Summary
Ribosomes elongate at a non-uniform rate during translation. Theoretical models and experiments disagree on the in vivo determinants of elongation rate and the mechanism by which elongation rate affects protein levels. To resolve this conflict, we measured transcriptome-wide ribosome occupancy under multiple conditions and used it to formulate a whole-cell model of translation in E. coli. Our model predicts that elongation rates at most codons during nutrient-rich growth are not limited by the intracellular concentrations of aminoacyl-tRNAs. However, elongation pausing during starvation for single amino acids is highly sensitive to the kinetics of tRNA aminoacylation. We further show that translation abortion upon pausing accounts for the observed ribosome occupancy along mRNAs during starvation. Abortion reduces global protein synthesis, but it enhances the translation of a subset of mRNAs. These results suggest a regulatory role for aminoacylation and abortion during stress, and our study provides an experimentally-constrained framework for modeling translation.
Introduction
Protein synthesis begins with initiation by ribosomes on an mRNA, and is followed by a sequence of elongation steps during which amino acids are added to the growing polypeptide chain. Initiation is the rate-limiting step for the translation of most mRNAs during nutrient-rich growth (Jacques and Dreyfus, 1990). However, stressful perturbations such as amino acid starvation or transgene overexpression can decrease the elongation rate of ribosomes and affect protein levels (Subramaniam et al., 2013a; Varenne et al., 1984; Welch et al., 2009). Biophysical modeling of translation can be used to infer the quantitative effect of these stressful perturbations on initiation rate, elongation rate, and the expression level of proteins (Shah et al., 2013). More generally, modeling integrates our biochemical knowledge of translation, and thus enables identification of novel regulatory processes when incorporation of known mechanisms is insufficient to recapitulate experimental measurements.
Biophysical modeling of translation has been greatly aided by the development of ribosome profiling which involves deep sequencing of ribosome-protected mRNA fragments to measure ribosome occupancy on mRNAs (Ingolia et al., 2009). Ribosome profiling has also produced surprising results that challenge two central assumptions in current theoretical models of translation. First, theoretical models assume that elongation rates at codons are directly proportional to the intracellular concentration of cognate aminoacyl-tRNAs (aa-tRNAs) during nutrient-rich growth (Shah et al., 2013; Tuller et al., 2010). However, this assumption is not supported by ribosome profiling – although concentrations of different tRNAs vary over greater than a 10-fold range in bacteria and yeast (Dong et al., 1996; Tuller et al., 2010), the measured ribosome occupancy at codons varies less than 2-fold in these organisms during nutrient-rich growth (Li et al., 2012; Qian et al., 2012). Differential aminoacylation of tRNAs is also unlikely to underlie this discrepancy since most tRNA species are >70% aminoacylated during nutrient-rich growth (Yegian et al., 1966). Second, most models assume that a decrease in ribosome elongation rate on an mRNA affects the level of the corresponding protein by causing a traffic jam of trailing ribosomes (Shah et al., 2013; Zhang et al., 1994). However, except for a few atypical mRNAs (Guydosh and Green, 2014; Li et al., 2012), traffic jams at ribosome pause sites have not been observed in vivo, and the effect of ribosome traffic jams on protein level remains unclear. Together, these results suggest that current theoretical models do not include the full set of mechanistic ingredients that is necessary for accurate modeling of in vivo translation, especially as it pertains to the elongation stage.
To identify mechanistic features of the elongation stage that enable accurate biophysical modeling of translation, we combined transcriptome-scale and reporter-based experiments with whole-cell computational modeling. This approach enabled us to decipher the contribution of various molecular processes to the elongation rate of ribosomes and the synthesis rate of proteins. We found that the differences in the intracellular concentration of tRNAs and the occurrence of ribosome traffic jams at pause sites, both of which have been key ingredients in previous theoretical models (Shah et al., 2013; Tuller et al., 2010), are insufficient to predict the measured ribosome occupancy in our experiments. We find that the molecular process of aminoacylation has a critical role in protein synthesis through modulation of ribosome elongation rates during stress. Further, we find that premature termination of translation before the synthesis of full length proteins (henceforth referred to as translation abortion) determines both the ribosome occupancy along mRNAs and protein expression during stress. More generally, our work illustrates the usefulness of integrating deep-sequencing experimental methods such as ribosome profiling together with quantitative whole-cell modeling to reveal the experimentally-relevant regimes of the a priori large parameter space in biophysical models of complex cellular processes.
Results
Changes in ribosome occupancy upon starvation for single amino acids
We previously found that during starvation for single amino acids in E. coli, the presence of certain codons cognate to the limiting amino acid can decrease the protein synthesis rate by up to 100-fold (Subramaniam et al., 2013a). Starvation for single amino acids decreases the concentration of the cognate aa-tRNAs (Dittmar et al., 2005; Sørensen, 2001; Sørensen et al., 2005), thus decreasing the elongation rate of ribosomes at cognate codons. Based on these observations, we used single amino acid starvation as an experimental condition to develop constraints for biophysical models of protein synthesis in an elongation-limited regime of translation.
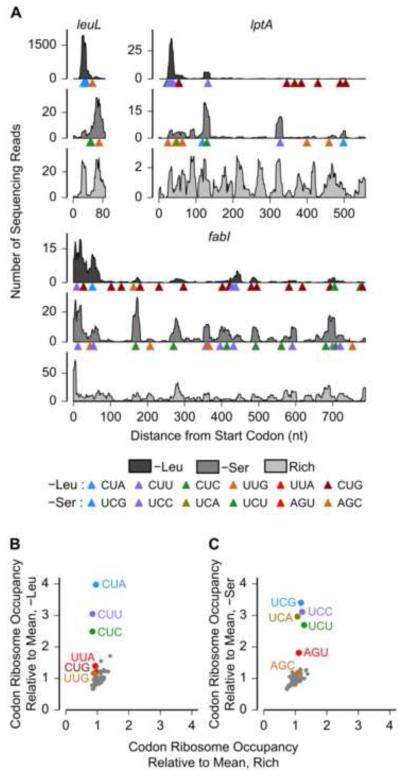
To characterize the effect of amino acid starvation on translation, we performed ribosome profiling in E. coli after 30 minutes of starvation for each of two amino acids – leucine and serine. For comparison with an initiation-limited regime of translation, we also performed ribosome profiling on cells grown in rich-defined medium with all twenty amino acids. The distribution of ribosome footprints along mRNAs was highly variable during nutrient-rich growth (Figure 1A), consistent with earlier observations (Oh et al., 2011). Starvation for either leucine or serine caused a pronounced change in the distribution of ribosome footprints along individual mRNAs (Figure 1A). Consistent with previous observations (Li et al., 2012), the transcriptome-averaged ribosome occupancy differed less than 2-fold across the 61 codons during nutrient-rich growth (Figures 1B, 1C, horizontal axis) and did not systematically vary with tRNA abundance (Figure S1A). Upon leucine or serine starvation, the average ribosome occupancy increased at leucine and serine codons, respectively, but this increase was not uniform (Figures 1B, 1C). The three leucine codons CUA, CUC and CUU had 2.5- to 4-fold higher ribosome occupancy than the average during leucine starvation, while the ribosome occupancy at the remaining three leucine codons CUG, UUA and UUG was comparable to that of non-cognate codons. Similarly, ribosome occupancy at the four serine codons UCA, UCC, UCG and UCU was markedly higher than at the two serine codons AGC and AGU during serine starvation. We attributed the increase in ribosome occupancy at leucine and serine codons to a decrease in the ribosome elongation rate, since we did not observe an increase in total mRNA density at these codons upon leucine or serine starvation (Figures S1B, S1C).
