SUMMARY
Mammalian transcriptomes display complex circadian rhythms with multiple phases of gene expression that cannot be accounted for by current models of the molecular clock. We have determined the underlying mechanisms by measuring nascent RNA transcription around the clock in mouse liver. Unbiased examination of eRNAs that cluster in specific circadian phases identified functional enhancers driven by distinct transcription factors (TFs). We further identify on a global scale the components of the TF cistromes that function to orchestrate circadian gene expression. Integrated genomic analyses also revealed novel mechanisms by which a single circadian factor controls opposing transcriptional phases. These findings shed new light on the diversity and specificity of TF function in the generation of multiple phases of circadian gene transcription in a mammalian organ.
INTRODUCTION
A substantial proportion of mammalian genes are expressed with a circadian rhythm driven by a cell autonomous molecular clock (Hughes et al., 2009; Miller et al., 2007; Panda et al., 2002). The clock mechanism involves a network of transcriptional-translational feedback loops comprised of core transcriptional activators BMAL1/CLOCK and two sets of repressors, PER/CRY (Reppert and Weaver, 2001; Takahashi et al., 2008) and Rev-erbs α and β (Bugge et al., 2012; Cho et al., 2012; Ripperger and Schibler, 2001). Under normal conditions, each cellular clock is synchronized by systemic cues and generates multiple phases of rhythmic output (Asher and Schibler, 2011; Dibner et al., 2010; Peek et al., 2012).
Although each circadian transcription factor (TF) binds DNA with genome-wide oscillation peaking at a specific time (Feng et al., 2011; Koike et al., 2012; Rey et al., 2011), binding of an individual circadian TF, e.g. BMAL1, has been reported at genes oscillating with a range of phases, many of which do not correlate with the circadian regulator’s binding phase (Menet et al., 2012). Moreover, genome-wide studies have revealed a substantial portion of circadian TF binding tens to hundreds of kilobases away from known transcription start sites (TSS) (Feng et al., 2011; Koike et al., 2012; Rey et al., 2011), and a high degree of overlap between core clock TFs with competing effects on circadian rhythms, such as BMAL1 and Rev-erbα (Cho et al., 2012; Koike et al., 2012). Furthermore, several clock output TFs have been suggested to generate transcriptional rhythms with delayed phase relative to BMAL1/CLOCK, but these mechanisms have not been explored genome-wide (Asher and Schibler, 2011). Thus, a fundamental question remains as to how the interaction of multiple regulators at the genome, particularly at distal enhancer elements, produces distinct phases of circadian transcriptional activity.
Here we applied Global Run-On sequencing (GRO-seq) (Core et al., 2008; Wang et al., 2011) to mouse liver collected at multiple times of day to measure the circadian activity of enhancer regions based on eRNA transcription (Hah et al., 2013; Kim et al., 2010). We identified thousands of oscillating enhancers with varying peak activity times, and in particular we found that specific phases of oscillation are associated with distinct regulatory motifs and TF binding patterns. Our data suggest for the first time that specific phases of enhancer activity in vivo are achieved by a dominant regulator at each site, determined in part by sequence content, in contrast to combinatorial regulation models based primarily on synthetic in vitro models (Ukai-Tadenuma et al., 2008). Furthermore, we show that eRNA oscillations are highly predictive of the rhythmicity and phase of transcription at nearby genes, demonstrating a large-scale and previously unexplored role for distal regulatory elements in the generation of transcriptional rhythms. By combining circadian enhancer maps, transcription factor cistromes, and genetic ablation of Rev-erbα and Clock we demonstrate that circadian eRNAs can be used to both identify the TFs coordinating specific phases of gene transcription and, importantly, uniquely distinguish the functional binding sites within a circadian TF cistrome. Thus, an integrative approach using multiple genomic techniques provides the most detailed and robust model to explain the generation and coordination of multiple phases of rhythm within a single tissue.
RESULTS
Circadian transcription in mouse liver
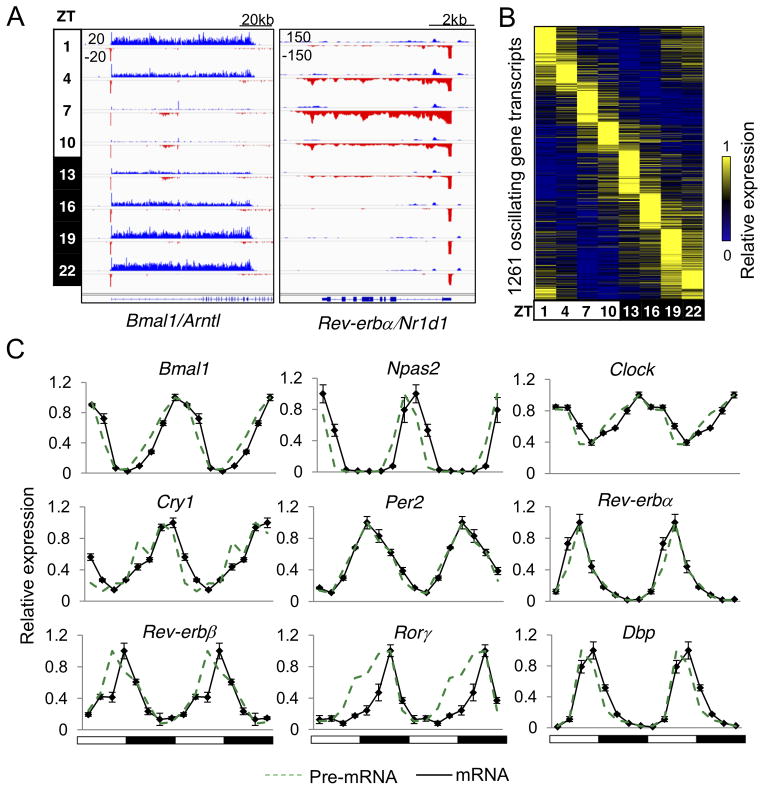
GRO-seq was performed on mouse liver nuclei collected every three hours throughout a 24-hr light-dark cycle. Transcription of known circadian genes showed robust oscillation patterns, exemplified by Bmal1 (Arntl) and Rev-erbα (Nr1d1) (Fig. 1A). 11,288 active gene transcripts were identified, of which 1,261 (11%) were transcribed with oscillating patterns (JTK_CYCLE (Hughes et al., 2010), p<0.01, 21≤ period (τ)≤24hr, peak to trough ratio>1.5) (Fig. 1B and Table S1A). Rhythmic mRNA expression of known circadian genes determined by RT-qPCR was associated with their nascent transcription (Fig. 1C), and biological replicates of GRO-seq samples at Zeitgeber Time (ZT) 10 and ZT22 showed a high degree of correlation (Pearson correlation coefficient, r=0.95) (Fig. S1A). In addition, genes oscillating in similar phases showed closely related biological functions (Fig. S1B and Table S1B). Together, these results demonstrate the robustness of our data.
Figure 1. Circadian transcription in mouse liver.
(A) Genome browser view of nascent transcripts at Bmal1/Arntl and Rev-erbα/Nr1d1 loci at 8 time points. GRO-seq signals on the + and- strand are illustrated in blue and red, respectively. Y-axis scale refers to the normalized tag count per 106 reads. (B) Heat map of the relative transcription of 1,261 oscillating genes sorted by oscillation phase. (C) Relative expression of pre-mRNA (green) and mRNA (black) determined by GRO-seq and RT-qPCR, respectively, throughout the day. Data are double plotted for better visualization. RT-qPCR data are expressed as the ±SEM (n=3–4 per time point) and normalized to the maximal expression of the day. See also Figure S1 and Table S1.
