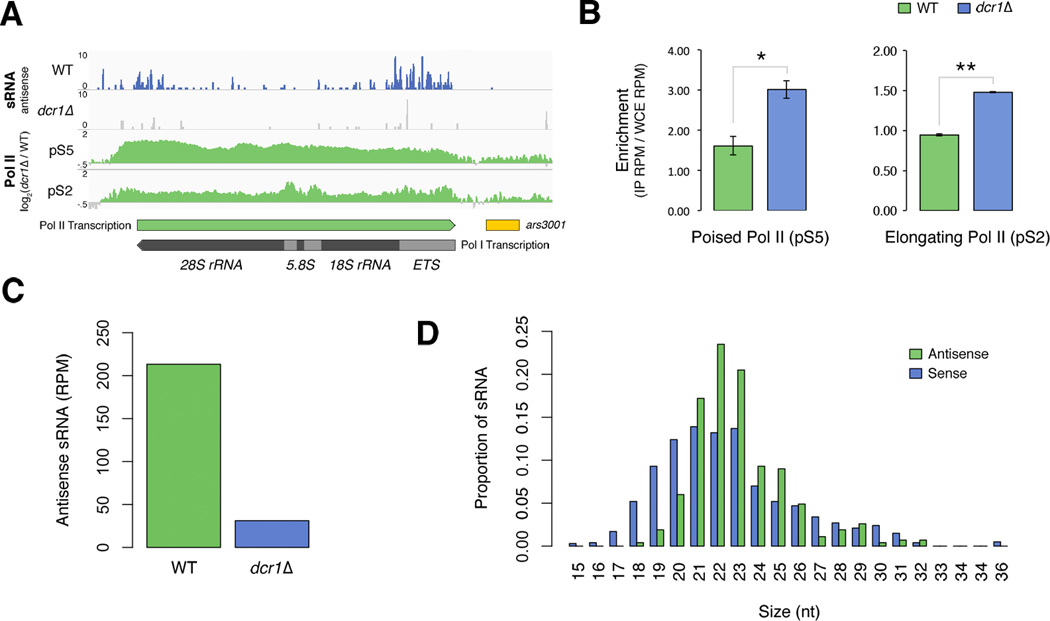
Figure 4. Dcr1 releases Pol II from antisense transcription at subtelomeric rDNA.
A) Distribution of normalized (RPM) sRNA reads mapping antisense to repetitive subtelomeric rRNA genes in WT (blue) and dcr1Δ (grey), and both poised and elongating Pol II accumulation in dcr1Δ vs WT (green). Direction of Pol I rRNA transcription and antisense Pol II transcription are indicated. Annotations for 18S, 5.8S, 28S rRNA genes, externally transcribed spacer (ETS) and replication origin containing region (ars3001) are shown. B) Quantification of poised (pS5) and elongating (pS2) Pol II enrichment within subtelomeric rDNA repeat regions containing sRNA for WT (green) and dcr1Δ (blue). Data are represented as mean ± SEM. The significance of differences across replicates is indicated (** = p < 0.01, * = p < 0.05). C) Normalized read counts of antisense sRNA mapping to rDNA in WT (green) and dcr1Δ (blue). D) Size distribution of antisense (green) and sense (purple) sRNA mapping to rDNA in WT cells.
The canonical RNAi pathway is not involved in Pol II release at novel Dcr1-terminated loci
Transcriptional gene silencing (TGS) in S. pombe occurs when sRNA generated by Dcr1 are loaded into Argonaute (Ago1) and direct H3K9me2 deposition at target loci (Castel and Martienssen, 2013). We tested the involvement of the RNAi pathway in transcriptional regulation by performing Pol II ChIP-qPCR in cells with a dcr1 RNase III catalytic dead allele (D937A, D1127A) unable to generate sRNA (Colmenares et al., 2007) as well as in ago1Δ cells alongside dcr1Δ and WT. We saw no increase in Pol II enrichment at tDNA or rDNA in either the catalytic dead dcr1 mutant or ago1Δ as compared to WT, unlike in dcr1Δ (Figure S2a). As expected all mutants showed centromeric Pol II accumulation vs WT due to H3K9me2 loss.
An indicator of RNAi mediated chromatin silencing is repressive H3K9 methylation at target sites, and both tDNA and rDNA are enriched for this mark in S. pombe (Figure S2b). We assessed the contribution of H3K9 methylation to transcriptional regulation by performing H3K9me2 ChIP-seq in WT and dcr1Δ cells. There was no decrease in H3K9me2 enrichment at either Dcr1-terminated protein coding genes, tDNA or rDNA in dcr1Δ, while a decrease at centromeric repeats was seen as expected (Figure S2b). In fact, there was a slight increase of H3K9me2 enrichment at novel Dcr1 targets that likely represents higher background levels in dcr1Δ samples due to the absence of centromeric heterochromatin. Thus Dcr1-dependent transcription termination at regions outside peri-centromeric heterochromatin can occur independently of the canonical RNAi pathway.
