SUMMARY
Mechanisms for human memory T cell differentiation and maintenance have largely been inferred from studies of peripheral blood, though the majority of T cells are found in lymphoid and mucosal sites. We present here a multidimensional, quantitative analysis of human T cell compartmentalization and maintenance over six decades of life in blood, lymphoid and mucosal tissues obtained from 56 individual organ donors. Our results reveal that the distribution and tissue residence of naïve, central and effector memory, and terminal effector subsets is contingent on both their differentiation state and tissue localization. Moreover, T cell homeostasis driven by cytokine or TCR-mediated signals is different in CD4+ or CD8+ T cell lineages, varies with their differentiation stage and tissue localization, and cannot be inferred from blood. Our data provide an unprecedented spatial and temporal map of human T cell compartmentalization and maintenance, supporting distinct pathways for human T cell fate determination and homeostasis.
INTRODUCTION
T lymphocytes, a critical component of the adaptive immune system, provide lifelong protection against pathogens by orchestrating immune responses at diverse sites of infection. Naïve T cells emerge from the thymus and populate lymphoid tissues sites, where they differentiate to effector T cells upon antigen encounter, and subsequently can develop into long-lived memory T cells. The complement of T cells within an individual is heterogeneous, consisting of naïve T cells, short-lived or terminally differentiated effector cells (also designated as TEMRA), and memory T cells that accumulate with successive antigen encounters and are the predominant T cell subset in adults (Farber et al., 2014; Saule et al., 2006). Memory T cells are comprised of multiple subsets defined by their migration capacities and tissue residence, including central (TCM) memory cells in circulation and lymphoid sites, effector memory (TEM) cells circulating through blood and peripheral sites (Sallusto et al., 2004), and a recently identified resident memory T cell (TRM) subset retained in tissues such as lungs, intestines, skin, liver and genital mucosa (Clark et al., 2006; Mueller et al., 2013; Purwar et al., 2011; Sathaliyawala et al., 2013; Turner et al., 2014a). Each of these subsets has specific roles in preserving immunity: maintenance of naïve T cells is important for responses to new antigens, and memory T cells mediate protection to diverse pathogens encountered at multiple anatomic locations. Identifying the pathways for memory generation and maintenance is therefore critical for designing effective ways to promote lifelong T cell-mediated immunity in humans, for which no strategies currently exist.
The development and maintenance of T cell subsets in humans remain poorly understood for several reasons. Primarily, most studies of human T cells are confined to sampling of peripheral blood, which contains less than 3% of the total T cells in the body (Ganusov and De Boer, 2007). There are few studies analyzing T cells in lymphoid tissue, where most immune responses are initiated, and only isolated studies in mucosal sites, where effector and memory T cells function and are maintained (Farber et al., 2014). This limited sampling in humans makes it virtually impossible to follow an immune response in vivo as in animal models, thus we lack essential insights into human T cell lineage and maintenance. Moreover, humans enjoy a long lifespan, with potential for dynamic changes in the T cell compartment due to increased antigen experience, decreased thymic output, and alterations in T cell homeostasis. However, most studies of human T cells examine cohorts of limited age range, while studies of aging and T cells compare young and aged cohorts in discrete, non-overlapping groups, rather than assessing how T cell subset composition may dynamically alter over the course of a lifetime. Defining the fundamental properties of human T cell subsets throughout the body can therefore provide an understanding of their lineage relationships and differentiation pathways in ways not previously possible.
Through an ongoing collaboration and research protocol with the New York Organ Donor Network (NYODN), we are studying human immunity by investigating immune cell subsets in multiple tissue sites obtained from individual organ donors. We previously demonstrated that obtaining blood, lymphoid and mucosal tissues during the time of organ acquisition for life-saving transplantation enables analysis of functional lymphocytes in situ (Sathaliyawala et al., 2013). Importantly, these initial studies revealed the potential to use this tissue resource as a “human model system” to address previously unanswerable questions in human T cell biology about differentiation and lifelong maintenance.
Here, we present a quantitative system-wide analysis of T cell differentiation, homeostasis and persistence in blood, lymphoid and mucosal tissues obtained from a highly diverse cohort of 56 organ donors aged 3–73 years. We incorporate an analysis of naïve, memory and effector T cells with functional markers of homeostasis, activation, and tissue residence along with quantification of in situ turnover and TCR clonal distribution to reveal distinct patterns of compartmentalization, maintenance and proliferation of human T cells. Notably, we show that mucosal sites are predominantly populated with TEM cells that bear features of TRM cells, which are discretely compartmentalized and stably maintained over life in mucosal tissues, while subsets in lymphoid sites undergo age-associated changes. Circulating naïve T cells decrease in lymphoid sites with a corresponding increase in TEM and TEMRA, while TCM cells persist at a steady frequency in circulation and tissues throughout life with a significant fraction continuously turning over. We identify key differences in memory CD4+ and CD8+ T cell homeostasis among lymphoid and mucosal tissues, with memory CD4+ T cell subsets responding to cytokine-mediated signals, and memory CD8+ T cells exhibiting cytokine-driven responses in lymphoid tissue and cognate signals in mucosal sites. Moreover, TCR sequencing of TEM cells in different sites revealed increased circulation and clonality of CD8+ TEM, while CD4+ TEM are primarily resting and compartmentalized. Our results reveal localization-dependent heterogeneity in T cell maintenance and homeostasis, implicating distinct pathways for peripheral differentiation of CD4+ and CD8+ T cell subsets.
RESULTS
Tissue compartmentalization of human T cell subsets is conserved among diverse individuals
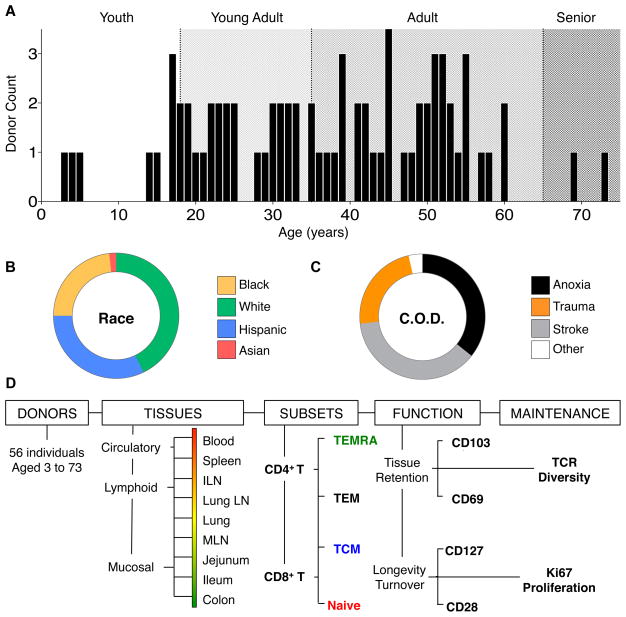
We have established a unique human tissue resource through a collaboration and research protocol with the NYODN, in which multiple lymphoid and mucosal tissues are obtained from research-consented human organ donors (Sathaliyawala et al., 2013). Here, we acquired tissues from donors across all ages of life for a unique assessment of human immune cell compartmentalization and maintenance. The 56 individuals used for this study are highly diverse, ranging in age from 3–73 yrs (median age 37.5, Figure 1A), and representing all major races/ethnicities (White, 43%; Hispanic, 32%; Black, 23%; and Asian, 2%; Figure 1B) and body types (Table S1). All donors were healthy (HIV−, HBV/HCV−, cancer-free, free of systemic infections) prior to brain death due to non-infectious traumas such as cerebrovascular stroke (37%), head trauma (23%), anoxia (36%), or other causes (4%) (Figure 1C and Table S1). The tissues analyzed from each donor sample circulation, lymphoid and mucosal sites and include peripheral blood, spleen, inguinal lymph node (ILN, draining the peripheral skin and muscle), lung-draining lymph node (LLN), lungs, mesenteric lymph node (MLN, draining the intestines), small intestine (jejunum and ileum), and colon.
Figure 1. Characteristics of the human donor population and analysis overview.