De novo identification of circadian liver enhancer RNAs
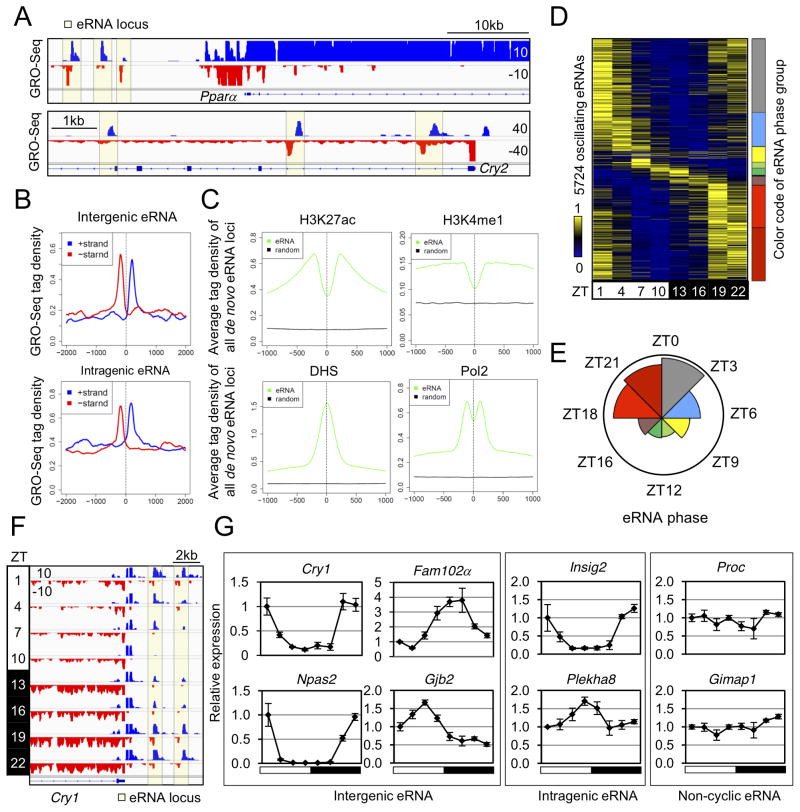
Analysis of the liver GRO-seq data revealed eRNA transcription in both inter- and intragenic regions, exemplified by highlighted regions in the vicinity of Pparα and Cry2, respectively (Fig. 2A). To globally identify eRNA loci, we developed a pipeline to search for genomic locations producing bi- and uni-directional short RNA transcripts (Extended Experimental Procedures), which identified 19,086 high confidence de novo eRNA loci (>300bp from TSS) (Table S2A). The average GRO-seq signal of de novo eRNAs showed a bimodal profile in both inter- and intragenic regions (Fig. 2B). Analysis of public ChIP-seq data (Table S2B) from mouse liver suggested that de novo eRNA loci were enriched for other epigenomic features including H3K27ac, H3K4me1, DNAse I hypersensitivity, and RNA polymerase II (Pol2) recruitment, consistent with the function of these sites as enhancers (Fig. 2C). eRNA signals correlated with Pol2 occupancy and histone acetylation but not histone methylation (Fig. S2A), consistent with earlier reports (Hah et al., 2013; Li et al., 2013; Wang et al., 2011) and in agreement with the notion that H3K4me1 and H3K27ac mark enhancer identity and activity, respectively (Creyghton et al., 2010).
Figure 2. De novo identification of circadian liver enhancer RNAs.
(A) Genome browser view of intergenic (upper panel) and intragenic (lower panel) eRNAs (yellow boxes). (B) GRO-seq tag densities in 4kb windows surrounding de novo intergenic (upper panel) and intragenic (lower panel) eRNA loci are shown for the plus (blue) and minus (red) strand. Y-axis shows average reads per 10 million reads (RPTM) per 10bp bin. (C) Average ChIP-seq tag densities of epigenetic marks in 2kb window surrounding all de novo eRNA loci (prior to the selection of high confidence eRNAs) and matched control regions. (D) Heat map of the relative transcription of oscillating eRNAs throughout the day. Color coding of eRNA population in 8 phase groups (from ZT0 to ZT24, at 3 hour intervals) is shown on the right. (E) Rose diagram showing the prevalence of eRNA loci in each phase group. For each wedge, the color corresponds to that in (D) and the area is proportional to the number of eRNAs in that group. (F) Genome browser view of oscillating eRNAs at Cry1 locus. (G) RT-qPCR validation of circadian transcription for intergenic, intragenic, and non-cyclic eRNAs at indicated gene loci. Data are expressed as mean±SEM (n=3–4 per time point) and normalized to the first time point. See also Figure S2 and Table S2.
To examine dynamics of eRNA transcription across the 24hr cycle, eRNA transcripts were quantified using GRO-seq tag counts within +/−500bp from the centers of eRNA loci. Remarkably, 5,724 (30%) of eRNAs were found to be transcribed in a circadian manner (JTK-CYCLE, p<0.05, 21≤ period (τ)≤24hr, peak to trough ratio>1.5) (Table S2C), and their relative expression peaked at different times of the day (Fig. 2D). Based on their peak expression time (hereafter referred to as “phase”), circadian eRNAs were divided into 8 groups (phase ZT0-ZT24, at 3hr intervals), represented by 8 colors in Fig. 2D. Interestingly, circadian eRNAs were not evenly distributed across the 8 phase groups. 71% of circadian eRNAs oscillated with a phase between ZT18 and ZT3, while 29% of circadian eRNAs oscillated in other phases (Fig. 2E and Table S2C). Examples of circadian eRNAs with phase ZT22 at the Cry1 locus are shown in Fig. 2F. eRNA transcripts oscillating in different phases were confirmed by RT-qPCR (Fig. 2G) at selected intergenic and intragenic eRNA loci (Fig. S2B). The unbalanced phase distribution of eRNAs agrees with the previous finding that histone acetylation, a reflection of enhancer activity, was globally high around ZT22 and low around ZT10 in the mouse liver (Feng et al., 2011). Moreover, the average H3K27ac level at 8 groups of eRNA loci showed the same oscillatory pattern as the circadian eRNAs within each group (Fig. S2C). Therefore, circadian eRNAs oscillate in diverse phases, suggesting that circadian enhancer activities are orchestrated by distinct mechanisms in liver.
Phase-specific transcription factors at circadian enhancers
We have shown that gene body and eRNA transcription occur in multiple phases. As previous studies suggested correlated transcription of eRNA and nearby target genes (Core et al., 2008; Hah et al., 2013; Kim et al., 2010), we examined whether eRNA oscillations are related to circadian gene transcription. The expression of genes mapped closest to oscillating eRNAs (within 200kb from TSS) showed rhythmic patterns in phase with eRNA expression (Fig. 3A). Among all genes mapped to circadian eRNAs, 423 (34%) circadian gene transcripts were mapped to 1,124 (20%) circadian enhancers and oscillation phases between each enhancer-gene pair were highly correlated (r=0.9) (Fig. S3A). This is likely an underestimate based on the stringent eRNA-gene mapping criteria and, indeed, if the analysis is not limited to the nearest gene, up to 76% of circadian genes in different phases have in-phase eRNAs (phase difference < 3hrs between gene and eRNA) located within 200kb of their TSSs. By contrast, for random genes this number is ~10% on average (hypergeometric test, p<0.001) (Fig. S3B). Together, these results suggest that circadian eRNAs predict rhythmic transcription of nearby genes and are likely to be functionally associated with circadian genes of the same phase.
Figure 3. Phase-specific transcription factors at circadian enhancers.
(A) Relative transcription of genes closest to oscillating eRNAs (within 200kb of TSS). (B) Motifs specifically enriched in each eRNA group are labeled in the clock diagram on the left. Position weight matrix (PWM) of each motif and its best enrichment p-value in assigned groups are shown in the table on the right. (C) Correlation of motif occurrence and TF binding in 8 eRNA phase groups. In each plot, the red dots represent the fraction of eRNA loci bound by the indicated TF (top 3,000 ChIP-seq peaks), and black bars represent the fraction of eRNA loci containing the corresponding motif. Correlation coefficient r is shown for phase-specific motifs. TFs recognizing different types of motifs are grouped in colored boxes corresponding to those used for eRNA phases. See also Figure S3 and Table S3.
Although gene body and eRNA transcription occur in multiple phases, the core clock oscillator in liver has only one peak and one trough in a 24 hour period (Koike et al., 2012). We considered the possibility that specific circadian TFs were responsible for the different phases of gene expression by driving the transcription of diversely phased eRNAs. To this end, we performed motif analysis on the 8 groups of circadian enhancers using 500bp windows centered on each eRNA locus (Fig. S3C). First, candidate phase-specific TFs with the most enriched motifs in each enhancer group were selected by de novo motif mining (Table S3). Then, annotated motifs of candidate TFs were used to quantify the motif enrichment in each enhancer group, revealing four major types of motifs specifically enriched in six enhancer groups (Fig. 3B). Specifically, an E-box motif was the most enriched at circadian eRNA loci in phase ZT6–9, coincident with the peak of BMAL1 binding to the genome (Koike et al., 2012; Rey et al., 2011; Ripperger and Schibler, 2006). However, although BMAL1/CLOCK has been previously linked to circadian gene regulation in liver, the ZT6–9 eRNAs comprised only ~6% of circadian enhancers, consistent with an earlier study in which only ~5% of total circadian genes were transcribed in phase with nearby BMAL1 binding (Menet et al., 2012).
We also discovered that a D-box motif, recognized by PAR-bZIP proteins including DBP, TEF, HLF, and E4BP4 (Cowell et al., 1992; Li and Hunger, 2001; Mitsui et al., 2001), was the most enriched motif at phase ZT9–15 eRNA loci (Fig. 3B), coinciding with the phase of known target genes for these TFs (Gachon et al., 2006). Moreover, the RevDR2 and RORE motifs, bound by Rev-erbαβ (Harding and Lazar, 1995) and RORα/γ (Giguere et al., 1994), were the top motifs at eRNA loci with the most common phase, ZT18–24 (Fig. 3B), coinciding with the trough of repression by Rev-erbα (Bugge et al., 2012; Feng et al., 2011). By contrast, motifs characteristic of ETS binding sites were highly enriched in the phase ZT0–3 enhancers, implying a potential role of ETS proteins in the circadian regulation of transcripts with this phase (Fig. 3B). In addition to these phase-specific motifs, constitutively enriched motifs in all enhancer groups were identified, most prominently the Forkhead and HNF4 motifs (Fig. 3B).
