Abstract
Aging of immune organs, termed as immunosenescence, is suspected to promote systemic inflammation and age-associated disease. The cause of immunosenescence and how it promotes disease, however, has remained unexplored. We report that the Drosophila fat body, a major immune organ, undergoes immunosenescence and mounts strong systemic inflammation that leads to de-regulation of immune deficiency (IMD) signaling in the midgut of old animals. Inflamed old fat bodies secrete circulating peptidoglycan recognition proteins that repress IMD activity in the midgut, thereby promoting gut hyperplasia. Further, fat body immunosenecence is caused by age-associated lamin-B reduction specifically in fat body cells, which then contributes to heterochromatin loss and de-repression of genes involved in immune responses. As lamin-associated heterochromatin domains are enriched for genes involved in immune response in both Drosophila and mammalian cells, our findings may provide insights into the cause and consequence of immunosenescence during aging.
Introduction
Much progress has been made in understanding the biology of aging, yet the mechanism that underlies many age-associated human diseases, such as cancer, remains poorly understood. While the majority of aging research focuses on cell/tissue autonomous mechanisms, chronic systemic inflammation in elderly humans, as revealed by elevated circulating pro-inflammatory cytokines, including interleukin 6 (IL-6), IL-1, and tumor necrosis factor α (TNFα), suggests that deterioration of one tissue could lead to disease manifestation in another (Ahmad et al., 2009; Fulop et al., 2011; Khatami, 2009). Indeed aging of immune organs, immunosenescence, is suspected to cause systemic inflammation and age-associated diseases (Day, 2010; Franceschi et al., 2000). However, the cell and molecular mechanisms that underlie the cause of immunosenescence upon aging and how immunosenescence leads to diseases are not understood.
It is known that circulating inflammatory cytokines can lead to NF-κB activation in cells, but such activation may either promote or limit tumorigenesis, depending on specific tumors and their tissue environment (Chen and Castranova, 2007; Grivennikov et al., 2010; Perkins, 2004). Therefore, to define the mechanism by which aging promotes immunosenescence and disease, it is critical to identify which aging immune organs promote systemic inflammation and how the circulating inflammatory cytokines affect the homeostasis of other tissues in the body. However, due to the complexity of organs, immune systems, and cell types, it is very challenging to decipher the cause and consequence of immunosenescence using vertebrates as model organisms.
Fortunately, simpler organisms such as Drosophila exhibit similar age-associated increase in tissue dysfunction and inflammation as those observed in mammals. Old Drosophila intestines exhibit increased proliferation and inappropriate differentiation of the intestinal stem cells (ISCs), which leads to gut hyperplasia, leakage, and animal death (Biteau et al., 2008; Biteau et al., 2010; Rera et al., 2012). Additionally, aging Drosophila up-regulates its innate immune response without apparent infection (Landis et al., 2004; Pletcher et al., 2002; Ramsden et al., 2008; Seroude et al., 2002; Zerofsky et al., 2005), but exactly which tissue(s) exhibits inflammation is unknown. Although Drosophila lacks an adaptive immunity, it has two main innate immune response pathways, called Toll and IMD (De Gregorio et al., 2002; Hoffmann and Reichhart, 2002), which are equivalent to the Toll-like receptor (TLR) and TNFα signaling pathways in mammals (Goto et al., 2008; Tanji and Ip, 2005), respectively. Upon microbial infection, both Toll and IMD pathways are strongly activated in the fat body, a major immune organ in Drosophila that is equivalent to mammalian fat and liver. This activation leads to enhanced secretion of antimicrobial peptides (AMPs) and PGRPs into the hemolymph (De Gregorio et al., 2002; Hoffmann and Reichhart, 2002). The circulating hemolymph (equivalent to human blood) in turn generates a systemic inflammatory response to aid in the killing of invading microbes. Therefore, the parallels of age-associated increase of immune response and tissue dysfunction make Drosophila an ideal model to investigate the cause and consequence of immunosenescence and systemic inflammation.
The anti-microbial function of several secreted PGRPs, such as PGRP-SCs and PGRP-LB, is based on their amidase activities that degrade microbial peptidoglycans. The reduction of peptidoglycans not only limits microbial growth but also decreases the amount of bacterial surface ligands that can activate the membrane associated pattern recognition receptors such as PGRP-LC. Since PGRP-LC activation leads to up-regulation of IMD pathway and inflammation, these secreted PGRPs have been recognized as both positive antimicrobial effectors and negative regulators of IMD signaling pathway (Bischoff et al., 2006; Zaidman-Remy et al., 2006).
Interestingly, previous studies have shown that the local intestinal IMD activity plays a critical role in maintaining gut homeostasis in Drosophila. Gut IMD activity functions to inhibit excessive ISC proliferation and maintains gut homeostasis (Buchon et al., 2009b; Wang et al., 2013). Since the local intestinal IMD activity is critical for gut homeostasis (Arthur et al., 2012; Buchon et al., 2009a; Buchon et al., 2009b; Ray, 2012; Wang et al., 2013), we reasoned that increased systemic inflammation in old flies could alter the local IMD activity in the gut, thereby leading to the age-associated gut hyperplasia. By analyzing age-associated changes in Drosophila fat body and midgut, we demonstrate that lamin-B loss in fat body cells leads to chronic systemic inflammation. The fat body-secreted PGRPs contribute to the down-regulation of intestinal IMD activity and hyperplasia. Our findings have important implications in deciphering immunosenescence and its role in systemic inflammation and age-associated diseases in mammals.
Results
Age-associated up-regulation of systemic immune response by fat body may repress IMD signaling in the midgut
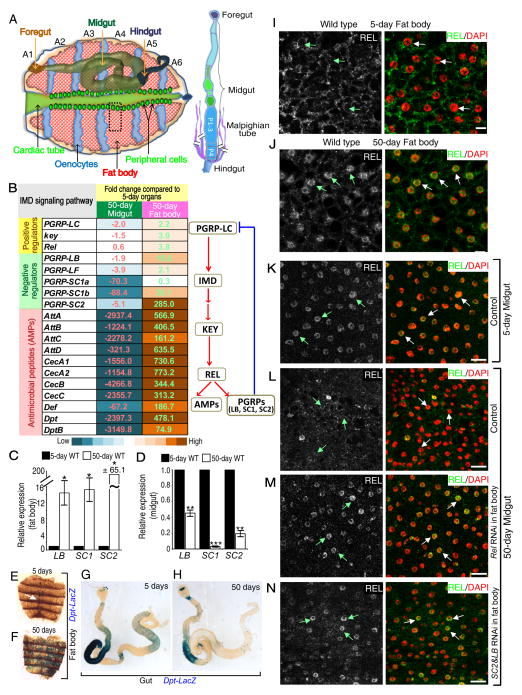
To understand whether systemic inflammation affects local tissue immune response, we performed RNA-sequencing (RNA-seq) on dissected fat bodies and midguts (Figure 1A) from wild-type 5-day and 50-day old flies. Compared to young flies, the expression of more than 2000 genes was altered by ≥2-fold in the old organs (Tables S1, S2). Many genes involved in immune response were strongly up regulated in old fat bodies, but these genes were significant down-regulation in aged midguts (Figure 1B, S1A–B, and Table S3). Thus, old Drosophila may exhibit systemic inflammation, while the midgut local immune response becomes repressed. We confirmed that both IMD regulators (PGRP-LC, receptor for IMD signaling; kenny, KEY, equivalent to mammalian IKKγ; and Relish, REL, equivalent to mammalian NFκB) and IMD effectors were up-regulated in aged fat bodies (Figure 1C, 1E–F, S1C–D). However, only the IMD downstream effectors were consistently down-regulated in old midguts (Figure 1D, 1G–H, S1E–F). The up- and down-regulation of the IMD signaling in aged fat body and midgut were further demonstrated by the increased or decreased nuclear REL accumulation, respectively, in the two organs compared to that of the young (Figures 1I–L).
Figure 1. Age-associated inflammation in fat body may repress IMD signaling in the midgut.
A. Adult Drosophila dorsal abdomen. Adult fat body cells (red) align the inner cuticle surface of abdominal segments (A1-6). The oenocytes (blue), cardiac tube with its peripheral cells (green), foregut, midgut, and hindgut are also shown. Gut with the indicated midgut segments (posterior P1-3 and P4) and Malpighian tubules (MT) attached to the posterior end of P4 are to the right. All immunofluorescence images of fat body and midgut in this manuscript are from A4 (black dashed box) and P4 (red box), respectively.
B. Selected up- or down-regulated genes involved in the IMD pathway upon aging in fat body or midgut, respectively. Numbers and color-codes indicate fold changes. A simplified IMD signaling pathway is shown to the right.
C–D. qRT-PCR analyses of PGRPs, LB, SC1, and SC2, in fat body (C) and midgut (D) from young and old wild-type (WT) flies. Expression in old organs was plotted relative to the young. Error bars, Standard error of the mean (SEM) from three independent experiments. Student’s t-tests, *p<0.05, **p<0.01, ***p<0.001.
E–H. Dpt-lacZ reporter assay of fat bodies (E, F) or midguts (G, H) from young (E, G) or old flies (F, H). Dpt-lacZ was only in peripheral cells in all (15) young fat bodies (white arrow in E), but was throughout 11 of 17 old fat bodies (F). 12 of 15 old midguts (H) had reduced Dpt-lacZ compared to that of the young (G, 12 analyzed).
I–J. REL (green or white) is mostly cytoplasmic in young (I) or nuclear (DAPI, red) in old (J) fat body cells.
K–N. Young control midguts (K) had nuclear REL, which was reduced by 50 days (L). Depletion of REL (M) or co-depletion of PGRP-SC2 plus PGRP-LB (N) by RNAi in fat bodies restored nuclear REL staining in old midguts.
Scale bars, 20 μm.
To test whether the de-repressed PGRPs in fat body could lead to IMD down-regulation in the aged midgut, we used different tissue-specific Gal4 lines to drive the repression of PGRPs. Depletion of REL or co-depletion of PGRP-LB plus PGRP-SC2 in fat bodies using Cg-Gal4 or r4-Gal4 (Figure S1G, S7D–I), but not in adult hemocytes using Hml.Δ-Gal4 or Pxn-Gal4 (Figure S1G and S7M–O), caused midgut IMD up-regulation as revealed by the nuclear accumulation of REL (compare Figure 1M–N to 1L) and Dpt up-regulation in the 50-day old midguts (Figure S1H–J). Thus systemic inflammation of the old fat body can inhibit the midgut IMD activity via PGRPs.