Lymphocyte frequency, distribution, function and maintenance was assessed in 9 tissue sites acquired from 56 deceased organ donors, aged 3–73. (A) Age distribution of analyzed donors grouped into four life stages: Youth (0–5 years, white), Young Adult (16–35 years, light gray), Adult (36–65 years, gray), and Senior (66+ years, dark gray). (B) Race distribution of the organ donor population: White (green), Hispanic (blue), Black (yellow), and Asian (red). (C) Cause of death distribution of the organ donor population: Cerebrovascular stroke (gray), Head Trauma (orange), Anoxia (black), other causes (white). Characteristics of each individual donor including age, gender, height, weight, cause of death and serology are detailed in Table S1. (D) Flow chart of different parameters investigated and the analyses performed for this study from 56 donors, with blood and 8 tissues from each donor, analyzing 8 distinct CD4+ and CD8+ T cell subsets in terms of their function, proliferation, TCR diversity and homeostasis.
We obtained viable lymphocyte populations from blood and 8 different lymphoid and mucosal tissue sites (see Experimental Procedures) that enabled unprecedented spatial and temporal phenotypic, functional and molecular analysis (Fig. 1D). The composition of CD4+ and CD8+ T cell subsets (naïve, TCM, TEM and TEMRA) was assessed in nine anatomic sites, and the resultant 72 populations were analyzed for the expression of functional markers of tissue residence, longevity, and turnover. We further investigated memory T cell maintenance and compartmentalization on the clonal level using TCR deep sequencing to quantify CD4+ and CD8+ T cell clonal distribution among multiple lymphoid sites (Fig 1D). We then statistically analyzed all these parameters to quantify age-, and tissue- associated changes (see extended methods). These multi-faceted analyses enabled an assessment of human T cell differentiation and maintenance in the body over life. Donors used for each aspect of this study are indicated in Table S2.
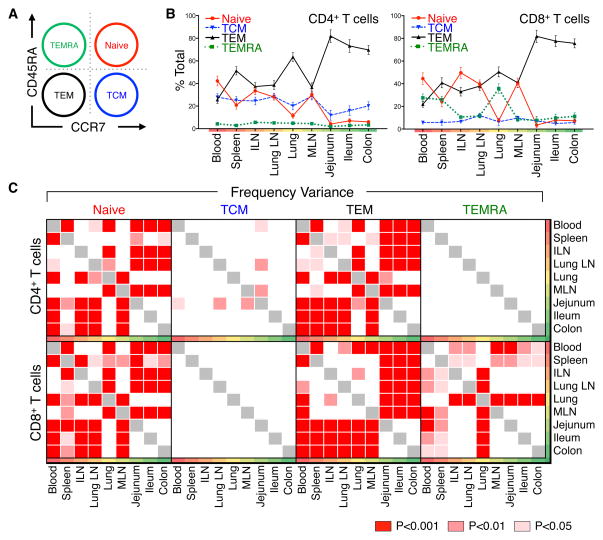
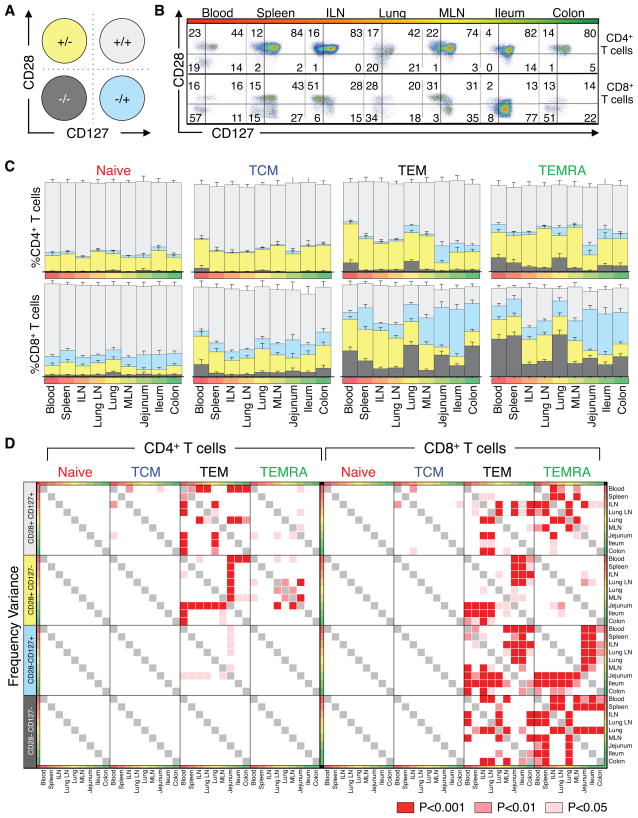
We assessed T cell subset composition in each tissue and variance among tissues and donors by flow cytometric analysis of well-established markers. CD3+CD19−CD4+ and CD8+ T cell subsets were further defined by coordinate expression of the CD45RA and chemokine receptor type 7 (CCR7), delineating naïve (CD45RA+/CCR7+), TCM (CD45RA−/CCR7+), TEM (CD45RA−/CCR7−), and TEMRA cells (CD45RA+/CCR7−) (Figure 2A). A representative staining profile of CD45RA and CCR7 expression by CD4+ and CD8+ T cells (Figure S1) and compiled results from 39 donors (Figure 2B) reveals a distinct pattern of subset composition in circulation (blood and spleen), lymph nodes, and mucosal tissues, that differs between CD4+ and CD8+ T cells. Both CD4+ and CD8+ T cells exhibited a similar distribution of TEM and naïve subsets, with TEM predominating in mucosal tissues and naïve cells only significantly present in blood and lymphoid tissues. However, the frequency and tissue localization of TCM and TEMRA cells differed between CD4+ and CD8+ T populations, with TCM cells more prevalent among CD4+ T cells, comprising 20–30% of total CD4 T cells in blood, lymph nodes and lungs, compared to <10% of CD8+ T cells in these sites (Figure 2B). Conversely, TEMRA cells were predominantly CD8+, and localized primarily to blood, spleen, and lung, with low frequencies (<10%) in lymphoid and mucosal tissues (Figure 2B, right). Remarkably, these tissue- and CD4+/CD8+-specific T cell subset distribution patterns were highly conserved among diverse individuals, as evidenced by the low variance in individual subset frequency at each site (Figure 2B and Table S3).
Figure 2. Tissue Distribution of naïve, memory and terminal effector subsets remains consistent between diverse donors.
Frequency of lymphocytes in 9 tissue sites isolated from organ donors was quantified by flow cytometry. (A) Gating strategy used to identify T cell subsets (CD3+ CD19−CD4+ or CD3+ CD19−CD8+ cells) based on CD45RA and CCR7 expression defining: CD45RA+/CCR7+ (Naïve, red), CD45RA−/CCR7+ (“Central Memory” or TCM, blue), CD45RA−/CCR7− (“effector-memory”, TEM, black), and CD45RA+/CCR7− (“Terminal effector” or TEMRA, green). Colors and abbreviations are used consistently throughout. (B) Mean frequency (±SEM) of each subset expressed as a percent of total CD4+ (left) and CD8+ (right) T cells in circulation, spleen, lymphoid tissue and mucosal sites (ordered from blood, red; to mucosal sites, green). Individual means and additional descriptive statistics are listed in Table S3. (C) Distribution variance between CD4+ and CD8+ T cell subset frequencies in 31 donors (Table S2) was assessed for each tissue by two-way ANOVA and adjusted for multiple comparisons by a Holm-Sidak correction. Significant frequency variances between tissue pairs are expressed as p-values with red- and pink-shaded boxes indicating significant differences (p<0.001, red; p<0.01, light red; p<0.05, pink) and white boxes indicating no significant difference in subset frequency between anatomic sites. Individual p-values are listed in Table S4.
To analyze differential T cell subset compartmentalization among blood, lymphoid and mucosal sites, we quantified the mean subset frequency variance in each tissue (see Methods) (Figure 2C and Table S4). Overall, the distribution of CD4+ and CD8+ naïve and TEM cells and CD8+ TEMRA cells varied significantly among blood, lymphoid and mucosal sites, while the frequency of TCM cells did not (Figure 2C). The distribution variance of CD4+ and CD8+ naïve and TEM cells is low between circulation and lymphoid sites, yet increases significantly between circulation/lymphoid and mucosal sites (Figure 2C). Moreover, the distribution variance of CD8+TEMRA is significant only in blood, spleen and lung compared to mucosal and lymphoid tissue. Together, these results demonstrate that naïve, TEM and CD8+TEMRA cells are compartmentalized in distinct patterns in circulation, lymphoid and mucosal tissues, which are consistent among diverse individuals. By contrast, TCM cells are not compartmentalized among tissues or individuals, suggesting either that they circulate among all sites, and/or represent an intermediate population in T cell differentiation that is maintained through continuous turnover.