We tested whether the motif enrichment in a given eRNA group was predictive of TF binding by overlapping each group of circadian eRNAs with TF cistromes determined by ChIP-seq. Specifically, we analyzed previously published cistrome data for core clock TFs (Feng et al., 2011; Koike et al., 2012), and performed additional ChIP-seq experiments for E4BP4 and RORα. To minimize the effects of variable ChIP-seq quality in different studies, only the 3,000 strongest ChIP-seq peaks for each TF were used in the analysis. Notably, the genomic binding sites of E-box-binding factors BMAL1, CLOCK, and NPAS2 were enriched at eRNAs with phase ZT6–9 (Fig. 3C), where de novo analysis implicated the E-box motif. Similarly, genomic binding of Reverbα and RORα was enriched at eRNAs whose transcription peaked at ZT21–24 (Fig. 3C), where the RevDR2 and RORE motifs were most prominent. Also consistent with the bioinformatic predictions, the D-box binding factor E4BP4 bound most commonly at eRNAs with phase ZT9–15 (Fig. 3C). By contrast, binding of FOXA1 and HNF4A, whose motifs were equally enriched in all eRNA groups, did not display a preference for eRNA loci of a specific phase (Fig. 3C). Thus, the regulatory activities of 6 TFs coincide with the rhythmic eRNA expression in the enhancer group at which they were enriched. These data strongly suggest that TFs bound specifically at each enhancer group are potential drivers of their circadian transcription and enhancer activities.
Phase-correlation between eRNA and gene body transcription marks functional enhancers of circadian genes
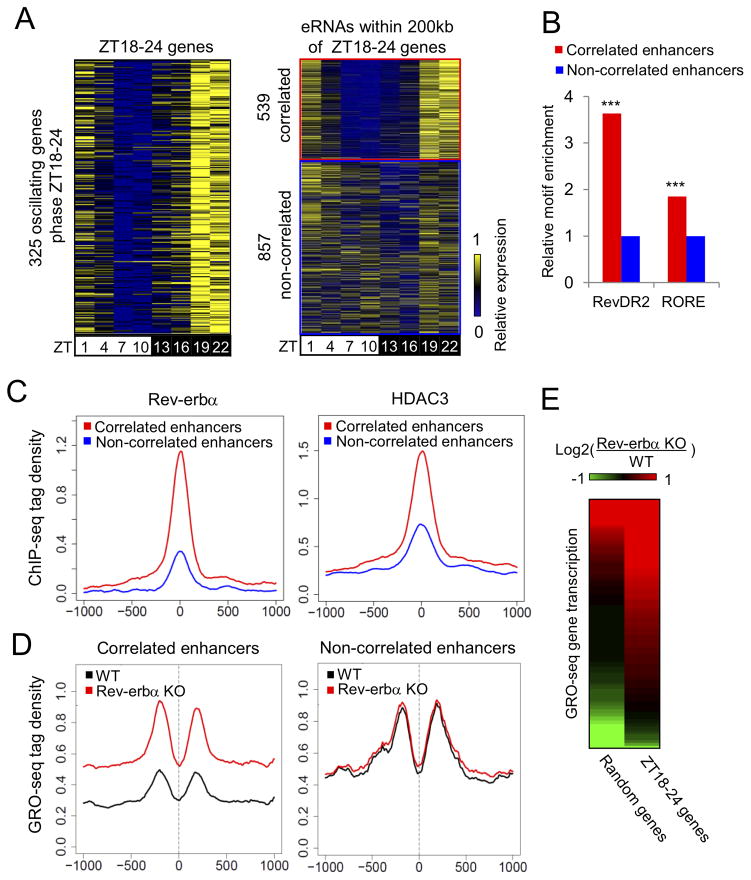
We next considered whether the specific TFs found to bind at circadian enhancers were driving transcription of nearby in-phase genes, focusing on the most common circadian enhancers (phase ZT18–24). Within 200kb of 325 circadian genes in phase ZT18–24, 539 neighboring eRNA loci showed circadian eRNA transcription in phase ZT18–24 (“correlated enhancers”), while 857 eRNA loci did not produce correlated eRNA transcription (“non-correlated enhancers”, eRNA expression ZT22/ZT10<1.5) (Fig. 4A).
Figure 4. Phase-correlation between eRNA and gene body transcription marks functional enhancers of circadian genes.
(A) Heat map of the relative transcription of 325 circadian genes in phase ZT18–24 (left panel) and their neighboring eRNAs (right panel). 539 eRNAs in correlated phase are shown in the red box while 857 non-correlated eRNAs are in the blue box. (B) Enrichment of RevDR2 and RORE motif in correlated eRNA loci relative to non-correlated eRNA loci (hypergeometric test, ***p<0.001). (C) ChIP-seq tag density of Rev-erbα (left) and HDAC3 (right) in 2kb windows surrounding correlated (red) and non-correlated eRNA loci (blue). y-axis shows the average tag count per 10bp bin normalized to 10 million total reads. (D) Comparison of GRO-seq tag density (RPTM per 10bp bin in 2kb window) surrounding correlated (left) and non-correlated (right) eRNA loci in WT and Rev-erbα −/− livers at ZT10. (E) Heat map of transcriptional changes between WT and Rev-erbα −/− livers at ZT10, for the 325 circadian genes in phase ZT18–24 (right column), compared to the same number of random genes (left column). Data are expressed as log2 fold change. See also Figure S4.
Correlated enhancers showed higher enrichment of the RevDR2 and RORE motifs in comparison to non-correlated enhancers (Fig. 4B). Notably, relative enrichment of the RevDR2 motif, which is a preferential binding site for Rev-erbα (Harding and Lazar, 1995; Zhao et al., 1998) was 2-fold higher than that of the RORE motif shared by Rev-erbα and RORα (Giguere et al., 1994), suggesting that Rev-erbα may play a more important role in regulating the correlated enhancers. ChIP-seq tag densities of Rev-erbα and its corepressor HDAC3 were dramatically stronger at correlated enhancers than at non-correlated enhancers (Fig. 4C), supporting the idea that the correlated enhancers in phase ZT18–24 were controlled by Rev-erbα. To test this hypothesis, GRO-seq was performed on livers from mice genetically lacking Rev-erbα Rev-erbα −/− at ZT10, when Rev-erbα levels normally peak and maximally repress histone acetylation and gene transcription (Feng et al., 2011). Indeed, eRNA signals at the correlated enhancers were markedly derepressed in Rev-erbα −/− mice, while no such change was seen at the non-correlated enhancers (Fig. 4D). Similar results were obtained at both inter- and intragenic enhancers (Fig. S4). Importantly, gene body transcription that normally peaked at ZT18–24 was also extensively derepressed in Rev-erbα −/− mice at ZT10 (Fig. 4E), indicating these genes are direct targets of Rev-erbα. Together, these results demonstrate that eRNAs in phase ZT18–24 mark functional Rev-erbα binding sites that regulate neighboring target genes with correlated phase. Conversely, non-correlated enhancers are not bound by Rev-erbα and do not control Rev-erbα target genes.