Figure 1.
Change in Ribosome Occupancy upon Starvation for a Single Amino Acid.
(A) Measured ribosome occupancy along three E. coli genes: leuL, lptA and fabI during leucine starvation (–Leu), serine starvation (–Ser) and amino acid rich growth (Rich). The horizontal axis extends from the start codon to the stop codon for each gene. Triangles indicate the positions of leucine (serine) codons along the coding sequence in the leucine (serine) starvation case. (B, C) Measured ribosome occupancy at the 61 sense codons averaged across the transcriptome. Start and stop codons are not shown. Standard errors of mean are smaller than data markers.
See also Figure S1.
A transcriptome-scale biophysical model of translation in E. coli
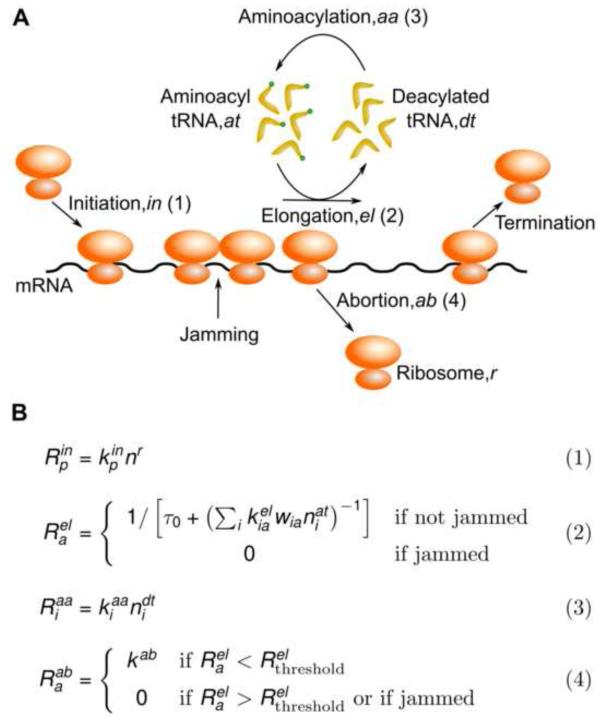
To systematically evaluate the consistency of different biophysical models with our measurements of ribosome occupancy, we formulated a generalized kinetic model of protein synthesis in E. coli that accounted for four different molecular processes we found to play a critical role in determining the ribosome occupancy and protein expression: initiation, elongation, aminoacylation and abortion (Figure 2A). Other molecular processes that are not expected to limit protein synthesis rate under the conditions of our study, such as termination at stop codons and ribosome recycling, were assumed to be instantaneous (rates set to infinity) for the sake of simplicity.
Figure 2.
A Transcriptome-scale Biophysical Model of Translation.
(A) Schematic of the four cellular processes modeled (initiation, elongation, aminoacylation, abortion) and the molecular species considered in the biophysical model. (B) Reaction rates, Ri, for the cellular processes shown in (A). The superscripts following Ri refer to the abbreviations for the cellular processes in (A). Subscript indices are used for distinct molecular species of the same kind (mRNA – p, tRNA – i, codon – a). The intracellular concentrations of molecular species and the values for rate constants in our whole-cell simulation are in Table S1.
See also Data S1.
The core features of our biophysical model of protein synthesis are summarized by the kinetic rate equations for the four molecular processes (Figure 2B). The full list of parameter values used in our biophysical model is given in Table S1. We implemented our model as a continuous-time, discrete-state process at the whole-cell level for E. coli by adapting the source code from a recent computational study in yeast (Shah et al., 2013). During balanced nutrient-rich growth, the availability of free ribosomes for initiation is rate-limiting for protein synthesis (Vind et al., 1993). We modeled this empirical observation in Eq. (1) by taking the initiation rate Rin to be proportional to nr, the number of free ribosomes in the cell (Figure 2B). The initiation rate constant kinp for each mRNA species p was estimated from our ribosome profiling measurements, and the macromolecular composition and synthesis rates that have been measured with high accuracy in E. coli (Bremer and Dennis, 2008) (Extended Experimental Procedures).
Intra-ribosomal events limit the rate of elongation during nutrient-rich growth
The elongation cycle is a multi-step process that begins with the delivery of the aa-tRNA (in ternary complex with Ef-Tu and GTP) to the ribosome. This step is followed by a series of intra-ribosomal events that include kinetic proofreading, peptidyl transfer, and translocation of the ribosome to the next codon (Wintermeyer et al., 2004). We considered a minimal model of elongation (Elf et al., 2003) which is composed of two effective steps [Eq. (2) in Figure 2B]: The first step accounts for the arrival of cognate aa-tRNAs at the ribosome and its rate is proportional to the intracellular concentration of aa-tRNA, nat. The proportionality factor kel is the second-order rate constant (kcat/KM) for association between the ribosome and the aa-tRNA. The second step accounts for all intra-ribosomal events that follow the arrival of the cognate aa-tRNA at the ribosome, and the cumulative rate of these events does not depend on the intracellular concentration of aa-tRNA. For simplicity, we set the total rate of this second step to a uniform value 1/τ0 for all codons. Finally, when translocation is prevented by the presence of a leading ribosome (henceforth referred to as jamming), the elongation rate is set to zero in our model.
If τ0 (the rate constant describing all intra-ribosomal events following arrival of the aa-tRNA) has the same value for all codons, the experimental observation that ribosome occupancies at codons do not vary inversely with the corresponding tRNA concentrations during nutrient-rich growth (Figure S1A) now implies a straightforward mechanistic constraint in our model – that the rate of intra-ribosomal events limits the overall rate of ribosome elongation under these conditions, i.e. τ0 » 1/kelnat in Eq. (2) of Figure 2B. Together, with the measured average elongation rate of ribosomes of ~20 codons per second (Bremer and Dennis, 2008), we can then infer that τ0 ≈ 0.05 s. In the more general case where τ0 is not uniform for all codons, most of the codons still need to have τ0 » 1/kelnat to recapitulate the observed lack of (inverse) proportionality between ribosome occupancy and tRNA concentration. Our conclusion that τ0 » 1/kelnat, based solely on ribosome occupancy measurements, is also consistent with the typical in vitro value of kel = 2 × 107 M−1s−1 (Bilgin and Ehrenberg, 1994; Pavlov and Ehrenberg, 1996) and the in vivo concentrations of tRNAs (nat) during nutrient-rich growth (Bremer and Dennis, 2008; Dong et al., 1996) (Data S1). Using these values of kel and nat, the median value of 1/kelnat for the 61 sense codons is 0.0036 s, and all codons had 1/kelnat < 0.013 s (« τ0 ≈ 0.05 s).