Dcr1 promotes termination of transcription at sites of replication stress
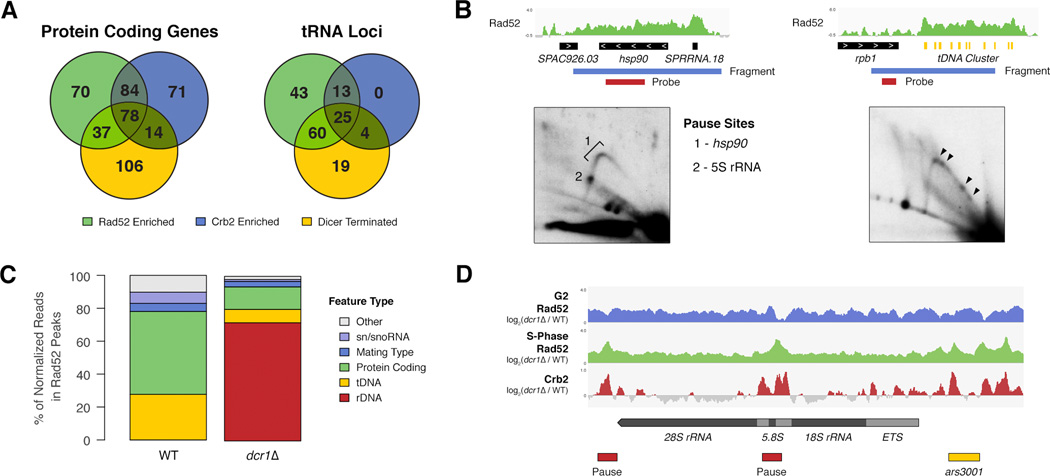
The novel Dcr1-regulated loci (actively transcribed genes, tDNA, and rDNA) that we have identified all represent ‘difficult to replicate’ regions because frequent passage of transcription complexes creates a barrier to DNA replication that can stall forks (Alzu et al., 2012; Sabouri et al., 2012). The homologous recombination (HR) protein Rad52 is recruited to stalled replication forks, which may eventually collapse leading to DNA damage and checkpoint (Chk1) activation via Crb2 (Nakamura et al., 2004). We performed both Rad52 and Crb2 ChIP-Seq to identify stalled and collapsed forks and to correlate these with Dcr1-terminated genes. We found a strong correlation between both Rad52 and Crb2 enriched genes and Dcr1-terminated genes in WT cells (Figure 5a), with over 55% of Dcr1-terminated genes enriched for at least one of these DNA damage signaling proteins. This correlation was also seen in dcr1Δ cells, however there was a large decrease in Rad52, while Crb2 remained similar (Figure S3a). We observed a similar correlation at tRNA genes, but unlike protein coding genes the number of Crb2-enriched tRNA genes increased by 30% in dcr1Δ. We validated the correlation between Rad52 enrichment and replication pausing by 2D gel electrophoresis at a protein-coding gene (hsp90) and a tDNA cluster, both of which showed significant Rad52 enrichment and Pol II accumulation (Figure 5b). We found that replication pausing occurs at these loci, indicating that sites of Rad52 enrichment detected by ChIP-seq are bona-fide difficult-to-replicate regions (Figure 5b).
Figure 5. Dcr1 releases Pol II at sites of replication stress.
A) Overlap between Rad52 enrichment, Crb2 enrichment and Dcr1 termination at either protein coding or tRNA genes in WT cells. B) Rad52 enrichment at hsp90 and adjacent 5S rRNA gene determined by ChIP-seq (log2(IP RPM / Input RPM)), and accompanying 2D gel of fragment containing both features. Sites of replication pausing, within the 5S rRNA gene and hsp90 are indicated. Rad52 enrichment in WT cells at the tDNA cluster in the left outer arm of centromere 2, and accompanying 2D gel of fragment containing the cluster. Arrowheads indicate sites of major pausing. C) Distribution of Rad52 by feature type in WT and dcr1Δ determined by normalized Rad52 ChIP-seq read counts in peaks. D) Rad52 accumulation in dcr1Δ vs WT at rDNA in both unsynchronized (G2, blue) and S-phase (green) cells, and Crb2 accumulation in dcr1Δ vs WT at rDNA in unsynchronized cells (red). rDNA annotations (black box), programmed pause sites (red box), and replication origin (yellow box) are indicated. See also Figure S3.