Systemic inflammation caused by old fat body leads to intestinal hyperplasia
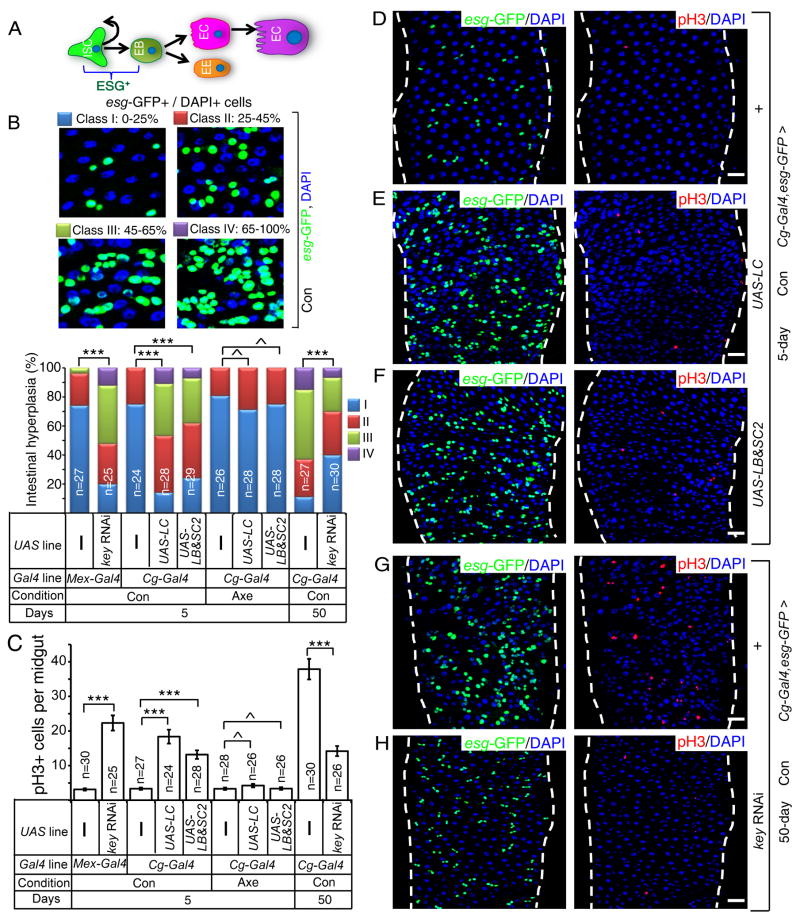
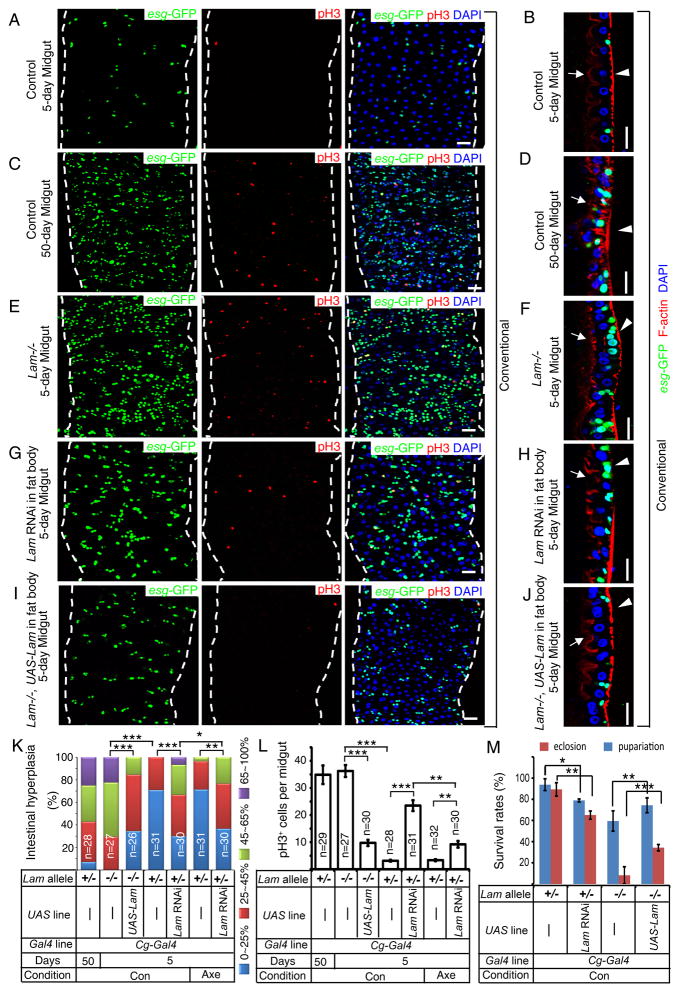
Previous studies show that age-associated midgut hyperplasia is caused by increased ISC proliferation and inappropriate differentiation as determined by the increased number of esg-GFP (escargot-GFP)+ cells that are ISCs, EBs, or mis-differentiated EBs (Biteau et al., 2008). Although inflammation is suspected as one of the culprits for hyperplasia in Drosophila midgut and human gastrointestinal system, whether it is the systemic or local inflammatory pathways that contribute to the disruption of tissue homeostasis remains unclear in either organism. Interestingly, reduction of midgut IMD activity has been shown to cause midgut hyperplasia (Buchon et al., 2009b; Wang et al., 2013). However, a recent study reports that it is the activation of gut IMD that leads to its hyperplasia in old flies (Guo et al., 2014a). Since the gut contains multiple subregions with distinct gene expression (Figure 1A) (Buchon et al., 2013; Marianes and Spradling, 2013), the discrepancy in these studies could be caused by not comparing proper regions of the gut. To address this discrepancy, we compared the equivalent whole midgut, P1-4 region or P4 region (for microscopy) of the midgut (Figure 1A). Midgut-specific Mex-Gal4 (Phillips and Thomas, 2006; Sieber and Thummel, 2009) and NP1-Gal4 (Buchon et al., 2009b; Jiang and Edgar, 2009) (also see Figure S1G and S7J–L) were used to deplete KEY by RNAi. The degree of midgut hyperplasia was determined using esg-GFP reporter (esgCB02017-GFP). Midguts were further labeled by an antibody to phosphorylated histone H3 (pH3) to mark mitotic ISCs (Figure S2A–B) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Intestinal hyperplasia was calculated as the percentage of esg-GFP+ cells of total DAPI+ nuclei in P1-4 regions (Figure 2B), whereas ISC proliferation was determined by counting the number of pH3+ cells in the whole midgut. Consistent with the two studies (Buchon et al., 2009b; Wang et al., 2013), we found that midgut IMD repression resulted in its hyperplasia (Figure 2B–C, S2A–B).
Figure 2. Systemic inflammation caused by old fat body results in midgut hyperplasia.
A. An intestinal stem cell (ISC) produces a new ISC and an enteroblast (EB). EBs differentiate into enteroendocrine cells (EE) or premature enterocytes (red). ECs further differentiate to mature polyploid ECs (purple). Both ISCs and EBs express ESCARGOT (ESG+).
B. Midgut hyperplasia based on esg-GFP in P1-4 region. The number of esg-GFP+ cells counted was divided by total cells counted based on DAPI staining. The percent of esg-GFP+ cell (% of intestinal hyperplasia) was grouped into four classes and plotted (bottom). ^p>0.05, **p<0.01, ***p<0.001, Wilcoxon two-sample test.
C. Midgut hyperplasia based on pH3 staining in the whole midgut. The average pH3+ cells/midgut was plotted. Error bars, SEM. ^p>0.05, ***p<0.001, Student’s t-tests.
D–H. Expression of PGRP-LC (LC, E) or PGRP-SC2 plus PGRP-LB (SC2&LB, F) in young fat bodies increased midgut esg-GFP+ (green) and pH3+ (red) cells compared to that of the control (D). Depletion of KEY in the fat body by RNAi (H) reduced the number of midgut esg-GFP+ and pH3+ cells in old midguts compared to that of control (G). DAPI (blue), nuclei. Scale bars, 20 μm.
Con and Axe, conventional and axenic conditions. n, numbers of midguts analyzed.
See also Figure S2.
To determine whether fat body IMD activation could lead to midgut IMD repression and hyperplasia, we overexpressed PGRP-LC (receptor for IMD) or co-overexpressed PGRP-LB plus PGRP-SC2 in the fat body of 5-day flies using Cg-Gal4 or r4-Gal4. A significant inhibition of Dpt expression in midguts was observed (Figure S2C), indicating midgut IMD repression. Conversely, reduction of IMD activity in old fat bodies (50 days) by depleting KEY (Cg-Gal4-driven RNAi), but not in hemocytes (Hml.Δ-Gal4-driven RNAi), resulted in a significant increase of Dpt expression in old midguts (Figure S2D–E). Therefore, increased IMD signaling or increased expression of secreted PGRPs in fat bodies due to natural aging or via genetic manipulations inhibits midgut IMD activity.
By comparing flies cultured under axenic (Axe, germ free) and conventional (Con, germ) conditions, we found that Midguts from axenic wild-type flies expressed significantly less Dpt than that of conventional ones as expected (Figure S2F). Expression of PGRP-LC or PGRP-SC2 plus PGRP-LB in adult fat body under axenic conditions resulted in higher midgut IMD activity than that of conventional ones (Figure S2F). However, the midgut IMD activity was lower in axenic flies with fat body expression of PGRP-LC or PGRP-SC2 plus PGRP-LB than those of axenic controls (Figure S2F). Under conventional conditions, the fat body-specific overexpression of PGRP-LC or co-overexpression of PGRP-LB plus PGRP-SC2 in young flies resulted in midgut hyperplasia (Figure 2B–F and Figure S2G–H), whereas depletion of KEY in fat bodies (but not in hemocytes) of old flies, significantly reduced the age-associated midgut hyperplasia (Figure 2B–C, 2G–H and Figure S2 G–H). Under axenic conditions, however, expression of PGRP-LC or PGRP-SC2 plus PGRP-LB in young adult fat body failed to induce midgut hyperplasia (Figure 2B–C and Figure S2I–J). Thus the fat body IMD activation represses the midgut IMD signaling in the bacteria dependent and independent manner, whereas the fat body IMD-induced midgut hyperplasia appears to be solely dependent on bacteria.
Age-associated lamin-B loss in fat body cells correlates with enhanced IMD signaling
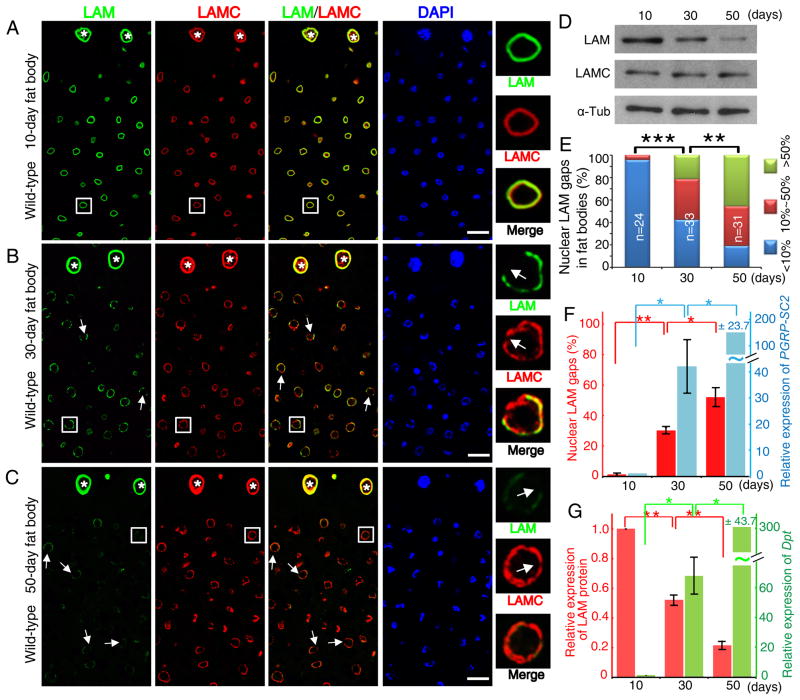
Senescence of mammalian fibroblasts is associated with lamin-B1 loss, increased secretion of inflammatory factors (also called senescence associated secretory phenotype, SASP), but it remains unknown whether lamin-B1 loss in vitro (Barascu et al., 2012; Dreesen et al., 2013; Freund et al., 2012; Shimi et al., 2011) or upon aging in vivo leads to SASP. Interestingly an immune effect has been observed in lamin mutant flies (Markovic et al., 2009). Since GO analyses of lamin-associated chromatin domains (LADs) in Drosophila Kc cells and four different mammalian cell types with mapped LADs showed a significant enrichment of genes involved in defense response to bacteria (Figure S3A) and since lamins can participate in gene repression (Kind and van Steensel, 2010), these nuclear proteins could limit the expression of inflammatory genes. Using antibodies to the two Drosophila lamins, LAM (B-type, lamin-B) and LAMC (A-type, lamin-C), we found similar LAM and LAMC staining in the gut (Figure S3B–C), the peripheral cells of the heart tube (white asterisks in Figure 3A–C), oenocytes and cardiomycytes (not shown) in young and old flies. However, LAM, but not LAMC, exhibited a gradual loss in the fat body cells (Figure 3A–D), which was accompanied by an increase in the percentage of nuclei containing LAM and LAMC staining gaps along the nuclear periphery (Figure 3A–C, 3E).
Figure 3. LAM loss in old fat body accompanies fat body inflammation.
A–C. Age-associated LAM reduction and appearance of nuclear LAM/LAMC gaps. Images (LAM, green; LAMC red; DAPI, blue) of a section of fat body next to the heart tube (see Figure 1A) from wild-type 10- (A), 30- (B), and 50-day (C) old flies. White asterisks mark the heart peripheral cells. Nuclei outlined by white squares were enlarged to the right. White arrows indicate LAM and LAMC gaps. Scale bars, 20 μm.
D. LAM and LAMC Western blotting analyses of 10-, 30-, and 50-day abdominal fat bodies. α-Tub, α-tubulin as loading control.
E. Quantification of LAM gaps. Nuclei with one or more LAM gaps were counted from confocal images from the fat body area in Figure 1A of 10-, 30-, and 50-day old flies. The percentages of nuclei with gap(s) per fat body were grouped and plotted. n, numbers of flies analyzed. **p<0.01, ***p<0.001, Wilcoxon two-sample test.