Circadian eRNAs reveal the functional Rev-erbα cistrome at oscillating genes
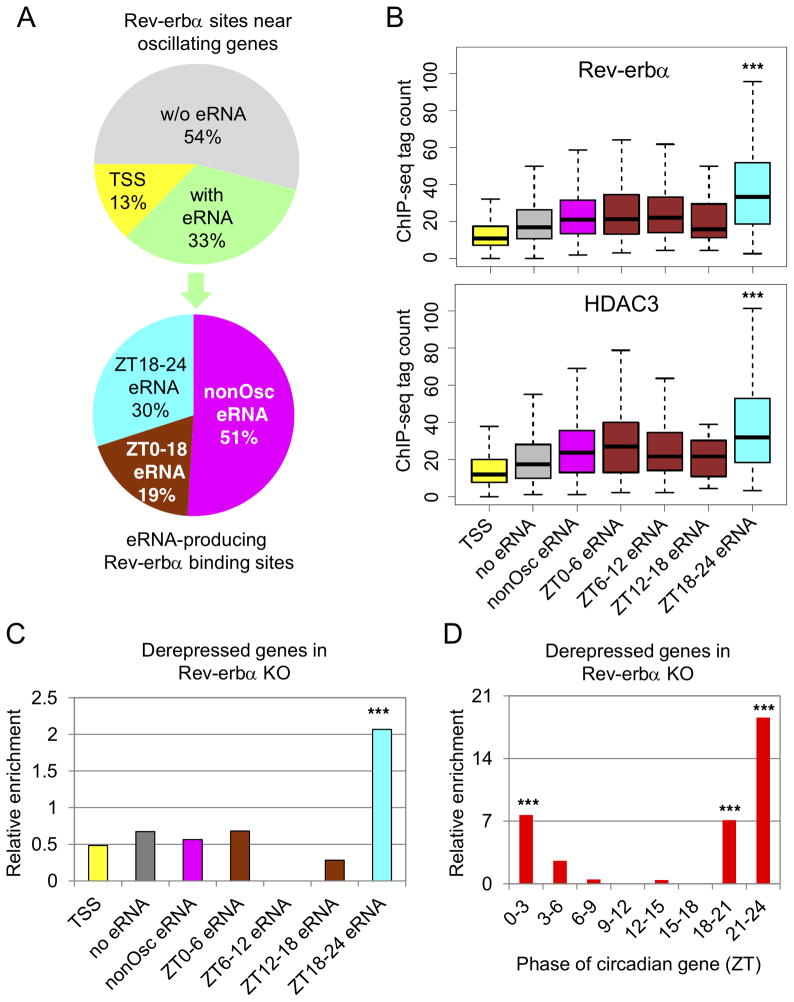
The findings to this point demonstrate that Rev-erbα regulates circadian genes in phase ZT18–24 via enhancers oscillating in phase with gene body transcription. However, these enhancers account for only a small fraction of the complete Rev-erbα cistrome (Feng et al., 2011). We therefore considered whether circadian eRNAs in phase ZT18–24 uniquely mark the functional subset of Rev-erbα binding sites controlling circadian genes in liver. To test this, Rev-erbα sites near circadian genes were divided into three groups, of which 887 (33%) overlapped de novo eRNA loci, 347 (13%) were found at TSSs of circadian genes (within 300bp), and the remaining 1,455 (54%) were not associated with detectable eRNA transcription (Fig. 5A). Of the eRNAs transcribed at Rev-erbα binding sites, 30% peaked at ZT18–24, while 19% peaked in other phases, and 51% were constitutively expressed eRNA and did not oscillate (Fig. 5A).
Figure 5. Circadian eRNAs reveal the function of the Rev-erb.
α cistrome at oscillating genes.
(A) Distribution of Rev-erbα ChIP-seq peaks near circadian genes (upper panel) and subdistribution of eRNA-producing Rev-erbα peaks near circadian genes (lower panel). (B) Boxplot showing Rev-erbα and HDAC3 peak height at binding sites from panel A. Y-axis indicates normalized tag count in each peak (RPTM) (***p<0.001, one-way ANOVA and Tukey’s test). (C) Enrichment of derepressed genes in Rev-erbα −/− mice at circadian genes bound by different Rev-erbα peaks from panel A relative to a random set of Rev-erbα peaks (hypergeometric test, ***p<0.001). (D) Enrichment of derepressed genes in Rev-erbα −/− mice in 8 groups of circadian genes with indicated phases relative to randomly selected genes (hypergeometric test, ***p<0.001). See also Figure S5 and Table S4.
Rev-erbα and its co-repressor HDAC3 bound more strongly at sites producing ZT18–24 eRNAs than at other types of binding sites (Fig. 5B), resulting in a marked decrease in histone H3K9 acetylation from ZT22 to ZT10 (Fig. S5). To directly assess the functionality of Rev-erbα binding on individual gene expression, we constructed a list of high confidence target genes whose nascent and mature transcripts were derepressed in Rev-erbα −/− livers at ZT10 compared to WT (Table. S4A-C). The enrichment of derepressed circadian genes in Rev-erbα −/− mice was >3-fold higher near Rev-erbα sites producing ZT18–24 eRNAs, compared to other Reverbα sites (Fig. 5C), suggesting that ZT18–24 eRNAs mark functional Rev-erbα sites. Moreover, circadian genes with phase around ZT21–24 were highly enriched for derepression in Reverbα −/− mice (Fig. 5D), consistent with the enrichment of circadian eRNAs in this phase. Together, these data strongly suggest that only a subset of the Rev-erbα cistrome associated with antiphase eRNAs is functional in controlling circadian gene transcription.
eRNA analysis identifies E4BP4 as a key mediator of gene activation by Rev-erbα
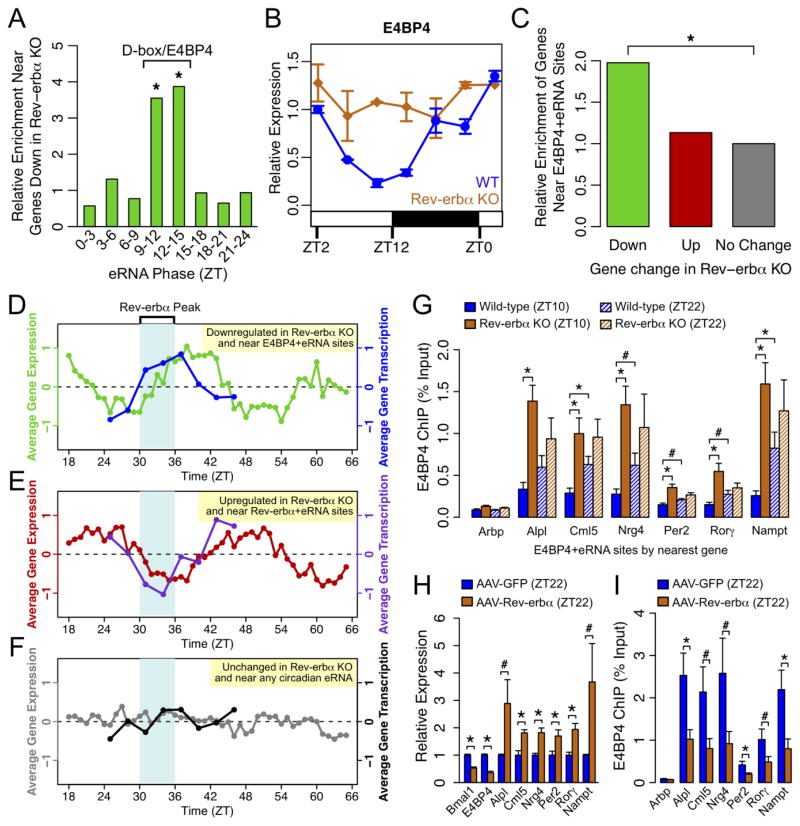
While eRNAs clearly delineate the functional Rev-erbα cistrome responsible for direct transcriptional repression, there remains a substantial set of genes paradoxically downregulated at ZT10 in Rev-erbα −/− mouse livers, which cannot be explained through direct regulation by Rev-erbα. To identify factors mediating this opposing effect on gene transcription, we constructed a list of high confidence target genes whose nascent and mature transcript levels were decreased in Reverbα −/− livers at ZT10 (Table. S4A–C). Profiling of eRNAs near genes that were downregulated in the Rev-erbα −/− livers revealed a marked and specific enrichment for phases between ZT9 and ZT15 (Fig. 6A), which were shown earlier to be enriched for the D-box motif and binding of the D-box repressor E4BP4 (Fig. 3C).
Figure 6. E4BP4 functions downstream of Rev-erbα.
(A) Enrichment of oscillating eRNAs in each phase group near genes downregulated in Rev-erbα −/− livers relative to control genes. Significantly enriched phases are noted as corresponding to D-box/E4BP4-enriched phase group. (hypergeometric test, *p<0.05). (B) mRNA expression of E4BP4/Nfil3 in WT and Reverbα −/− livers measured by RT-qPCR throughout the day. Data are expressed as mean±SEM (n=2 per time point and genotype) normalized to the first WT time point. (C) Enrichment of E4BP4+eRNA bound genes among those downregulated (green) or upregulated (red) in Reverbα −/− livers relative to unchanged genes (grey) (hypergeometric test, *p<0.05). (D-F) Average circadian expression profiles in WT mouse livers (Hughes et al., 2009) and corresponding transcription profiles by GRO-seq for (D) genes downregulated in Rev-erbα −/− livers within 200kb of E4BP4 binding at ZT9–15 circadian eRNAs, (E) genes upregulated in Rev-erbα −/− livers within 200kb of Rev-erbα binding at ZT18–24 circadian eRNAs, and (F) non-regulated control genes (expressed in liver within 200kb of near circadian eRNA in any phase). (G) ChIP-qPCR of E4BP4 binding at genes downregulated in Rev-erbα −/− livers at ZT10. Binding is shown at ZT10 (solid bars) and ZT22 (hashed bars) in WT (blue) and Rev-erbα −/− (orange) livers. Data are expressed as mean±SEM (One-way ANOVA, *p<0.05, #p<0.1, n=3–4 per group). (H) mRNA expression measured by RT-qPCR in liver overexpressing Rev-erbα (mice injected with AAV-Tbg-Rev-erbα) or control liver (mice injected with AAV-Tbg-GFP) at ZT22. Data are expressed as mean±SEM (One-way ANOVA, *p<0.05, #p<0.1; n=6 per group). (I) ChIP-qPCR of E4BP4 binding at same sites as (G) in liver overexpressing Rev-erbα (blue) or control liver (orange). Data are expressed as mean±SEM (One-way ANOVA, *p<0.05, #p<0.1, n=5–6 per group). See also Figure S6.