The loss of Rad52 from many of the Dcr1-terminated genes in dcr1Δ was somewhat surprising, but analysis of sequencing read distribution clearly showed that in dcr1Δ the bulk (72%) of Rad52 is localized to the rDNA repeats (Figure 5c). Indeed we have previously observed a substantial increase in Rad52 foci in dcr1Δ cells as compared to WT (Zaratiegui et al., 2011), however the total level of Rad52 remains unchanged (data not shown), indicating that the Rad52 pool is limited. Rad52 nucleation occurs at sites of DNA damage during S-phase and subsequently spreads from the stall site (Zhou et al., 2013). To determine the precise location of replication stalling within rDNA we synchronized cells and performed Rad52 ChIP in S-phase. During S-phase Rad52 accumulation in dcr1Δ vs WT peaked over programmed replication pause sites (Sanchez et al., 1998) and replication origins within rDNA repeats (Figure 5d). We also saw overlapping peaks of Crb2 accumulation in dcr1Δ vs WT, indicating fork collapse at these loci (Figure 5d).
RNA:DNA hybrids can occur at stall sites resulting from transcription and replication collision (Aguilera and García-Muse, 2012; Alzu et al., 2012; Bermejo et al., 2012). These hybrids are themselves highly recombinogenic, and recruit Rad52 (Wahba et al., 2013). We hypothesized that hybrids might form within rDNA repeats due to failed Pol II release in dcr1Δ. We performed DNA:RNA Immunoprecipitation (DRIP) (Ginno et al., 2012) to assess hybrid formation at rDNA and found an increase in hybrid levels in dcr1Δ cells (Figure S3b) that was significant across replicates in the 18S and 28S regions (Figure S3c). Conversely, hybrids that are Dcr1-dependent have been reported at centromeric repeats (Nakama et al., 2012) and our DRIP-seq results support this (Figure S3d). We did however observe an increase in hybrids within regions containing replication origins, again suggesting hybrid formation as a consequence of replication and transcription collision.
Dcr1 is required for copy number maintenance of rDNA repeats
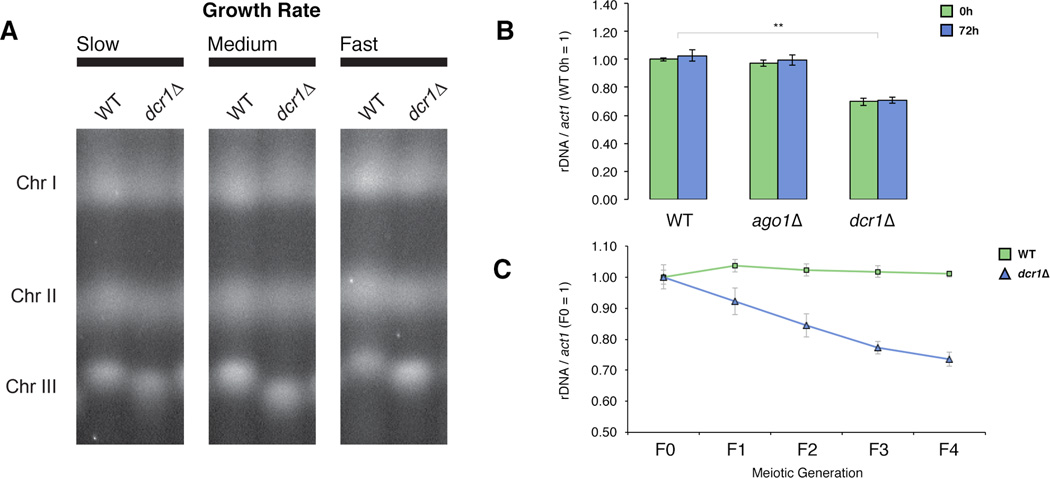
The dramatic increase in Rad52 enrichment, and the presence of RNA:DNA hybrids at rDNA in dcr1Δ suggested that recombination within the repeats could result in genomic instability. We isolated whole chromosomes from individual WT and dcr1Δ colonies of varying growth rates using pulsed-field gel electrophoresis (PFGE). Consistently dcr1Δ cells showed a significant reduction in chromosome III size suggesting a loss of subtelomeric rDNA repeats (Figure 6a). To understand the dynamics of rDNA loss we created de novo dcr1 deletion strains from WT cells and assessed rDNA copy number using quantitative polymerase chain reaction (qPCR). Freshly transformed dcr1Δ cells showed a 30% reduction in rDNA copy number, which remained stable through 72h of continuous mitotic division (Figure 6b). As expected this loss was not seen in either the catalytic dead dcr1 mutant (Figure S4a), or ago1Δ cells, again indicating a function outside of canonical RNAi. We then tested rDNA stability through meiosis, by assessing copy number through four generations of progeny. Strikingly, rDNA repeats were progressively lost at each meiosis in dcr1Δ (Figure 6c), while being maintained in WT and ago1Δ cells (Figure S4b).
Figure 6. Dcr1 is required for copy number maintenance of rDNA repeats.