F–G. The increase of nuclei with LAM gaps (F, left) or decrease of LAM protein (G, left) upon aging is accompanied by the increase of PGRP-SC2 (F, right) and Dpt (G, right) expression in fat bodies. The percentage of nuclear LAM gap was calculated by dividing the number of nuclei with one or more gaps by the total nuclei analyzed. LAM amount and PGRP-SC2, Dpt mRNAs measured from Western blots or qRT-PCR, respectively, were plotted relative to 10-day old fat bodies. Error bars, SEM, from three independent experiments. *p<0.05, **p< 0.01, Student’s t-tests.
All flies were raised in conventional condition.
See also Figure S3.
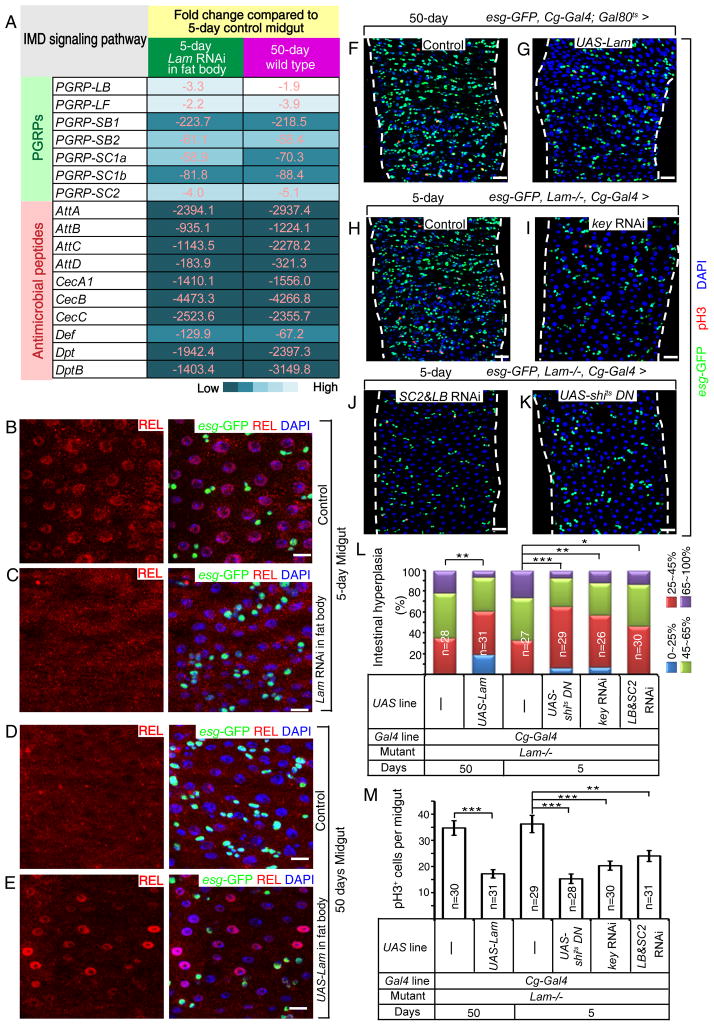
Both our RNA-seq and RT-qPCR analyses revealed no changes of LAM or LAMC transcription (Figure S3D) upon fat body aging, indicating a post transcriptional LAM reduction during aging. By quantifying LAM gaps or LAM protein levels (based on Western blotting analyses) together with mRNA levels of PRGP-SC2 or Dpt, we found that the age-associated LAM loss in fat body correlated with increased IMD signaling (Figure 3F–G). Therefore fat body-specific LAM loss upon aging could lead to de-repression of immune responsive genes in this organ.
Lamin-B represses immune response in fat bodies
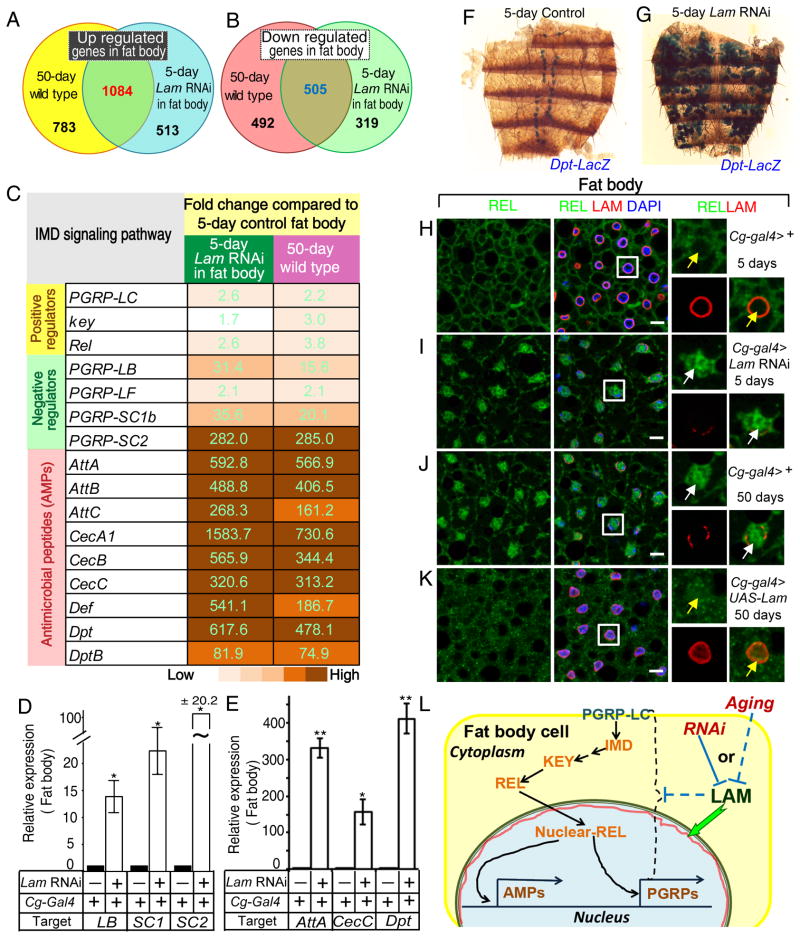
RNA-seq of young (5 day) fat bodies dissected from either control flies (Cg-Gal4/+;tub-Gal80ts/+) or flies depleted of Lam in adult fat bodies (Cg-Gal4/+;tub-Gal80ts/Lam RNAi, Figure S4A) revealed a large number of genes whose expression was altered by ≥2-fold upon Lam depletion (Tables S4). This altered gene set overlapped with those observed in the fat body upon aging (Figure 4A–B). Importantly, most (86.4%) of the up-regulated genes involved in immune response in old fat bodies were also up-regulated to similar degrees in the young fat bodies upon Lam depletion (Figure 4C, Table S5). We further verified that the increased IMD activation upon LAM depletion in young fat bodies (Figure 4D–I, S4B–E) to be similar to those observed in old fat bodies (see Figure 1, S1). Conversely, induced expression of LAM in aging flies (Cg-Gal4/+;tub-Gal80ts/UAS-Lam) significantly reduced the nuclear accumulation of REL in 50-day old fat body cells compared to controls (Figure 4J–K). Induced expression of LAM in adult fat bodies, but not in hemocytes, also resulted in the reduction of Dpt expression in aging fat bodies (Figure S4F–H). Thus lamin-B loss in aging fat bodies leads to elevated fat body IMD signaling (Figure 4L).
Figure 4. LAM depletion in fat body causes its inflammation.
A–B. Graphs of the overlap of up- (A) or down-regulated (B) genes between old wild-type fat bodies and young fat bodies depleted of LAM.
C. A list of selected up-regulated genes of IMD pathway from the old wild-type fat bodies or the young fat bodies depleted of LAM. Numbers and colors indicate similar fold increases.
D–E. qRT-PCR of selected PGRPs (D) and AMPs (E) in control or Lam depleted young fat bodies. The increase of expression upon Lam RNAi was plotted relative to controls. Error bars, SEM from three independent experiments. Student’s t-tests, *p<0.05, **p<0.01.
F–G. Lam RNAi in fat bodies causes increased expression of Dpt-lacZ (F, a representative of 13 out of 15 fat bodies) compared to the control (G, 12 fat bodies analyzed).
H–K. Increased nuclear REL localization in old fat body (J, white arrows) compared to young (H, yellow arrows) was reduced by forced LAM expression in old fat bodies (K, yellow arrows) or mimicked by LAM depletion in young fat bodies (I, white arrows). REL, green; LAM, red; DAPI, blue. Nuclei boxed by white squares were enlarged to the right. Scale bars, 10 μm.
L. LAM loss upon aging or by RNAi causes up regulation of genes involved in the IMD signaling in fat bodies. Red line, nuclear LAM.
All flies were raised in conventional conditions.
Lamin-B in fat body represses intestinal hyperplasia and promotes survival
Since enhanced IMD signaling in fat body induces hyperplasia in midgut in aging flies, we asked whether forced LAM depletion in fat body would disrupt gut homeostasis and reduce animal survival. A previous report showed increased cell proliferation in the gut of Lam mutant (null) pupae (Osouda et al., 2005). Since ~8% Lam mutant pupae survive to adulthood (Chen et al., 2013), we examined midguts of 5-day old wild-type and Lam-mutant flies. Severe midgut hyperplasia was indeed present in the adult Lam mutant flies as judged by numerous pH3+ and esg-GFP+ cells (Figure 5A–B, 5E–F, 5K–L). This gut phenotype is reminiscent of that of 50-day old wild-type flies (Figure 5C–D, 5K–L).
Figure 5. LAM loss in fat bodies causes midgut hyperplasia and reduces survival.
A–J. Compared to control young midguts (A, B), midguts from young Lam mutant flies (E, F), flies with fat body-specific Lam depletion (G, H), or old control flies (C, D) exhibited hyperplasia. esg-GFP, green; pH3, red. Fat body-specific LAM expression in young Lam mutant flies significantly reduced hyperplasia (I, J). Images show the face on view (A, C, E, G and I) or cross-section view (B, D, F, H and J, revealing the basally localized esg-GFP+ ISCs and inappropriately differentiated EBs) of midguts. Arrows and arrowheads indicate the luminal and muscular surfaces, respectively, as revealed by phalloidin staining of actin (red). DAPI (blue), nuclei. All flies were kept in conventional condition. Scale bars, 20 μm.
K. Midgut hyperplasia based on esg-GFP. The percent of esg-GFP+ cells (% intestinal hyperplasia) was grouped into four classes and plotted. *p<0.05, **p<0.01, ***p<0.001, Wilcoxon two-sample test.
L. Midgut cell proliferation based on pH3 staining. The average number of pH3+ cells was counted from the whole midgut and plotted. Error bars, SEM. Student’s t-tests, **p<0.01, ***p<0.001.
M. Survival rate of Lam-mutant and flies depleted of LAM in fat body. Error bars, SEM based on three independent experiments performed with 200 larvae per experiment. Student’s t-tests, **p<0.05, **p<0.01, ***p<0.001.
Con or Axe, conventional or axenic conditions. n, numbers of midguts analyzed.
See also Figure S5.
Next, we depleted LAM by RNAi in midgut, fat body, or hemocytes using respective Gal4 lines (see Figure S1G). Only depleting LAM in the fat body resulted in midgut hyperplasia (Figures 5G–H, 5K–L, S5A–B). To further confirm that lamin-B functions in fat bodies to prevent intestinal hyperplasia, we expressed LAM only in the fat body of Lam mutant animals using Cg-Gal4-driven UAS-Lam cDNA. This fat body-specific LAM expression not only significantly reduced intestinal hyperplasia (Figure 5I–L) but also increased the eclosion rate of Lam mutant flies from ~8% to ~40% (Figure 5M). Thus lamin-B functions in the fat body to maintain intestine homeostasis and promote animal survival to adulthood.
To determine the role of bacteria in the intestinal hyperplasia induced by LAM depletion in fat body, we raised flies in axenic condition and found a weaker intestinal hyperplasia in both Lam null flies and flies with fat body LAM depletion compared to those raised in conventional condition (Figure 5K–L, S5C–F). However, when compared to axenic control flies, the axenic Lam null flies and flies with fat body LAM depletion still exhibited higher gut hyperplasia (Figure 5K–L, S5C–F). Moreover, axenic culturing enhanced the survival of the Lam null flies as measured by the increase of eclosion rate from ~8% to ~30% (Figure S5G). Thus lamin-B functions in the fat body to maintain intestinal homeostasis and to promote animal survival through both bacteria dependent and independent pathways.