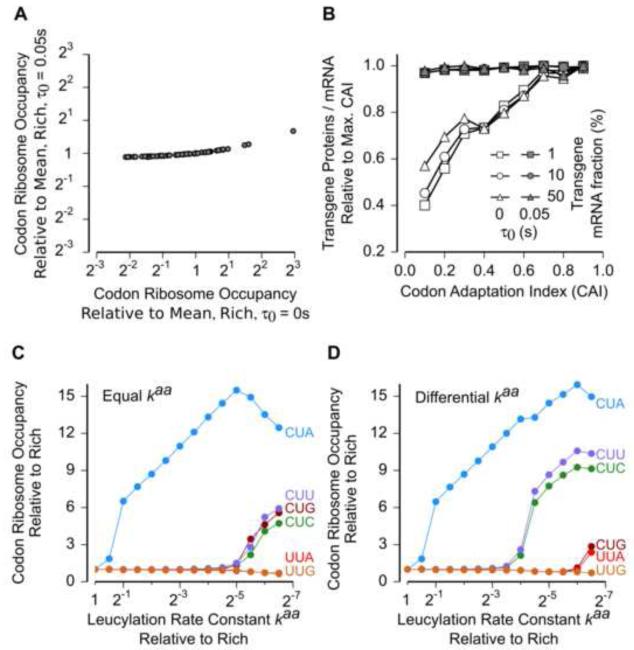
To test the consistency of our whole-cell model with ribosome profiling measurements, we simulated our model with the above-constrained rate constants for elongation. With τ0 = 0.05 s and kel = 2 × 107 M−1s−1, we observed less than 2-fold variation (s.d./mean = 10%) in ribosome occupancy across the 61 codons (Figure 3A, vertical axis). By contrast, when we set the timescale of intra-ribosomal events to be zero (τ0 = 0 s), as assumed in previous work (Shah et al., 2013; Tuller et al., 2010), ribosome occupancy varied 34-fold (s.d./mean = 105%) across the 61 codons (Figure 3A, horizontal axis), even though for both values of τ0 (0.05 s and 0 s), the average elongation rate (Rel) was ~ 20 codons s−1. Thus, we conclude that ribosome occupancy at codons during nutrient-rich growth of E. coli, as measured using ribosome profiling, is consistent with a model in which intra-ribosomal events, rather than the arrival of aa-tRNA to the ribosome A-site, are the slowest steps in the elongation cycle for most codons.
Figure 3.
Elongation and Aminoacylation Kinetics Determine Ribosome Occupancy at Codons.
(A) Mean ribosome occupancy at the 61 sense codons averaged across the transcriptome during nutrient-rich growth calculated from whole-cell model. Simulations were run with the time for intra-ribosomal events at a single codon, τ0, set to either 0 s (horizontal) or 0.05 s (vertical). The value of kel was chosen such that the mean elongation rate of ribosomes Rel was approximately equal to the experimentally measured value of 20 s−1 in both cases.
(B) Amount of transgene proteins produced per mRNA upon overexpression during nutrient-rich growth calculated from whole-cell model as a function of codon adaptation index (CAI) and the transgene fraction. All data points corresponding to a single transgene mRNA fraction were normalized by the data point at CAI = 0.9.
(C, D) Mean ribosome occupancy at the six leucine codons as a function of leucylation rate constant calculated from whole-cell model. The leucylation rate constants of the five leucine tRNA isoacceptors were set either equal (C) or different (D). In the differential case (D), the leucylation rate constants were in the proportion 1.5: 0.5: 1: 0.5: 0.5 (Leu1 through Leu5).
See also Figure S2.
Highly-expressed genes in E. coli, such as those coding for the translation machinery, display a characteristic codon bias (called the major codon bias) towards codons that are decoded by tRNA isoacceptors with high intracellular concentration (Andersson and Kurland, 1990). Motivated by this observation, biophysical models often predict that optimizing the codon usage of an overexpressed transgene by enriching for codons decoded by abundant tRNA isoacceptors can significantly improve the yield of its protein (Shah et al., 2013; Zhang et al., 1994). Since this question is of significant interest in biotechnological applications (Gustafsson et al., 2004), we re-examined it in the light of our evidence that intra-ribosomal events, rather than the arrival of aa-tRNA, limit the rate of elongation during nutrient-rich growth.
We simulated the expression of three transgenes at different fractions of the cellular transcriptome and with various codon bias (Extended Experimental Procedures, Data S1). When we assumed that the elongation rate at codons is proportional to the concentration of aa-tRNAs [by setting τ0 = 0 s in Eq. (2) of Figure 2B], we found that increasing codon bias can improve protein expression from the transgenes by over 2-fold (Figure 3B, open markers). However, this effect of codon bias was essentially absent when elongation rates at codons were limited by intra-ribosomal events (Figure 3B, filled markers). We observed a similar effect of intra-ribosomal events when we simulated a model for yeast translation (Shah et al., 2013) (Figure S2A). The lack of effect of codon bias on transgene overexpression is consistent with measurements using synthetic gene libraries in E. coli, which detected little correlation between yield of overexpressed proteins and the codon adaptation index (Kudla et al., 2009; Welch et al., 2009). Thus, based on the elongation kinetics inferred from ribosome profiling, we suggest that the major codon bias is not a significant determinant of protein yield during transgene overexpression in E. coli.
Differential aminoacylation determines the specificity of elongation pausing
Having formulated our biophysical model during nutrient-rich growth when initiation is rate-limiting for translation, we sought to test our model in an elongation-limited regime during starvation for single amino acids. To simulate starvation for a single amino acid, we reduced the aminoacylation rate constant of the corresponding tRNA isoacceptors [kaa in Eq.(3) in Figure 2B] while keeping all other parameters identical to those during nutrient-rich growth. Reducing the leucylation rate caused a non-uniform increase in ribosome occupancy at both leucine and serine codons (Figure 3C, S2B). However, the hierarchy predicted by the model significantly deviated from the measured hierarchy among these codons both during leucine and serine starvation (Figure 1B).