A) Whole chromosomes isolated by Contour Clamped Homogenous Electric Field Pulsed Field Gel Electrophoresis (CHEF-PFGE) from individual WT and dcr1Δ colonies of varying growth rates (slow, medium, fast) run side-by-side for comparison. Intervening lanes have been removed for ease of viewing. B) rDNA copy number determined by qPCR of genomic DNA from 6 colonies of WT and freshly transformed dcr1 or ago1 knockout cells, and cells after 72h (~25–30 generations) of mitotic division. Copy number is normalized to WT 0h. Data are represented as mean ± SEM. C) rDNA copy number of WT and freshly transformed dcr1 knockout cells (F0) and subsequent meiotic generations (Fn). Copy number is normalized to F0 of each strain. Data are represented as mean ± SEM. The significance of differences is indicated (** = p < 0.01, * = p < 0.05). See also Figure S4.
Dcr1 is required in the face of replicative stress at rDNA
Programmed replication fork pausing facilitates the directional replication of DNA, preventing the collision of replication forks with transcription complexes. The histone demethylase Lsd1 is required for replication fork pausing within rDNA (Holmes et al., 2012), and is enriched at tDNA (Lan et al., 2007) where it may also play the same role, as H3K9me2 spreads across tRNA boundaries in lsd1 mutants (Lan et al., 2007) and depends on association of CLRC with the replisome (Li et al., 2011; Zaratiegui et al., 2011). We hypothesized that in the absence of programmed fork pausing, collisions between Pol II and replication forks would increase, and that Dcr1 would be required to resolve these. lsd1 single mutants are slow growing but viable, however we found that dcr1 and lsd1 are synthetically lethal, supporting our hypothesis (Figure S4c). Increased activity of Dcr1 in the face of replicative stress at rDNA should result in higher sRNA levels. The helicase Pfh1 is required for replication fork progression through rDNA, and in its absence stalling occurs (Sabouri et al., 2012). We sequenced sRNA from temperature sensitive pfh1-R23 cells (Tanaka et al., 2002) at a semi-permissive temperature (Figure S5), to induce replication stress in rDNA without arresting growth. As predicted, there was a Dcr1-dependent increase in antisense sRNA originating from rDNA in pfh1 cells relative to WT, supporting increased Dcr1 activity in the face of replicative stress (Figure S4d). Importantly this increase was not due to rDNA repeat expansion, since rDNA copy number in pfh1-R23 was not significantly different from WT (Figure S4e). Intriguingly, the pfh1-R23 dcr1Δ double mutant showed a further reduction in rDNA copy number as compared to both parents suggesting that increased replication stress in the absence of Dcr1 results in an enhancement of rDNA loss.
Discussion
A Dcr1-specific role in transcriptional termination
Previous genome-wide studies aimed at identifying targets of RNAi in S. pombe have focused on RNA transcript levels and histone modification, but have failed to identify a consensus group of targets outside of heterochromatin (Gullerova et al., 2011; Hansen et al., 2005; Woolcock et al., 2012; 2011; Yamanaka et al., 2012). Because of the well-established role of RNAi in transcriptional silencing we interrogated Pol II directly at the chromatin level using ChIP-seq in WT and dcr1Δ cells. With this robust approach we identified a comprehensive set of loci that showed a significant increase of Pol II in dcr1Δ, suggesting transcriptional regulation by Dcr1. These diverse loci included highly transcribed protein coding genes, tDNA, and rDNA in addition to pericentromeric repeats.
At these loci Pol II accumulation was most striking at the 3’ end of the transcription unit, suggesting a termination defect in dcr1Δ cells, and we present several lines of evidence indicating that Dcr1 promotes transcriptional termination. Canonical termination involves two steps, the first being Pol II pausing, and the second being Pol II release (Kuehner et al., 2011; Park et al., 2004; Yang and Roberts, 1989). We saw no evidence of read-through transcription, which is indicative of a pausing defect, by either Pol II ChIP-seq or RNA-seq. Instead, Pol II peaked just upstream of the transcription stop site, and RNA-seq showed reduced transcript levels of most Dcr1-terminated genes in dcr1Δ cells, consistent with a release defect. In some instances we observed an increase in Pol II enrichment that extended upstream of the 3’ end. A failure to remove stalled Pol II at the 3’ end has been shown to result in an upstream Pol II “pile-up” (Hanawalt and Spivak, 2008; Trautinger et al., 2005), and could explain this observation. Taken together, these results support a role for Dcr1 in releasing Pol II.
We found strong evidence for direct Dcr1 activity at regions of Pol II accumulation in the form of Dcr1-dependent sRNA that matched the expected size distribution. As an RNase III enzyme Dcr1’s substrate, an RNA duplex, can be generated in two ways. First, the presence of antisense sRNA from tDNA and rDNA, and their persistence in rdp1Δ cells (Yu et al., 2013), strongly suggests that antisense transcription by Pol II is occurring at these loci, providing the potential for dsRNA. Alternatively, secondary structures in Pol II transcribed RNA molecules themselves might produce hairpins. This latter pathway is more likely at protein coding genes since sRNA arise almost exclusively from the sense strand at the 3’ end, where Pol II accumulated consistent with a failure to release. Predicting secondary RNA structure at an example Dcr1-terminated gene revealed a highly stable hairpin corresponding to sRNA peaks, consistent with this model.