Age dependence of T cell subset distribution in lymphoid but not mucosal sites
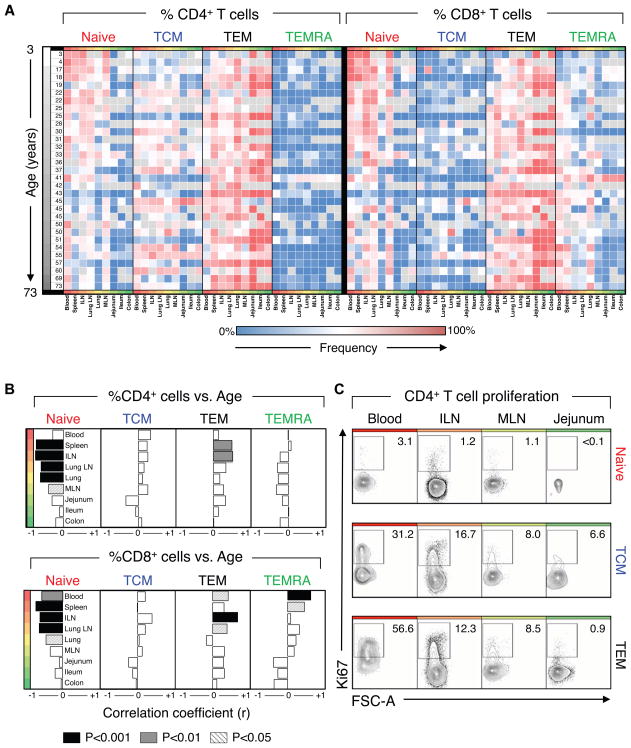
By analyzing donors of ages that span the majority of the human lifespan over six decades, we were able to assess whether T cell subset tissue distribution changes as a function of age. We plotted the relationship between age and subset distribution as a heat map of individual subset frequencies among blood and 8 tissue sites ordered by increasing age to generate a spatial and temporal atlas of human T cell subset composition (Figure 3A). The differential compartmentalization of naïve, TEM and TEMRA subsets is conserved between individuals, with TEM subsets predominating in the mucosa, naïve T cells localized to blood and lymphoid tissue, and CD8+ TEMRA predominant in blood, spleen and lung independent of donor age. (Figure 3A). The low frequency of CD4+ TEMRA and CD8+ TCM subsets is also conserved in all individuals of all ages (Figure 3A).
Figure 3. Temporal dependence of T cell distribution varies by subset and compartment.
Lymphocyte frequency was quantified in 9 tissues from donors aged 3 to 73. (A) Heatmap of CD4+ and CD8+ T cell subset frequency (ranging from 0% (blue) to 100%, (red)) in individual donors from youngest (white) to oldest (black), from blood to lymphoid to mucosal sites. (B) Spearman correlation analysis of changes in lymphocyte tissue distribution with increasing age shows temporal changes in subset distribution in circulating and lymphoid sites. Correlation strength and directionality (r=−1, left to r=+1, right) is shown for each tissue ordered from blood to lymphoid to mucosal tissues. Correlation significance for each tissue is denoted by shading (p<0.05, striped; p<0.01, gray; p<0.001, black). Individual r- and p-values are listed in Table S5.) (C) Ex vivo analysis of proliferation of naïve, TCM, and TEM CD4 T cell subsets in different tissue sites. Ki67 expression by CD4+ naïve (red), TCM (blue), and TEM (black) CD3+ T cell subsets in blood, ILN, MLN, and jejunum from Donor 93. Compiled Ki67 data from multiple donors are in Figure S3B.
To quantitatively probe the age dependence of subset distribution, we calculated the correlation between the subset frequencies in each tissue as a function of donor age. Our analyses revealed significant age-dependent changes in distribution of naïve, TEM and TEMRA subsets in blood and lymphoid tissue while their composition in mucosal tissue remain unchanged (Figure 3B). CD4+ and CD8+ naïve T cell frequency decreased in lymphoid tissues with increasing age, though only the proportion of naïve CD8+ T cells decreased significantly in blood (Figure 3B, right). This depletion of naïve T cells corresponded with a compensatory increase in lymphoid CD4+ TEM in spleen and lymph nodes, CD8+TEM in blood and lymph nodes, and CD8+ TEMRA in blood and spleen (Figure 3B). The TCM subset (i.e., CD4 TCM), did not exhibit significant age-associated changes in any of the tissues (Figure 3B). These results demonstrate that age-associated changes in T cell composition only occurred with certain subsets and in specific sites, with lymph nodes undergoing dynamic changes and mucosal sites remaining stable.
The change in specific subset frequency could be due to either antigen-activation and/or homeostatic turnover. We investigated the contribution of ongoing homeostatic turnover to alterations in subset composition by examining Ki67 expression, upregulated by cycling cells. In blood, lymph nodes and mucosal tissue of a representative donor, differences in Ki67 expression were observed between subsets and tissues: TCM and TEM had a higher proportion of Ki67+ cells compared to naïve T cells in all sites. Among CD4+ memory subsets, the highest frequency of Ki67+ cells was found in blood (30–56%) followed by lymph nodes (8–17%) with few Ki67+ cells in mucosal sites (1–7%) (Figure 3C). Though the overall percentage of Ki67+ cells varied between donors, TCM and TEM subsets consistently had a higher proportion of cycling cells compared to naïve T cells, particulatly among blood TEM cells (Figure S2A). Together, these results indicate that TEM and TCM subsets in lymphoid tissues and blood are dynamically altered throughout life, likely due to their continuous homeostatic turnover, while TEM cells in mucosal sites remains consistent over life with lower levels of in situ turnover. Loss of naïve T cells, by contrast, is not associated with turnover, but with conversion to TEM/TEMRA cells, likely through antigen-mediated activation.
Markers of tissue residence are predominant among TEM cells in tissues throughout life
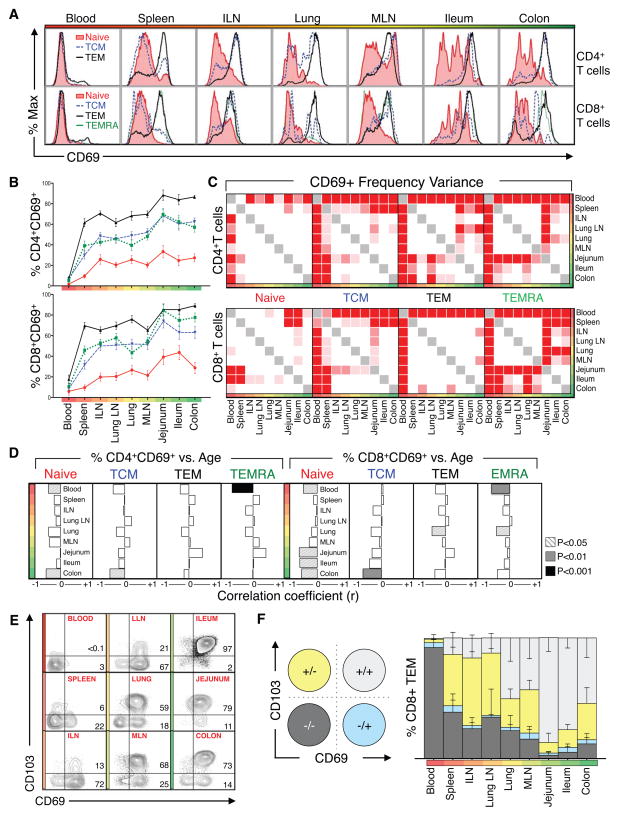
To assess the residence and tissue retention properties of different T cell subsets in each site, we examined tissue-, subset- and age-dependent expression of CD69, an early activation marker for T cells, recently established as a marker for TRM due to its constitutive expression by mucosal and skin TRM in mice and humans (Farber et al., 2014; Mueller et al., 2013; Sathaliyawala et al., 2013). Our analysis from blood and tissues of 29 donors reveals that CD69 expression is predominant among TEM in tissues, is established early, and remains stably maintained through life (Figure 4). T cell subsets in tissues are predominantly CD69-, while in tissues CD69 expression is stratified among different subsets with 60–90% TEM, 30–60% TCM and TEMRA, and <25% of naïve T cells expressing CD69, consistent between diverse individuals (Figure 4B). Moreover, the highest frequency of CD69+ TEM cells is observed in mucosal sites, with lower frequencies in lymphoid tissue (Figure 4B). Variance analyses reveal significant differences in CD69 expression by TCM, TEM and TEMRA subsets between blood and tissues (Figure 4C). Correlation analysis reveals no significant change in CD69 expression by memory subsets in specific sites (Figure 4D). CD69 expression by T cells in tissues thus appears primarily dependent on differentiation, with TEM having the highest propensity for CD69 expression that is enhanced in mucosal sites.
