SUMMARY
Fighting viral infections is hampered by the scarcity of viral targets and their variability resulting in development of resistance. Viruses depend on cellular molecules for their life cycle, which are attractive alternative targets, provided that they are dispensable for normal cell functions. Using the model organism Drosophila melanogaster, we identify the ribosomal protein RACK1 as a cellular factor required for infection by internal ribosome entry site (IRES)-containing viruses. We further show that RACK1 is an essential determinant for hepatitis C virus translation and infection indicating that its function is conserved for distantly related human and fly viruses. Inhibition of RACK1 does not affect Drosophila or human cell viability and proliferation, and RACK1-silenced adult flies are viable, indicating that this protein is not essential for general translation. Our findings demonstrate a specific function for RACK1 in selective mRNA translation and uncover a new target for the development of broad antiviral intervention.
INTRODUCTION
Viral infections are a significant threat for all living organisms. In humans, acute and chronic viral infections cause a wide spectrum of diseases, including life-threatening inflammation and cancer. A major challenge for the control of viral infections is that viruses, due to the small size of their genomes, offer few intrinsic targets either for recognition by the immune system or for inhibition by antiviral effector molecules. Furthermore, the error-prone viral polymerases allow RNA viruses to rapidly escape detection by the immune system and to resist the adverse effects of directly acting antiviral molecules. Significantly, viruses rely on numerous host factors for essential functions during their life cycle. These are not subject to rapid sequence changes and hence provide good alternative targets for antiviral therapy. Therefore, a central challenge is to identify cellular factors required for viral replication but dispensable for normal cell function.
RNA replication, transcription and translation are critical steps in the life cycle of RNA viruses, which involve interactions with host-cell molecules. In the model organism Drosophila melanogaster, the small interfering (si) RNA pathway targets viral RNAs (reviewed in (Ding, 2010)). In order to better characterize the contribution of the three core components of this pathway, Dicer-2, R2D2 and AGO2, we performed a proteomic analysis of the complexes assembling around these molecules in infected Drosophila cells (in preparation). One protein copurifying with R2D2 and AGO2 in cells infected with the picorna-like Drosophila C virus (DCV) was the evolutionarily conserved ribosomal protein RACK1. The RACK1 protein has been extensively studied during the last two decades, and shown to be involved in different aspects of cell regulation. RACK1 is an adapter protein, interacting with a variety of signaling molecules (e.g. PKC, Src, MAPK) (Belozerov et al., 2014; Gibson, 2012; Long et al., 2014), and is a component of the 40S subunit of the ribosome (Coyle et al., 2009; Sengupta et al., 2004). RACK1 is thus ideally suited to connect signal transduction pathways to the regulation of translation (Nilsson et al., 2004). Indeed, RACK1 was found to interact with the initiation factor eIF6, which associates with the 60S subunit of the ribosome, and prevents its association with the 40S subunit. eIF6 phosphorylation by RACK1-assisted PKC triggers its release from the 60S subunit, thus promoting the formation of 80S active ribosomes (Ceci et al., 2003).
Here, we show that RACK1 is mandatory for DCV replication, but largely dispensable for cell viability and proliferation. We further demonstrate that RACK1 is required for internal ribosome entry site (IRES)-dependent translation in Drosophila, and in human hepatocytes, where this factor is an essential determinant of hepatitis C virus infection. By contrast, RACK1 is not required for 5′ cap-dependent translation. Collectively, our data unravel a specific function for ribosomal protein RACK1 in selective mRNA translation of fly and human viruses and uncover a previously undiscovered target for the development of broad antiviral intervention.
RESULTS
RACK1 is required for Dicistroviridae infection in Drosophila
In a proteomic analysis of the interactome of Dicer-2, R2D2 and AGO2 in virus infected cells, to be reported elsewhere, we identified 16 ribosomal proteins. To address the functional relevance of this finding, we systematically depleted these ribosomal proteins from S2 cells by RNAi, and tested DCV replication. Knockdown of most ribosomal genes affected cell viability or proliferation and did not yield interpretable results with regards to DCV infection (Figure 1A, B). Indeed, silencing of these genes may result in decreased ability of the cells either to support viral replication, or to control the infection. By contrast, depletion of RACK1 (Figure S1A) did not affect cell viability or proliferation in S2 cells (Figure 1B, C) or in two other cell lines (Figure S1B). However, it resulted in a significant decrease of DCV titer in infected cells (Figure 1A). Furthermore, RACK1 silencing did not affect replication of either FHV or VSV (Figure 2A, B), indicating that the RACK1-depleted cells are not only viable and able to proliferate, but can also support replication of other viruses. To test whether the effect of RACK1 was specific to DCV, or to the family to which it belongs, we infected S2 cells with Cricket Paralysis Virus (CrPV), another member of the Dicistroviridae family. Replication of CrPV was also strongly impaired when RACK1 was depleted (Figure 2B).
Figure 1. RACK1 is required for DCV replication, but not for viability or proliferation in Drosophila cells.
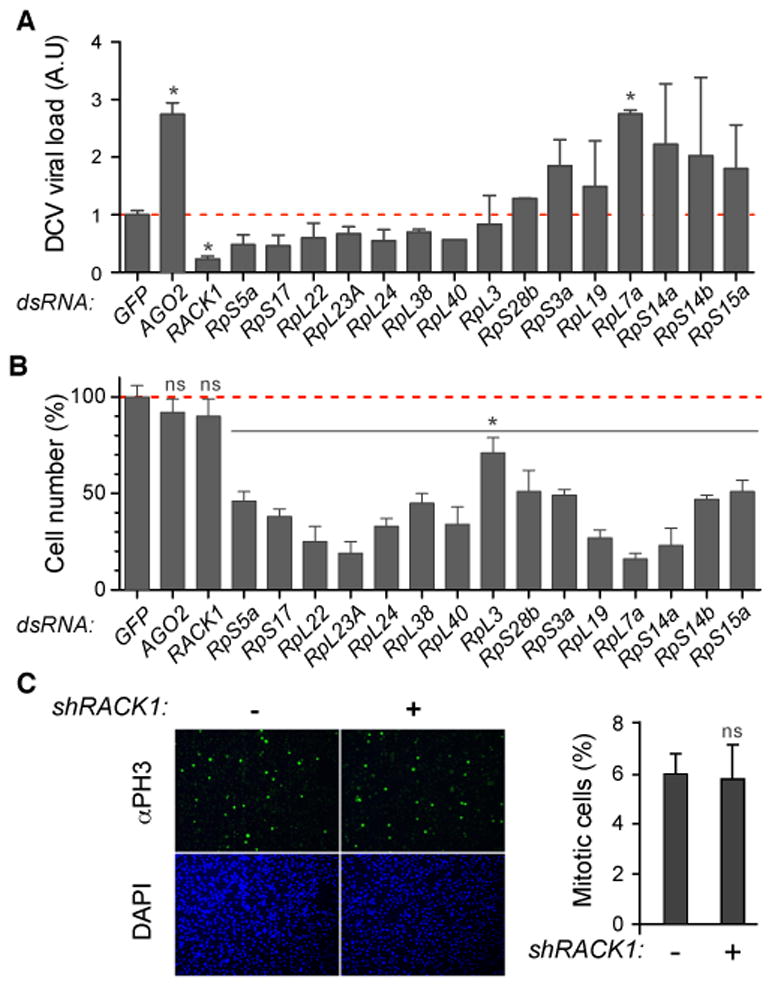
(A, B) Quantification of DCV viral RNA levels by qRT-PCR (A) and of cell numbers as estimated by DAPI staining (B) in cells treated with the indicated dsRNAs to induce silencing. Cells treated with a dsRNA corresponding to GFP and AGO2 sequences are used as a reference and a control, respectively. (C) S2 cells stably transfected with a metallothionein promoter driven vector expressing a shRNA targeting the 5′ UTR from the RACK1 gene were treated or not with CuSO4 for three days, stained with DAPI and an anti-phospho H3 antibody (left panels) and counted (right panel). Data represent the mean and s.e.m. of at least three independent experiments. ns: non significant; * p<0.05. See also Figure S1 and Table S1.