Our findings suggest that while a RNA duplex is present at target sites and could be required for recruitment, Dcr1 mediated termination does not rely on its catalytic activity or its sRNA products. Importantly, we did not see Pol II accumulation in RNase III catalytic dead Dicer mutant nor in ago1Δ cells at tDNA or rDNA, and there was no reduction in rDNA copy number. Supporting this, the TRAMP complex containing the poly(A) polymerase Cid14 targets sRNA arising from tDNA and rDNA for degradation by the exosome, and prevents their loading into Ago1 (Bühler et al., 2008). Furthermore, overexpression of Dcr1 results in an increase of sRNA mapping antisense to tDNA and rDNA (Yu et al., 2013). Taken in light of our results these sRNA are likely by-products of Dcr1’s role in transcriptional termination, and because Ago1 is similarly not required for this process, the TRAMP complex ensures that they do not enter the RNAi pathway. If TRAMP-targeted sRNA from Dcr1 are not degraded, such as in an exosome mutant, they are able to increase H3K9 methylation at rDNA (Marasovic et al., 2013). Yet we did not observe a decrease of H3K9me2 in the absence of Dcr1-dependent sRNA, supporting that these sRNA are targeted for degradation and not loaded into Ago1. Consequently, they would be highly unstable, which may explain their low abundance compared to centromeric siRNA.
While Dcr1 mediated termination does not rely on its well-known function in sRNA biogenesis, the presence of Dcr1-dependent sRNA at termination loci strongly supports a direct effect of Dcr1 at these loci. For example, it could bind to transcripts to be terminated and serve as a platform for the timely recruitment of effectors for fork protection and Pol II release. Such a structural role for Dcr1 in H3K9 methylation, that goes beyond the sRNA biogenesis function has recently been reported at the centromere (Yu et al., 2013).
Dcr1 acts in the unique context of transcription and replication collision
Why should Dcr1 promote transcriptional termination at some loci and not others? Our results suggest that in S. pombe this regulation occurs specifically at sites where collision between transcription and replication occurs. Head on collisions first occur at the 3’ end of transcribed regions and result in stalled Pol II, corresponding to sites of Dcr1 activity. Such collisions result in stalled replication forks (Azvolinsky et al., 2009) and the recruitment of Rad52 (Lambert et al., 2010). If stalled forks are not resolved they will collapse, leading to γH2A deposition and Crb2 recruitment (Rozenzhak et al., 2010). We performed both Rad52 and Crb2 ChIP-seq and found a strong correlation between Dcr1-terminated loci and peaks of both proteins in WT cells. This suggests that these loci are “natural” sites of replication stress and pausing. Indeed highly transcribed RNA Pol II genes, tDNA, and rDNA all constitute “difficult-to-replicate” regions in S. pombe (Sabouri et al., 2012). The presence of Rad52 in WT cells at Dcr1-terminated loci suggests that collision, fork stalling, and Rad52 localization occur upstream of Dcr1 termination. A similar function has been proposed for the RNA/DNA helicase Sen1, which terminates transcription of non-polyadenylated transcripts, and has other functions in replication fork progression (Bermejo et al., 2012; Mischo et al., 2011). Loss of Sen1 also results in stalled replication forks, and recombination at Pol II transcribed genes (Alzu et al., 2012).
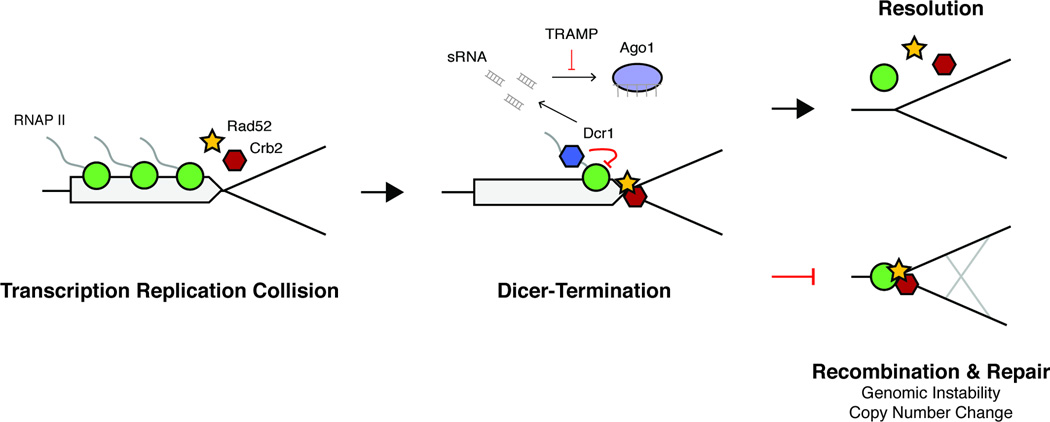
These findings suggest a model whereby Dcr1 terminates transcription by releasing stalled Pol II specifically at transcription-replication collisions (Figure 7). This explains why read-through transcription is not observed at Dcr1 targets, as collision with the replisome would presumably prevent further transcription. In this model Dcr1 does not prevent collisions from occurring, but through termination it does resolve them. Without Dcr1 termination, Rad52 and Crb2 persist at stall sites, which ultimately must be restarted by homologous recombination for replication to proceed (Lambert et al., 2010). It’s possible that Dcr1 is specifically recruited to stalled forks through a pathway not yet understood. Supporting this Neurospora Rad52 is required for the generation of aberrant RNA (aRNA) from rDNA repeats by HU-induced replication fork stalling, which are then processed by Dicer into qiRNA (QDE-2 interacting) (Zhang et al., 2013). Of all the loci we detected, only the pericentromeric repeats were enriched for H3K9me2 in a Dcr1-dependent manner, consistent with the idea that CLRC is recruited to heterochromatin, but not to euchromatin, for spreading via the replisome (Chen et al., 2008; Li et al., 2011; Zaratiegui et al., 2011).