Figure 4. CD69 expression marks T cell compartmentalization in tissue sites.
(A) Representative flow cytometry histograms showing CD69 expression by CD4+ (top) and CD8+ (bottom) naïve (red), TCM (blue), TEM (black), and TEMRA (green) cell subsets from donor #101. (B) Mean frequency (+− SEM) of CD69+ CD4+ and CD8+ T naïve, TCM, TEM, and TEMRA cell subsets among blood, lymphoid and mucosal tissues. Data shown are compiled from 29 donors (Table S2). (C) Distribution variance of CD69+CD4+ (top) and CD8+ (bottom) naïve, TCM, TEM, and TEMRA subsets in blood, lymphoid and mucosal tissues from (B). (D) Spearman correlation analysis of changes in CD69+ subset tissue distribution with increasing age. Correlation strength and directionality (r=−1, left to r=+1, right) is shown for each tissue. Correlation significance for each tissue is denoted by shading (p<0.05, striped; p<0.01, gray; p<0.001, black; not significant, white). Individual r- and p-values are listed in Table S5) (E) Representative flow cytometry plots showing CD103/CD69 expression patterns by CD8+ TEM subsets in circulation, lymphoid, and mucosal tissues. Number in upper and lower right quadrants indicate %CD69+CD103+ and %CD69+CD103− cells, respectively. (F) Overall distribution of TEM subpopulations delineated by CD69 and CD103 expression: CD69+CD103+, light gray; CD69+CD103−, blue; CD69− CD103+, yellow; and CD69− CD103−, dark gray. Graphs shows mean frequencies (±SEM) of these subsets among CD8+ TEM from nine tissues compiled from 10 donors with each tissue representing 5 or more donors. Individual donors used are indicated in Table S2.
As CD69 expression can also connote activation, we examined expression of the αE integrin CD103, another key marker for CD8+ TRM cells (Mueller et al., 2013; Turner et al., 2014a) also known to promote T cell retention in mucosal sites in mice (Schon et al., 1999; Strauch et al., 2001). As shown in representative flow cytometry plots (Figure 4E) and data compiled from multiple donors (Figure 4F), CD103 is co-expressed by CD69+ CD8+ TEM in mucosal tissues (lung, intestines) and intestinal-draining MLN (Figure 4E), indicating a canonical TRM phenotype. In peripheral lymphoid sites, including spleen, ILN and LLN, there is minimal expression of CD103 by CD69+ CD8+ TEM, raising the question of whether CD8+ TEM in lymphoid sites are resident or migratory. Because mouse and human memory CD4 T cells do not appreciably express CD103 (Sathaliyawala et al., 2013; Turner et al., 2014a), the migration and residence of lymphoid CD69+ CD4+ TEM, which undergo turnover and alterations with age, requires further investigation (see below).
Coordinate expression of CD28 and CD127 identifies tissue- and subset-specific T cell homeostasis
To assess potential mechanisms for the distinct dynamics of memory T cells in lymphoid and mucosal sites, we analyzed functional markers of activation/homeostasis including the costimulatory receptor CD28 and the homeostatic cytokine IL-7 receptor CD127 (IL-7R). CD28 expression is downregulated by human T cells in response to TCR-mediated activation, proliferation and replicative senescence (Lo et al., 2011; Vallejo et al., 1999), while IL-7R expression is downregulated following responses to IL-7 (Mazzucchelli and Durum, 2007) and by activated effector T cells (Kiazyk and Fowke, 2008). When examined separately, CD28 and CD127 expression by CD4 and CD8 T cell subsets in blood and tissues is variably expressed by TEM (and CD8 TEMRA subset) in mucosal sites (Figure S3). Coordinate expression of CD28/CD127 delineated four subsets (Figure 5A): CD28+CD127+, resting or unstimulated; CD28+CD127−, receiving IL-7 signals; CD28−CD127+, indicative of previous TCR-driven responses; and CD28−CD127−, a phenotype of antigen-activated effector cells. While naïve and TCM subsets exhibited a predominant resting CD28+/CD127+ phenotype, CD28/CD127 expression by TEM cells (and CD8+ TEMRA cells) varied between CD4+ and CD8+ T cells and lymphoid versus mucosal sites, shown in a representative flow cytometry plot (Figure 5B) and compiled analysis from 23 donors (Figure 5C). Specifically, the majority of CD4+ TEM (50–80%) were CD28+CD127+ in all tissues, with a significant fraction (30–40%) exhibiting a cytokine-responding CD28+CD127− phenotype in lymph nodes, spleen and blood, and low frequency (10–18%) TCR-responding CD28−CD127+ TEM in lung and intestines (Figure 5B,C and Table S3). By contrast, CD8+ TEM and TEMRA cells were predominantly CD28+CD127− in lymphoid tissues and CD28−CD127+ in mucosal sites, with only low frequency (<20–40%) CD28+CD127+ resting populations (Figure 5B, C and Table S3). The prevailing CD28− profile of CD8+ TEM in mucosal sites occurred within the CD69+ TRM fraction (Figure S4A, B), suggesting in situ activation of mucosal resident T cells. Additionally, in blood, spleen and lung, significant proportions (23–26%) of CD8+ TEM and TEMRA cells (and 10% CD4+TEM) were CD28−CD127−, indicative of circulating activated effector cells (Figure 5C and Table S3).
Figure 5. CD28 and CD127 expression reveal distinct homeostatic patterns in lymphoid and mucosal tissues.
Coordinate expression of CD28 and CD127 by CD4+ and CD8+ naïve, TCM, TEM, and TEMRA subsets was analyzed. (A) Diagram showing the 4 subpopulations delineated by CD28/CD127 expression: CD28+ CD127+, light gray; CD28− CD127+, blue; CD28+ CD127−, yellow; and CD28− CD127−, dark gray T cells. (B) Representative flow cytometry plots showing pattern of CD28 and CD127 expression by CD4+ (top) and CD8+ (bottom) TEM cells. (C) Frequency (±SEM) of these functional subsets in (B) within CD4+ (top) and CD8+ (bottom) naïve, TCM, TEM, and TEMRA subsets. Individual frequencies from each of 23 donors are in Table S3. (D) Variance between CD4+ (left) and CD8+ (right) CD127/CD28 subset frequencies within naïve, TCM, TEM, and TEMRA subsets was assessed for each tissue by two-way ANOVA and corrected by Holm-Sidak for multiple comparisons. Significant variation among tissues is denoted by colored squares (p<0.001, red; p<0.01, light red; p<0.05, pink). Individual p-values for each tissue are listed in Table S4.
Variance analyses of population distribution frequencies revealed significant differences in CD28/CD127 co-expression by TEM and TEMRA subsets between blood and lymphoid or blood and mucosal sites, while naïve and TCM populations did not vary in CD28/CD127 expression between diverse sites (Figure 5D). CD4+ TEM differed significantly in CD28+CD127+ and CD28+CD127− cell frequency in blood and lymphoid tissue compared to mucosal sites while CD8+ TEM and TEMRA subsets significantly varied in CD28+CD127+, CD28−CD127+, and CD28−CD127− population distribution among circulation, lymphoid and mucosal sites (Figure 5D), indicating compartmentalized maintenance of TEM cells in specific sites. Naïve and TCM cells showed no tissue-specific expression differences, indicating that their maintenance is not compartmentalized.