The discrepancy between model and experiments in the hierarchy of ribosome occupancy at codons during starvation could arise from two sources. One possibility is that the uniform rate constants for codon-tRNA interaction (kel) could differ between tRNA isoacceptors since we used only average estimates in our model. However, when we replaced the average estimates with in vitro measurements of kel, which vary over a 3-fold range for the leucine family (Sørensen et al., 2005), it had little effect on the predicted hierarchy of ribosome occupancy at codons (Figure S2C). A second explanation for the above discrepancy is that the rate constants for aminoacylation (kaa) might be unequal between the different tRNA isoaccceptors. Indeed, when we allowed the aminoacylation rate constants for tRNA isoacceptors to differ over a 3-fold range (Figure S2D), the model could largely recapitulate the experimentally-measured hierarchy among codons during starvation (Figures 3D, S2E). Our modeling of differential aminoacylation kinetics is consistent with previous in vitro measurements which found that tRNA isoacceptors could differ 2-20 fold in their aminoacylation rate constants (Fender et al., 2004; Harris and Marashi, 1980; Myers et al., 1971).
Accounting for aminoacylation itself has a critical effect on protein expression during amino acid starvation. A recent modeling study arrived at the conclusion that protein expression can be rescued during amino acid starvation by reducing the number of ribosomes in the cell (Shah et al., 2013). This conclusion was based on lowering the total number of tRNAs to simulate amino acid starvation without considering aminoacylation (Figure S2F). However, there is little evidence that the total concentration of tRNAs significantly changes upon amino acid starvation, while the canonical effect of amino acid starvation is the reduction in aminoacyl-tRNA concentration due to lower aminoacylation rate (Dittmar et al., 2005; Sørensen, 2001; Sørensen et al., 2005). When we simulated starvation by reducing the levels of aminoacyl-tRNA rather than that of total tRNA using the model of (Shah et al., 2013), we did not observe rescue of protein expression upon decreasing the number of ribosomes (Figures S2G). Thus, accounting for aminoacylation qualitatively alters the prediction from biophysical models of translation in the elongation-limited regime of amino acid starvation.
Ribosome traffic jams at ribosome pause sites during amino acid starvation
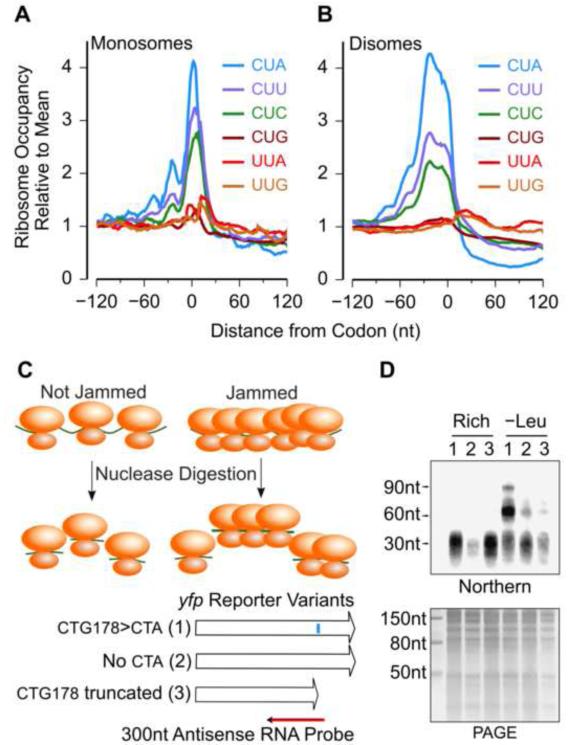
Single mRNAs are often simultaneously translated by several ribosomes. As a result, if ribosomes pause for a sufficiently long duration during elongation, a traffic jam of trailing ribosomes can occur in the 5’ region of the ribosome pause site. To detect traffic jams in our ribosome profiling measurement, we calculated the average ribosome occupancy across the transcriptome in the 120 nt region on either side of the leucine codons during leucine starvation (Figure 4A). We observed an increase in ribosome occupancy at three of the leucine codons CUA, CUC and CUU, which is consistent with ribosomes pausing at these codons during elongation. In addition, we observed smaller peaks in ribosome density centered approximately −28 nt and −56 nt upstream of these three codons (Figure 4A). These peaks are consistent with a traffic jam of one and two ribosomes, behind the paused ribosome. We observed similar, but smaller peaks at −28 nt for four of the six serine codons during serine starvation (Figure S3A). Notably, we did not observe smaller peaks upstream of Shine-Dalgarno like codons which also cause an increase in ribosome occupancy during nutrient-rich growth (Li et al., 2012) (Figure S3B).
Figure 4.
Ribosome Traffic Jams at Ribosome Pause Sites.
(A) Measured monosome occupancy from −120 nt to +120 nt around the six leucine codons during leucine starvation. The monosome occupancy was averaged across all occurrences of each codon in the transcriptome.
(B) Measured disome occupancy from −120 nt to +120 nt around the six leucine codons during leucine starvation. The disome occupancy was averaged across all occurrences of each codon in the transcriptome.
(C) Nuclease footprinting assay for detecting ribosome traffic jams on yfp reporter mRNAs. The blue vertical bar along the first variant indicates the location of the CTG200>CTA substitution. Northern blotting was performed using a 32P-labeled antisense RNA complementary to the 300 nt mRNA region from −250 nt to +50 nt of the CTG200>CTA substitution.
(D) Upper panel: Northern blot of nuclease-digested polysomes for the three yfp variants; Lower panel: Polyacrylamide gel corresponding to the Northern blot. Numbers above individual lanes correspond to the three yfp variants in (C). The size markers on the left of the Northern blot were inferred by aligning it to the polyacrylamide gel image. The arrows at 30 nt, 60 nt and 90 nt indicate the approximate locations of monosomes, disomes and trisomes respectively.
See also Figure S3.
Previous work has shown that nuclease treatment of polysomes with ribosome traffic jams can result in longer mRNA footprints that are protected by multiple ribosomes (Guydosh and Green, 2014; Wolin and Walter, 1988). To test whether longer mRNA footprints occur during amino acid starvation in E. coli, we measured the size of nuclease-protected mRNA fragments using three yellow fluorescent protein (yfp) reporter variants (Figure 4C). We observed a ~30 nt fragment corresponding to the monosome for all yfp variants both during rich growth and during leucine starvation (Figure 4D). In addition, we observed a prominent ~60 nt fragment and a weaker ~90 nt fragment in the yfp variant with the CUA codon during leucine starvation. These longer fragments were either faint or absent in the control variants without CUA codons and during nutrient-rich growth, which is consistent with a traffic jam of multiple ribosomes caused by pausing of ribosomes at the CUA codon during leucine starvation. These results also suggest that the standard ribosome profiling method, in which only monosome-protected fragments are sequenced, underestimates the actual in vivo magnitude of ribosome occupancy 5’ to the pause site in the presence of ribosome traffic jams. Deep-sequencing of longer mRNA fragments (50-80 nt) that were protected by two ribosomes (disomes) from nuclease digestion revealed an increase in average disome occupancy up to 90 nt upstream of CUA codons during leucine starvation (Figure 4B), which is consistent with a traffic jam of between 2-3 ribosomes behind the paused ribosome. The length of this region with increased footprint density was similar to that obtained using the standard ribosome profiling method in which only short (~30 nt) monosome-protected mRNA fragments were sequenced (Figure 4A), which suggests that both the paused, leading ribosome as well as the jammed, trailing ribosomes have an equal likelihood of occurring either as monosomes or as disomes upon nuclease treatment.