We hypothesized that, by controlling the circadian expression of E4BP4, Rev-erbα indirectly dictated the circadian expression of a large set of genes controlled by D-box enhancers whose expression would thus be in phase with Rev-erbα. Indeed, E4BP4 gene expression was circadian in WT mouse livers but constitutively elevated in Rev-erbα −/− mice (Fig. 6B), consistent with a previous report (Duez et al., 2008). Furthermore, Rev-erbα bound along with its NCoR-HDAC3 corepressor complex to several sites at the E4BP4 (Nfil3) locus, suggesting that E4BP4 expression is directly controlled by Rev-erbα (Fig. S6A). By contrast, there were weaker changes in hepatic expression of D-box activating factors Dbp, Tef, and Hlf in livers of Rev-erbα −/− mice, and the expression of these factors remained circadian with similar phases (Fig. S6B).
To identify putative functional E4BP4 sites, we analyzed the complete set of E4BP4 ChIP-seq peaks for those with higher eRNA levels at ZT9–15 (ZT10/ZT22 > 3 or ZT13/ZT1 > 3). These sites, which we refer to as “E4BP4+eRNA” sites, were enriched 2-fold around genes downregulated in Rev-erbα −/− mice (Fig. 6C), demonstrating a significant association between E4BP4 binding and gene regulation downstream of Rev-erbα. Transcriptome profiles from livers of WT mice (Hughes et al., 2009) confirmed that putative E4BP4 target genes (downregulated in Rev-erbα −/− livers and near E4BP4+eRNA sites) were generally circadian with average peak and trough expression in phase with Rev-erbα and E4BP4 levels, respectively (Fig. 6D, green line). The average GRO-seq transcription profile for this same group of genes showed a similar pattern over a 24-hour cycle (Fig. 6D, blue line). Both patterns are consistent with direct repression by E4BP4 leading to circadian oscillation in phase with Rev-erbα protein levels. In contrast, Rev-erbα target genes (upregulated in Rev-erbα −/− livers and near Rev-erbα sites overlapping ZT18–24 eRNAs) were on average antiphase to Rev-erbα expression in WT livers, consistent with direct transcriptional repression by Rev-erbα (Fig. 6E). As a control, genes that were expressed near oscillating eRNAs, but unchanged in the Rev-erbα −/− livers, were not systematically phased relative to Rev-erbα or E4BP4 levels (Fig. 6F).
These findings support a model in which Rev-erbα indirectly activates genes in phase ZT9–15 by repressing the D-box repressor E4BP4. Such a model predicts that E4BP4 target genes would be constitutively downregulated in Rev-erbα −/− livers, with increased E4BP4 binding at nearby functional sites. Indeed, expression profiling over a 24hr cycle revealed that genes near E4BP4+eRNA sites showed attenuated rhythmic expression in Rev-erbα −/− livers (Fig. S6C). Furthermore, E4BP4 genomic binding was increased at ZT10 and no longer circadian at these sites in Rev-erbα −/− livers (Fig. 6G).
We also tested the effect of ectopic expression of Rev-erbα in mouse livers on E4BP4 expression and function. Interrogation of data from a previously published experiment (Kornmann et al., 2007) revealed upregulation of the genes putatively controlled by E4BP4 in livers constitutively expressing Rev-erbα, particularly at the physiological peak time of E4BP4 expression (Fig. S6D). Indeed, while constitutive expression of Rev-erbα in mouse liver repressed its direct targets such as Bmal1 and E4BP4/Nfil3, it upregulated E4BP4 target genes at ZT22 (Fig. 6H). This effect was much less apparent at ZT10 when E4BP4 is already at physiologically low levels (Fig. S6E). Importantly, E4BP4 binding at putative functional sites near these genes was reduced at ZT22, consistent with loss of repression by E4BP4 at the implicated D-box elements (Fig. 6I). These results strongly suggest that E4BP4 functions downstream of Rev-erbα, via sites transcribing eRNA in phase ZT9–15, to repress the genes that are downregulated in Rev-erbα −/− livers and upregulated when Rev-erbα is overexpressed.
Circadian eRNAs define functional cistromes that distinguish CLOCK and Reverbα target genes
CLOCK and Rev-erbα have opposite effects on gene transcription, however their maximal binding to the genome occur in roughly the same time window (ZT8-ZT10) (Cho et al., 2012; Feng et al., 2011; Koike et al., 2012). ChIP-seq results suggest that 80% of genes bound by CLOCK within 200kb of TSS were also bound by Rev-erbα (Fig. S7A), resulting in 15–35% of circadian genes in different phases cobound by these two factors (Fig. S7B). The question as to how co-occurrence of CLOCK and Rev-erbα binding affects rhythmic gene transcription remains unsolved (Zhao et al., 2014).
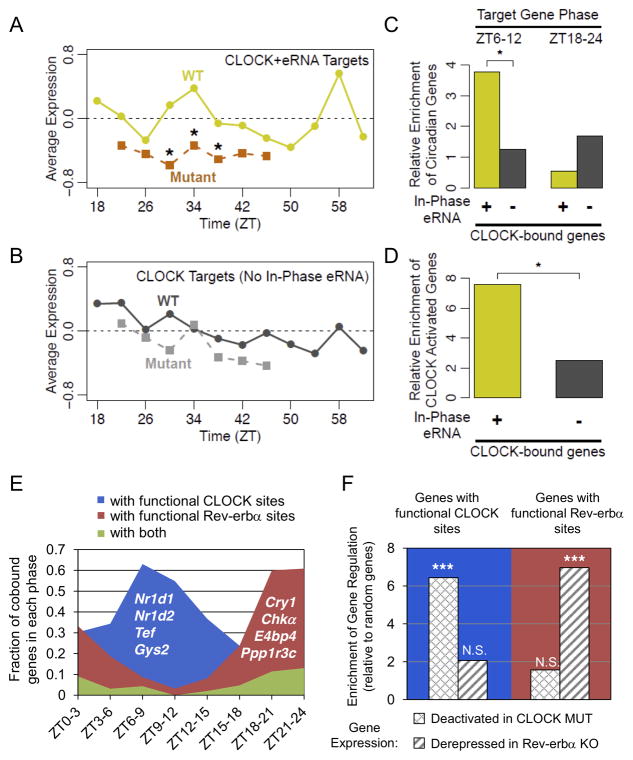
Having demonstrated that functional Rev-erbα sites marked by ZT18–24 eRNAs correlated with target gene phase (Figs. 4 and 5), we tested whether eRNAs oscillating in other phases could identify the functional cistromes of other clock components. To this end, we analyzed published microarray data measuring gene expression in livers of WT and Clock mutant mice (Miller et al., 2007). We first noted that genes downregulated in the Clock mutant mice were significantly enriched for circadian eRNAs in the phase ZT6–9 compared to control genes (Fig. S7C), corresponding to the enrichment of E-box motif and CLOCK binding. We then selected putatively functional CLOCK sites (Koike et al., 2012) producing eRNAs in phase with CLOCK binding (Table S5, eRNA level ZT7/ZT19 > 3 or ZT10/ZT22 > 3) and correlated with nearby gene transcription, and compared these sites to the remainder of the CLOCK cistrome.
Target genes within 200kb of putatively functional CLOCK sites showed rhythmic mRNA expression in WT mice (Miller et al., 2007), peaking at the time point corresponding to ZT10 in our studies (Fig. 7A, yellow line). These genes also showed reduced expression overall in Clock mutant mice, particularly at time points corresponding to ZT6 and ZT10 (Fig. 7A, orange line). By comparison, genes near other CLOCK sites showed weaker average rhythm, and weaker average reduction in Clock mutant mice (Fig. 7B). Further confirming that CLOCK sites marked by in phase eRNA represent the functional subset of the CLOCK cistrome, target genes near these sites are significantly enriched for circadian genes specifically in phases ZT6–12, but not opposing phases (Fig. 7C) and are also significantly enriched for genes downregulated >1.5 fold in Clock mutants (Fig. 7D). The fact that some CLOCK target gene mRNA levels cycle in phases ZT9–12 is likely due to delays in the phase of mature mRNA oscillations relative to nascent transcription, as noted in previous studies (Menet et al., 2012). Taken together, these results demonstrate that CLOCK sites marked by in phase eRNAs represent the functional component of the total cistrome.