Figure 7. Dcr1-termination of Pol II transcription at stalled replication forks maintains genomic stability.
Transcription by RNA Pol II (green circle) and DNA replication collide producing stalled replication forks that recruit Rad52 (yellow star) and Crb2 (red hexagon). Dcr1 (blue hexagon) acts at these sites to release Pol II and facilitate replication. The TRAMP complex prevents sRNA produced by Dcr1 during termination from being loaded into Ago1 (purple oval) by targeting them for degradation. Without Dcr1 homologous recombination is necessary to restart the replication fork and results in genome instability and copy number changes.
Dcr1 is required for genome stability at rDNA
The subtelomeric rDNA repeats are a suitable locus to study the necessity of Dcr1 termination at collision sites because of their well-known replication dynamics, and tolerance of copy number change. In the absence of Dcr1 there was a dramatic accumulation of Pol II and Rad52 within rDNA repeats, which was accompanied by a reduction in rDNA copy number likely occurring through homologous recombination. After an initial loss, copy number subsequently remained stable for 72h of mitotic division, however further loss occurred in subsequent meiotic generations. Recombination pathways are hyper activated as part of the normal meiotic progression, and it is possible that without Dcr1 this leads to an enhancement of rDNA loss. Similarly, RNAi prevents detrimental recombination at the centromeres during meiosis (Ellermeier et al., 2010).
The direction of DNA replication within rDNA repeats is tightly controlled to prevent collisions with transcribing Pol I that would result in stalled forks. However, the presence of antisense sRNA at rDNA and patterns of poised and elongating Pol II enrichment suggest that Pol II transcription occurs antisense to Pol I and would therefore collide with replication. Our results show that Dcr1-termination is required at these collision sites to prevent recombination and thus maintain genome stability. We demonstrated this by increasing replication stress within rDNA using a partial loss-of-function pfh1 allele, and by observing an increase in Dcr1-dependent sRNA in the pfh1-R23 single mutant, as well as an enhancement of rDNA loss in the double pfh1-R23 dcr1Δ mutant. Furthermore, in the absence of programmed fork pausing, Pol II release by Dcr1 is essential, revealed by the synthetic lethality of lsd1 and dcr1Δ We also detected increased RNA:DNA hybrids in dcr1Δ at rDNA that form when ssDNA is exposed as a result of fork stalling outside of programmed pause sites, providing further evidence of collision. Similar hybrids occur within S. cerivisiae rDNA in the absence of a master repressor of transcription, sin3, which leads to Rad52 recruitment and genome instability (Gottlieb and Esposito, 1989; Wahba et al., 2011).
The presence of antisense sRNA and enrichment of Pol II suggest that antisense transcription of rDNA occurs even in WT cells. In S. cerevisiae Pol II transcription of the intergenic spacer region stimulates recombination and copy number change, thought to be mediated by loss of cohesin localization (Kobayashi and Ganley, 2005). Similar to what we describe, within rDNA Pol II is released by the exosome in budding yeast (Vasiljeva et al., 2008). Pol II transcription is negatively regulated by the silencing protein Sir2 (Smith and Boeke, 1997), and a balance between transcription and silencing therefore regulates copy number. A similar mechanism may exist in S. pombe, whereby some level of Pol II transcription is required to promote basal recombination that maintains copy number. This would presumably lead to transcription-replication collisions that are resolved by Dcr1. Similarly to Sir2, Dcr1 is required for cohesin localization at some loci (Gullerova and Proudfoot, 2008), and may also suppress recombination in rDNA by this mechanism.