Lamin-B maintains midgut IMD signaling and homeostasis by inhibiting systemic inflammation in fat bodies
RNA-seq in young (5 day) midguts dissected from control flies or flies depleted of LAM in the fat body revealed that LAM depletion in fat bodies resulted the change of expression of a large number of genes (≥2 fold) in the midgut (Table S6). This set of genes exhibited a striking overlap to those found in the old midgut (Figure S6A–B). Importantly, among the shared repressed genes, many have known functions in immune response (Table S7), including those involved in IMD signaling (Figure 6A, see the similar degree of repression). Further qRT-PCR analyses of selected downstream targets of IMD signaling confirmed that LAM depletion in fat bodies leads to repression of midgut IMD signaling (Figure S6C–D).
Figure 6. LAM maintains gut homeostasis by inhibiting systemic inflammation caused by the fat body.
A. A list of selected down-regulated IMD pathway genes in midguts from old wild-type flies or young flies raised in conventional condition with fat body-specific LAM depletion. Numbers and color shades indicate similar fold decrease.
B–E. In conventional condition, age-associated repression of IMD signaling in midgut (D) was reversed by fat body-specific expression of LAM in old flies (E) to levels similar to those of control young flies (B) or mimicked by fat body-specific Lam RNAi in young flies (C). REL, red; esg-GFP, green. DAPI (blue), nuclei. Scale bars, 10 μm.
F–K. Midgut hyperplasia in old flies (F) was inhibited by fat body-specific expression of LAM (G). Midgut hyperplasia in young Lam-mutant flies (H) was inhibited by fat body-specific RNAi of KEY (I), co-depletion of PGRP-SC2 & PGRP-LB (SC2 & LB, J), or fat body-specific expression of a dominant negative form of dynamin (shits) (K) in young Lam-mutant flies. Flies were raised in conventional condition. esg-GFP, green; pH3, red; DAPI, blue. Scale bars, 20 μm.
L. Midgut hyperplasia based on esg-GFP. The % of esg-GFP+ cells (% of intestinal hyperplasia) was grouped into four classes and plotted. *p<0.05, **p<0.01, ***p<0.001, Wilcoxon two-sample test.
M. Midgut cell proliferation based on pH3 staining. The average number of pH3+ cells was determined in whole midguts and plotted. Student’s t-tests, **p<0.01, ***p<0.001.
n, numbers of midguts analyzed. Error bars, SEM.
By analyzing nuclear REL, we found that depletion of LAM in 5-day old fat bodies (Cg-Gal4,esg-GFP/+;tub-Gal80ts/Lam RNAi) resulted in reduced nuclear REL immunostaining in the midgut compared to controls (Cg-Gal4,esg-GFP/+;tub-Gal80ts/+) (Figure 6B–C). Conversely, forced expression of LAM in old (50 days) fat body (Cg-Gal4,esg-GFP/+;tub-Gal80ts/UAS-Lam) significantly increased nuclear REL in old midguts compared to controls (Cg-Gal4,esg-GFP/+;tub-Gal80ts/+) (Figure 6D–E), which was accompanied by a reduction of intestinal hyperplasia (Figure 6F–G, 6L–M). LAM expression in old fat bodies also significantly increased midgut Dpt expression compared to controls (Figure S6E). Additionally, depletion of KEY or co-depletion of PGRP-LB plus PGRP-SC2 in fat bodies of Lam mutant flies resulted in decreased midgut hyperplasia and increased midgut IMD signaling compared to controls (Figure 6H–J, 6L–M, S6F). Inhibition of secretion using an allele of dynamin (shits) (Kitamoto, 2001) revealed a significant increase in midgut IMD signaling and reduction in gut hyperplasia in Lam mutant flies (Figure 6H, 6K, 6L–M, S6F). These show that elevated PGRPs secreted from old fat bodies contribute toward midgut hyperplasia. Finally, by comparing flies raised in axenic and conventional conditions, we found midgut IMD repression caused by LAM depletion in fat body to be mediated by bacteria dependent and independent pathways (Figure S6G).
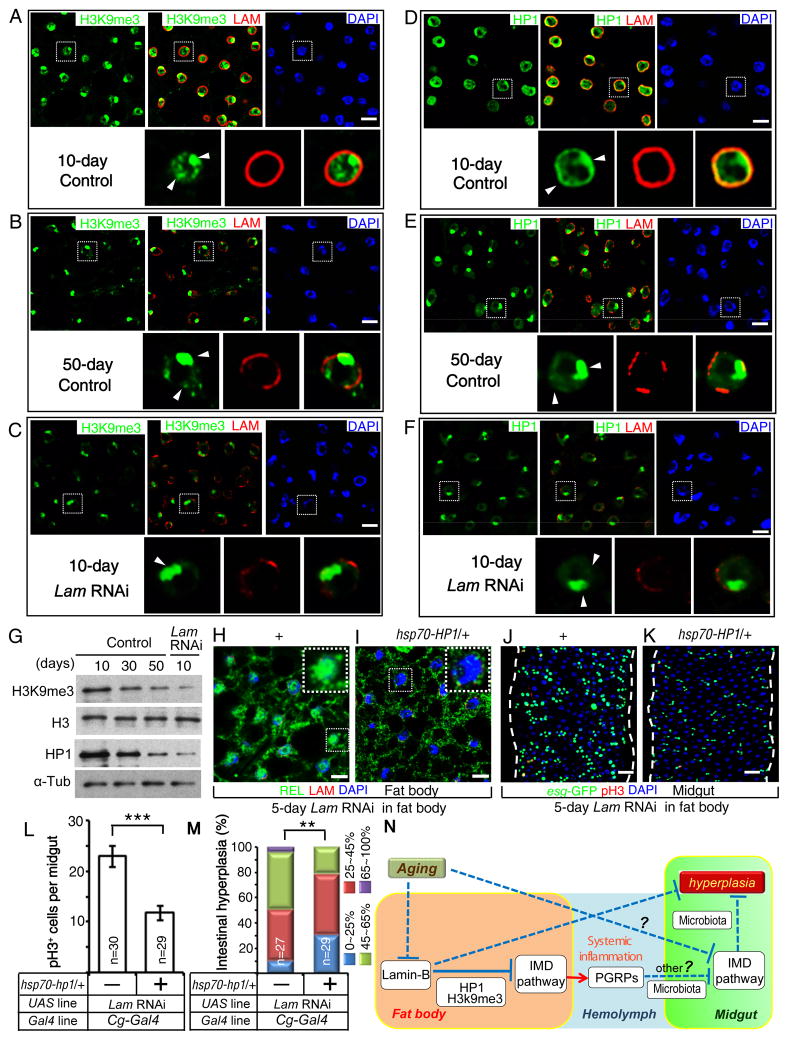
Lamin-B inhibits systemic inflammation by maintaining heterochromatin in the fat body
Drosophila aging is associated with heterochromatin changes in some tissues (Larson et al., 2012; Wood et al., 2010). Since lamin-B is known to associate with heterochromatin and to contribute to heterochromatin formation and/or maintenance (Bank and Gruenbaum, 2011; Dechat et al., 2008), we examined the heterochromatin and found a clear reduction of H3K9me3 in old fat bodies (Figure 7A–B), similar to the H3K9me3 reduction in young (10 day) fat bodies depleted of LAM (Figure 7C, 7G). Consistent with the global reduction of heterochromatin upon LAM loss, we found a clear reduction of HP1, a protein involved in heterochromatin formation (Danzer and Wallrath, 2004), in both wild-type old and LAM-depleted young fat bodies (Figure 7D–G). Since introducing one copy of Hsp70-HP1 could extend fly lifespan (Larson et al., 2012), we used the same strategy to enhance heterochromatin formation. We found that an extra copy of HP1 resulted in a significant reduction of IMD signaling in fat bodies depleted of LAM, and a corresponding decrease and increase of midgut hyperplasia and IMD activity, respectively (Figure 7H–M, S7A–B). Finally, we analyzed H3K9me3 using chromatin immunoprecipitation (ChIP)-qPCR of the fat body and found that LAM depletion resulted in the decrease of H3K9me3 modification on PGRP-LC and key (Figure S7C). Thus, lamin-B prevents persistent IMD response in fat bodies in the absence of infection by maintaining heterochromatin states (Figure 7N).
Figure 7. LAM represses systemic inflammation by maintaining fat body heterochromatin.
A–F. Compared to young wild-type fat bodies (A, D), reduction of H3K9me3 (B) and HP1 (E) in old wild-type fat bodies was mimicked by fat body specific LAM RNAi in young flies (C, F) under conventional condition. LAM, red; H3K9me3, green; HP1, green; DAPI, blue. Nuclei outlined by dashed boxes were enlarged at the bottom of each panel. Arrowheads, H3K9me3 or HP1 staining. Scale bars, 10 μm.
G. Western blotting of fat bodies revealed a reduction of H3K9me3 and HP1 upon aging or by fat-body specific LAM RNAi under conventional condition. Loading controls, Histone 3 (H3), α-tubulin (α-Tub).
H–I. Enhanced fat body IMD signaling (increased nuclear REL) in flies depleted of fat body LAM (H) was reversed by HP1 expression from the basal activity of Hsp70 promoter (I) under conventional condition. REL, green; LAM, red (missing due to RNAi); DAPI, blue. Nuclei boxed by dashed lines were enlarged to highlight the high (H) or low (I) nuclear REL. Scale bars, 10 μm.
J–K. Enhanced midgut hyperplasia in flies depleted of fat body LAM (J) was reversed by low level of HP1 expression (K). esg-GFP, green; pH3, red; DAPI, blue. Scale bars, 20 μm.
L. HP1-mediated reduction of midgut cell proliferation. The average number of pH3+ cells in the whole midgut was counted. n, number of midguts analyzed. Error bars, SEM. Student’s t-tests, ***p<0.001.
M. HP1-mediated reduction of midgut hyperplasia. The percent of esg-GFP+ cells (% of intestinal hyperplasia) was grouped into four classes and plotted. n, number of midguts analyzed. **p<0.01, Wilcoxon two-sample test.
N. A model. Aging associated lamin-B loss in fat body causes the loss of heterochromatin and de-repression of genes involved in fat body IMD signaling. This leads to increased fat body secretion of PGRPs and midgut IMD repression, which in turn causes gut hyperplasia. The effect of midgut IMD signaling by fat body PGRPs is mediated by microbiota dependent and independent pathways. Lamin-B loss in old fat body also causes midgut hyperplasia in microbiota and IMD dependent and independent pathways. Our studies do not rule out the possibility that aging fat body could cause midgut hyperplasia independent of lamin-B loss.
See also Figure S7.
Discussion
By analyzing gene expression changes upon aging in fat bodies and midguts, we show that an increase of immune response in the fat body is accompanied by a striking reduction in the midgut. Specifically, we demonstrate that the age-associated increase in IMD signaling in fat bodies leads to reduction of IMD activity in the midgut, which in turn causes midgut hyperplasia. This fat body to midgut effect requires PGRPs secreted from fat body cells and is mediated by both bacteria dependent and independent pathways. Therefore, fat body aging contributes to systemic inflammation, which leads to the disruption of gut homeostasis (Figure 7N). Importantly, we show that the age-associated lamin-B loss in fat body cells contributes to the de-repression of a large number of immune responsive genes, thereby resulting in fat body-based systemic inflammation.
B-type lamins have long been suggested to have a role in maintaining heterochromatin and gene repression. Consistently, our global analyses of fat body depleted of lamin-B revealed a loss of heterochromatin and de-repression of a large number of immune responsive genes. This is further supported by our ChIP-qPCR analyses of H3K9me3 on specific IMD regulators. Recent studies in different cell types show that tethering genes to nuclear lamins do not always lead to their repression (Finlan et al., 2008; Kumaran and Spector, 2008). Deleting lamins in mouse ES cells or trophectdoderm cells does not result in de-repression of all genes in LADs (Kim et al., 2011; Kim et al., 2013). In light of these studies, we suggest that the transcriptional repression function of lamin-B could be gene and cell type dependent. Interestingly, our GO analyses revealed a significant enrichment of immune responsive genes in LADs in four different mammalian cell types and Drosophila Kc cells. Since the large-scale pattern of LADs is conserved in different cell types in mammals (Meuleman et al., 2013; Peric-Hupkes et al., 2010), it is possible that the immune-responsive genes are also enriched in LADs in the fly fat body cells. Supporting this notion, key, which is one of the two de-repressed IMD regulators and we found to exhibit H3K9me3 reduction and gene activation, is localized to LADs in Kc cells. We speculate that lamin-B might play an evolutionarily conserved role in repressing a subset of inflammatory genes in certain tissues, such as the immune organs, in the absence of infection or injury. Consistently, senescence-associated lamin-B1 loss in mammalian fibroblasts is correlated with SASP. Although the in vivo relevance of fibroblast SASP in chronic inflammation and aging-associated diseases in mammals remains to be established, our findings in Drosophila provide insights and impetus to investigate the role of lamins in immunosenescence and systemic inflammation in mammals.