Figure 2. RACK1 is required for replication of DCV and CrPV, but not FHV and VSV.
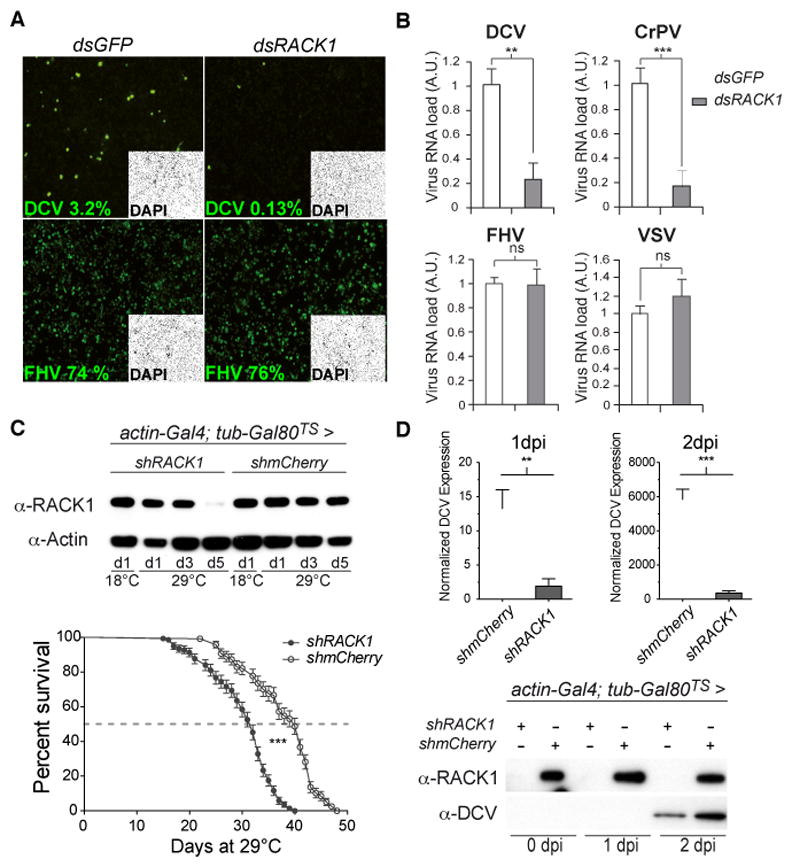
(A, B) S2 cells were treated with either control (GFP) or RACK1 dsRNA for 4 days, before challenge with DCV, FHV, VSV or CrPV. Viral infection was monitored by immunofluorescence (A) and qRT-PCR (B) 16h or, in the case of VSV, 48h later using antibodies recognizing capsid proteins. The percentage of infected cells is indicated for each virus in panel A. (C) Silencing of RACK1 expression in transgenic flies expressing a shRNA targeting the 5′ UTR from the RACK1 gene, using the Gal4-UAS system and the broadly expressed actin-Gal4 driver controlled by the thermosensitive (TS) tub-Gal80 repressor. A shRNA targeting the mCherry protein was used as a control. The life span of RACK1 depleted flies is shown in the bottom graph. (D) RACK1 silenced flies infected by DCV after 5 days at 29°C show a decrease of the viral RNA and protein, as indicated by qRT-PCR (upper panel) and western blot. Data represent the mean and s.e.m. from at least three independent experiments. ns: non significant; dpi: days post-infection; * p<0.05; ** p<0.01; *** p<0.001. See also Figures S1, S2.
We next confirmed these findings in vivo. RACK1 null mutant flies are not viable, indicating that RACK1 exerts developmental functions (Kadrmas et al., 2007). In agreement with this finding, silencing RACK1 expression with a small hairpin (sh) RNA driven by the broadly active actin5C promoter was embryonic lethal. When the thermosensitive Gal80 system was used to express the shRNA only in adult flies, development occurred normally and the adult flies expressed significantly reduced levels of RACK1 at the permissive temperature of 29°C (Figure 2C). The reduced levels of RACK1 did not affect the viability of the flies, although it reduced longevity by 20% at this temperature. In addition, the eggs laid by RACK1-silenced females showed a phenotype similar to that of RACK1 mutants (Figure S1C) (Kadrmas et al., 2007). Thus, even though RACK1 is required during development, it appears to be largely dispensable in adult flies. As expected, when these flies were challenged with DCV, both viral RNA and capsid protein levels were markedly reduced at 1 and 2 days post-infection compared to controls (Figure 2D). Overall, our data indicate that replication of the Dicistroviridae DCV and CrPV requires the ribosomal factor RACK1, which is otherwise dispensable for the viability of S2 cells and adult flies.
RACK1 is required for viral IRES-dependent translation
Our data indicate that RACK1 is required for a step of viral replication specific to Dicistroviridae. Whereas FHV and VSV use a canonical strategy of cap-dependent initiation of translation, DCV and CrPV RNA recruits the 40S ribosomal subunit through IRES sequences to initiate translation (Figure S2A). Furthermore, although initially identified as a scaffolding protein involved in protein kinase C signaling, RACK1 is now recognized as a component of the 40S subunit of the ribosome. This suggested to us that RACK1 was required for viral translation. We first verified that RACK1 is indeed required at the ribosome level for CrPV replication. We silenced RACK1 expression in a stable cell line using an shRNA targeting the 5′ untranslated region (Figure S1D), and observed a marked decrease in CrPV replication (Figure S1E). Transfection of a vector expressing wild-type RACK1 restored CrPV replication in these cells (Figure 3A). By contrast, expression of mutant proteins unable to interact with either RpS17 (D108Y) (Kuroha et al., 2010) or 18S rRNA (R38D/K40E) (Coyle et al., 2009) did not rescue CrPV replication (Figure 3A). We conclude that RACK1 is required in the 40S ribosomal subunit for CrPV replication.