Figure 7. Circadian eRNAs define functional cistromes that distinguish CLOCK and Reverbα target genes.
(A) Average expression of genes within 200kb of CLOCK binding sites producing eRNA in phase with CLOCK binding and target gene expression in WT (yellow line) and Clock mutant (orange line) mouse livers from (Miller et al., 2007) (*Wilcoxon test of gene fold-change distribution versus matching time points in (B), *p<0.05). (B) Average expression of genes within 200kb of CLOCK binding sites lacking in phase eRNA in WT (dark grey line) and Clock mutant (light grey line) mouse livers from (Miller et al., 2007). (C) Enrichment of circadian genes expressed in phase with CLOCK binding (ZT6–12) or anti-phase to CLOCK binding (ZT18–24) for the gene groups used in panel A (yellow) and panel B (grey) relative to random genes (hypergeometric test, *p<0.05). (D) Enrichment of genes downregulated in Clock mutant livers among the gene groups used in panels A-C relative to random genes (hypergeometric test, *p<0.05). (E) Fraction of oscillating genes co-bound by CLOCK and Rev-erbα that are within 200kb of TF binding sites producing rhythmic eRNA in phase with CLOCK activation (blue), Rev-erbα repression (red), or both (green). Oscillating genes are divided according to their phases. Representative genes are noted in each group. (F) Enrichment of CLOCK and Rev-erbα regulatedgenes (expression fold change in mutant >95% of random genes) in those with eRNA predicted functional binding sites in panel E, relative to random genes (hypergeometric test, ***p<0.001, N.S. p>0.05). See also Figure S7 and Table S5.
To examine whether CLOCK and Rev-erbα are both functional at co-bound circadian genes, functional binding sites of each factor were mapped to their closest circadian genes. CLOCK binding sites at TSS were included in this analysis as they are also enriched at genes downregulated in Clock mutant mouse livers (Fig. S7D), consistent with previous studies (Rey et al., 2011). Remarkably, the majority of co-bound circadian genes contained functional binding sites of only one factor but not both, with genes around phase ZT6–9 and ZT18–24 most enriched for functional CLOCK and Rev-erbα sites, respectively (Fig. 7E). These findings suggest exclusive functions of either CLOCK or Rev-erbα at most co-bound genes. Consistent with this notion, expression profiling showed that co-bound genes exclusively carrying functional CLOCK sites, such as Nr1d1, Nr1d2, and Tef, are deactivated in Clock mutant mice, while those only carrying functional Rev-erbα sites, such as Cry1 and E4BP4, are derepressed in Rev-erbα −/− mice (Fig. 7F and Fig. S7E). Therefore, despite frequent colocalization of their binding, CLOCK and Rev-erbα coordinate distinct sets of circadian genes that can be predicted from their regulation of eRNAs.
DISCUSSION
Unbiased analysis of the nascent transcription of over 5,000 circadian eRNAs and the TF motifs at these sites has allowed us to identify the direct genomic targets of multiple circadian regulators in mouse liver. Circadian eRNA loci are enriched for enhancer marks, the phase of eRNA oscillation correlated with that of nearby genes, and knockout studies demonstrated the causal relationship between TF binding and the transcriptional regulation at enhancers and the genes they control. These results informed the comparison of cistromes with gene expression, and thus revealed the functional cistromes of multiple TFs that bind at thousands of genomic sites in liver.
Previous genomic studies of circadian gene regulation have focused primarily on the core clock components BMAL1/CLOCK, which bind DNA with a uniform genome-wide phase peaking at ZT6–9 (Hatanaka et al., 2010; Koike et al., 2012; Menet et al., 2012; Rey et al., 2011; Yoshitane et al., 2014), yet only a small fraction of circadian gene transcription is in this phase. Our data suggest that only the genes with phase ZT6–9 are the true BMAL1/CLOCK targets, while many other genes are bound, but not controlled, by BMAL1/CLOCK possibly due to inactive binding or long distance looping to different genes. Moreover, despite extensive binding region overlap with Rev-erbα (Cho et al., 2012), whose repressive activity would conflict with activation by BMAL1/CLOCK, our results demonstrate on a genome-wide scale that enhancer activity is primarily controlled by one factor or the other.
Importantly, our unbiased identification of enhancers revealed not only the ZT6–9 enhancers marked by E-box motifs and bound by BMAL1, NPAS2, and CLOCK but also more abundant sets of enhancers in other phases. Those peaking at ZT0–3, ZT9–15, and ZT18–24 were enriched for ETS, D-box, and RevDR2/RORE motifs, respectively. The ETS motif is recognized by a large family of TFs (Hollenhorst et al., 2011), some of which have recently been implicated in circadian biology and will be focus of future research (Anafi et al., 2014; Ciarleglio et al., 2014). Moreover, by integration of enhancer sites with cistromic data, E4BP4 emerged as a key regulator of the ZT9–15 D-box enhancers in normal liver, as well in the Rev-erbα −/− livers, and Rev-erbα was clearly a strong antiphase repressor bound to RevDR2/RORE sites at ZT18–24 enhancers.
Interestingly, the phase of circadian enhancers exhibited an uneven distribution, with 42% of circadian eRNAs peaking during the late night (ZT18–24), while rhythmic gene transcription was more evenly distributed across all phases. A possible explanation is that the regulation of genes whose transcription peaks in the light cycle might be primarily regulated at promoters. For example, BMAL1 controls gene transcription at both promoters and enhancers (Rey et al., 2011), whereas Rev-erbα, the main controller of the ZT18–24 phase, binds mainly intergenically (Feng et al., 2011; Lam et al., 2013). The overabundance of enhancers in phase ZT18–24 is surprising, yet remarkably consistent with the previously unexplained finding of Koike et al that the global peak of initiated Pol2 occurs at ~ZT22–24 (Koike et al., 2012).
Analysis of oscillating eRNAs in mice fed normal chow ad libitum did not reveal the motifs for TFs previously suggested to entrain liver circadian gene expression to feeding/fasting cycles, such as CREB, SREBP, PPARs, and FOXO1 (Adamovich et al., 2014; Eckel-Mahan et al., 2013; Vollmers et al., 2009). Some of these TFs, such as CREB and SREBP, bind preferentially to promoters of target genes (Everett et al., 2013; Gilardi et al., 2014; Seo et al., 2009), which would not be captured by analysis of eRNAs. Phase-specific enrichment could also have been masked by motifs bound by constitutive liver TFs, such as HNF4A and FOXA1, that bind at enhancers in all phases. It will be interesting to profile eRNAs under altered dietary conditions in future studies to examine the interplay between metabolic cues and circadian rhythms at enhancers.
Rev-erbα expression and repressive function peaks at ZT10 in liver, thereby orchestrating circadian transcription in the opposing phase (ZT22) (Feng et al., 2011). Consistent with this, recruitment of Rev-erbα and its corepressor was strongest at sites of ZT18–24 eRNA transcription. It should be noted that the entire Rev-erbα cistrome in liver includes thousands of other binding sites, with <10% characterized by rhythmic eRNAs antiphase to Rev-erbα binding. Deletion of Rev-erbα specifically activated transcription of these eRNAs, as well as the genes they control, thus clearly delineating the functional component of the Rev-erbα cistrome.
In addition to the direct regulation of circadian genes antiphase to Rev-erbα expression, we uncovered a large set of in-phase circadian transcripts that were downregulated in the absence of Rev-erbα, contrary to its powerful repressive function. Functional enhancer analysis suggested that the downregulated genes in Rev-erbα −/− mice were mediated by D-box factors, including E4BP4, a direct target of Rev-erbα. While the direct regulation of E4BP4 by Rev-erbα has been recognized (Duez et al., 2008), a relatively small number of E4BP4 target genes have been identified in liver, based primarily on in vitro studies of proximal promoter constructs (Tong et al., 2010; Ueda et al., 2005). Our study includes the first ChIP-seq study of E4BP4 in liver, and our integrative analysis demonstrates the extensive, genome-wide effects of this pathway, revealing how a single TF, such as Rev-erbα, can regulate opposing phases of circadian gene expression by its direct and indirect actions.
Together, the present studies reveal mechanisms for generating and coordinating multiple phases of circadian transcription in a single organ. They also demonstrate that the unbiased analysis of enhancer activity and correlated gene expression is a powerful method of discovering relevant TFs and their specific functional cistromes, which can be more generally applied to understanding the transcriptional regulation of physiology and disease states.