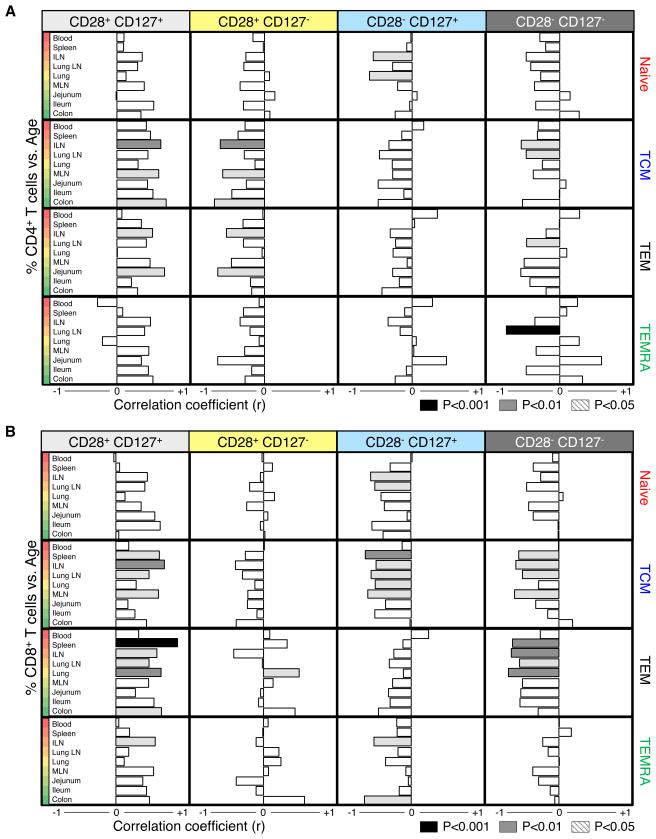
We further quantified the relationship between subset CD28/CD127 co-expression in individual tissues and age through correlation analyses (Figure 6, individual data in Table S5). Significant changes in CD28/CD127 expression occurred primarily in lymphoid tissues, while the CD28/CD127 profile in mucosal sites remained stable over life-- another measure of mucosal TEM stability. Notably, both CD4+ and CD8+ TEM cells (and CD4+ TCM cells) increased in CD28+CD127+ cell frequency with increasing age in lymphoid tissue, particularly in spleen, ILN and LLN, with a corresponding decrease in CD28+CD127− cells (cytokine-responding) for CD4+ T cells, and CD28−CD127+ or CD28− CD127− cells (TCR-stimulated) for CD8+ T cells (Figure 6). This specific population loss and proportional accumulation of resting phenotype CD28+CD127+ cells suggests homeostatic turnover in the depleted (CD127− or CD28−) populations. Increased turnover of the CD4+CD127− compared to CD127+ subset in lymphoid and mucosal tissues is further supported by increased Ki67 expression (Figure S2B). Together, our analyses of functional markers of homeostasis and residence reveal stability in mucosal sites, dynamic changes in lymphoid tissue and different patterns of homeostasis between CD4+ and CD8+ TEM.
Figure 6. Temporal dependence of T cell subset maintenance is site-specific.
Correlation between increasing age and frequency of (A) CD4+ and (B) CD8+ naïve, TCM, TEM, and TEMRA subsets divided by expression of CD127 and CD28 was determined in each tissue. Correlation degree (bar length) and directionality (left, negative; right, positive) are shown for each population and subset: CD28+CD127+ (light gray), CD28+CD127− (yellow), CD28−CD127+ (blue), and CD28−CD127− (dark gray). Significant correlation is denoted by bar shading (p<0.001=black, p<0.01=gray, p<0.05=light gray). Individual r- and p-values for each tissue and subset are shown in Table S5.
TCR sequencing reveals distinct compartmentalization and maintenance of CD4+ and CD8+ TEM
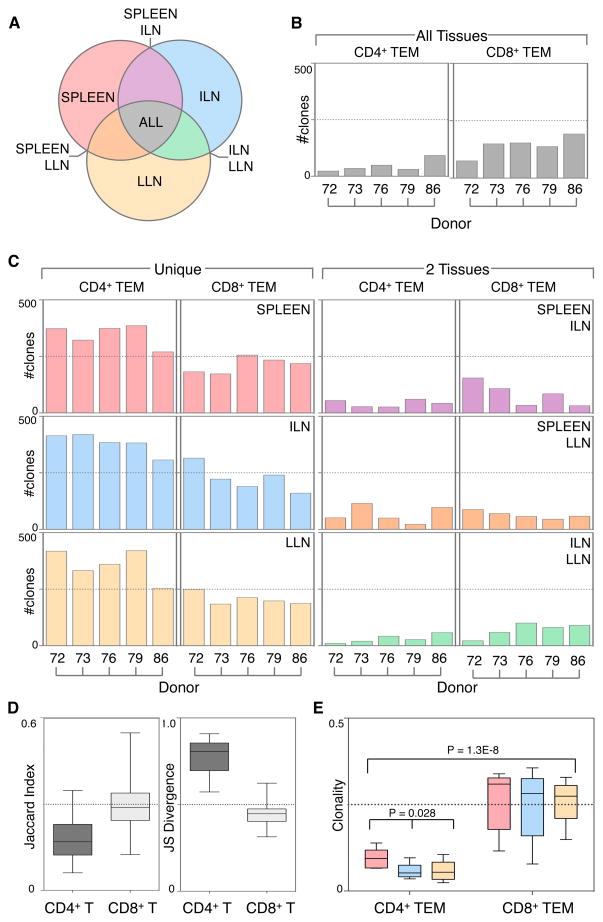
We further investigated the mechanisms for the differential homeostasis and maintenance of CD4+ and CD8+ TEM cells by TCR sequencing to quantify the distribution and expansion of individual T cell clones within and between multiple tissues of an individual. Because TCR sequencing identifies all TCR clones within a site and can be affected by sampling errors, we sorted CD4+ and CD8+ TEM cells from spleen and whole lymph nodes (ILN, LLN) to enable analysis of a full complement of tissue-associated T cell subsets. We compared clonal composition of circulating, lymphoid, and mucosal-draining T cells from five donors with ages spanning four decades (Table S6). TCRβ deep sequencing was performed on comparable numbers of TEM cells from each site (Table S6) using the ImmunoSEQ platform (Robins, 2013; Robins et al., 2009). Analysis of nucleotide sequences enabled assessment of clonal overlap and TCR repertoire diversity between tissue sites and T cell subsets (Figure 7 and extended methods). Of the total clones identified, the proportion shared between sites was different for CD4+ and CD8+ TEM (Figure S5). To eliminate potential sampling issues of rare clones, we assessed the clonal overlap between tissues of the top 500 clones in each site (Figure 7A–C). Compared to CD4+ TEM, CD8+ TEM had more clones common to all three tissues, consistent in all donors (Figure 7B). Conversely, the majority (>80%) of CD4+TEM clones were unique to each site, while <50% of CD8+TEM clones were unique, and few TEM clones were shared by only two sites (Figure 7C, left), indicating no specific bias of clonal distribution between sites (Figure 7B and C).
Figure 7. Differential clonal compartmentalization and TCR diversity between CD4+ and CD8+ TEM cells.
(A) TCR clonal distribution among and between three tissue sites from five individual donors. Venn diagram schematic showing how TCR clones were grouped according to their tissue distribution indicated by colored shading: TCR sequences unique to spleen (pink), ILN (blue), and LLN (yellow); sequences shared by spleen and LLN but not ILN (orange), spleen and ILN but not LLN (purple), ILN and LLN but not spleen (green); sequences shared by all three tissues (spleen, ILN and LLN) indicated by gray shading. (B) Clonal TCR sequence overlap of CD4+ and CD8+ TEM in all 3 tissues (spleen, ILN, and LLN) from 5 donors aged 24–55 (Table S1) is shown for the top 500 TCR sequences. (C) Number of unique TCR clones in each site (left graphs), and clones that were shared specifically between two tissues indicated (right graphs) for CD4+ and CD8+ TEM from five individual donors. TCR sequence data are listed in Table S6. (D) Left: Jaccard index values of CD4+ (dark gray) and CD8+ (light gray) TEM clones from combined spleen, ILN, and LLN are shown for 5 donors to quantify population similarity. Right: Jensen-Shannon (JS) divergence indices quantify population diversity of CD4+ (dark gray) and CD8+ (light gray) TEM from combined spleen, ILN, and LLN of 5 donors. Individual donor and tissue Jaccard indices and Jensen-Shannon indices are shown in Figure S6A. (E) Clonality of TCR sequences is shown for CD4+ and CD8+ TEM from spleen (pink), ILN (blue), and LLN (yellow) among 5 donors. Box plots denote 25th to 75th percentile and whiskers indicate minimum and maximum values.