Translation abortion determines the distribution of ribosomes along mRNAs during amino acid starvation
The strong ribosome pausing that we observed during starvation for single amino acids enabled us to test whether the measured ribosome occupancy along mRNAs is quantitatively consistent with a biophysical model of ribosome traffic jams. During nutrient-rich growth, except for a peak at the start codon, ribosome occupancy was uniformly distributed across the entire length of the mRNA (Figure 5A, grey line), which is similar to previous observations (Oh et al., 2011). By contrast, during starvation for leucine, the distribution of ribosome occupancy was highly non-uniform, varying over a 10-fold range within the first 900 nt (Figure 5A, black line). Ribosome occupancy increased sharply to a maximum at around 30 nt from the start codon, and then decreased towards the 3’ end of the mRNA. Starvation for serine produced a similar, but less skewed distribution of ribosome occupancy along mRNAs (Figure S4A). By comparison, the distribution of total mRNA density was not significantly skewed during leucine starvation (Figure S4B).
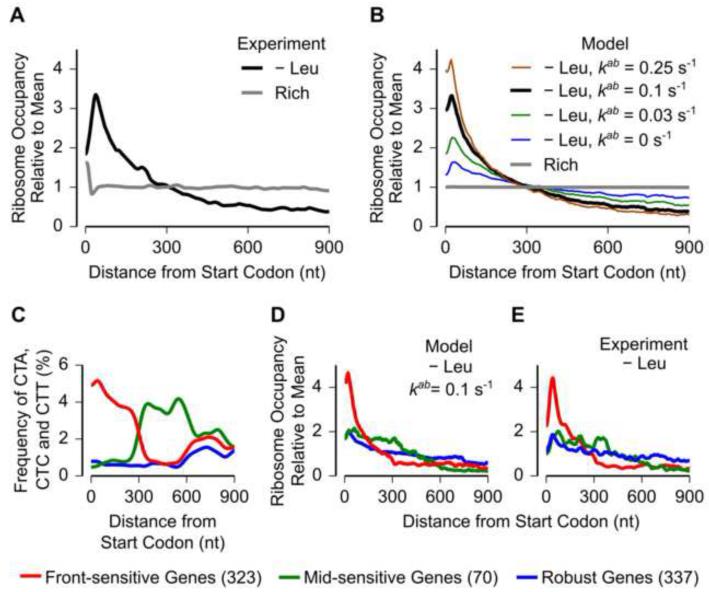
Figure 5.
Translation abortion determines the distribution of ribosomes along mRNAs during amino acid starvation.
(A) Measured ribosome occupancy along mRNAs averaged across the transcriptome (1518 genes).
(B) Ribosome occupancy along mRNAs averaged across the transcriptome (1518 genes) calculated from the whole cell model. The abortion rate constant kab was varied. Leucine starvation was modeled as a constant 100-fold reduction in the leucylation rate constant, kaa, Leu.
(C) Codon frequency of the three leucine codons, CTA, CTC and CTT, in three sets of genes (red, green, blue) with different intragenic distributions of these codons. The number of genes in each class is shown in parentheses in the legend. The codon frequency distribution was smoothed using a Gaussian window of 30 nt width.
(D) Ribosome occupancy averaged across the three sets of genes during leucine starvation calculated from the whole-cell model.
(E) Measured ribosome occupancy averaged across the three sets of genes during leucine starvation.
Ribosome occupancy profiles in all panels were smoothed using a sliding window of 30 nt. Each ribosome occupancy profile was normalized to have a mean value of 1.
See also Figure S4.
We then used the distribution of measured ribosome occupancy along mRNAs to evaluate our biophysical model of translation. Our model reproduced the uniform distribution of measured ribosome occupancy during nutrient-rich growth (Figure 5B, grey line), which is consistent with initiation rate being rate-limiting for translation under these conditions (Li et al., 2014). However, when we simulated leucine starvation by a 100-fold reduction in the leucylation rate, a model that included only ribosome traffic jams predicted a more uniform distribution of ribosome occupancy than the one observed experimentally (Figure 5B, blue line, compared to black line Figure 5A). This disagreement between the model and our measurements could not be offset by varying the leucylation rate, the only remaining free parameter in our model, over a 1000-fold range (Figure S4C), and was also observed for serine starvation (Figure S4D).
The discrepancy in the distribution of ribosome occupancy between ribosome profiling measurements and our model that included only ribosome traffic jams led us to consider other molecular processes that occur in response to ribosome pausing. Translation abortion is a widespread mechanism of ribosome rescue that has been observed both in bacteria and eukaryotes in response to ribosome pausing (Shoemaker and Green, 2012). Inactivation of translation abortion factors can decrease the growth rate of cells during stressful perturbations (Keiler and Feaga, 2014), indicating the importance of abortion during stress. We modeled translation abortion [Eq. (4) in Figure 2B] as occurring at a constant rate kab whenever the elongation rate of ribosomes falls below a threshold, Relthreshold. This threshold elongation rate was set below the elongation rate of all codons under nutrient-rich growth to account for the experimental observation that the abortion is selective for paused ribosomes (Moore and Sauer, 2005). In our model, increasing the rate of translation abortion (kab) produced higher ribosome occupancy at the 5’ end of mRNAs during starvation (Figures 5B, S4D). An in vivo abortion rate (kab) of ~0.1 s−1 in our model predicted an average distribution of ribosome occupancy that matched the experimentally observed distribution both during leucine and serine starvation (Figures 5B, S4D, black lines).
To test the role of translation abortion at ribosome pause sites in determining the distribution of measured ribosome occupancy along mRNAs, we analyzed three subsets of E. coli genes with varying codon usage patterns (Figure 5C). These gene sets had different intragenic locations of the three leucine codons CUA, CUC and CUU that result in ribosome pausing during leucine starvation (Figure 1B). These three codons were present at a high frequency within the first 300 nt in the first set (Figure 5C, red, 323 genes), between 300 nt and 600 nt in the second set (Figure 5C, green, 70 genes), and at a low frequency throughout the third set (Figure 5C, blue, 337 genes). The whole-cell model incorporating abortion predicted a decreased ribosome occupancy following the mRNA region where the CUA, CUC and CUU codons were present at high frequency in these gene sets (Figure 5D). This prediction matched the measured distribution of ribosome occupancy for all the three gene sets (Figure 5E), indicating the validity of the abortion model. By contrast, a model that incorporated only ribosome traffic jams, did not recapitulate the measured distribution of ribosome occupancy in the three gene sets (Figure S4E). Together, these observations indicate that incorporating abortion at ribosome pause sites in our whole-cell model of translation is necessary for quantitative consistency with the measured ribosome occupancy along mRNAs during starvation for single amino acids in E. coli.