Figure 3. The ribosomal protein RACK1 is required for IRES-mediated translation.
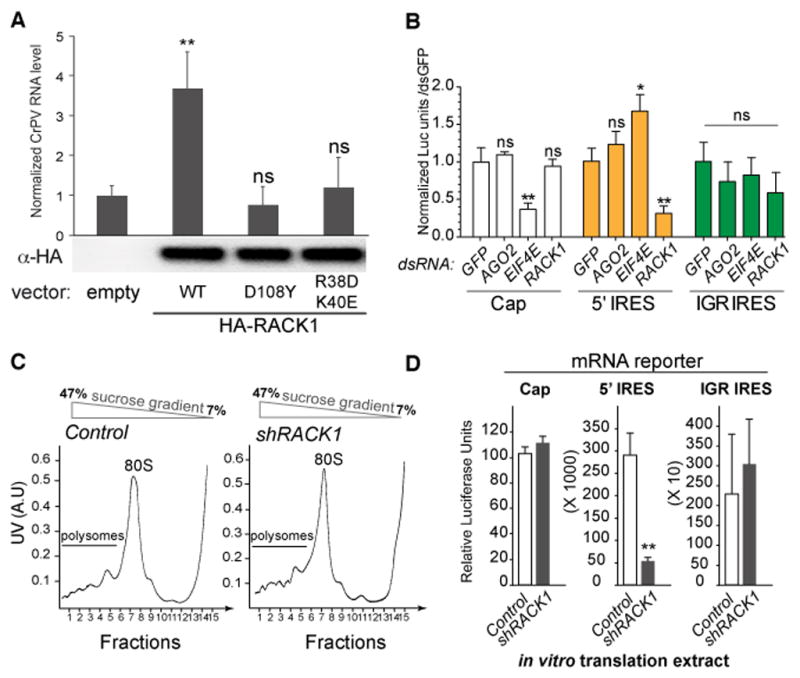
(A) Stable S2 transformants expressing a shRNA targeting the 5′ UTR of RACK1 were transfected with vectors expressing three versions of RACK1 (WT, D108Y or R38D/K40E). Expression of the transfected RACK1 was monitored by western blot using an antibody recognizing the N-terminal tag HA. The cells were infected with CrPV for 16h, and viral RNA loads were determined by qRT-PCR. Data represent the mean and s.e.m. from three independent experiments. (B) RACK1 is required for translation regulated by the 5′ IRES, but not the intergenic (IGR) IRES, of CrPV. S2 cells were treated with dsRNAs corresponding to GFP (control), AGO2, eIF4E or RACK1 for 3 days, before transfection of the indicated Luciferase reporters (5′CAP, IRESCrPV-IGR or IRESCrPV-5′; see Fig. S2). Luciferase activity was monitored 48h later. The ratio of the activity of the IRES-dependent luciferase and the 5′ cap-dependent luciferase is plotted and normalized to the control for the three reporters. Data represent the mean and s.d. from six independent experiments. (C) Polysome profiles from S2 cells expressing or not a shRNA targeting the 5′ UTR of RACK1. The position of the peaks corresponding to the 80S ribosomes and the polysomes are indicated. (D) In vitro translation of capped and IRES-dependent reporters using cell free extracts prepared from control or RACK1-silenced S2 cells. Data represent the mean and s.d. from three independent experiments. ns: non significant, * p<0.05, ** p<0.01. See also Figures S2, S3.
To confirm that RACK1 is involved in translation from Dicistroviridae RNAs, we tested whether its depletion affected translation of luciferase reporters placed under the control of the two IRES elements from CrPV (Figure 3B and Figure S2C). Translation of a 5′ cap-dependent RNA was not affected in the absence of RACK1, although it was affected when expression of eIF4E was knocked down. Translation from the CrPV 5′ IRES reporter was not reduced, and was even slightly increased, when eIF4E was silenced, suggesting that the 5′ IRES drives non canonical translation. Interestingly, a significant reduction of luciferase production was observed for the 5′ IRES reporter in RACK1 silenced cells (Figure 3B). Silencing of RACK1 did not affect the amount of the 5′ IRES reporter luciferase mRNA in the cells, indicating that RACK1 affects translation, rather than RNA stability (Figure S3). By contrast, translation driven by the intergenic (IGR) IRES (Jan and Sarnow, 2002; Spahn et al., 2004) was not affected by the level of RACK1 in the cells (Figure 3B). Polysome profiles from S2 cells and RACK1-silenced stable derivatives of these cells (Figure S1D) were similar, confirming that RACK1 does not affect significantly general translation (Figure 3C). Finally, we prepared cell-free translation extracts from control and RACK1-depleted S2 cells, and used them to monitor translation of in vitro transcribed, capped and IRES-dependent RNAs. Translation of the 5′ IRES reporter RNA was strongly reduced in the RACK1 depleted extract. By contrast, translation of the 5′ CAP and IGR IRES dependent reporters was not inhibited and was even slightly stimulated (Figure 3D). Overall, our data indicate that ribosomal RACK1 is required for IRES-dependent translation of Dicistroviridae both ex vivo and in vitro.
RACK1 is an essential host factor for HCV infection
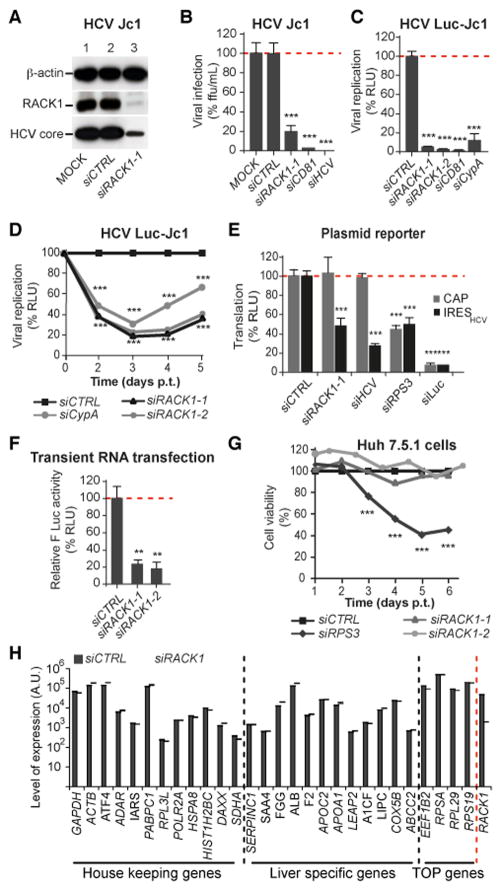
RACK1 is an evolutionarily strongly conserved factor, and we asked whether it plays a role in the translation driven by the IRES of a mammalian virus. Hepatitis C virus (HCV), a major cause of liver disease and hepatocellular carcinoma, is a positive strand RNA virus member of the Flaviviridae family depending on a highly structured IRES for its translation (Figure S2) (Spahn et al., 2001). Transfection of an siRNA targeting RACK1 markedly reduced expression of the protein in Huh7.5.1 cells (Figure 4A), a human hepatocyte-derived cell line highly permissive for HCV infection (Lindenbach et al., 2005; Wakita et al., 2005). Infection of RACK1-depleted Huh7.5.1 cells by cell culture-derived HCV (Jc1 strain) was strongly and significantly reduced, as revealed both by immunodetection of the viral core protein (Figure 4A) and the focus forming assay performed by infection of naïve Huh7.5.1 cells with supernatants from infected and treated cells (Figure 4B). A similar inhibition of infection was observed for HCV Luc-Jc1 (Figure 4C), a well-characterized recombinant virus expressing a luciferase reporter (Figure S2B). Inhibition of RACK1 expression was as efficient as the silencing of the key HCV host factors CD81 (Koutsoudakis et al., 2007) and Cyclophilin A (CypA) (Kaul et al., 2009) (Figure 4A, B, C). We next transiently depleted RACK1 in Huh7.5.1 cells replicating the reporter virus HCV Luc-Jc1, and observed a marked impairment of HCV replication (Figure 4D), demonstrating that RACK1 is required for HCV translation/replication rather than entry. HCV replication rebound observed after day 4 was due to progressive loss of RACK1 silencing leading to neosynthesis of RACK1 (Figure S4A).