The extent of repertoire sharing by each subset was determined by computing the Jaccard index of CD4 and CD8 TEM subsets across all sites for clones greater than 0.05% in frequency, and showed a greater overlap among the CD8+ TEM compared to the CD4+ TEM repertoire (Figure 7D, Figure S6). The Jensen-Shannon (JS) divergence metric quantified differences between repertoire distributions, and revealed that the CD4+ TEM repertoire had increased JS divergence indicating increased TCR diversity compared to CD8+ TEM across all donors and tissues (Figure 7D, Figure S6). Together, this clonal overlap, repertoire sharing and distribution analysis shows that CD8+ TEM clones are broadly distributed among tissues while CD4+TEM clones remain more compartmentalized.
We next determined whether the increased sharing of CD8+ TEM clones between tissues was due to increased clonal expansion. The degree of clonality was measured for all TCR clones in each tissue site by normalizing the entropy values of each sample to the number of unique TCR sequences, resulting in a value ranging from 0 (most diverse) to 1 (least diverse). CD8+ TEM had higher clonality in all tissue sites in all donors compared to CD4+ TEM (Figure 7E), indicating increased clonal expansion by CD8+ TEM. Between sites, the clonality of spleen CD4+ TEM was significantly greater than that of both ILN and LLN CD4+ TEM, suggesting that circulating CD4+ TEM are more clonally expanded than those in lymph nodes, while CD8+ TEM clonality did not significantly differ between tissues (Figure 7C). Together these results indicate profound and reproducible tissue-specific distinctions between CD4+ and CD8+ TEM clonal diversity. Notably, we found more clonal expansion, less TCR diversity and a broader clonal distribution of CD8+ compared to CD4+ TEM consistent in diverse donors of different ages.
DISCUSSION
Manipulating human T cell immunity for vaccines and immunotherapies requires a fundamental understanding of the processes driving T cell differentiation and maintenance. Most knowledge of in vivo T cell responses in lymphoid and mucosal tissues derives from studies in mouse models, and translating results to humans has been limited to analysis of peripheral blood. Here, we investigate how human T cells are distributed and maintained in the body using a multi-dimensional, quantitative analysis of T cell subset differentiation, residence, turnover and clonal distribution in blood, lymphoid and mucosal tissues derived from 56 human organ donors spanning over 60 years of life. Our results provide insights into T cell migration, differentiation and maintenance that cannot be extrapolated from studies of T cell subsets in blood. We reveal that the tissue distribution and residence of heterogeneous T cell subsets is contingent on both subset differentiation and tissue-intrinsic factors, and that control of T cell homeostasis depends on tissue localization and CD4+ versus CD8+ lineage. Our elucidation of the human T cell composition in the body and pathways for T cell maintenance over life suggest distinct mechanisms for memory CD4+ and CD8+ T cell differentiation.
Human T cell heterogeneity in blood was initially delineated based on CCR7 expression into two major memory T cell subsets, with CCR7+ TCM cells migrating primarily to lymphoid sites and CCR7− TEM cells freely circulating in blood, peripheral and non-lymphoid tissue sites (Sallusto et al., 2004; Sallusto et al., 1999). These homing properties for human TCM and TEM cells were mainly extrapolated from ex vivo migration studies and limited analysis of tissue isolates (Campbell et al., 1998; Campbell et al., 2001). When the tissue distribution of these subsets is directly examined among lymphoid and mucosal sites within an individual and compared among many individuals as accomplished here, a far more complex picture emerges. We found that CCR7+ TCM cells comprise a similar proportion of CD4+ T cells in blood, spleen, three different lymph nodes, colon and ileum, which did not alter with age, and were present in similar proportions to TEM cells in LN. Moreover, loss of CCR7 expression did not reliably indicate exclusion from lymphoid sites: both TEM and TEMRA cells lack CCR7 expression, yet they differed in tissue distribution. TEM represented a significant fraction (>20–50%) of T cells in lymphoid tissues, while TEMRA cells (exclusively CD8+) were found only in blood, spleen and lung, suggesting a distinct circulatory niche for this subset. Recent studies in mice showing that memory T cell migration is not driven by CCR7 expression (Vander Lugt et al., 2013), and that tissue-homing of CD8+ T cells during allograft rejection occurs independent of chemokine receptor signaling (Walch et al., 2013), further suggest that memory T cell localization is not driven by CCR7 expression. In addition, few CCR7+ TCM cells were found among CD8+ T populations in multiple lymph nodes from over 50 individuals studied, indicating that lymphoid CD8+ T cell memory does not persist as canonical TCM cells.
Additional insights into maintenance and homeostasis of T cell subsets were obtained by examining changes in tissue subset composition within this donor population as a function of age. While it is not possible to directly follow T cell responses in tissues within the same individual over time, the broad and continuous age distribution of our donors, and the consistent tissue T cell subset composition between donors enabled a quantitative analysis of temporal changes in subset composition over decades of life and allowed us to interpolate homeostasis mechanisms and lineage relationships of T cell subsets. We found that lymphoid tissue and blood exhibited the most dynamic changes in subset composition (decreases in naïve, increases in TEM subsets) while mucosal sites maintained stable T cell subset frequencies and phenotypes. Ki67 expression showed significant proliferative turnover of TEM and TCM subsets in lymphoid tissue compared to mucosal sites, while naïve T cells exhibited minimal turnover, consistent with human studies demonstrating higher turnover of TCM and TEM compared to naïve T cells in human blood through D2O incorporation (De Boer and Perelson, 2013; Macallan et al., 2004) and with in vivo mouse studies in mice showing increased turnover of memory T cells in lymphoid compared to peripheral tissues (Wang et al., 2003). Interestingly, the highest spontaneous turnover of T cell subsets was found in blood, suggesting that the most activated and/or cycling subsets leave the tissues and enter circulation. Though the frequency of TCM cells did not alter with age in blood or any tissue, proliferation of this subset suggests that TCM generation is equally counterbalanced by their turnover. Because loss of naïve T cells in lymphoid sites was not associated with turnover, we conclude that loss of naïve T cells over time is due to antigen-driven conversion to short-lived effector cells or long-lived memory T cells.
The acquisition of multiple tissues from individual donors enabled a detailed assessment of T cell tissue residence and stability over life not previously possible through studies of blood or isolated tissue biopsies. In particular, we were able to assess the frequency and stability of TRM cells, recently identified as non-circulating, tissue-resident memory T cells marked by CD69 and CD103 expression (Clark, 2010; Farber et al., 2014; Masopust et al., 2006; Mueller et al., 2013; Sathaliyawala et al., 2013). In mouse models, CD69 expression by TRM mediates retention in tissues through degradation of the sphingosine-1-phosphate receptor (S1PR1) (Bankovich et al., 2010; Shiow et al., 2006; Skon et al., 2013). Here, we show that CD69 expression is subset- and tissue-dependent, with human TEM cells in all tissues exhibiting the highest frequency of CD69+ cells while TEM cells in blood and all naïve T cells were largely CD69− (Figure 4). In individuals at all life stages, the highest proportion of CD69+ TEM cells was found in intestines, suggesting that TRM may be more readily generated and/or stably maintained in mucosal compared to lymphoid sites. The co-expression of CD69 and CD103 by CD8+ TEM in mucosal but not lymphoid tissue further confirms that CD8+ TRM predominate in these sites. Though the precise signals regulating CD69 and CD103 expression by TRM are not known, they likely involve cognate TCR and/or cytokine signals that induce CD69 upregulation (Turner et al., 2014b), and TCR and/or TGF-β signals that trigger CD103 expression (Casey et al., 2012; Wang et al., 2004). Our data further suggest that the signals perceived by TEM in mucosal sites induce molecules controlling long-term retention in situ, and that mucosal TEM/TRM do not readily enter circulation as assessed by an absence of CD69- and CD103- expressing TEM in blood.