Translation abortion and its effectors during amino acid starvation
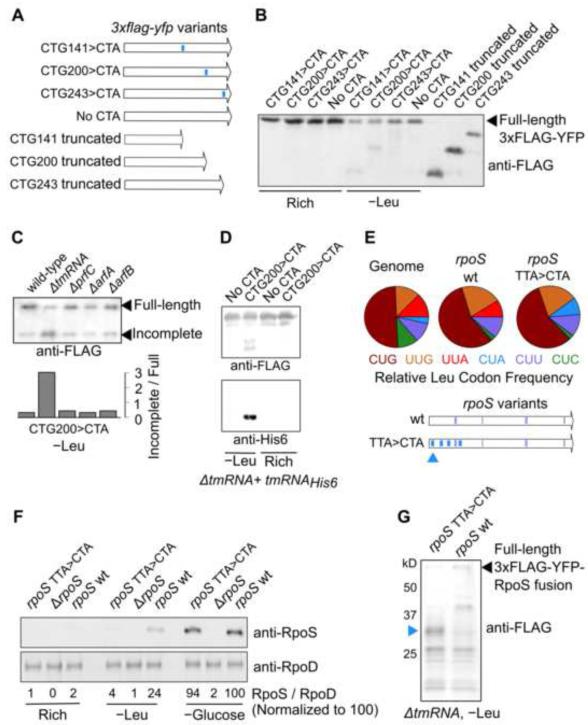
Based on the prediction of translation abortion from our analysis of the measured ribosome occupancy, we looked for abortion events at ribosome pause sites using a yfp reporter system. We constructed synonymous variants of yfp that encoded a 3xFLAG epitope at the N-terminus for detection of incomplete polypeptides, and that had a single leucine starvation-sensitive CTA codon at one of three different locations along 3xflag-yfp (Figure 6A). During leucine starvation, we detected shorter length polypeptide fragments whose size was consistent with translation abortion at the CUA codon (Figure 6B).
Figure 6.
Translation abortion and its effectors during amino acid starvation.
(A) Schematic of 3xflag-yfp reporter variants with either single CTG>CTA substitutions (indicated in blue) or truncated at one of three locations.
(B) Western blot using Anti-FLAG antibody for the 3xflag-yfp variants shown in (A).
(C) Western blot with Anti-FLAG antibody of the CTG200>CTA variant of yfp during leucine starvation in strains with deletion of one of four different genes encoding factors that mediate translation abortion (tmRNA, prfC, arfA, arfB). ‘Wild-type’ refers to the parent leucine auxotroph strain. The lower panel indicates the densitometric ratio of these two bands.
(D) Upper panel – Immunoprecipitation with Anti-FLAG antibody of CTG200>CTA yfp variant expressed during leucine starvation in a ΔtmRNA strain with a tmRNAHis6 mutant. Lower panel – tmRNAHis6 activity detected with an Anti-His6 antibody.
(E) Pie charts – Relative frequency of the six leucine codons across all coding sequences in the genome, in the rpoS wild-type coding sequence, and in the rpoS synonymous variant. Four TTA codons were replaced by CTA codons in the rpoS synonymous variant at the locations indicated by thick blue bars. The thin blue and green bars correspond to the location of the CTC and CTT codons in the rpoS wt and synonymous variant. Blue triangle indicates the location of the first ribosome pause site encoded by the CTA codon during leucine starvation.
(F) Western blot against the RpoS protein (upper panel) and RpoD protein (lower panel) during nutrient-rich growth, leucine starvation and glucose starvation. The rpoS wild-type coding sequence at the native chromosomal locus was either deleted (ΔrpoS) or replaced by the rpoS TTA>CTA synonymous variant without additional selection markers. Numbers between the two panels indicate the normalized densitometric ratio of the RpoS and RpoD bands for each lane.
(G) Western blot with Anti-FLAG antibody against 3XFLAG-YFP-RpoS fusion proteins during leucine starvation. Approximate molecular weight in kilodaltons (kD) was estimated using a protein ladder. Blue triangle corresponds to the approximate location of the expected truncated peptide caused by ribosome abortion at the first pause site in the rpoS TTA>CTA synonymous variant [indicated as a blue triangle in (E)].
Paused ribosomes with an empty A-site are stable in vitro (Ivanova et al., 2005), which suggests a role for a trans-acting factor in mediating abortion at ribosome pause sites during leucine starvation. Hence we tested the role of four known abortion-mediating factors in E. coli, tmRNA (Keiler et al., 1996), RF3 (Zaher and Green, 2011), ArfA (Chadani et al., 2010), and ArfB (Chadani et al., 2011), by deleting the genes for each of these factors individually (Figure 6C). Deletion of the gene encoding tmRNA, a chimeric transfer-messenger RNA which releases paused ribosomes and tags the incomplete polypeptide for proteolysis (Keiler et al., 1996), caused a significant increase in the ratio of the incomplete to the full length YFP (Figure 6C). Further, complementation with a tmRNAHis6 mutant (Roche and Sauer, 2001), which adds a modified His6 proteolysis-resistant tag but still causes ribosome release, revealed an incomplete YFP polypeptide with a His6 epitope upon immunoprecipitation (Figure 6D). Together, these experiments suggest that tmRNA mediates abortion at ribosome pause sites during leucine starvation. This conclusion is consistent with a previously-ascribed role for tmRNA during amino acid starvation (Garza-Sánchez et al., 2008; Li et al., 2008). However, in contrast to these studies and previous in vitro experiments (Ivanova et al., 2004), we did not find evidence for significant mRNA cleavage near the pause site during amino acid starvation (Figure S5A). Furthermore, the measured total mRNA density around the pause site did not indicate appreciable degradation of the region 3’ to the pause site (Figure S5B). Finally, ribosome profiling in a ΔtmRNA strain showed only a slight decrease in ribosome occupancy at the 5’ end of mRNAs compared to a tmRNA+ strain during leucine starvation (Figure S5C). This observation suggests that abortion is still efficient in the absence of tmRNA and is likely mediated by the abortion-mediating factor ArfA. Such an auxillary role for ArfA in the absence of tmRNA is supported by earlier studies, which found that both the arfA mRNA and a truncated, but functional form of the ArfA protein are highly stabilized by the deletion of tmRNA (Garza-Sánchez et al., 2011), and that double knockout of arfA and tmRNA is synthetic lethal (Chadani et al., 2010).