Figure 4. RACK1 is a specific host-factor required for IRES-mediated translation of HCV.
(A–C) Huh7.5.1 cells were transfected with siRNAs either control (siCTRL) or targeting RACK1 (siRACK1-1 or -2), CD81 (siCD81), Cyclophylin A (siCypA), or HCV IRES (siHCV) before infection three days later with HCV Jc1 (A, B), or HCV Luc-Jc1 (C). Viral infection was monitored 3 days post-infection, by immunoblotting using antibodies recognizing HCV core protein (A); by counting foci forming units (ffu/ml) (B); or by quantifying luciferase activity (C). (D) HCV Luc-Jc1 replicating cells were transfected with siCTRL, two different siRNAs targeting RACK1 or siCypA, and replication was monitored during 5 days by luciferase activity quantification. (E) Huh7.5.1. cell lines stably expressing an IRES (IRESHCV-Luc) or a 5′ cap (CTRL-Luc) dependent luciferase reporter gene were transfected with siCTRL, siRACK1, siHCV, siRPS3 or siLuc. Translation was monitored 72h later by luciferase activity quantification. (F) Huh7.5.1 cells were transfected with the indicated siRNAs and, 72 h later with in vitro transcribed IRESHCV or 5′ cap dependent luciferase mRNAs. Luciferase activity was monitored 5h later. (G) Cell viability of Huh7.5.1 cells silenced with the indicated siRNAs was measured during 5 days using MTT assay. ** p<0.01; *** p<0.005. (H) Quantification of representative mRNAs in polysomes prepared from Huh7.5.1 cells transfected with siCTRL or siRACK1. Gene expression levels, shown in arbitrary units, was determined by hybridization on genome wide microarrays, and represent the mean +/− s.d. of 4 individual samples. Each sample was analyzed individually. See also Figures S2, S4 and Table S2.
To confirm that the inhibition of HCV replication is indeed mediated by the effect of RACK1 on IRES-mediated translation, we established stable cell lines expressing an IRESHCV-luciferase reporter construct or a classical capped reporter gene (Figure S2C), and transfected these cells with RACK1-specific siRNAs. Silencing of RACK1 markedly and specifically decreased IRESHCV-dependent translation, to a similar extent as an antiviral siRNA directed against the IRESHCV (Figure 4E). By contrast, silencing of ribosomal protein RPS3 inhibited translation from both IRES- and 5′ cap-dependent reporter constructs (Figure 4E). Similar results were obtained when in vitro transcribed reporter mRNAs were transfected into Huh7.5.1 cells, ruling out an effect of RACK1 on transcription of the IRESHCV-luciferase reporter gene (Figure 4F).
Importantly, RACK1-specific siRNAs did not affect cell proliferation (Figure S4B) or viability, in contrast to silencing of the ribosomal protein RPS3 (Figure 4G). A genome-wide microarray analysis of polysomes prepared from control or RACK1-silenced human Huh7.5.1 cells revealed that the amount in polysomes of mRNAs for most genes, including house keeping genes and important hepatocyte specific genes such as albumin or lipoproteins, was not affected by RACK1 depletion (Figure 4H). Of note, silencing of RACK1 also did not affect the presence of 5′ terminal oligopyrimidine tract (TOP) mRNAs in polysomes (for details, see Supplemental information). This result suggests that translation of the large majority of mRNAs is not affected by the absence of RACK1 in human hepatocytes under normal culture conditions and confirms the results obtained in the model organism Drosophila.
The effect of RACK1 on viral translation is independent of the miRNA pathway
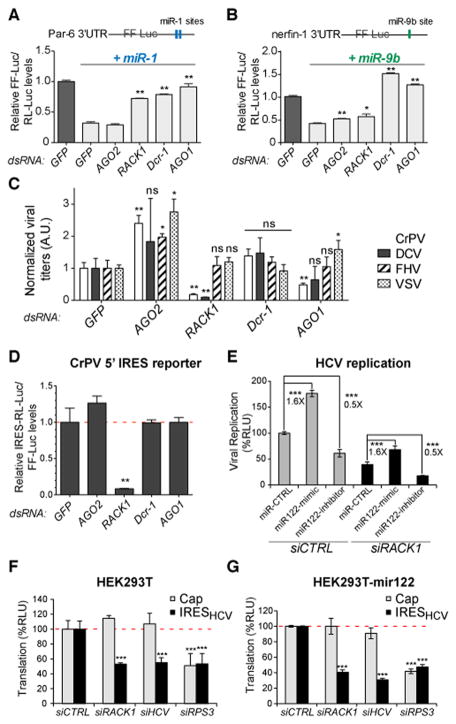
While this work was in progress, a role for RACK1 in miRNA function was reported in the plant Arabidopsis thaliana (Speth et al., 2013), the model organism Caenorhabditis elegans (Chu et al., 2014; Jannot et al., 2011) and humans (Otsuka et al., 2011). In light of the important impact of the cellular microRNA miR122 on HCV replication (Jopling et al., 2005), this suggested that RACK1 might operate on viral translation through the miRNA pathway. We first verified that RACK1 affects the miRNA pathway in Drosophila. Expression in S2 cells of two previously described miRNA reporters, Par-6 and nerfin-1 (Eulalio et al., 2007), was derepressed when RACK1 was silenced, indicating that in Drosophila as well, RACK1 is involved in miRNA function (Figure 5A, B). We note however that the derepression is much stronger for the miR1 reporter than for the miR9b reporter, suggesting that the role of RACK1 may be specific of a subset of miRNAs. By contrast, silencing of Dcr-1 or AGO1 derepressed equally well the two miR reporters (Figure 5A, B). We next tested whether miRNAs play a role in viral replication, by monitoring accumulation of viral RNAs in cells depleted of Dcr-1 or AGO1. Silencing of Dcr-1 had no effect on the viral RNA load of the four viruses tested (Figure 5C). Silencing of AGO1 did reduce to some extent CrPV and DCV RNA load. However, this reduction was variable in the case of DCV, and not to the extent of the reduction observed when RACK1 was silenced for DCV and CrPV (Figure 5C). Thus, although the miRNA pathway may have a contribution in the replication of Dicistroviridae, our data suggest that the strong effect of RACK1 cannot be accounted for only by its effect on miRNA function. This was confirmed by the observation that silencing of Dcr-1 or AGO1 had no effect on translation driven by the IRESCrPV-5′, unlike silencing of RACK1 (Figure 5D).