EXPERIMENTAL PROCEDURES
Mice
Wild-type (WT) C57Bl/6 mice were purchased from the Jackson Laboratories. The Reverbα −/− mice were obtained from B. Vennström, and backcrossed ≥7 generations with C57Bl/6 mice. 10–12 week old WT and mutant male mice were housed under standard 12h-light/12h-dark cycles, with lights on (ZT0) at 7AM and lights off (ZT12) at 7PM, and euthanized at indicated times. All animal care and procedures followed the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Antibodies
E4BP4 antibodies (Santa Cruz sc-9550 and sc-9549) were mixed in 1:1 ratio for ChIP. RORα antibody was purchased from Santa Cruz (sc-6062).
Global Run On Sequencing (GRO-Seq)
The GRO-seq was performed as previously described (Core et al., 2008; Step et al., 2014; Wang et al., 2011). Raw data are available in GEO (GSE59486). See also Extended Experimental Procedures.
De novo identification of eRNAs
A pipeline was constructed for genome wide de novo identification of eRNA loci. See also Extended Experimental Procedures.
Analysis of oscillating gene transcripts and eRNAs
RPKTM values across all time points for each transcript and eRNA feature were analyzed for significant circadian oscillations using JTK_CYCLE (Hughes et al., 2010). Motif mining at oscillating eRNAs was performed by applying HOMER to the 500bp window centered on each locus. See also Extended Experimental Procedures.
Gene and eRNA expression analysis
Total RNA was extracted from liver using the RNeasy Mini Kit (Qiagen) and treated with DNase (Qiagen). RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed with Power SYBR Green PCR Mastermix on the PRISM 7500 (Applied Biosystems), and analyzed by the standard curve method. Gene or eRNA expression was normalized to mRNA levels of housekeeping gene 36B4 (Arbp). Primer sequences can be found in Table S1D.
Microarray analysis
Microarray analysis of WT and Rev-erbα −/− livers (n=5) was performed by the Penn Microarray Core. Raw data are available from GEO (GSE59460). See also Extended Experimental Procedures.
Chromatin Immunoprecipitation (ChIP)
ChIP-qPCR and ChIP-seq experiments were performed as described (Feng et al., 2011) with minor changes. Raw data for RORα and E4BP4 ChIP-seq are available in GEO (GSE59486). See also Extended Experimental Procedures.
ChIP-seq data analysis
Sequenced reads were aligned to the mouse reference genome (mm9) and peak calling was performed with HOMER (Heinz et al., 2010). Sources of public ChIP-seq data analyzed are listed in Table S2B. See also Extended Experimental Procedures.
Liver-specific gene expression
Flag-Rev-erbα and GFP cDNAs were subcloned into hepatocyte-specific AAV vector AAV8-Tbg (Bell et al., 2011) and tail veins were injected with 1 x 1012 genome copies per mouse. Livers were harvested 2 weeks after injection.
Supplementary Material
(A) Consistent gene transcription levels (Pearson correlation coefficient r=0.95) were observed for biological replicates at both ZT10 (red) and ZT22 (black). Gene transcription is expressed as reads per kb in ten million tags (RPKTM). (B) KEGG pathways that are most highly enriched (P-value cutoff 0.05) in circadian genes grouped by their phases. 8 groups of genes are labeled in different colors.
Table S1. Oscillating gene transcripts identified by GRO-seq. Related to Figure 1.
Table S2. De novo and oscillating eRNAs identified by GRO-seq. Related to Figure 2.
Table S3. Motif mining in oscillating eRNA loci. Related to Figure 3.
(A) Correlation between eRNA signal and other epigenomic marks at intergenic bi-directional eRNA loci. ChIP-seq signal of histone modifications and Pol2, expressed as log tag count (RPTM) on the x-axis, was compared with GRO-seq data at ZT22 on the y-axis (log RPTM). Pearson correlation coefficient was shown in each panel. (B) Genome browser view of eRNAs examined by RT-qPCR in Fig 2G. eRNA loci are highlighted in yellow boxes. (C) ChIP-seq tag density (RPTM per 10bp bin) of H3K27ac at each group of circadian eRNA loci defined in Fig. 2E is shown in separated panels. In each panel, the average tag densities at 6 time points of the day are illustrated in different colors.
(A) Phase correlation between oscillating eRNAs and closest oscillating genes. Each row contains a unique pair of eRNA (red dot) and gene (blue dot), and is double plotted for better visualization. (B) Fraction of oscillating genes containing eRNA sites within 200kb of TSS in comparison with equivalent number of random genes. In-phase eRNAs refers to oscillating eRNAs having similar phases with nearby genes (difference <3hr). In-trend eRNAs refer to eRNAs with 2-fold or higher expression at the in-phase time point than at the anti-phase time point. (C) Protocol for identifying phase-specific TFs at eRNA loci. Detailed description can be found in Extended Experimental Procedures.
GRO-seq tag density (RPTM per 10bp bin) in 2kb window surrounding correlated (left) and non-correlated (right) eRNA loci from Fig. 4D are shown. Inter- and intragenic eRNAs were analyzed separately in the upper and lower panel, respectively. In each figure, GRO-seq tag densities in WT (black) and Rev-erbα −/− mice (red) were compared.
Changes in H3K9ac from ZT10 to ZT22 at Rev-erbα binding sites in Fig. 5B, expressed as ChIP-seq tag count (RPTM) normalized by random eRNA sites and TSSs. (*** p<0.001, one-way ANOVA followed by Tukey’s test).
(A) ChIP-seq binding profiles of Rev-erbα, NCoR, and HDAC3 around Nfil3 locus coding for E4BP4. (B) RT-qPCR measurements of D-box activators Dbp, Tef, and Hlf in WT (blue) and Rev-erbα −/− (orange) livers at multiple circadian time points. Data are represented as mean±SEM (n=2 per genotype per time point). (C) RT-qPCR measurements of E4BP4-bound gene expression in Rev-erbα −/− livers (orange) versus WT (blue). Data are represented as mean±SEM (n=2 per genotype per time point). (D) Average profile of genes from previously published microarray experiment (Kornmann et al., 2007) comparing Rev-erbα over-expressing livers (orange) versus control livers (blue) at multiple time points. Average profiles are shown for genes downregulated in Rev-erbα −/− livers within 200kb of E4BP4 binding at ZT9–15 circadian eRNAs (top), genes upregulated in Rev-erbα −/− livers within 200kb of Rev-erbα binding at ZT18–24 circadian eRNAs (middle), and control genes (unchanged in Rev-erbα −/− livers within 200kb of any circadian eRNA; bottom). (E) Gene expression changes in AAV-Tbg-Rev-erbα treated mouse livers (orange) compared to control AAV-Tbg-GFP treated livers (blue) at ZT10, when E4BP4 expression is normally low (see for comparison ZT22 in Fig. 6H). Data are represented as mean±SEM (One-way ANOVA, *p<0.05, #p<0.1, n=6 per group).
(A) Overlap of genes near CLOCK and Rev-erbα ChIP-seq peaks (within 200kb of TSS). (B) Fraction of oscillating genes in each phase that are bound by both CLOCK and Rev-erbαcompared with 300 random genes (closest mapping within 200kb of TSS). (C) Enrichment of circadian eRNA around genes down-regulated at CT34 in Clock mutant livers (Miller et al., 2007) relative to all oscillating eRNA (hypergeometric test vs all oscillating eRNA, **p<0.01). (D) Enrichment of CLOCK activated genes relative to random genes for: circadian genes bound by CLOCK only at TSS (yellow), and circadian genes bound by CLOCK only at distal sites (>300bp from TSS) not producing in-trend eRNAs (ZT7/ZT19>3 or ZT10/ZT22>3) (grey) (hypergeometric test, **p<0.01). (E) Expression changes in Clock mutant livers (left) and Rev-erbα −/− livers (right) for genes from Fig. 7E: those within 200kb of binding sites for CLOCK (blue) or Rev-erbα (red) with eRNA rhythm indicating functional activation or repression respectively. Expression changes for random genes shown for comparison (grey) (student’s t-test, ***p<0.001).
Table S4. List of differentially expressed genes identified in Rev-erbα −/− livers. Related to Figure 5.
Table S5. Relative eRNA level at CLOCK binding sites. Related to Figure 7.
HIGHLIGHTS.
Enhancer activities predict circadian gene transcription in vivo.
Distinct transcription factors control multiple phases of circadian gene expression.
Circadian eRNAs reveal the functional component of transcription factor cistromes.
A single circadian transcription factor controls opposing transcriptional phases.