We demonstrate that lamin-B gradually decreases in fat body cells of aging flies, whereas lamin-C amount remains the same. Since we have recently shown that the assembly of an even and dense nuclear lamina is dependent on the total lamin concentration (Guo et al., 2014b), the age-associated appearance of lamin-B and lamin-C gaps around the nuclear periphery of fat body cells is likely caused by the drop of the lamin-B level. How aging triggers lamin-B loss is unknown, but it appears to be post-transcriptional, because lamin-B transcripts in fat bodies remain unchanged upon aging. Interestingly, among the tissues examined, we found no changes of lamin-B and lamin-C proteins in cells in the heart tube, oenocytes, or gut epithelia in old flies. Therefore, the age-associated lamin-B loss does not occur in all cell types in vivo. A systematic survey to establish the cell/tissue types that undergo age-associated reduction of lamins in both flies and mammals should provide clues to the cause of loss. Deciphering how advanced age leads to lamin loss should open the door to further investigate the cellular mechanism that contributes to chronic systemic inflammation and how it in turn promotes age-associated diseases in humans.
Old Drosophila gut is known to exhibit increased microbial load (Broderick and Lemaitre, 2012; Buchon et al., 2009a; Guo et al., 2014a), which would cause increased stress response and activation of tissue repair, thereby leading to midgut hyperplasia. We show that systemic inflammation caused by lamin-B loss in fat body leads to repression of local midgut IMD signaling. We note that Guo et al (Guo et al., 2014a) recently reported the up-regulation of targets of IMD in the aged whole gut, while we observed a down-regulation of target genes in our analyses of the midgut. Since we also found a similar up-regulation of the genes reported by Guo et al when performing RNA-sequencing of the whole gut (Table S2), the differences were likely due to the use of whole gut versus midgut in the analyses of gene expresion.
Our studies reveal an involvement of bacteria in the repression of midgut IMD signaling by the PGRPs secreted from the fat body. How PGRPs from the fat body repress midgut IMD is still unknown. One possibility is that the body cavity bacteria contribute to the maintenance of midgut IMD activity, and the increased circulating PGRPs limit these bacteria. The circulating PGRPs may also reduce midgut IMD activity indirectly by affecting other tissues. Our evidence suggests that lamin-B loss could also contribute to midgut hyperplasia independent of the IMD pathway. While it will be important to further address these possibilities, our findings have revealed a fat body mediated inflammatory pathway that can lead to reduced migut IMD, increased gut microbial accumulation, and midgut hyperplasia upon aging (Figure 7N).
Interestingly, microbiota changes also occur in aging human intestine and have been linked to altered intestinal inflammatory states and diseases (Biagi et al., 2013; Cheng et al., 2013; Guinane and Cotter, 2013; Hopkins et al., 2002; Lozupone et al., 2012). Although, much effort has been devoted to understand how local changes in aging mammalian intestines affect gut microbial community, the cause remains unclear. Our findings in Drosophila reveal the importance of understanding the impact of immunosenescence and systemic inflammation on gut microbial homeostasis. Indeed, if increased circulating inflammatory cytokines perturb the ability of local intestine epithelium and the gut-associated lymphoid tissue to maintain a balanced microbial community, the unfavorable microbiota in the old intestine would cause chronic stress response and tissue repair, thereby leading to uncontrolled cell growth as observed in age-associated cancers.
Experimental procedures
Fly stocks, culture, and manipulations
All flies were maintained on standard cornmeal/molasses/yeast fly food. Fly stocks were cultured at 25°C on light/dark cycles. For all experiments using Gal4-UAS system, the crosses were performed at 19°C. Female flies were used for this study. Detailed genetic information for all stocks, crosses, and culturing conditions are described in the Supplemental Experimental Procedures.
Immunofluorescence and β-galactosidase histochemistry
Adult fat bodies with attached dorsal cuticles and intact adult guts were dissected in PBS and xed either in paraformaldehyde for immunofluorescence or in glutaraldehyde for β-galactosidase histochemistry. Detailed procedures, antibodies used, and imaging were described in the Supplemental Experimental Procedures.
qRT-PCR and Western blotting analyses
Adult fat bodies were dissected from dorsal abdomen from 50 flies. 30 adult midguts were dissected from whole guts by carefully removing foreguts, hindguts, trachea, and Malpighian tubules. Total RNA or protein was prepared from the dissected tissues for qRT-PCR or Western blotting analyses, respectively. Arcturus PicoPure RNA isolation kit (Life Technologies # KIT0204) was used to prepare total RNA. The RNA samples were processed using iScript One-Step RT-PCR Kit and SYBR Green kit (Bio-Rad #170-8892) along with 2 pmol relevant primers in a 20 μl reaction for qPCR in a C1000 touch thermal cycler (Bio-Rad) with Bio-Rad CFX96 real-time system. The amount of mRNA detected was quantified by comparison with a standard curve and normalized to control rp49 mRNA values. The 2−Δ ΔCT method (Livak and Schmittgen, 2001) was used for evaluating the relative quantity of a given mRNA. The qRT-PCR primers used can be found in the Supplemental Experimental Procedures.
Whole-Transcriptome Shotgun Sequencing (RNA-Seq)
Dissected adult fat bodies, midguts, or whole guts (see Supplemental Experimental Procedures) were used to prepare RNA using Arcturus PicoPure RNA isolation kit (Life Technologies # KIT0204). Poly-A selected mRNA was purified and sequencing libraries were built using Illumina TruSeq RNA sample prep kit V2 (Illumina # RS-122-2001). Libraries were sequenced by single end 50-bp reads on Illumina HiSeq 2000.
Bioinformatics
For RNA-seq, low-quality reads (quality score below 20) were trimmed and reads shorter than 36 bp after trimming were filtered out. The remaining reads were further mapped. The mapped LADs in four mammalian cells and Drosophila Kc cells were used to perform GO analyses (for details see Supplemental Experimental Procedures).
Supplementary Material
A and B. Gene Ontology (GO, by DAVID Bioinformatics Resources 6.7) analyses of up- or down-regulated (≥2 fold) genes upon aging (50-day versus 5-day old flies) in fat body (A) or midgut (B), respectively. Genes involved in the GO term ‘immune response’ (highlighted in red), which includes the innate immune response genes, are significantly enriched in old fat body and midgut, revealing opposite immune responses in the two organs upon aging.
C and D. qRT-PCR analyses of selected IMD regulatory components (PGRPs, key, and Rel, C) or IMD-induced AMPs (D) found to be up-regulated by RNA-seq in old (50 day) wild-type fat bodies. The fold expression change of 50-day fat bodies was plotted relative to 5-day fat bodies, which was set to 1. Error bars, standard error of the mean (SEM) based on three independent experiments. *p<0.05, Student’s t tests.
E and F. qRT-PCR analyses of selected IMD regulatory components (PGRP-LC, key, and Rel, E) or IMD-induced AMPs (F) found to be down regulated by RNA-seq in old (50 day) wild-type midgut. The fold expression change of 50-day midgut was plotted relative to that of 5-day midguts, which was set to 1. Only the IMD pathway downstream effectors, including PGRP-LF and AMPs were confirmed to exhibit reduction. Error bars, SEM, based on three independent experiments. ^p>0.05, *p<0.05, ***p<0.001, Student’s t-tests.
G. Expression pattern of Cg-Gal4, r4-Gal4, Mex-Gal4, NP1-Gal4, and Hml. Δ-Gal4 in adult fat body, midgut, and circulating hemocytes as tested by the UAS-GFPS65T reporter. UAS-eGFP was used as a reporter for Pxn-Gal4::UAS-eGFP line. Cg-Gal4, Pxn-Gal4, and Hml. Δ-Gal4 are expressed in a few cells localized to the basal side of the gut. Since these cells are not ISC, EB, EE (identified by marker expression) or EC (identified by cell size and morphology), their identity remains unknown. The GFP fluorescence images of the three tissues or cell types can be found in Figure S7D–O.
H. qRT-PCR analysis of Dpt in midgut from 5-day or 50-day old Gal4 control flies (Cg-Gal4/+;UAS-Dicer-2/+), UAS control flies (Rel RNAi/+ or PGRP-LB RNAi/+;PGRP-SC2 RNAi/+), flies depleted of REL by Cg-Gal4 (Cg-Gal4/Rel RNAi;UAS-Dicer-2/+), or flies co-depleted of PGRP-LB plus PGRP-SC2 by Cg-Gal4 (PGRP-LB RNAi/+;Cg-Gal4/+;UAS-Dicer-2/PGRP-SC2 RNAi). The fold change of Dpt expression was determined relative to the 5-day Gal4 control flies (Cg-Gal4/+;UAS-Dicer-2/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
I. qRT-PCR analysis of Dpt in midgut from 5-day or 50-day old Gal4 control flies (UAS-Dicer-2/+;r4-Gal4/+), UAS control flies (Rel RNAi/+ or PGRP-LB RNAi/+;PGRP-SC2 RNAi/+), flies depleted of REL by r4-Gal4 (UAS-Dicer-2/Rel RNAi;r4-Gal4/+), or flies co-depleted of PGRP-LB plus PGRP-SC2 by r4-Gal4 (PGRP-LB RNAi/+;UAS-Dicer-2/+;r4-Gal4/PGRP-SC2 RNAi). The fold change of Dpt expression was determined relative to the 5-day Gal4 control flies, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
J. qRT-PCR analysis of Dpt in midgut from 5-day or 50-day old Gal4 control flies (UAS-Dicer-2/+;Hml. Δ-Gal4/+), UAS control flies (Rel RNAi/+ or PGRP-LB RNAi/+;PGRP-SC2 RNAi/+), flies depleted of REL in the circulating hemocytes by Hml. Δ-Gal4 (UAS-Dicer-2/Rel RNAi;Hml. Δ-Gal4/+), or flies co-depleted of PGRP-LB plus PGRP-SC2 in hemocytes by Hml. Δ-Gal4 (PGRP-LB RNAi/+;UAS-Dicer-2/+;Hml. Δ-Gal4/PGRP-SC2 RNAi). The fold change of Dpt expression was determined relative to the 5-day Gal4 control flies (UAS-Dicer-2/+;Hml. Δ-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
List A shows the changed genes in 50-day wild-type midgut compared to the 5-day wild-type midgut. List B shows changed genes in 40-day wild-type whole gut compared to the 5-day wild-type whole gut. Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
Gene symbols, description, and fold changes are indicated.
Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
Gene symbols, description, and fold changes are indicated.
Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
A and B. Representative images showing that compared to the control (A, Mex-Gal4,esg-GFP/+;tub-Gal80ts/+), midgut-specific depletion of KEY by Mex-Gal4-mediated RNAi in 5-day old animals led to gut hyperplasia (B, Mex-Gal4,esg-GFP/+;tub-Gal80ts/key RNAi) as judged by esg-GFP reporter (green) and pH3 staining (red). DAPI labels nuclei (blue). Flies were raised under the conventional (germ) conditions (Con). Scale bars, 20 μm.
C. qRT-PCR analysis of Dpt in midguts from 5-day old Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+ or r4-Gal4/+;tub-Gal80ts/+), 5-day old UAS control flies (UAS-PGRP-LC/+ or UAS-PGRP-SC2/+;UAS-PGRP-LB/+), 5-day old flies expressing PGRP-LC cDNA or PGRP-LB plus PGRP-SC2 cDNAs in fat bodies by Cg-Gal4 or r4-Gal4. The fold change of Dpt expression was plotted relative to the 5-day controls, which was set to 1. Error bars, SEM, based on three independent experiments. ***p<0.001, Student’s t-tests.