Figure 5. The effect of RACK1 on viral translation is independent of the miRNA pathway.
(A, B) RACK1 is required for miR1 and miR9b silencing. The structure of the Par-6 3′ UTR and nerfin-1 3′ UTR reporter constructs is represented on top, and the luciferase activity in cells silenced for the indicated genes is shown below. (C) Effect of the depletion of AGO1, Dcr-1 and RACK1 on replication in Drosophila S2 cells of CrPV, DCV, FHV and VSV. Cells were transfected with the indicated dsRNAs, and infected four days later. Viral RNA was extracted 24hpi, and quantified by qRT-PCR. (D) Silencing of AGO1 or Dcr-1 does not affect the activity of a Luciferase reporter gene controlled by the IRESCrPV-5′ in Drosophila S2 cells. (E) A miR122 mimic and a miR122 inhibitor affect HCV replication similarly in control or RACK1-silenced Huh7.5.1 hepatocytes. (F–G) Silencing of RACK1 affects the activity of the IRESHCV-luciferase reporter in miR122 deficient (F) and stably transfected miR122 expressing (G) HEK-293T cells, respectively. Data represent the mean and s.e.m. of at least three independent experiments. ns: non significant; * p<0.05; ** p<0.01, *** p<0.001. See also Figure S5.
In mammalian hepatocytes, HCV translation depends on AGO2 and miR122 (Conrad et al., 2013; Roberts et al., 2011). As expected, transfection of Huh7.5.1 cells with a miR122 mimic increased HCV replication, while transfection of a miR122 inhibitor led to decreased viral replication (Figure 5E). Importantly, the impact of the miR122 mimic and the miR122 inhibitor on HCV replication did not depend on RACK1 (Figure 5E). To unambiguously determine whether the contribution of RACK1 to HCV translation was dependent on miR122, we used HEK-293T cells, which do not express miR122 ((Da Costa et al., 2012), Figure S5). Silencing of RACK1 expression efficiently repressed translation driven by the IRESHCV in these cells (Figure 5F). Finally, transduction of HEK-293T cells with an expression vector for miR122 did not affect the impact of RACK1 on HCV translation (Figure 5G), although miR122 was expressed and functional in these cells (Figure S5A, B). Collectively, these results indicate that RACK1 and miR122 regulate HCV translation by different mechanisms.
The eIF3j subunit is dispensable for cell viability, but important for CrPV and HCV replication
We next attempted to gain mechanistic insight on the role of RACK1 in viral translation. Previous cryo-electron microscopy studies have highlighted the interaction of the 40S subunit with the HCV IRES and, in spite of their low resolution, have suggested that binding of the HCV IRES triggers a pronounced conformational change in the small subunit of the ribosome (Spahn et al., 2001; 2004). HCV IRES has been also visualized on the 80S human ribosome and RACK1 localized in its vicinity (Boehringer et al., 2005; Sengupta et al., 2004). The recently elucidated crystal structure of the small subunit of the ribosome at 3.9Å (Rabl et al., 2011) allows to fit the crystal structure in the cryo-electron microscopy density. The picture obtained suggests that RACK1 is located in close proximity to the IRES of HCV in the region affected by the conformational change triggered upon IRESHCV binding (Figure S6A). By contrast, the IRESCrPV-IGR, which does not depend on RACK1 (Figure 2C), interacts with a distinct site of the 40S subunit, directly contacting RpS25 (Figure S6B) (Fernandez et al., 2014; Koh et al., 2014; Schuler et al., 2006; Spahn et al., 2004). Although no direct contacts between RACK1 and IRESHCV could be observed, a recent study indicates that a peripheral domain of the translation initiation factor eIF3, which is required for IRESHCV-dependent translation (Kieft, 2008), is in contact with RACK1 (Figure S6C) (Hashem et al., 2013a; Sun et al., 2013). This domain may be the functional link between RACK1 and IRESHCV-dependent translation.
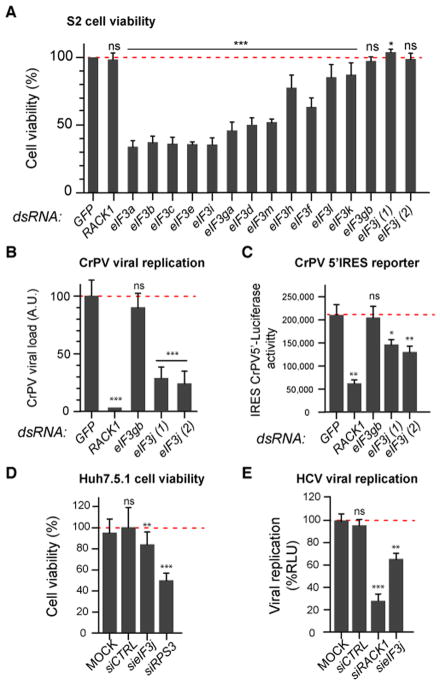
We asked whether some subunits of eIF3, such as eIF3c, which has been shown to interact with RACK1 in yeast, may be specifically involved in IRES-dependent translation, like RACK1. We first tested in Drosophila S2 cells whether some subunits of the eIF3 complex are dispensable for cell viability in normal culture conditions. Out of the 14 genes encoding eIF3 components (the Drosophila genome contains two eIF3g paralogues, CG8636 (eIF3ga) and CG10881 (eIF3gb)), only two were not required for cell viability or proliferation (Figure 6A). One of these genes is CG10881, encoding eIF3gb, which is expressed specifically in testis (Chintapalli et al., 2007) and thus provides a useful negative control. The second gene is the Drosophila orthologue of eIF3j (Figure 6A). We next monitored CrPV replication in cells silenced for eIF3j or eIF3gb (CG10881). Although silencing of eIF3gb did not affect CrPV replication, silencing of eIF3j resulted in a significant reduction of CrPV replication (Figure 6B). Silencing of eIF3j, but not of eIF3gb, also affected translation of the IRESCrPV5′-luciferase reporter, although not as strongly as silencing of RACK1 (Figure 6C). In Huh7.5.1 cells, silencing of eIF3c affected cell viability. By contrast, silencing of eIF3j only marginally affected cell viability (Figure 6D and (Wagner et al., 2014)). Interestingly however, it resulted in a moderate but significant decrease of HCV replication (Figure 6E). Altogether, these results suggest that the eIF3j subunit might participate in the observed effects of RACK1 on translation.