We then tested whether ribosome pausing and abortion also affects the expression level of an endogenous E. coli protein, the sigma factor RpoS, that is highly up-regulated at the transcriptional level in response to leucine starvation (Subramaniam et al., 2013a). Ribosome pause sites encoded by the two leucine codons CTA and CTC during leucine starvation are underrepresented in the rpoS protein coding sequence in comparison to their average frequency in the genome (Figure 6E), which suggests that ribosome pause sites during leucine starvation are selected against in the rpoS coding sequence due to their adverse effect on the expression of this critical stress response protein. To test the effect of ribosome pause sites on RpoS expression, we substituted, at the chromosomal rpoS locus, four leucine TTA codons with the synonymous CTA codons which encode ribosome pause sites (Figure 6E, thick blue bars). Western blotting indicated that the expression level of RpoS decreased ~6-fold during leucine starvation due to the presence of the CTA codons in the rpoS coding sequence (Figure 6F). This decrease was specific to leucine starvation since the synonymous substitutions did not significantly affect the expression of RpoS during glucose starvation which also strongly up-regulates RpoS (Figure 6F). By fusing 3XFLAG-YFP to the N-terminus of RpoS, we detected a truncated peptide (Figure 6G, blue triangle) whose size is consistent with translation abortion at the first ribosome pause site in the rpoS synonymous variant during leucine starvation (Figure 6E, blue triangle).
Effect of translation abortion on protein expression during amino acid starvation
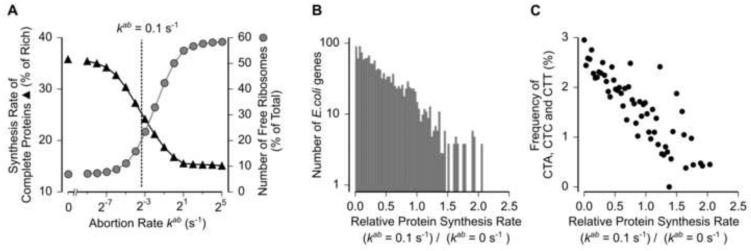
Previous studies have suggested that the primary function of abortion mediated by factors such as tmRNA is to rescue inactive mRNA-bound ribosomes during stress and thereby increase the translational capacity in the cell (Moore and Sauer, 2007). To quantify this effect of abortion on ribosome rescue and translational capacity, we tracked the number of free ribosomes and the global synthesis rate of proteins as a function of the abortion rate in our whole-cell model (Figure 7A, S6). Based on earlier work (Bremer and Dennis, 2008), we assumed that 15% of the ribosomes in a cell are free (not bound to mRNAs) during nutrient-rich growth. In the absence of abortion (kab = 0 s−1), starvation for leucine decreased the fraction of free ribosomes in the cell to 7% (Figure 7A, leftmost circle). Increasing the rate of abortion during leucine starvation gradually rescued the fraction of free ribosomes to a maximum of ~60% in our model (Figure 7A, rightmost circle). We then examined the effect of abortion on global protein expression using our whole-cell model. In the absence of abortion (kab = 0 s−1), leucine starvation decreased the synthesis rate of complete proteins in the cell to 35% of its value during nutrient-rich growth (Figure 7A, leftmost triangle). Increasing the rate of abortion during leucine starvation further decreased the global synthesis rate to a minimum of ~15% (Figure 7A, rightmost triangle). Notably, increasing the abortion rate had widely different effects on protein expression from individual mRNAs during leucine starvation (Figure 7B). Proteins with higher expression level upon increasing the abortion rate also had a lower frequency of ribosome pause sites in the corresponding mRNAs (Figure 7C). As a result, protein expression from these mRNAs is less susceptible to abortion, while their translation initiation rate is increased by the released ribosomes due to abortion from other mRNAs. Thus our model predicts that abortion enhances the translation of proteins in a selective manner even though it can have a deleterious effect on global protein expression during amino acid starvation.
Figure 7.
Effect of Translation Abortion on Protein Expression.
(A) Effect of varying abortion rate constant (kab) on the number of free ribosomes in the cell (grey circles) and the global synthesis rate of complete proteins (black triangles) during leucine starvation calculated from whole-cell model. The value kab = 0.1 s−1 that fits the measured ribosome occupancy (black line in Figure 5B) is indicated as a dashed line.
(B) Effect of non-zero abortion rate constant on the synthesis rate of individual E. coli proteins during leucine starvation calculated from the whole-cell model.
(C) Average frequency of the three leucine codons CTA, CTC and CTT for genes in each of the histogram bins in (B). Only genes with greater than ten leucine codons were considered in (B) and (C).
See also Figure S6.
Discussion
Here we formulated a biophysical model of translation in E. coli by leveraging the near single-codon resolution of the ribosome profiling method. Notably, we constrained our model not only in the initiation-limited regime of nutrient-rich growth, but also in the regime of amino acid starvation, during which the elongation rate of ribosomes has a large effect on the measured ribosome occupancy. By contrast, previous computational models of translation used data solely from initiation-limited regimes, with the consequence that mechanistic features of the elongation stage of translation were not fully constrained (Shah et al., 2013; Tuller et al., 2010). As our results generally illustrate, the exact mechanistic features assumed for the elongation stage have a critical role in models of translation, and changing these features qualitatively alters several of the model predictions.
Consistency between ribosome profiling measurements and our model suggests that the concentration of aminoacyl-tRNAs does not limit the elongation rate of ribosomes at most codons during nutrient-rich growth. This conclusion relies on our assumption that biases in the generation and analysis of ribosome footprinting data are sufficiently small such that the measured ribosome occupancy at codons reflects the residence times of ribosomes with an empty A-site at these codons. This assumption is partly supported by our observation that differences in ribosome occupancy at codons during starvation for their cognate amino acid are consistent with their corresponding effect on protein expression. A more direct test of our conclusion will be to characterize the effect of overexpressing low abundance tRNA isoacceptors on the measured ribosome occupancy during nutrient-rich growth. Our model still predicts a small variation in ribosome occupancy at codons that is correlated with tRNA abundance, the exact magnitude of which depends on the kcat/KM for ribosome-tRNA association (Figure 3A, vertical axis). While we predict that this small variation neither limits the ribosome elongation rate at most codons nor affects protein levels during nutrient-rich growth on the physiological time scale of protein synthesis, it can nevertheless leave an evolutionary signature on protein coding sequences and thus underlie the widely-observed correlation between tRNA abundance and codon frequency in microorganisms (Andersson and Kurland, 1990; Drummond and Wilke, 2008; Wallace et al., 2013).