Dicer’s regulation of rDNA appears to be conserved across those eukaryotes with Dicer family proteins. Drosophila DCR-2 is required to maintain stability, as well as K9 methylation at rDNA repeats (Peng and Karpen, 2007). In Neurospora Dicer produces sRNA from rDNA repeats and is similarly required for their stability (Cecere and Cogoni, 2009). Dicer physically localizes to rDNA repeats in mouse ES cells (Sinkkonen et al., 2010), and associates with both rDNA and tDNA chromatin in human cells (White et al., 2014). Our research suggests that this conserved role of Dicer at rDNA is related to Pol II regulation.
Aberrant expression of Dicer occurs frequently in human cancers and is significantly correlated with clinical outcome (Bahubeshi et al., 2011; Merritt et al., 2008). In many tumor types Dicer acts as a tumor suppressor (Bahubeshi et al., 2011), a function which is supported by studies in mouse models (Kumar et al., 2009; Lambertz et al., 2010), and could be related to its role in maintaining genome integrity that we describe here. Indeed, rDNA restructuring through recombination is one of the most common chromosomal alterations in adult tumors (Stults et al., 2009) and it is well-known that rDNA instability is linked to cellular senescence (Ganley and Kobayashi, 2014).
Experimental Procedures
Yeast Strains and Growth
Strains used in this study are listed in Table S5. Standard media (YEA) and genetic protocols for fission yeast were used. Cells were harvested in mid-log phase. Cells for pfh1-R23 ts allele sRNA-seq experiment were grown at the semi-permissive temperature as determined by plate assay (Figure S5). All strains used in this study are listed in Table S5.
Cell Cycle Synchronization
Cells were arrested with HU and released as previously described (Zaratiegui, 2011). Synchrony was measured using septation index. Samples for Rad52 ChIP-seq were taken from the first S-phase, occurring approximately 90 minutes after release.
ChIP
ChIP was performed as previously described (Zaratiegui, 2011). The following antibodies were used: Pol II pS2 - Abcam ab5095, Pol II pS5 - Abcam ab5131, H3K9me2 – Milipore 07-441, Myc - Invitrogen R950, GFP - Abcam ab290, RNA:DNA hybrids - S9.6.
ChIP-Seq Analysis
ChIP peaks versus appropriate inputs (whole cell extract) were called using MACS v1.4. When replicates were performed only peaks found in all replicates were considered. MEDIPS v1.12.9 was used to compare Pol II enrichment genome wide between WT and dcr1Δ experiments. Differential coverage in MEDIPS was calculated using EdgeR and a cutoff of FDR < 0.01. Genome browser tracks and meta-analysis were created using enrichment (IP reads per million (RPM) / input rpm) of representative replicates. Enrichment at individual loci was calculated as IP RPM / input RPM within the genomic interval and significance was calculated using a two-tailed heteroscedastic T-test.
Illumina Sequencing (DNA and sRNA)
Genomic DNA libraries were created using either the standard Illumina protocol (Pol II pS5, pS2 and H3K9me2), or with the Nugen Ovation Ultralow DR kit (0330, all others). Small RNA libraries were created using the NEBNext Small RNA kit (E7300). Sequencing was performed on Illumina GA II, Illumina HiSeq, or Illumina MiSeq platforms depending on the experiment. A full list of all libraries used in this study is listed in Table S7.
Illumina Read Processing and Alignment (DNA and sRNA Libraries)
Illumina reads were quality filtered using Trimmomatic and aligned to the S. pombe genome assembly ASM294v2.21 using Bowtie v2.1.0 and local alignment, with multi-mappers randomly distributed. For genomic DNA libraries all duplicate reads were discarded. Read counts were normalized to reads per million (RPM), using total library size.
Boxplot Representation
Boxes represent lower and upper quartiles surrounding the median (line), outliers are hidden for ease of viewing. Whiskers represent minimum and maximum values within 1.5× of the inner quartile range. The significance of differences between groups was calculated using a Wilcoxon Rank Sum test.
Supplementary Material
Acknowledgements
We thank M. O’Connell, and B. Arcangioli for strains, advice, and discussion, F. Chedin for advice on DRIP-seq, and M.T.A. Donoghue for discussion and advice relating to statistical analysis. S.C. was supported by a fellowship from the Natural Sciences and Engineering Research Council of Canada (PGSD) and a Cashin Scholarship from the Watson School of Biological Sciences. This work was supported by grants BFU2001-28804 and CSD2007-00015 from the Spanish Ministerio de Economía y Competitividad to F. A., and by a grant from the National Institutes of Health (GM076396) to R.A.M.. R.A.M is also supported by the Howard Hughes Medical Institute-Gordon and Betty Moore Foundation (GMBF3033) and acknowledges a Chaire Blaise Pascal (Region Ile-de-France) at Institut Biologie École Normale Superieure, Paris. The authors acknowledge assistance from the Cold Spring Harbor Laboratory Shared Resources, which are funded in part by the Cancer Center Support Grant (5PP30CA045508).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The GEO accession number for the raw sequencing data reported in this paper is XXXX.