Figure 6. eIF3j is required for CrPV and HCV replication, but not for cell viability.
(A) Quantification by the MTS assay of the number of viable cells 5 days after treatment of S2 cells with the indicated dsRNAs. Two different dsRNA preparations, targeting different regions of the gene, were used for eIF3j. (B) Quantification by qRT-PCR of CrPV viral RNA levels in S2 cells treated with the indicated dsRNAs. (C) Activity of the IRESCrPV5′ in S2 cells silenced for the indicated genes. (D) Quantification of Huh7.5.1 cell viability after silencing of the indicated genes. (E) Quantification of HCV replication in Huh7.5.1 cells transfected with the indicated siRNAs. See also Figure S6.
DISCUSSION
A new function for RACK1 in IRES-dependent translation
Our data reveal a new function for RACK1 in specific mRNA translation. Indeed, silencing RACK1 expression does not affect viability of Drosophila S2 or human Huh7.5.1 cells in tissue culture, indicating that formation of active ribosomes is not strictly dependent on RACK1. In vivo as well, translation can occur in the absence of RACK1, as lethality in RACK1 mutant animals does not occur before larval stages for Drosophila and gastrulation in mice (Kadrmas et al., 2007; Volta et al., 2013). In agreement with this observation, translation of a 5′ cap-dependent reporter was not affected in the absence of RACK1 in Drosophila and human cells. Nevertheless, the fact that RACK1 mutant animals cannot complete their development suggests that this protein is required for the translation of some cellular mRNAs, in addition to viral IRES-containing RNAs. Interestingly, previous studies have highlighted the role of another protein from the 40S subunit of the ribosome, RpS25, in IRES-dependent translation (Landry et al., 2009). Performed on yeast and mammalian tissue-culture cells with IRES reporter assays, these experiments concluded that RpS25 is essential for the activity of two viral IRES, IRESHCV and IRESCrPV-IGR. The mechanism used by RpS25 and RACK1 to promote translation is probably different because (i) RpS25 is required for IRESCrPV-IGR, unlike RACK1; and (ii) structural data place RpS25 at a distance from RACK1 on the 40S subunit of the ribosome, providing an explanation for its importance on the activity of the IRESCrPV-IGR. Several other ribosomal proteins (e.g. RpL38, RpL40) were recently proposed to be involved in specific translation of some 5′ cap-dependent mRNAs (Kondrashov et al., 2011; Lee et al., 2013), indicating that transcript-specific regulation can occur in the absence of IRES elements. Our data lend support to an evolving picture of the eukaryotic ribosome, which includes structurally peripheral components such as RACK1 involved in the modulation of translation of specific mRNAs (reviewed in (Xue and Barna, 2012)). They have implications for the development of new antivirals, and raise questions on the mechanism underlying the role of RACK1 in IRES-dependent translation.
RACK1 as a target for broad antiviral intervention
Our results open interesting therapeutic perspectives for a broad range of viral infections including chronic hepatitis C, a major cause of liver cirrhosis and cancer. Because HCV translation initiates viral genome neosynthesis via the formation of the replication complex, RACK1-mediated translation is a crucial step in virus propagation. Thus, RACK1 is a novel host target for antiviral therapy, which is complementary to interferon-based therapies or direct-acting antivirals (DAAs). DAAs have achieved high response rates with cure in late-stage clinical trials, but high costs will limit their broad access. In addition, certain patient groups (e.g. genotype 3, renal failure, hepatic decompensation, liver transplantation) will need complementary approaches (Chung and Baumert, 2014; Liang and Ghany, 2013).
The low variability of host factors targeted by host-targeted antivirals (HTAs) results in a high genetic barrier to resistance (Nathan, 2012). Indeed, HTAs effectively inhibit HCV escape variants (Fofana et al., 2010; Lupberger et al., 2011), as well as DAA-resistant virus (Xiao et al., 2014a). Furthermore, their complementary mechanism of action results in synergy with DAAs (Xiao et al., 2014b). Given that HTAs interfere with host targets, one theoretical caveat is the possibly greater risk of cellular toxicity as compared to DAAs. Interestingly, our data obtained in cell culture models did not reveal any major toxicity linked to RACK1 inhibition. Thus, our proof-of-concept studies in state-of-the-art cell culture models open a highly attractive and innovative perspective to develop small molecules targeting RACK1. RACK1 inhibitors may also be of interest for treatment of infection of many other human or animal viruses using 5′ cap-independent mechanisms for the translation of their RNAs.
Mechanistic insight on the role of RACK1 in IRES-dependent translation
While this work was in preparation, several reports described a role for RACK1 in miRNA function. However, our data in Drosophila and human cells indicate that the role of RACK1 in IRES-dependent translation does not involve small regulatory RNAs. Nevertheless, the connection between RACK1 and AGO proteins is intriguing, and suggests that RACK1 may participate in a checkpoint for the control of the translation of specific mRNAs by miRNAs or siRNAs.
The ribosome code or filter hypothesis posits that some ribosomal proteins have evolved to mediate translation of specific mRNAs (Mauro and Edelman, 2002; Topisirovic and Sonenberg, 2011; Xue and Barna, 2012). A central unresolved issue of this hypothesis is the nature of the cis-acting elements defining a possible “ribosome code”. In the case of RACK1, these cis-acting elements include viral IRES. Interestingly, the IRESCrPV-IGR is active in the absence of RACK1, unlike the IRESCrPV-5′ or the IRESHCV. This IRESCrPV-IGR (class I IRES) is capable on its own, without any initiation factors, of binding directly the 40S subunit and of recruiting the 60S subunit to form an active 80S ribosome, thus bypassing the loading of the initiator methionyl-tRNAi (Jan and Sarnow, 2002; Pestova et al., 2004). By contrast, the function of IRESHCV (class II IRES) requires two canonical eIFs, eIF2 and 3, as well as Met-tRNAi (Kieft, 2008). This suggests that the effect of RACK1 on translation initiation may require one of these factors. Interestingly, the eIF3 complex binds to the 40S ribosomal subunit, and to the IRESHCV (e.g. (Kieft et al., 2001)). Furthermore, RACK1 was shown to associate with one of the eIF3 subunits in order to assemble a translation pre-initiation complex in yeast (Hashem et al., 2013a; Kouba et al., 2012).
Although our understanding of the molecular structure of the core of the 13 subunits eIF3 complex has progressed remarkably in recent years (e.g.(Hashem et al., 2013b; Sun et al., 2011)), the role of the non-core subunits remains essentially untested in animals. Interestingly, the subunit eIF3e in the yeast Schizosaccharomyces pombe is involved in translation of a selected set of RNAs (Sha et al., 2009; Zhou et al., 2005). More recently, one of the two eIF3h genes present in zebrafish, eIF3ha, was shown to encode a factor specifically targeting crystalline isoform mRNAs for translation during lens development (Choudhuri et al., 2013). Our data indicate that, like RACK1, the subunit eIF3j is not required for cell viability in Drosophila, but is required for CrPV replication and IRESCrPV5′ driven translation. This raises the possibility that RACK1 and eIF3j act together in translation of a specific subset of mRNAs.