D. qRT-PCR analyses of Dpt in midguts from 5- or 50-day old Gal4 control (Cg-Gal4/+;UAS-Dicer-2/+), UAS control (key RNAi/+), or flies depleted of key in fat body by Cg-Gal4 (Cg-Gal4/+;UAS-Dicer-2/key RNAi). The fold change of Dpt expression was plotted relative to the 5-day Gal4 control, which was set to 1. Con, conventional condition. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
E. qRT-PCR analyses of Dpt in midguts from 5- or 50-day old Gal4 control flies (UAS-Dicer-2/+;Hml. Δ-Gal4), or flies depleted of key in hemocytes by Hml. Δ-Gal4 (UAS-Dicer-2/+;Hml. Δ-Gal4/key RNAi). The fold change of Dpt expression was plotted relative to the 5-day Gal4 control (UAS-Dicer-2/+;Hml. Δ-Gal4), which was set to 1. Con, conventional condition. Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
F. qRT-PCR analysis of Dpt in midguts from 5-day old Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), 5-day old UAS control flies (UAS-PGRP-LC/+ or UAS-PGRP-SC2/+;UAS-PGRP-LB/+), 5-day old flies with PGRP-LC cDNA expressed in fat bodies, or 5-day old flies with co-expression of PGRP-LB plus PGRP-SC2 cDNAs in fat bodies from axenic (Axe) or conventional (Con) flies. The fold change of Dpt expression was plotted relative to the 5-day Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+) cultured under conventional condition, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, **p<0.01, ***p<0.001, Student’s t-tests.
G. Midgut hyperplasia based on esg-GFP. The number of esg-GFP+ cells counted from P1-4 regions of midguts was divided by total cells counted from DAPI staining and used to define the percentages of intestinal hyperplasia. This percentage was grouped into four classes and plotted as colored bars. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, **P<0.01, ***P<0.001, Wilcoxon two-sample test.
H. Midgut hyperplasia based on pH3 staining. The average number of pH3+ cells was determined from the whole midguts and plotted. Error bars, SEM. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, ***P<0.001, Student’s t-tests.
I and J. Representative images showing that under axenic conditions (Axe), the expression of PGRP-LC (I, Cg-Gal4, esg-GFP/+;tub-Gal80ts/UAS-PGRP-LC) in 5-day old fat bodies did not increase the number of midgut esg-GFP+ (green) and pH3+ (red) cells compared to that of the Gal4 control (J, Cg-Gal4,esg-GFP/+;tub-Gal80ts/+). Scale bars, 20 μm.
A. The GO analyses using DAVID Bioinformatics Resources 6.7 revealed an enrichment of defense responsive genes, including those involved in immune response, in LADs in mouse ES cells (ESC), mouse astrocytes (Astro), mouse neural progenitor cells (NPC), mouse embryonic fibroblasts (MEF), and in Drosophila Kc cells based on the published LADs’ maps.
B and C. LAM signal remained the same in both 10- (B) and 50-day (C) old midguts as revealed by LAM (red) staining. esg-GFP reporter (green) and DAPI labels are also shown. Boxed areas are enlarged to the right of each set of images. Flies were raised in conventional condition. Scale bars, 20 μm.
D. qRT-PCR analyses of Lam and LamC in fat body from 10- or 50-day old wild-type flies showed no change of their transcription. The expression level of old fat body was plotted relative to that of the young, which was set to 1. Error bars, SEM, based on three independent experiments. Flies were raised in conventional condition. ^P>0.05, Student’s t-tests.
A. qRT-PCR analyses of Lam in fat body from 5-day old control flies (Lam RNAi/+) or flies with fat body-specific LAM depletion (Cg-Gal4/+;tub-Gal80ts/Lam RNAi or tub-Gal80ts/+;r4-Gal4/Lam RNAi). The expression change was relative to the 5-day fat body control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, **p<0.01, Student’s t-tests.
B. qRT-PCR analyses of selected genes of IMD signaling components in fat body from 5-day old Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (Lam RNAi/+) or flies with Cg-Gal4-driven fat body-specific LAM depletion (Cg-Gal4/+;tub-Gal80ts/Lam RNAi). The expression change was relative to the 5-day Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
C. qRT-PCR analyses of selected PGRP genes in fat body from 5-day old Gal4 control flies (tub-Gal80ts/+;r4-Gal4/+) or flies with r4-Gal4-driven fat body-specific LAM depletion (tub-Gal80ts/+;r4-Gal4/Lam RNAi). The expression change was relative to the 5-day fat body Gal4 control (tub-Gal80ts/+;r4-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
D. qRT-PCR analyses of selected AMP genes in fat body from 5-day old control flies (tub-Gal80ts/+;r4-Gal4/+ or Lam RNAi/+) or flies with r4-Gal4 driven fat body-specific LAM depletion (tub-Gal80ts/+;r4-Gal4/Lam RNAi). The expression change was relative to the 5-day fat body Gal4 control (tub-Gal80ts/+;r4-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
E. qRT-PCR analyses of selected genes of IMD signaling pathway in fat body from 5-day old Gal4 control flies (tub-Gal80ts/+;Hml. Δ-Gal4/+) or flies with Hml. Δ-Gal4 driven hemocyte-specific LAM depletion (tub-Gal80ts/+;Hml. Δ-Gal4/Lam RNAi). The expression change was relative to the 5-day Gal4 control (tub-Gal80ts/+; Hml. Δ-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
F. qRT-PCR analysis of Dpt in fat bodies from 5- or 50-day Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (UAS-Lam/+) or flies with Cg-Gal4 driven fat body-specific expression of LAM (Cg-Gal4/+;tub-Gal80ts/UAS-Lam). Dpt expression change was relative to the 5-day Gal4 control (Cg-Gal4/+; tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
G. qRT-PCR analysis of Dpt in fat bodies from 5- or 50-day Gal4 control flies (tub-Gal80ts/+;r4-Gal4/+), UAS control flies (UAS-Lam/+), or flies with r4-Gal4 driven fat body-specific expression of LAM (tub-Gal80ts/+;r4-Gal4/UAS-Lam). Dpt expression change was relative to the 5-day fat body Gal4 control (tub-Gal80ts/+;r4-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
H. qRT-PCR analysis of Dpt in fat bodies from 5- or 50-day Gal4 control flies (tub-Gal80ts/+;Hml. Δ-Gal4/+), UAS control flies (UAS-Lam/+) or flies with Hml. Δ-Gal4 driven hemocyte-specific expression of LAM (tub-Gal80ts/+;Hml. Δ-Gal4/UAS-Lam). Dpt expression change was relative to the 5-day hemocyte Gal4 control (tub-Gal80ts/+; Hml. Δ-Gal4/+), which was set to 1. Pxn-Gal4 (another hemocyte Gal4 line) gave similar results (not shown). Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
A. Midgut hyperplasia of conventional flies based on pH3 staining. The average number of pH3+ cells was determined from the whole midgut and plotted. Error bars, SEM. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, ***P<0.001, Student’s t-tests.
B. Midgut hyperplasia of conventional flies based on esg-GFP. The number of esg-GFP+ cells counted from P1-4 regions of midguts was divided by total cells counted from DAPI staining and used to define the percentages of intestinal hyperplasia. This percentage was grouped into four classes and plotted as colored bars. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, ***P<0.001, Wilcoxon two-sample test.
C and D. Representative images of axenic Lam mutant flies (D, LamD395/Lamk2,esg-GFP) that exhibit stronger midgut hyperplasia than the axenic Lam heterozygous control flies (C, Lamk2,esg-GFP/+) as judged by esg-GFP reporter expression (green) and pH3 (red) staining in 5-day old flies. Scale bars, 20 μm.
E. Midgut hyperplasia of axenic and conventional flies based on pH3 staining. The average number of pH3+ cells was determined from the whole midguts and plotted. Error bars, SEM. The number (n) of midguts analyzed in each group is indicated. **P<0.01, ***P<0.001, Student’s t-tests.
F. Midgut hyperplasia of axenic and conventional flies based on esg-GFP. The number of esg-GFP+ cells counted from P1-4 regions of midguts was divided by total cells counted from DAPI staining and used to define the percentages of intestinal hyperplasia. This percentage was grouped into four classes as indicated by the colored bars and plotted. The number (n) of midguts analyzed in each group is indicated. **P<0.01, ***P<0.001, Wilcoxon two-sample test.
G. Eclosion and pupariation rates. The axenic (Axe) Lam mutants exhibited increased eclosion and pupariation rates compared to conventional (Con) Lam mutants. Error bars, SEM based on three independent experiments performed with 200 larvae per experiment. Student’s t-tests, **p<0.01, ***p<0.001.
A and B. Graphs showing the overlap of down- (A) or up-regulated (B) genes between midguts from wild-type old (50 days) flies and midguts from young (5 days) flies depleted of LAM in the fat body by Cg-Gal4;tub-Gal80ts driven LAM RNAi.
C and D. qRT-PCR confirmation of selected PGRPs (C, PGRP-LB, LB; PGRP-LF, LF; PGRP-SB1, SB1; PGRP-SB2, SB2; PGRP-SC1, SC1; PGRP-SC2, SC2) and AMPs (D) that were found to be down-regulated in the midgut upon fat body-specific LAM RNAi in 5-day old flies by RNA-seq. The expression changes were plotted relative to the 5-day Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. **p<0.01, ***p<0.001, Student’s t-tests.
E. qRT-PCR analysis of Dpt in midgut from 5- or 50-day Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (UAS-Lam/+) or flies with fat body-specific expression of LAM cDNA (Cg-Gal4/+;tub-Gal80ts/UAS-Lam). Dpt expressions in midgut were plotted relative to 5-day Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
F. qRT-PCR analyses of Dpt in midgut from 5-day old Lam-mutant control flies (Cg-Gal4,LamD395/Lamk2), or 5-day old Lam-mutant flies with dynamin inhibition (UAS-shits), RNAi of key, or RNAi of PGRP-SC2 plus PGRP-LB (SC2&LB) in fat bodies. The Dpt expression change was plotted relative to the control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
G. qRT-PCR analysis of Dpt in midgut from 5-day conventional (Con) or axenic (Axe) Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (Lam RNAi/+) or flies with fat body-specific deletion of Lam (Cg-Gal4/+;tub-Gal80ts/Lam RNAi). Dpt expression in midgut were plotted relative to 5-day conventional Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, **p<0.01, ***p<0.001, Student’s t-tests.
A. qRT-PCR analyses of Dpt in fat body from 5-day old control (Cg-Gal4/+;tub-Gal80ts/+) flies, 5-day old flies depleted of LAM in fat body (Cg-Gal4/+;tub-Gal80ts/Lam RNAi), or the 5-day LAM depleted flies carrying one copy of hsp70-HP1 transgene (Cg-Gal4/hsp70-HP1;tub-Gal80ts/Lam RNAi). Dpt expression was plotted relative to the control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
B. qRT-PCR analyses of Dpt in midgut from 5-day old control flies (Cg-Gal4/+;tub-Gal80ts/+), 5-day old flies depleted of fat body LAM (Cg-Gal4/+;tub-Gal80ts/Lam), or the 5-day LAM depleted flies carrying one copy of hsp70-HP1 transgene (Cg-Gal4/hsp70-HP1;tub-Gal80ts/Lam RNAi). Dpt expression was plotted relative to the control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
C. Chromatin immunoprecipitation (ChIP) analysis of selected IMD signaling components of adult fat body from 5-day wild type (w1118) and Lam mutant (LamD395/Lamk2) flies. Chromatin was immunoprecipitated with antibodies to H3K9me3 and control IgG. Primers corresponding to PGRP-LC, IMD, key, Rel, and rp49 (negative control) were used to amplify precipitated DNA. ChIP samples were normalized to 5% of input DNA. Error bars, SEM, based on three independent experiments. ^p>0.05, *p<0.05, **p<0.01, Student’s t-tests.
D–O. Expression patterns of different Gal4 lines as reported by UAS-GFPS65T in adult fat body (D, G, J, M), adult gut (E, H, K, N), and adult circulating hemocytes (F, I, L, O). DAPI labels the nuclei (blue). Phalloidin stains of actin (red) in F, I, L and O. Yellow asterisks mark the heart peripheral cells. Whole guts and hemocytes are outlined by white dashed lines. Scale bars: D, F, G, I, J, L, M, and O (20 μm); E, H, K, and N (200μm).
Gene symbols, description, and fold changes are indicated.
Highlights.
Old flies exhibit fat body inflammation and midgut immune repression.
Age-associated lamin-B loss in fat bodies causes uncontrolled systemic inflammation.
Lamin-B loss in the old fat body triggers midgut immune repression and hyperplasia.
PGRPs secreted by the old fat body inhibit midgut immunity and trigger hyperplasia.