Acknowledgments
We thank the Functional Genomics Core (J. Schug and K. Kaestner) and Viral Vector Core (J. Johnston and A. Sandhu) of the Penn Diabetes Research Center (P30 DK19525) for next generation sequencing and AAV production, respectively. We also thank the Penn Microarray Core for microarray analysis. We thank Dr. Ken Zaret for critical reading of the manuscript. This work was supported by NIH grants R01 DK45586 (MAL), F32 DK095526 (LJE), K99 DK099443 (ZS), and F32 DK095563 (ZGH), and the JPB Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
BF, LJE, JJ, and MAL conceived the project design and wrote the manuscript. BF performed analysis of GRO-seq data. BF, LJE, and AR performed integrative genomic analyses. JJ performed GRO-seq experiments. EB and LJE performed E4BP4 ChIP-seq. DF performed RORα ChIP-seq. JJ, EB, and ZGH performed RT-qPCR and ChIP-qPCR experiments. SMA and ZS performed AAV over-expression experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, Asher G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anafi RC, Lee Y, Sato TK, Venkataraman A, Ramanathan C, Kavakli IH, Hughes ME, Baggs JE, Growe J, Liu AC, et al. Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 2014;12:e1001840. doi: 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bell P, Gao G, Haskins ME, Wang L, Sleeper M, Wang H, Calcedo R, Vandenberghe LH, Chen SJ, Weisse C, et al. Evaluation of adeno-associated viral vectors for liver-directed gene transfer in dogs. Hum Gene Ther. 2011;22:985–997. doi: 10.1089/hum.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarleglio CM, Resuehr HE, Axley JC, Deneris ES, McMahon DG. Pet-1 deficiency alters the circadian clock and its temporal organization of behavior. PLoS One. 2014;9:e97412. doi: 10.1371/journal.pone.0097412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Skinner A, Hurst HC. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Bauge E, Havinga R, Bloks VW, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LJ, Le Lay J, Lukovac S, Bernstein D, Steger DJ, Lazar MA, Kaestner KH. Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics. 2013;14:337. doi: 10.1186/1471-2164-14-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- Gilardi F, Migliavacca E, Naldi A, Baruchet M, Canella D, Le Martelot G, Guex N, Desvergne B. Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals. PLoS Genet. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hunger SP. The DBP transcriptional activation domain is highly homologous to that of HLF and TEF and is not responsible for the tissue type-specific transcriptional activity of DBP. Gene. 2001;263:239–245. doi: 10.1016/s0378-1119(00)00565-5. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Peek CB, Ramsey KM, Marcheva B, Bass J. Nutrient sensing and the circadian clock. Trends Endocrinol Metab. 2012;23:312–318. doi: 10.1016/j.tem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Circadian regulation of gene expression in animals. Curr Opin Cell Biol. 2001;13:357–362. doi: 10.1016/s0955-0674(00)00220-9. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Seo YK, Chong HK, Infante AM, Im SS, Xie X, Osborne TF. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc Natl Acad Sci U S A. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Step SE, Lim HW, Marinis JM, Prokesch A, Steger DJ, You SH, Won KJ, Lazar MA. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARgamma-driven enhancers. Genes Dev. 2014;28:1018–1028. doi: 10.1101/gad.237628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Muchnik M, Chen Z, Patel M, Wu N, Joshi S, Rui L, Lazar MA, Yin L. Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. J Biol Chem. 2010;285:36401–36409. doi: 10.1074/jbc.M110.172866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitane H, Ozaki H, Terajima H, Du NH, Suzuki Y, Fujimori T, Kosaka N, Shimba S, Sugano S, Takagi T, et al. CLOCK-controlled polyphonic regulations of circadian rhythms through canonical and non-canonical E-boxes. Mol Cell Biol. 2014 doi: 10.1128/MCB.01465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Khorasanizadeh S, Miyoshi Y, Lazar MA, Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell. 1998;1:849–861. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- Zhao X, Cho H, Yu RT, Atkins AR, Downes M, Evans RM. Nuclear receptors rock around the clock. EMBO Rep. 2014;15:518–528. doi: 10.1002/embr.201338271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Consistent gene transcription levels (Pearson correlation coefficient r=0.95) were observed for biological replicates at both ZT10 (red) and ZT22 (black). Gene transcription is expressed as reads per kb in ten million tags (RPKTM). (B) KEGG pathways that are most highly enriched (P-value cutoff 0.05) in circadian genes grouped by their phases. 8 groups of genes are labeled in different colors.
Table S1. Oscillating gene transcripts identified by GRO-seq. Related to Figure 1.
Table S2. De novo and oscillating eRNAs identified by GRO-seq. Related to Figure 2.
Table S3. Motif mining in oscillating eRNA loci. Related to Figure 3.
(A) Correlation between eRNA signal and other epigenomic marks at intergenic bi-directional eRNA loci. ChIP-seq signal of histone modifications and Pol2, expressed as log tag count (RPTM) on the x-axis, was compared with GRO-seq data at ZT22 on the y-axis (log RPTM). Pearson correlation coefficient was shown in each panel. (B) Genome browser view of eRNAs examined by RT-qPCR in Fig 2G. eRNA loci are highlighted in yellow boxes. (C) ChIP-seq tag density (RPTM per 10bp bin) of H3K27ac at each group of circadian eRNA loci defined in Fig. 2E is shown in separated panels. In each panel, the average tag densities at 6 time points of the day are illustrated in different colors.
(A) Phase correlation between oscillating eRNAs and closest oscillating genes. Each row contains a unique pair of eRNA (red dot) and gene (blue dot), and is double plotted for better visualization. (B) Fraction of oscillating genes containing eRNA sites within 200kb of TSS in comparison with equivalent number of random genes. In-phase eRNAs refers to oscillating eRNAs having similar phases with nearby genes (difference <3hr). In-trend eRNAs refer to eRNAs with 2-fold or higher expression at the in-phase time point than at the anti-phase time point. (C) Protocol for identifying phase-specific TFs at eRNA loci. Detailed description can be found in Extended Experimental Procedures.
GRO-seq tag density (RPTM per 10bp bin) in 2kb window surrounding correlated (left) and non-correlated (right) eRNA loci from Fig. 4D are shown. Inter- and intragenic eRNAs were analyzed separately in the upper and lower panel, respectively. In each figure, GRO-seq tag densities in WT (black) and Rev-erbα −/− mice (red) were compared.
Changes in H3K9ac from ZT10 to ZT22 at Rev-erbα binding sites in Fig. 5B, expressed as ChIP-seq tag count (RPTM) normalized by random eRNA sites and TSSs. (*** p<0.001, one-way ANOVA followed by Tukey’s test).
(A) ChIP-seq binding profiles of Rev-erbα, NCoR, and HDAC3 around Nfil3 locus coding for E4BP4. (B) RT-qPCR measurements of D-box activators Dbp, Tef, and Hlf in WT (blue) and Rev-erbα −/− (orange) livers at multiple circadian time points. Data are represented as mean±SEM (n=2 per genotype per time point). (C) RT-qPCR measurements of E4BP4-bound gene expression in Rev-erbα −/− livers (orange) versus WT (blue). Data are represented as mean±SEM (n=2 per genotype per time point). (D) Average profile of genes from previously published microarray experiment (Kornmann et al., 2007) comparing Rev-erbα over-expressing livers (orange) versus control livers (blue) at multiple time points. Average profiles are shown for genes downregulated in Rev-erbα −/− livers within 200kb of E4BP4 binding at ZT9–15 circadian eRNAs (top), genes upregulated in Rev-erbα −/− livers within 200kb of Rev-erbα binding at ZT18–24 circadian eRNAs (middle), and control genes (unchanged in Rev-erbα −/− livers within 200kb of any circadian eRNA; bottom). (E) Gene expression changes in AAV-Tbg-Rev-erbα treated mouse livers (orange) compared to control AAV-Tbg-GFP treated livers (blue) at ZT10, when E4BP4 expression is normally low (see for comparison ZT22 in Fig. 6H). Data are represented as mean±SEM (One-way ANOVA, *p<0.05, #p<0.1, n=6 per group).
(A) Overlap of genes near CLOCK and Rev-erbα ChIP-seq peaks (within 200kb of TSS). (B) Fraction of oscillating genes in each phase that are bound by both CLOCK and Rev-erbαcompared with 300 random genes (closest mapping within 200kb of TSS). (C) Enrichment of circadian eRNA around genes down-regulated at CT34 in Clock mutant livers (Miller et al., 2007) relative to all oscillating eRNA (hypergeometric test vs all oscillating eRNA, **p<0.01). (D) Enrichment of CLOCK activated genes relative to random genes for: circadian genes bound by CLOCK only at TSS (yellow), and circadian genes bound by CLOCK only at distal sites (>300bp from TSS) not producing in-trend eRNAs (ZT7/ZT19>3 or ZT10/ZT22>3) (grey) (hypergeometric test, **p<0.01). (E) Expression changes in Clock mutant livers (left) and Rev-erbα −/− livers (right) for genes from Fig. 7E: those within 200kb of binding sites for CLOCK (blue) or Rev-erbα (red) with eRNA rhythm indicating functional activation or repression respectively. Expression changes for random genes shown for comparison (grey) (student’s t-test, ***p<0.001).
Table S4. List of differentially expressed genes identified in Rev-erbα −/− livers. Related to Figure 5.
Table S5. Relative eRNA level at CLOCK binding sites. Related to Figure 7.