Mechanisms for lifelong human T cell maintenance have been difficult to elucidate due to limitations in sampling, although mouse studies demonstrated that homeostatic cytokines such as IL-7 and IL-15 promote memory CD4+ and CD8+ T cell homeostasis (Purton et al., 2007; Surh and Sprent, 2008). Our analysis of CD28/CD127 expression by T cell subsets as functional markers of TCR- or IL-7-mediated signaling, respectively, (Lo et al., 2011; Mazzucchelli and Durum, 2007; Rethi et al., 2005; Warrington et al., 2003) provide evidence for distinct in situ control of human CD4+ and CD8+ T cell maintenance. While naïve and TCM persisted as predominant resting (CD28+CD127+) populations, CD4+ TEM contained significant CD28+CD127− (cytokine-responding) populations in all tissues, while CD8+ TEM had predominant CD28+CD127− populations in lymphoid sites and CD28−CD127+ (TCR-responding) populations in mucosal sites. These cytokine-regulated and TCR-stimulated (CD28−) CD8+ and CD4+ TEM cells, respectively, decreased with age specifically in lymphoid tissue, as another indication of homeostatic turnover occurring in lymphoid sites. The CD28−CD8+ TEM subset in mucosal sites is analogous to CD28− TEM previously identified in specific inflammatory sites (Martens et al., 1997; Schmidt et al., 1996), and its persistence suggests continuous in situ influences on T cell activation state. It is also notable that blood contained the highest proportion of CD4+ and CD8+ TEM exhibiting an activated (CD28−CD127−) profile, providing further evidence that the most activated, cycling T cells enter circulation.
We further investigated the different dynamics of CD4+ and CD8+ TEM homeostasis and tissue compartmentalization by T cell clonal analysis using TCRβ deep sequencing. Direct analysis of the individual TCR clones, as defined by a unique nucleotide sequences in the CDR3 region, enabled us to determine whether specific T cell clones represented in one tissue site were also found in others and whether specific clonal expansion occurred in certain subsets and/or tissues. Moreover, our ability to obtain TEM from pooled whole lymph nodes and large segments of spleen reduced sampling bias. Notably, we found increased overlap of TCR clones among CD8+ TEM in mucosal and peripheral=-draining lymph nodes and spleen compared to CD4+ TEM, consistent among five donors spanning four decades of life (Figure 7). Clonal analysis of TCR sequences further revealed increased clonality of CD8+ compared to CD4+ TEM, consistent with their increased activation and homeostasis based on CD28/CD127 expression, and the increased overlap of CD8+ compared to CD4+ TEM clones between tissue sites. This increased clonality and TCR-driven maintenance of human memory CD8+ T cells may be a result of ongoing responses to chronic viruses regulated by this subset. CD4+ TEM, however, are less clonally expanded and maintain more clonal diversity and compartmentalization, and therefore may be poised to respond to acute infections.
Current models or T cell differentiation based on studies in mice and human peripheral blood posit either progressive differentiation from naïve to memory subsets, and ultimately to terminal effector cells (Restifo and Gattinoni, 2013), or divergent generation of effector and memory subsets as separate lineages (Ahmed et al., 2009; Chang et al., 2007). Our results provide evidence for distinct pathways controlling differentiation and maintenance of human memory CD4+ and CD8+ T cell subsets, with naïve CD4+ T cells undergoing progressive differentiation into TCM and TEM cells (Figure S7A), while naïve CD8+ T cells divergently generate TEMRA and TEM subsets that migrate through lymphoid tissue and ultimately reside in mucosal sites as resident subsets (Figure S7B). TEM in circulation, lymphoid tissue and mucosal sites exhibit distinct properties and differentiation states, and TEM which enter tissues are capable of becoming TRM through upregulation of CD69 expression and other retention molecules. Lymphoid TEM and TCM cells undergo the highest degree of turnover, likely replenishing terminal TRM cells lost to attrition (Figure S7).
The findings obtained from our quantitative assessment of T cell immunity in multiple anatomic sites of a diverse population over six decades of human life, have broad implications for improving immune monitoring, vaccines and immunotherapies. Our study makes important advances in demonstrating how peripheral blood T cells relate to in situ tissue responses, with peripheral blood containing more activated and cycling cells compared to tissues. We also demonstrate that CD8 T cells have distinct differentiation fates and maintenance mechanisms compared to CD4 T cells, which is of great relevance for designing vaccines and monitoring their efficacy. Finally, our analyses provide a dynamic map of T cell composition in healthy tissues over life, and establish a standard from which to define immune health and homeostasis and better understand tissue-specific and immune-associated pathologies.
EXPERIMENTAL PROCEDURES
Acquisition of human tissues
Human tissues were obtained from deceased organ donors at the time of organ acquisition for clinical transplantation through an approved research protocol and MTA with the NYODN. All donors were free of chronic disease and cancer, were Hepatitis B, C, and HIV-negative. Tissues were collected after the donor organs were flushed with cold preservation solution and clinical procurement process was completed. The study does not qualify as “human subjects” research, as confirmed by the Columbia University IRB as tissue samples were obtained from brain-dead (deceased) individuals.
Lymphocyte isolation from human lymphoid and non-lymphoid tissues
Tissue samples were maintained in cold saline and brought to the laboratory within 2–4hrs of organ procurement. Samples were rapidly processed using enzymatic and mechanical digestion resulting in high yields of live lymphocytes, as described (Sathaliyawala et al., 2013) with additional optimizations detailed in extended methods.
Flow Cytometry Analysis
The following flurochrome-conjugated antibodies were used: anti-human CD3 (Brilliant Violet 650, OKT3, BioLegend, San Diego, CA), CD4 (PeCy7, SK3, BioLegend), CD8 (APC-Cy7, SK1, BD Biosciences, San Jose, CA), CD19 (PE Texas Red, SJ25-C1, Life Technologies, Carlsbad, CA), CD28 (PE, CD28.2, eBioscience, San Diego, CA), CD45RA (Brilliant Violet 605, HI100, BioLegend), CD45RO (PerCpFl710, UCHL1, eBioscience), CD69 (Brilliant Violet 421, FN50, BioLegend), CD103 (Alexa Fluor 647, Ber-Act8, Biolegend), CD127 (Brilliant Violet 711, BioLegend), and CCR7 (Alexa Fluor 488, TG8, BioLegend). Stained cells were acquired on a 6-laser LSRII analytical flow cytometry (BD Biosciences, San Jose, CA). Control samples included unstained and single fluorochrome-stained compensation beads (OneComp ebeads, eBioscience) for accurate compensation and subsequent data analysis. Flow cytometry data were analyzed using FlowJo software (Treestar, Ashland, OR).
Cell proliferation analysis
To monitor cell proliferation, cells were stained for surface markers followed by fixation and permeabilization with in specific buffers (eBioscience), and stained with Ki67 (PE, B56, BD Biosciences) and analyzed by flow cytometry.
Statistical analysis and data visualization
Descriptive statistics (percent means, standard deviations, counts) were calculated for each T cell subset and tissue in Microsoft Excel. Frequency variance was determined for each subset and tissue by Holm-Sidak post-hoc multiple comparison following two-way ANOVA to exclude subset-dependent effects in GraphPad PRISM (Graphpad software, Inc., La Jolla, CA). Frequency comparison p-values for each tissue versus each remaining tissue were graphed using Microsoft Excel. Correlation analysis of subset distribution vs. age was calculated in GraphPad PRISM by nonparametric Spearman correlation for non-Gaussian distributions. Resulting two-tailed p-values and r-values were graphed in Microsoft Excel and using GraphPad Prism.
T cell receptor sequencing
TEM-phenotype (CD45RA−CCR7−) CD4+ and CD8+ T cells were sorted from 2–3 whole lymph nodes for ILN and LLN and large pieces (7cm2) of human spleen obtained from five individual donors (donors 72, 73, 76, 79, 86; Tables S1 and S2) using a BD-influx cell sorter (BD Biosciences). Purified populations were sent to Adaptive Biotechnologies (Seattle, WA) for TCRβ deep sequencing using the ImmunoSEQ™ platform (Robins et al., 2009). Cell number and productive reads for each population from each donor are presented in Table S6. Analysis of nucleotide sequences for repertoire overlap, TCR diversity and clonality were calculated using R-based approaches as detailed in the extended methods in supplementary information.