Author Contributions
S.C. and J.R. contributed equally to this work. S.C., J.R., S.B., A-Y.C., M.S., and A.V. performed experiments, and S.C. analyzed the data. R.A.M. and P.A. designed experiments, and R.A.M. and S.C. and J.R. wrote the manuscript.
The authors declare no competing financial interests.
References
- Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly Transcribed RNA Polymerase II Genes Are Impediments to Replication Fork Progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahubeshi A, Tischkowitz M, Foulkes WD. miRNA processing and human cancer: DICER1 cuts the mustard. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002493. 111ps46. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Bühler M, Gasser SM. Silent chromatin at the middle and ends: lessons from yeasts. Embo J. 2009;28:2149–2161. doi: 10.1038/emboj.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Cogoni C. Quelling targets the rDNA locus and functions in rDNA copy number control. BMC Microbiol. 2009;9:44. doi: 10.1186/1471-2180-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Sardo Lo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Bühler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci USA. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley ARD, Kobayashi T. Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 2014;14:49–59. doi: 10.1111/1567-1364.12133. [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto DB, Nakayama J-I. RNA and epigenetic silencing: insight from fission yeast. Dev. Growth Differ. 2012;54:129–141. doi: 10.1111/j.1440-169X.2011.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Moazed D, Proudfoot NJ. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 2011;25:556–568. doi: 10.1101/gad.618611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bähler J, Thon G. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Roseaulin L, Schurra C, Waxin H, Lambert S, Zaratiegui M, Martienssen RA, Arcangioli B. Lsd1 and Lsd2 Control Programmed Replication Fork Pauses and Imprinting in Fission Yeast. Cell Reports. 2012;2:1513–1520. doi: 10.1016/j.celrep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley ARD. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Fréon K, Murray JM, Carr AM, Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell. 2010;39:346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, Marine J-C. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Zaratiegui M, Villén J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA, et al. S. pombe LSD1 Homologs Regulate Heterochromatin Propagation and Euchromatic Gene Transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Bayne EH, Allshire RC. On the Connection between RNAi and Heterochromatin at Centromeres. Cold Spring Harbor Symposia on Quantitative Biology. 2011 doi: 10.1101/sqb.2010.75.024. [DOI] [PubMed] [Google Scholar]
- Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Bakht S, Dean C. Cotranscriptional role for Arabidopsis DICERLIKE 4 in transcription termination. Science. 2012;335:1621–1623. doi: 10.1126/science.1214402. [DOI] [PubMed] [Google Scholar]
- Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell. 2013;52:173–183. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng J-F, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gómez-González B, Grzechnik P, Rondón AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells. 2012;17:218–233. doi: 10.1111/j.1365-2443.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Du L-L, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- Park NJ, Tsao DC, Martinson HG. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol Cell Biol. 2004;24:4092–4103. doi: 10.1128/MCB.24.10.4092-4103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenzhak S, Mejía-Ramírez E, Williams JS, Schaffer L, Hammond JA, Head SR, Russell P. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 2010;6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev. 2012;26:581–593. doi: 10.1101/gad.184697.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JA, Kim SM, Huberman JA. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- Schlackow M, Marguerat S, Proudfoot NJ, Bähler J, Erban R, Gullerova M. Genome-wide analysis of poly(A) site selection in Schizosaccharomyces pombe. Rna. 2013;19:1617–1631. doi: 10.1261/rna.040675.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Mello CC. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012 doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS ONE. 2010;5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009;69:9096–9104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ryu G-H, Seo Y-S, Tanaka K, Okayama H, MacNeill SA, Yuasa Y. The fission yeast pfh1(+) gene encodes an essential 5” to 3” DNA helicase required for the completion of S-phase. Nucleic Acids Res. 2002;30:4728–4739. doi: 10.1093/nar/gkf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ. Transcriptional termination enhances protein expression in human cells. Mol Cell. 2009;33:354–364. doi: 10.1016/j.molcel.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Schlackow M, Kamieniarz-Gdula K, Proudfoot NJ, Gullerova M. Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock KJ, Stunnenberg R, Gaidatzis D, Hotz HR, Emmerth S, Barraud P, Buhler M. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 2012 doi: 10.1101/gad.186866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Bühler M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol. 2011;18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SIS. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2012 doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Roberts JW. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc Natl Acad Sci USA. 1989;86:5301–5305. doi: 10.1073/pnas.86.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Jih G, Iglesias N, Moazed D. Determinants of Heterochromatic siRNA Biogenesis and Function. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marín L, Chang A-Y, Goto D, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chang S-S, Zhang Z, Xue Z, Zhang H, Li S, Liu Y. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes Dev. 2013;27:145–150. doi: 10.1101/gad.209494.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z-X, Zhang M-J, Peng X, Takayama Y, Xu X-Y, Huang L-Z, Du L-L. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Research. 2013;23:705–715. doi: 10.1101/gr.146357.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.