Acknowledgments
We thank Dr. Allan Spradling and his lab for valuable help, Drs. Allan Spradling, Mitchell Dushay, Bruno Lemaitre, Lori Wallrath, Georg Krohne, and Erin Zeituni for fly strains and antibodies, Matthew Sieber, Alexis Marianes, and the Zheng lab for helpful discussions. Supported by a Senior Scholar Award to YZ from the Ellison Medical Foundation, GM056312 and GM106023 (YZ).
Footnotes
Author Contribution Section: Yixian Zheng and Haiyang Chen conceived the idea. Haiyang Chen performed all the experiments. Yixian Zheng and Haiyang Chen interpreted the data and wrote the paper. Xiaobin Zheng analyzed and interpreted all the RNA-sequencing data and performed GO term analyses of transcriptionally altered genes and genes in LADs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad A, Banerjee S, Wang Z, Kong D, Majumdar AP, Sarkar FH. Aging and inflammation: etiological culprits of cancer. Curr Aging Sci. 2009;2:174–186. doi: 10.2174/1874609810902030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank EM, Gruenbaum Y. The nuclear lamina and heterochromatin: a complex relationship. Biochem Soc Trans. 2011;39:1705–1709. doi: 10.1042/BST20110603. [DOI] [PubMed] [Google Scholar]
- Barascu A, Le Chalony C, Pennarun G, Genet D, Imam N, Lopez B, Bertrand P. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. EMBO J. 2012;31:1080–1094. doi: 10.1038/emboj.2011.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E, Candela M, Turroni S, Garagnani P, Franceschi C, Brigidi P. Ageing and gut microbes: perspectives for health maintenance and longevity. Pharmacol Res. 2013;69:11–20. doi: 10.1016/j.phrs.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009a;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009b;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Chen F, Castranova V. Nuclear factor-kappaB, an unappreciated tumor suppressor. Cancer Res. 2007;67:11093–11098. doi: 10.1158/0008-5472.CAN-07-1576. [DOI] [PubMed] [Google Scholar]
- Chen H, Chen X, Zheng Y. The nuclear lamina regulates germline stem cell niche organization via modulation of EGFR signaling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Palva AM, de Vos WM, Satokari R. Contribution of the intestinal microbiota to human health: from birth to 100 years of age. Curr Top Microbiol Immunol. 2013;358:323–346. doi: 10.1007/82_2011_189. [DOI] [PubMed] [Google Scholar]
- Danzer JR, Wallrath LL. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 2004;131:3571–3580. doi: 10.1242/dev.01223. [DOI] [PubMed] [Google Scholar]
- Day MJ. Ageing, immunosenescence and inflammageing in the dog and cat. J Comp Pathol. 2010;142(Suppl 1):S60–69. doi: 10.1016/j.jcpa.2009.10.011. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O, Chojnowski A, Ong PF, Zhao TY, Common JE, Lunny D, Lane EB, Lee SJ, Vardy LA, Stewart CL, et al. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J Cell Biol. 2013;200:605–617. doi: 10.1083/jcb.201206121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Freund A, Laberge RM, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop T, Larbi A, Kotb R, de Angelis F, Pawelec G. Aging, immunity, and cancer. Discov Med. 2011;11:537–550. [PubMed] [Google Scholar]
- Goto A, Matsushita K, Gesellchen V, El Chamy L, Kuttenkeuler D, Takeuchi O, Hoffmann JA, Akira S, Boutros M, Reichhart JM. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in drosophila and mice. Nat Immunol. 2008;9:97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol. 2013;6:295–308. doi: 10.1177/1756283X13482996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014a;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell. 2014b doi: 10.1091/mbc.E13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34(Suppl 2):S12–18. doi: 10.1016/s1590-8658(02)80157-8. [DOI] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development. 2009;136:483–493. doi: 10.1242/dev.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatami M. Inflammation, aging, and cancer: tumoricidal versus tumorigenesis of immunity: a common denominator mapping chronic diseases. Cell Biochem Biophys. 2009;55:55–79. doi: 10.1007/s12013-009-9059-2. [DOI] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res. 2013;23:1420–1423. doi: 10.1038/cr.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, Tavare S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianes A, Spradling AC. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife. 2013;2:e00886. doi: 10.7554/eLife.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic MP, Kylsten P, Dushay MS. Drosophila lamin mutations cause melanotic mass formation and lamellocyte differentiation. Mol Immunol. 2009;46:3245–3250. doi: 10.1016/j.molimm.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Osouda S, Nakamura Y, de Saint Phalle B, McConnell M, Horigome T, Sugiyama S, Fisher PA, Furukawa K. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev Biol. 2005;284:219–232. doi: 10.1016/j.ydbio.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. NF-kappaB: tumor promoter or suppressor? Trends Cell Biol. 2004;14:64–69. doi: 10.1016/j.tcb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Phillips MD, Thomas GH. Brush border spectrin is required for early endosome recycling in Drosophila. J Cell Sci. 2006;119:1361–1370. doi: 10.1242/jcs.02839. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Ramsden S, Cheung YY, Seroude L. Functional analysis of the Drosophila immune response during aging. Aging Cell. 2008;7:225–236. doi: 10.1111/j.1474-9726.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- Ray K. Gut microbiota: Colorectal cancer-driven by inflammation and gut bacteria? Nat Rev Gastroenterol Hepatol. 2012;9:558. doi: 10.1038/nrgastro.2012.172. [DOI] [PubMed] [Google Scholar]
- Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroude L, Brummel T, Kapahi P, Benzer S. Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging Cell. 2002;1:47–56. doi: 10.1046/j.1474-9728.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T, Ip YT. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005;26:193–198. doi: 10.1016/j.it.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hang S, Purdy AE, Watnick PI. Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. MBio. 2013;4:e00337–00313. doi: 10.1128/mBio.00337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Hillenmeyer S, Lawrence C, Chang C, Hosier S, Lightfoot W, Mukherjee E, Jiang N, Schorl C, Brodsky AS, et al. Chromatin remodeling in the aging genome of Drosophila. Aging Cell. 2010;9:971–978. doi: 10.1111/j.1474-9726.2010.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging Cell. 2005;4:103–108. doi: 10.1111/j.1474-9728.2005.00147.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A and B. Gene Ontology (GO, by DAVID Bioinformatics Resources 6.7) analyses of up- or down-regulated (≥2 fold) genes upon aging (50-day versus 5-day old flies) in fat body (A) or midgut (B), respectively. Genes involved in the GO term ‘immune response’ (highlighted in red), which includes the innate immune response genes, are significantly enriched in old fat body and midgut, revealing opposite immune responses in the two organs upon aging.
C and D. qRT-PCR analyses of selected IMD regulatory components (PGRPs, key, and Rel, C) or IMD-induced AMPs (D) found to be up-regulated by RNA-seq in old (50 day) wild-type fat bodies. The fold expression change of 50-day fat bodies was plotted relative to 5-day fat bodies, which was set to 1. Error bars, standard error of the mean (SEM) based on three independent experiments. *p<0.05, Student’s t tests.
E and F. qRT-PCR analyses of selected IMD regulatory components (PGRP-LC, key, and Rel, E) or IMD-induced AMPs (F) found to be down regulated by RNA-seq in old (50 day) wild-type midgut. The fold expression change of 50-day midgut was plotted relative to that of 5-day midguts, which was set to 1. Only the IMD pathway downstream effectors, including PGRP-LF and AMPs were confirmed to exhibit reduction. Error bars, SEM, based on three independent experiments. ^p>0.05, *p<0.05, ***p<0.001, Student’s t-tests.
G. Expression pattern of Cg-Gal4, r4-Gal4, Mex-Gal4, NP1-Gal4, and Hml. Δ-Gal4 in adult fat body, midgut, and circulating hemocytes as tested by the UAS-GFPS65T reporter. UAS-eGFP was used as a reporter for Pxn-Gal4::UAS-eGFP line. Cg-Gal4, Pxn-Gal4, and Hml. Δ-Gal4 are expressed in a few cells localized to the basal side of the gut. Since these cells are not ISC, EB, EE (identified by marker expression) or EC (identified by cell size and morphology), their identity remains unknown. The GFP fluorescence images of the three tissues or cell types can be found in Figure S7D–O.
H. qRT-PCR analysis of Dpt in midgut from 5-day or 50-day old Gal4 control flies (Cg-Gal4/+;UAS-Dicer-2/+), UAS control flies (Rel RNAi/+ or PGRP-LB RNAi/+;PGRP-SC2 RNAi/+), flies depleted of REL by Cg-Gal4 (Cg-Gal4/Rel RNAi;UAS-Dicer-2/+), or flies co-depleted of PGRP-LB plus PGRP-SC2 by Cg-Gal4 (PGRP-LB RNAi/+;Cg-Gal4/+;UAS-Dicer-2/PGRP-SC2 RNAi). The fold change of Dpt expression was determined relative to the 5-day Gal4 control flies (Cg-Gal4/+;UAS-Dicer-2/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
I. qRT-PCR analysis of Dpt in midgut from 5-day or 50-day old Gal4 control flies (UAS-Dicer-2/+;r4-Gal4/+), UAS control flies (Rel RNAi/+ or PGRP-LB RNAi/+;PGRP-SC2 RNAi/+), flies depleted of REL by r4-Gal4 (UAS-Dicer-2/Rel RNAi;r4-Gal4/+), or flies co-depleted of PGRP-LB plus PGRP-SC2 by r4-Gal4 (PGRP-LB RNAi/+;UAS-Dicer-2/+;r4-Gal4/PGRP-SC2 RNAi). The fold change of Dpt expression was determined relative to the 5-day Gal4 control flies, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
J. qRT-PCR analysis of Dpt in midgut from 5-day or 50-day old Gal4 control flies (UAS-Dicer-2/+;Hml. Δ-Gal4/+), UAS control flies (Rel RNAi/+ or PGRP-LB RNAi/+;PGRP-SC2 RNAi/+), flies depleted of REL in the circulating hemocytes by Hml. Δ-Gal4 (UAS-Dicer-2/Rel RNAi;Hml. Δ-Gal4/+), or flies co-depleted of PGRP-LB plus PGRP-SC2 in hemocytes by Hml. Δ-Gal4 (PGRP-LB RNAi/+;UAS-Dicer-2/+;Hml. Δ-Gal4/PGRP-SC2 RNAi). The fold change of Dpt expression was determined relative to the 5-day Gal4 control flies (UAS-Dicer-2/+;Hml. Δ-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
List A shows the changed genes in 50-day wild-type midgut compared to the 5-day wild-type midgut. List B shows changed genes in 40-day wild-type whole gut compared to the 5-day wild-type whole gut. Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
Gene symbols, description, and fold changes are indicated.
Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
Gene symbols, description, and fold changes are indicated.
Gene symbol, gene location, fold change (logFC, log2 of the fold change), P values (by hypergeometric test), and FDR (False Discover Rate) are indicated. Flies were raised in conventional condition.
A and B. Representative images showing that compared to the control (A, Mex-Gal4,esg-GFP/+;tub-Gal80ts/+), midgut-specific depletion of KEY by Mex-Gal4-mediated RNAi in 5-day old animals led to gut hyperplasia (B, Mex-Gal4,esg-GFP/+;tub-Gal80ts/key RNAi) as judged by esg-GFP reporter (green) and pH3 staining (red). DAPI labels nuclei (blue). Flies were raised under the conventional (germ) conditions (Con). Scale bars, 20 μm.
C. qRT-PCR analysis of Dpt in midguts from 5-day old Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+ or r4-Gal4/+;tub-Gal80ts/+), 5-day old UAS control flies (UAS-PGRP-LC/+ or UAS-PGRP-SC2/+;UAS-PGRP-LB/+), 5-day old flies expressing PGRP-LC cDNA or PGRP-LB plus PGRP-SC2 cDNAs in fat bodies by Cg-Gal4 or r4-Gal4. The fold change of Dpt expression was plotted relative to the 5-day controls, which was set to 1. Error bars, SEM, based on three independent experiments. ***p<0.001, Student’s t-tests.