Supplementary Material
Acknowledgments
This work was supported by NIH P01AI06697 awarded to D.L.F. J.J.T is supported by NIH F31AG047003 and a BD Bioscience Immunology Research Grant. These studies were performed in the CCTI Flow Cytometry Core funded in part through an S10 Shared Instrumentation Grant, 1S10RR027050. We wish to gratefully acknowledge the generosity of the donor families and the outstanding efforts of the NYODN transplant coordinators and staff for making this study possible. We also wish to thank Dr. Claire Gordon and Mr. Sam Mickel for assistance with tissue processing, and Drs. Uri Hershberg and Claire Gordon for critical reading of this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
JJT and NY contributed equally to this work. JJT processed donor tissues, performed flow cytometry, collected the data, and wrote the paper; NY performed statistical analyses, visualized the data, created models and wrote the paper. YO, TS, and MK acquired donor tissues and helped with flow cytometry. HL coordinated tissue donation and acquisition. B.G. and Y.S. analyzed the TCR sequencing data and calculated the different parameters for diversity, clonality and normalized entropy. DLF planned experiments, coordinated tissue acquisition and data acquisition/analysis, analyzed data, and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed R, Bevan MJ, Reiner SL, Fearon DT. The precursors of memory: models and controversies. Nat Rev Immunol. 2009;9:662–668. doi: 10.1038/nri2619. [DOI] [PubMed] [Google Scholar]
- Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1 -phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J Biol Chem. 2010;285:22328–22337. doi: 10.1074/jbc.M110.123299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JJ, Bowman EP, Murphy K, Youngman KR, Siani MA, Thompson DA, Wu L, Zlotnik A, Butcher EC. 6-C-kine (SLC), a lymphocyte adhesion-triggering chemokine expressed by high endothelium, is an agonist for the MIP-3beta receptor CCR7. J Cell Biol. 1998;141:1053–1059. doi: 10.1083/jcb.141.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JJ, Murphy KE, Kunkel EJ, Brightling CE, Soler D, Shen Z, Boisvert J, Greenberg HB, Vierra MA, Goodman SB, et al. CCR7 expression and memory T cell diversity in humans. J Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. Antigen-Independent Differentiation and Maintenance of Effector-like Resident Memory T Cells in Tissues. J Immunol. 2012;188:4866–4875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- Clark RA. Skin-resident T cells: the ups and downs of on site immunity. J Invest Dermatol. 2010;130:362–370. doi: 10.1038/jid.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RA, Chong B, Mirchandani N, Brinster NK, Yamanaka K, Dowgiert RK, Kupper TS. The vast majority of CLA+ T cells are resident in normal skin. J Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- De Boer RJ, Perelson AS. Quantifying T lymphocyte turnover. J Theor Biol. 2013;327:45–87. doi: 10.1016/j.jtbi.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusov VV, De Boer RJ. Do most lymphocytes in humans really reside in the gut? Trends Immunol. 2007;28:514–518. doi: 10.1016/j.it.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Kiazyk SA, Fowke KR. Loss of CD127 expression links immune activation and CD4(+) T cell loss in HIV infection. Trends Microbiol. 2008;16:567–573. doi: 10.1016/j.tim.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Lo DJ, Weaver TA, Stempora L, Mehta AK, Ford ML, Larsen CP, Kirk AD. Selective targeting of human alloresponsive CD8+ effector memory T cells based on CD2 expression. Am J Transplant. 2011;11:22–33. doi: 10.1111/j.1600-6143.2010.03317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macallan DC, Wallace D, Zhang Y, De Lara C, Worth AT, Ghattas H, Griffin GE, Beverley PC, Tough DF. Rapid turnover of effector-memory CD4(+) T cells in healthy humans. J Exp Med. 2004;200:255–260. doi: 10.1084/jem.20040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens PB, Goronzy JJ, Schaid D, Weyand CM. Expansion of unusual CD4+ T cells in severe rheumatoid arthritis. Arthritis Rheum. 1997;40:1106–1114. doi: 10.1002/art.1780400615. [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol. 2006;176:2079–2083. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- Purton JF, Tan JT, Rubinstein MP, Kim DM, Sprent J, Surh CD. Antiviral CD4+ memory T cells are IL-15 dependent. J Exp Med. 2007;204:951–961. doi: 10.1084/jem.20061805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo NP, Gattinoni L. Lineage relationship of effector and memory T cells. Curr Opin Immunol. 2013;25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethi B, Fluur C, Atlas A, Krzyzowska M, Mowafi F, Grutzmeier S, De Milito A, Bellocco R, Falk KI, Rajnavolgyi E, et al. Loss of IL-7Ralpha is associated with CD4 T-cell depletion, high interleukin-7 levels and CD28 down-regulation in HIV infected patients. AIDS. 2005;19:2077–2086. doi: 10.1097/01.aids.0000189848.75699.0f. [DOI] [PubMed] [Google Scholar]
- Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions [see comments] Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, Bickham KL, Lerner H, Goldstein M, Sykes M, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saule P, Trauet J, Dutriez V, Lekeux V, Dessaint JP, Labalette M. Accumulation of memory T cells from childhood to old age: central and effector memory cells in CD4(+) versus effector memory and terminally differentiated memory cells in CD8( +) compartment. Mech Ageing Dev. 2006;127:274–281. doi: 10.1016/j.mad.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Goronzy JJ, Weyand CM. CD4+ CD7- CD28- T cells are expanded in rheumatoid arthritis and are characterized by autoreactivity. J Clin Invest. 1996;97:2027–2037. doi: 10.1172/JCI118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, Marsal J, Donohue JP, Her H, Beier DR, et al. Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. J Immunol. 1999;162:6641–6649. [PubMed] [Google Scholar]
- Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. 2013;14:1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch UG, Mueller RC, Li XY, Cernadas M, Higgins JM, Binion DG, Parker CM. Integrin alpha E(CD103)beta 7 mediates adhesion to intestinal microvascular endothelial cell lines via an E-cadherin-independent interaction. J Immunol. 2001;166:3506–3514. doi: 10.4049/jimmunol.166.5.3506. [DOI] [PubMed] [Google Scholar]
- Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, Farber DL. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014a;7:501–510. doi: 10.1038/mi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Gordon CL, Farber DL. Tissue-resident T cells, in situ immunity and transplantation. Immunol Rev. 2014b;258:150–166. doi: 10.1111/imr.12149. [DOI] [PubMed] [Google Scholar]
- Vallejo AN, Brandes JC, Weyand CM, Goronzy JJ. Modulation of CD28 expression: distinct regulatory pathways during activation and replicative senescence. J Immunol. 1999;162:6572–6579. [PubMed] [Google Scholar]
- Vander Lugt B, Tubo NJ, Nizza ST, Boes M, Malissen B, Fuhlbrigge RC, Kupper TS, Campbell JJ. CCR7 plays no appreciable role in trafficking of central memory CD4 T cells to lymph nodes. J Immunol. 2013;191:3119–3127. doi: 10.4049/jimmunol.1200938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch JM, Zeng Q, Li Q, Oberbarnscheidt MH, Hoffman RA, Williams AL, Rothstein DM, Shlomchik WD, Kim JV, Camirand G, et al. Cognate antigen directs CD8+ T cell migration to vascularized transplants. J Clin Invest. 2013;123:2663–2671. doi: 10.1172/JCI66722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yuan R, Feng Y, El-Asady R, Farber DL, Gress RE, Lucas PJ, Hadley GA. Regulation of CD103 expression by CD8+ T cells responding to renal allografts. J Immunol. 2004;172:214–221. doi: 10.4049/jimmunol.172.1.214. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Stepp SE, Brehm MA, Chen HD, Selin LK, Welsh RM. Virus -specific CD8 T cells in peripheral tissues are more resistant to apoptosis than those in lympho id organs. Immunity. 2003;18:631–642. doi: 10.1016/s1074-7613(03)00116-x. [DOI] [PubMed] [Google Scholar]
- Warrington KJ, Vallejo AN, Weyand CM, Goronzy JJ. CD28 loss in senescent CD4+ T cells: reversal by interleukin-12 stimulation. Blood. 2003;101:3543–3549. doi: 10.1182/blood-2002-08-2574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.