Several observations support a role for eIF3j in selective mRNA translation. First, it is located in the decoding center of the 40S ribosomal subunit, where it can regulate access to the mRNA binding cleft (Fraser et al., 2007; 2009). Second, it is located at the periphery of the eIF3 complex, often in sub-stoichiometric quantities, indicating that it can undergo regulated cycles of association and dissociation (Hinnebusch, 2006; Miyamoto et al., 2005; Sha et al., 2009). Third, experiments in S. pombe and human cells indicate that it can be regulated post-translationally by phosphorylation (Sha et al., 2009) or caspase-mediated C-terminal truncation (Bushell et al., 2000). Altogether, this suggests that RACK1 may act as a scaffold recruiting an enzyme modifying eIF3j in order to allow access of the entry channel of the 40S subunit to IRES-containing mRNAs. In a way, such a scenario would be reminiscent of the recently described role of another eIF3 subunit, eIF3e, which controls the recruitment of the kinase Mnk1 to phosphorylate eIF4E, thus promoting selective mRNA translation in human cells (Walsh and Mohr, 2014).
EXPERIMENTAL PROCEDURES
Silencing candidate gene expression by RNAi and screening
DsRNAs targeting the candidate genes were designed using the E-RNAi algorithm (http://www.dkfz.de/signaling/e-rnai3/). Knock-down in Drosophila S2 cells was performed in 96-well plates using the bathing method, and cells were challenged with virus 4 days later. Viral load was determined by qRT-PCR. Alternatively, infected cells were fixed and labeled with anti-capsid antibodies for immunofluorescence analysis using the InCELL1000 Analyzer workstation (GE LifeSciences). Image data processing was performed using the InCELL Analyzer software. See Extended Experimental procedures in Supplemental material for more details.
Preparation of cell-free extract for in vitro translation
In vitro translation competent extracts were prepared from control or RACK1-silenced S2 cells as described in (Wakiyama et al., 2005). Briefly, cells were resuspended in lysis solution [40 mM Hepes–KOH (pH 8), 100 mM potassium acetate, 1 mM magnesium acetate, and 1 mM dithiothreitol] at a cell density of approximately 109 mL−1 and were placed in the Cell Disruption Bomb (Parr Instrument Company). The homogenate produced upon the pressure release was cleared by centrifugations at 4°C, and creatine kinase was added at 0.24 mg.mL −1 of lysate, before storage in aliquotes at −80°C. Reporter mRNAs were synthesized by transcription in vitro using recombinant T7 RNA polymerase. A non-functional cap (ApppG) (New England Biolabs) was added at the 5′ end of the IRES monocistronic reporter mRNAs to protect them from degradation. Cap-dependent translation was measured with a Renilla Luciferase reporter mRNA that was capped with the ScriptCap m7G capping system (Epicentre Biotechnologies). In vitro translation was performed as previously described (Wakiyama et al., 2005) and under sub-saturating conditions to avoid substrate titration.
HCV infection and replication assays
Huh7.5.1 human hepatoma cells were infected with cell culture-derived HCV (HCVcc strains Jc1 and Luc-Jc1, half-maximal tissue culture infectious dose (TCID50 104 mL−1 for both viruses)) as described (Lupberger et al., 2011; Pietschmann et al., 2006). Two days before infection, gene silencing was performed by reverse transfection with 10 nM of siRNA (Silence®Select siRNA, Ambion) specific for RACK1, CD81, Cyclophilin A, HCV IRES or a nonspecific control siRNA. Viral infection and RACK1 depletion were analyzed by western blotting and quantified by counting of focus forming units (ffu)/mL following immunostaining using a HCV core-specific antibody (mAbC7-50, Affinity BioReagents, CO) or by luciferase reporter gene expression in cell lysates 3 days post infection. For HCV replication experiments, Huh7.5.1 cells were electroporated with HCV Luc-Jc1 RNA (Koutsoudakis et al., 2007). Three days later, cells were reverse transfected with siRNAs.
Supplementary Material
Acknowledgments
This work was supported by CNRS, Inserm, and grants from NIH (PO1 AI070167, to J.A.H. and J.-L.I.); Investissement d’Avenir programs (HepSys ANR-10-LAB-28, to T.B. and NetRNA ANR-10-LABX-36; I2MC ANR-11-EQPX-0022, to J.-L.I. and S.M.; IDEX W13RATCS to C.S.), ANR (ANR-09-MIEN-006-01 to J.-L.I. and ANR-11-SVSE802501 to F.M.), ARC (IHU201301187 to T.B.), ANRS (2010-307/2011-415, to C.S.), FRM (to C.M.). Financial support from the TGE FT-ICR, the ESPCI ParisTech and the Fonds pour la Recherche et la Technologie of the French Ministry of Research for the LTQ FT acquisition are gratefully acknowledged. M.L.H., K.M. and A.F. were supported by fellowships from ANRS, CNRS/Région Alsace, and the IDEX program from the University of Strasbourg respectively. We thank S. Pellegrino and M. Yusupov (IGBMC, Strasbourg) for advice and assistance in the preparation of translation competent cell free extracts; R. Bartenschlager (University of Heidelberg, Heidelberg, Germany) for providing Jc1 and Luc-Jc1 expression vectors; F.V. Chisari (TSRI, La Jolla, CA) for Huh7.5.1 cells; M. Beckerle (University of Utah) for providing RACK1 antiserum; S. Thompson (Birmingham, AL) for sharing the plasmid pSRT208; the transgenic RNAi Project (TRiP) at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks. We also thank E. Santiago, N. Ftaich and M. Parnot for technical assistance; B. Fischer and D. Dembele (siRNA screening and microarray platforms at IGBMC) for the InCell1000 and the transcriptomic analysis; and E. Westhof and W. Filipowicz for discussions and critical reading of the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
KM and LMH performed and analyzed most experiments in drosophila and Huh7.5.1 cells, respectively. CM and AG designed, performed or supervised, and analyzed the data for the experiments in flies and in Fig. 6. SM prepared Fig. S6, and AF analyzed the polysome microarray expression data. YV and JV identified RACK1 as a protein associating with AGO2 and R2D2 in DCV infected cells. FM designed, performed or supervised, and analyzed the data for the polysome and in vitro translation experiments. JAH, TFB, CS and JLI designed the experiments, analyzed the data and wrote the article.