D. qRT-PCR analyses of Dpt in midguts from 5- or 50-day old Gal4 control (Cg-Gal4/+;UAS-Dicer-2/+), UAS control (key RNAi/+), or flies depleted of key in fat body by Cg-Gal4 (Cg-Gal4/+;UAS-Dicer-2/key RNAi). The fold change of Dpt expression was plotted relative to the 5-day Gal4 control, which was set to 1. Con, conventional condition. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
E. qRT-PCR analyses of Dpt in midguts from 5- or 50-day old Gal4 control flies (UAS-Dicer-2/+;Hml. Δ-Gal4), or flies depleted of key in hemocytes by Hml. Δ-Gal4 (UAS-Dicer-2/+;Hml. Δ-Gal4/key RNAi). The fold change of Dpt expression was plotted relative to the 5-day Gal4 control (UAS-Dicer-2/+;Hml. Δ-Gal4), which was set to 1. Con, conventional condition. Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
F. qRT-PCR analysis of Dpt in midguts from 5-day old Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), 5-day old UAS control flies (UAS-PGRP-LC/+ or UAS-PGRP-SC2/+;UAS-PGRP-LB/+), 5-day old flies with PGRP-LC cDNA expressed in fat bodies, or 5-day old flies with co-expression of PGRP-LB plus PGRP-SC2 cDNAs in fat bodies from axenic (Axe) or conventional (Con) flies. The fold change of Dpt expression was plotted relative to the 5-day Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+) cultured under conventional condition, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, **p<0.01, ***p<0.001, Student’s t-tests.
G. Midgut hyperplasia based on esg-GFP. The number of esg-GFP+ cells counted from P1-4 regions of midguts was divided by total cells counted from DAPI staining and used to define the percentages of intestinal hyperplasia. This percentage was grouped into four classes and plotted as colored bars. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, **P<0.01, ***P<0.001, Wilcoxon two-sample test.
H. Midgut hyperplasia based on pH3 staining. The average number of pH3+ cells was determined from the whole midguts and plotted. Error bars, SEM. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, ***P<0.001, Student’s t-tests.
I and J. Representative images showing that under axenic conditions (Axe), the expression of PGRP-LC (I, Cg-Gal4, esg-GFP/+;tub-Gal80ts/UAS-PGRP-LC) in 5-day old fat bodies did not increase the number of midgut esg-GFP+ (green) and pH3+ (red) cells compared to that of the Gal4 control (J, Cg-Gal4,esg-GFP/+;tub-Gal80ts/+). Scale bars, 20 μm.
A. The GO analyses using DAVID Bioinformatics Resources 6.7 revealed an enrichment of defense responsive genes, including those involved in immune response, in LADs in mouse ES cells (ESC), mouse astrocytes (Astro), mouse neural progenitor cells (NPC), mouse embryonic fibroblasts (MEF), and in Drosophila Kc cells based on the published LADs’ maps.
B and C. LAM signal remained the same in both 10- (B) and 50-day (C) old midguts as revealed by LAM (red) staining. esg-GFP reporter (green) and DAPI labels are also shown. Boxed areas are enlarged to the right of each set of images. Flies were raised in conventional condition. Scale bars, 20 μm.
D. qRT-PCR analyses of Lam and LamC in fat body from 10- or 50-day old wild-type flies showed no change of their transcription. The expression level of old fat body was plotted relative to that of the young, which was set to 1. Error bars, SEM, based on three independent experiments. Flies were raised in conventional condition. ^P>0.05, Student’s t-tests.
A. qRT-PCR analyses of Lam in fat body from 5-day old control flies (Lam RNAi/+) or flies with fat body-specific LAM depletion (Cg-Gal4/+;tub-Gal80ts/Lam RNAi or tub-Gal80ts/+;r4-Gal4/Lam RNAi). The expression change was relative to the 5-day fat body control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, **p<0.01, Student’s t-tests.
B. qRT-PCR analyses of selected genes of IMD signaling components in fat body from 5-day old Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (Lam RNAi/+) or flies with Cg-Gal4-driven fat body-specific LAM depletion (Cg-Gal4/+;tub-Gal80ts/Lam RNAi). The expression change was relative to the 5-day Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
C. qRT-PCR analyses of selected PGRP genes in fat body from 5-day old Gal4 control flies (tub-Gal80ts/+;r4-Gal4/+) or flies with r4-Gal4-driven fat body-specific LAM depletion (tub-Gal80ts/+;r4-Gal4/Lam RNAi). The expression change was relative to the 5-day fat body Gal4 control (tub-Gal80ts/+;r4-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
D. qRT-PCR analyses of selected AMP genes in fat body from 5-day old control flies (tub-Gal80ts/+;r4-Gal4/+ or Lam RNAi/+) or flies with r4-Gal4 driven fat body-specific LAM depletion (tub-Gal80ts/+;r4-Gal4/Lam RNAi). The expression change was relative to the 5-day fat body Gal4 control (tub-Gal80ts/+;r4-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
E. qRT-PCR analyses of selected genes of IMD signaling pathway in fat body from 5-day old Gal4 control flies (tub-Gal80ts/+;Hml. Δ-Gal4/+) or flies with Hml. Δ-Gal4 driven hemocyte-specific LAM depletion (tub-Gal80ts/+;Hml. Δ-Gal4/Lam RNAi). The expression change was relative to the 5-day Gal4 control (tub-Gal80ts/+; Hml. Δ-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
F. qRT-PCR analysis of Dpt in fat bodies from 5- or 50-day Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (UAS-Lam/+) or flies with Cg-Gal4 driven fat body-specific expression of LAM (Cg-Gal4/+;tub-Gal80ts/UAS-Lam). Dpt expression change was relative to the 5-day Gal4 control (Cg-Gal4/+; tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
G. qRT-PCR analysis of Dpt in fat bodies from 5- or 50-day Gal4 control flies (tub-Gal80ts/+;r4-Gal4/+), UAS control flies (UAS-Lam/+), or flies with r4-Gal4 driven fat body-specific expression of LAM (tub-Gal80ts/+;r4-Gal4/UAS-Lam). Dpt expression change was relative to the 5-day fat body Gal4 control (tub-Gal80ts/+;r4-Gal4/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
H. qRT-PCR analysis of Dpt in fat bodies from 5- or 50-day Gal4 control flies (tub-Gal80ts/+;Hml. Δ-Gal4/+), UAS control flies (UAS-Lam/+) or flies with Hml. Δ-Gal4 driven hemocyte-specific expression of LAM (tub-Gal80ts/+;Hml. Δ-Gal4/UAS-Lam). Dpt expression change was relative to the 5-day hemocyte Gal4 control (tub-Gal80ts/+; Hml. Δ-Gal4/+), which was set to 1. Pxn-Gal4 (another hemocyte Gal4 line) gave similar results (not shown). Error bars, SEM, based on three independent experiments. ^p>0.05, Student’s t-tests.
A. Midgut hyperplasia of conventional flies based on pH3 staining. The average number of pH3+ cells was determined from the whole midgut and plotted. Error bars, SEM. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, ***P<0.001, Student’s t-tests.
B. Midgut hyperplasia of conventional flies based on esg-GFP. The number of esg-GFP+ cells counted from P1-4 regions of midguts was divided by total cells counted from DAPI staining and used to define the percentages of intestinal hyperplasia. This percentage was grouped into four classes and plotted as colored bars. The number (n) of midguts analyzed in each group is indicated. ^P>0.05, ***P<0.001, Wilcoxon two-sample test.
C and D. Representative images of axenic Lam mutant flies (D, LamD395/Lamk2,esg-GFP) that exhibit stronger midgut hyperplasia than the axenic Lam heterozygous control flies (C, Lamk2,esg-GFP/+) as judged by esg-GFP reporter expression (green) and pH3 (red) staining in 5-day old flies. Scale bars, 20 μm.
E. Midgut hyperplasia of axenic and conventional flies based on pH3 staining. The average number of pH3+ cells was determined from the whole midguts and plotted. Error bars, SEM. The number (n) of midguts analyzed in each group is indicated. **P<0.01, ***P<0.001, Student’s t-tests.
F. Midgut hyperplasia of axenic and conventional flies based on esg-GFP. The number of esg-GFP+ cells counted from P1-4 regions of midguts was divided by total cells counted from DAPI staining and used to define the percentages of intestinal hyperplasia. This percentage was grouped into four classes as indicated by the colored bars and plotted. The number (n) of midguts analyzed in each group is indicated. **P<0.01, ***P<0.001, Wilcoxon two-sample test.
G. Eclosion and pupariation rates. The axenic (Axe) Lam mutants exhibited increased eclosion and pupariation rates compared to conventional (Con) Lam mutants. Error bars, SEM based on three independent experiments performed with 200 larvae per experiment. Student’s t-tests, **p<0.01, ***p<0.001.
A and B. Graphs showing the overlap of down- (A) or up-regulated (B) genes between midguts from wild-type old (50 days) flies and midguts from young (5 days) flies depleted of LAM in the fat body by Cg-Gal4;tub-Gal80ts driven LAM RNAi.
C and D. qRT-PCR confirmation of selected PGRPs (C, PGRP-LB, LB; PGRP-LF, LF; PGRP-SB1, SB1; PGRP-SB2, SB2; PGRP-SC1, SC1; PGRP-SC2, SC2) and AMPs (D) that were found to be down-regulated in the midgut upon fat body-specific LAM RNAi in 5-day old flies by RNA-seq. The expression changes were plotted relative to the 5-day Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. **p<0.01, ***p<0.001, Student’s t-tests.
E. qRT-PCR analysis of Dpt in midgut from 5- or 50-day Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (UAS-Lam/+) or flies with fat body-specific expression of LAM cDNA (Cg-Gal4/+;tub-Gal80ts/UAS-Lam). Dpt expressions in midgut were plotted relative to 5-day Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
F. qRT-PCR analyses of Dpt in midgut from 5-day old Lam-mutant control flies (Cg-Gal4,LamD395/Lamk2), or 5-day old Lam-mutant flies with dynamin inhibition (UAS-shits), RNAi of key, or RNAi of PGRP-SC2 plus PGRP-LB (SC2&LB) in fat bodies. The Dpt expression change was plotted relative to the control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
G. qRT-PCR analysis of Dpt in midgut from 5-day conventional (Con) or axenic (Axe) Gal4 control flies (Cg-Gal4/+;tub-Gal80ts/+), UAS control flies (Lam RNAi/+) or flies with fat body-specific deletion of Lam (Cg-Gal4/+;tub-Gal80ts/Lam RNAi). Dpt expression in midgut were plotted relative to 5-day conventional Gal4 control (Cg-Gal4/+;tub-Gal80ts/+), which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, **p<0.01, ***p<0.001, Student’s t-tests.
A. qRT-PCR analyses of Dpt in fat body from 5-day old control (Cg-Gal4/+;tub-Gal80ts/+) flies, 5-day old flies depleted of LAM in fat body (Cg-Gal4/+;tub-Gal80ts/Lam RNAi), or the 5-day LAM depleted flies carrying one copy of hsp70-HP1 transgene (Cg-Gal4/hsp70-HP1;tub-Gal80ts/Lam RNAi). Dpt expression was plotted relative to the control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
B. qRT-PCR analyses of Dpt in midgut from 5-day old control flies (Cg-Gal4/+;tub-Gal80ts/+), 5-day old flies depleted of fat body LAM (Cg-Gal4/+;tub-Gal80ts/Lam), or the 5-day LAM depleted flies carrying one copy of hsp70-HP1 transgene (Cg-Gal4/hsp70-HP1;tub-Gal80ts/Lam RNAi). Dpt expression was plotted relative to the control, which was set to 1. Error bars, SEM, based on three independent experiments. *p<0.05, Student’s t-tests.
C. Chromatin immunoprecipitation (ChIP) analysis of selected IMD signaling components of adult fat body from 5-day wild type (w1118) and Lam mutant (LamD395/Lamk2) flies. Chromatin was immunoprecipitated with antibodies to H3K9me3 and control IgG. Primers corresponding to PGRP-LC, IMD, key, Rel, and rp49 (negative control) were used to amplify precipitated DNA. ChIP samples were normalized to 5% of input DNA. Error bars, SEM, based on three independent experiments. ^p>0.05, *p<0.05, **p<0.01, Student’s t-tests.
D–O. Expression patterns of different Gal4 lines as reported by UAS-GFPS65T in adult fat body (D, G, J, M), adult gut (E, H, K, N), and adult circulating hemocytes (F, I, L, O). DAPI labels the nuclei (blue). Phalloidin stains of actin (red) in F, I, L and O. Yellow asterisks mark the heart peripheral cells. Whole guts and hemocytes are outlined by white dashed lines. Scale bars: D, F, G, I, J, L, M, and O (20 μm); E, H, K, and N (200μm).
Gene symbols, description, and fold changes are indicated.