Supplemental information includes Extended Experimental procedures, six Supplementary Figures and two Supplementary Tables. The microarray data presented in Figure 4H have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database under the accession number GSE60374.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belozerov VE, Ratkovic S, McNeill H, Hilliker AJ, McDermott JC. In Vivo Interaction Proteomics Reveal a Novel p38 Mitogen-Activated Protein Kinase/Rack1 Pathway Regulating Proteostasis in Drosophila Muscle. Mol Cell Biol. 2014;34:474–484. doi: 10.1128/MCB.00824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Bushell M, Wood W, Clemens MJ, Morley SJ. Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur J Biochem. 2000;267:1083–1091. doi: 10.1046/j.1432-1327.2000.01101.x. [DOI] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. Release of eIF6 (p27(BBP)) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Choudhuri A, Maitra U, Evans T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc Natl Acad Sci USA. 2013;110:9818–9823. doi: 10.1073/pnas.1302934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YD, Wang WC, Chen SAA, Hsu YT, Yeh MW, Slack FJ, Chan SP. RACK-1 regulates let-7 microRNA expression and terminal cell differentiation in Caenorhabditis elegans. Cell Cycle. 2014;13:1995–2009. doi: 10.4161/cc.29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RT, Baumert TF. Curing chronic hepatitis C--the arc of a medical triumph. N Engl J Med. 2014;370:1576–1578. doi: 10.1056/NEJMp1400986. [DOI] [PubMed] [Google Scholar]
- Conrad KD, Giering F, Erfurth C, Neumann A, Fehr C, Meister G, Niepmann M. microRNA-122 Dependent Binding of Ago2 Protein to Hepatitis C Virus RNA Is Associated with Enhanced RNA Stability and Translation Stimulation. PLoS One. 2013;8:e56272. doi: 10.1371/journal.pone.0056272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, Doudna JA. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa DD, Turek MM, Felmlee DJD, Girardi EE, Pfeffer SS, Long GG, Bartenschlager RR, Zeisel MBM, Baumert TFT. Reconstitution of the entire hepatitis C virus life cycle in nonhepatic cells. J Virol. 2012;86:11919–11925. doi: 10.1128/JVI.01066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SHW, Ramakrishnan V. Initiation of Translation by Cricket Paralysis Virus IRES Requires Its Translocation in the Ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ, et al. Monoclonal Anti-Claudin 1 Antibodies Prevent Hepatitis C Virus Infection of Primary Human Hepatocytes. Gastroenterology. 2010;139:953–U339. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JWB, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Molecular Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Fraser CS, Hershey JWB, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson TJ. RACK1 research – ships passing in the night? FEBS Letters. 2012;586:2787–2789. doi: 10.1016/j.febslet.2012.04.048. [DOI] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CUT, Pestova TV, Frank J. Structure of the Mammalian Ribosomal 43S Preinitiation Complex Bound to the Scanning Factor DHX29. Cell. 2013a;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CUT, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013b;503:539. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends in Biochemical Sciences. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Jan EE, Sarnow PP. Factorless Ribosome Assembly on the Internal Ribosome Entry Site of Cricket Paralysis Virus. J Mol Biol. 2002;324:14–14. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Jannot G, Bajan S, Giguere NJ, Bouasker S, Banville IH, Piquet S, Hutvagner G, Simard MJ. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011;12:581–586. doi: 10.1038/embor.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi MK, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Smith MA, Pronovost SM, Beckerle MC. Characterization of RACK1 function in Drosophila development. Dev Dyn. 2007;236:2207–2215. doi: 10.1002/dvdy.21217. [DOI] [PubMed] [Google Scholar]
- Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathogens. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou KH, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis CIRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends in Biochemical Sciences. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, Korostelev AA. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc Natl Acad Sci USA. 2014;111:9139–9144. doi: 10.1073/pnas.1406335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T, Rutkai E, Karaskova M, Valasek LS. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucl Acids Res. 2012;40:2683–2699. doi: 10.1093/nar/gkr1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Burdeinick-Kerr R, Whelan SPJ. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Long L, Deng Y, Yao F, Guan D, Feng Y, Jiang H, Li X, Hu P, Lu X, Wang H, et al. Recruitment of Phosphatase PP2A by RACK1 Adaptor Protein Deactivates Transcription Factor IRF3 and Limits Type I Interferon Signaling. Immunity. 2014;40:515–529. doi: 10.1016/j.immuni.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Patel P, Hershey J. Changes in ribosomal binding activity of eIF3 correlate with increased translation rates during activation of T lymphocytes. J Biol Chem. 2005;280:28251–28264. doi: 10.1074/jbc.M414129200. [DOI] [PubMed] [Google Scholar]
- Nathan C. Fresh Approaches to Anti-Infective Therapies. Sci Transl Med. 2012;4:140sr2. doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Takata A, Yoshikawa T, Kojima K, Kishikawa T, Shibata C, Takekawa M, Yoshida H, Omata M, Koike K. Receptor for Activated Protein Kinase C: Requirement for Efficient MicroRNA Function and Reduced Expression in Hepatocellular Carcinoma. PLoS One. 2011;6:e24359. doi: 10.1371/journal.pone.0024359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen C. Position of the CrPVIRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci USA. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Roberts APE, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucl Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA. The elF3 Interactome Reveals the Translasome, a Supercomplex Linking Protein Synthesis and Degradation Machineries. Molecular Cell. 2009;36:141–152. doi: 10.1016/j.molcel.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Speth C, Willing EM, Rausch S, Schneeberger K, Laubinger S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013;76:433–445. doi: 10.1111/tpj.12308. [DOI] [PubMed] [Google Scholar]
- Sun C, Querol-Audí J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JHD. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucl Acids Res. 2013;41:7512–7521. doi: 10.1093/nar/gkt510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audí J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3) Proc Natl Acad Sci USA. 2011;108:20473–20478. doi: 10.1073/pnas.1116821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. Translational control by the eukaryotic ribosome. Cell. 2011;145:333–334. doi: 10.1016/j.cell.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Volta V, Beugnet A, Gallo S, Magri L, Brina D, Pesce E, Calamita P, Sanvito F, Biffo S. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell Mol Life Sci. 2013;70:1439–1450. doi: 10.1007/s00018-012-1215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannová A, Malík R, Peclinovská L, Valasek LS. Functional and biochemical characterization of human eIF3 in living cells. Mol Cell Biol. 2014;34:3041–3052. doi: 10.1128/MCB.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Kaitsu Y, Yokoyama S. Cell-free translation system from Drosophila S2 cells that recapitulates RNAi. Biochemical and Biophysical Research Communications. 2005;343:1067–1071. doi: 10.1016/j.bbrc.2006.03.076. [DOI] [PubMed] [Google Scholar]
- Walsh D, Mohr I. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev. 2014;28:835–840. doi: 10.1101/gad.236752.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Fofana I, Heydmann L, Barth H, Soulier E, Habersetzer F, Doffoël M, Bukh J, Patel AH, Zeisel MB, et al. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathogens. 2014a;10:e1004128. doi: 10.1371/journal.ppat.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Fofana I, Thumann C, Mailly L, Alles R, Robinet E, Meyer N, Schaeffer M, Habersetzer F, Doffoël M, et al. Synergy of entry inhibitors with direct-acting antivirals uncovers novel combinations for prevention and treatment of hepatitis C. Gut. 2014b doi: 10.1136/gutjnl-2013-306155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CS, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.