Our analysis of aminoacylation kinetics raises the interesting possibility that cells might utilize differential aminoacylation rates of tRNA isoacceptors as a mechanism to regulate the elongation rate of codons specifically during nutrient stress. Further, small differences in aminoacylation rate between tRNA isoacceptors might also modulate mis-aminocylation rates during other stresses (Netzer et al., 2009), while having little or no deleterious effect on translation during nutrient-rich growth when aminoacylation is not limiting for elongation.
Our work provides in vivo evidence for the widespread occurrence of ribosome traffic jams in response to pausing of a leading ribosome. However, we found that translation abortion at ribosome pause sites is the primary determinant of ribosome occupancy along mRNAs during amino acid starvation. While abortion from non-stop mRNAs is thought to increase global protein expression during nutrient-rich growth (Moore and Sauer, 2007), our whole-cell modeling suggests that abortion of paused ribosomes during nutrient stress might have the function of enabling selective translation from specific mRNAs. Additionally, releasing nascent polypeptides from paused ribosomes by abortion might facilitate their proteolysis and prevent protein misfolding during stress. However, previous studies have found that eliminating the ability of the abortion-mediating factor tmRNA to target nascent polypeptides to proteolysis does not significantly alter the cellular response to stress (Abo et al., 2002; Huang et al., 2000), suggesting that prevention of protein misfolding is not the primary cellular function of abortion - mediating factors during stress.
The four processes that we modeled in our study (Figure 2) are an essential part of protein synthesis in both bacteria and eukaryotes. Hence our analysis in E. coli can be readily extended to eukaryotes, where ribosome profiling has revealed that a variety of stresses result in a slow elongation rate (Liu et al., 2013; Shalgi et al., 2013). It will also be useful to integrate our whole-cell model of translation with quantitative models of other cellular processes such as transcription (Brewster et al., 2014) and metabolism (Bordbar et al., 2014). Such an integrated model can shed light on the complex interplay between metabolism and gene expression that occurs during environmental changes (Subramaniam et al., 2013b).
Experimental Procedures
Construction of all strains and plasmids (listed in Table S2), Western blots and Northern blots were performed using standard molecular biology techniques (Extended Experimental Procedures).
Ribosome Profiling
Ribosome profiling was carried out as described previously (Li et al., 2012; Oh et al., 2011) with the following modifications. To accurately capture the ribosome occupancy on mRNAs with single-codon resolution, we flash-froze the cells immediately upon harvesting and stabilized ribosomes with the translation inhibitor chloramphenicol only at the lysis stage. Cells were lysed using glass beads (G1277, Sigma, vortex 10 × 30 s at 4°C with 60 s cooling on ice in between). Micrococcal nuclease digestion was carried out with 1 U Worthington Biochemicals MNase per μg of nucleic acid as measured by A260. Monosome-protected mRNA footprints between 20 nt and 40 nt were size-selected by polyacrylamide gel electrophoresis for monosome sequencing. For disome sequencing, the disome peak was collected from the MNase-treated polysomes after sucrose-gradient fractionation, and fragments of size between 50 nt and 80 nt were used for sequencing. For total mRNA sequencing, the Microbe Express kit (Ambion) was used for subtracting rRNA from total mRNA and then fragmented using a bicarbonate buffer (Ingolia et al., 2009) for 20 min. For library construction, polyA-tailing (Ingolia et al., 2009) was used instead of linker ligation.
High-throughput Sequencing Data Analysis
Analysis steps were similar to that in previous ribosome profiling studies (Li et al., 2012; Subramaniam et al., 2013b), and were implemented using Python and Bash programming languages. Full programming code for generating the final figures in our paper starting from raw sequencing data is provided both as an interactive IPython notebook (Perez and Granger, 2007) and as a static HTML file (sequencing_data_analysis.html in Data S1). Briefly, single-end reads were polyA-trimmed and then aligned to the E. coli genome (NC_000913.3 build) using Bowtie (Langmead et al., 2009). Aligned reads were trimmed by 8 nt on each side. Each genomic position corresponding to the trimmed read was assigned ribosome occupancy equal to the inverse of the read length. The transcriptome-averaged ribosome occupancy at individual codons for each sample (Figures 1B, 1C) was computed by first averaging the ribosome occupancy at the first nucleotide position of the codon across all occurrences of that codon within each coding sequence, and then by averaging across all coding sequences that had a minimum average occupancy of 1 read/codon. Since the start codon and the three stop codons have a high ribosome density both during nutrient rich growth and during starvation, they were excluded in the plots showing all 61 sense codons (Figures 1, S1) for clarity. The ribosome occupancy profiles around codons (Figures 5A, 5B) were calculated using the same procedure, but for the 120 nt region on each side of the codon rather than just the first nucleotide position of the codon. Ribosome occupancy along mRNAs (Figure 6A) was computed by first normalizing the ribosome occupancy at each position of a coding sequence by the aggregated ribosome occupancy for the full coding sequence, and then by averaging this quantity at each position beginning from the start codon across all coding sequences that had a minimum average occupancy of 1 read/codon.
Whole-Cell Model of E. coli translation
Our whole-cell model for translation (Figure 2) was implemented using the stochastic Gillespie algorithm (Gillespie, 1977). The source code for implementing the kinetic model in Figure 2 was adapted from (Shah et al., 2013) with modifications as shown in Data S1. Full programming code for running the simulation and for the subsequent data analysis to generate the final figures in our paper is provided both as an interactive IPython notebook (Perez and Granger, 2007) and as a static HTML file (simulation.html and simulation_data_analysis.html in Data S1). Standard errors of mean for model predictions in all figures are smaller than data markers. Our whole-cell model tracked the state of 44,000 ribosomes, 408,000 tRNA molecules (38 distinct tRNA species) and ~7500 mRNA molecules (1518 distinct mRNA species) in the E. coli cell (Table S1). The transition rates between different states of each of these molecules are determined by the four rate equations in Figure 2B. The key parameters that control the predictions from our model are the rate constants k for the four processes in Figure 2B. Based on the analysis presented in the main text, we chose a default set of parameters for all our simulations. Table S1 lists these choices, together with the corresponding references and a footnote explaining the choice (see also Extended Experimental Procedures). We chose parameters corresponding to a cell doubling time of 30 min that we measured during nutrient-rich growth in our experiments.
Supplementary Material
Acknowledgements
We thank J. Calarco, P. Cluzel, V. Denic, A. Murray, and C. Shoemaker for discussions; G. W. Li and E. Oh for advice on ribosome profiling; G. L. Chew and A. Gutu for advice on Northern blotting; H. Aiba for the XL001 strain; P. Shah for clarifications on the yeast model; K. Amarnath, A. Buskirk, S. Chandrasekaran, A. Darnell, G. W. Li, W. Moebius, J. Moffitt, P. Shah and E. Wallace for comments on the manuscript. The computations in this paper were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. This research was supported by an NIH K99 Pathway to Independence Award GM107113 (A.R.S.) and by the Howard Hughes Medical Institute (E.K.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequencing data is publicly available from Gene Expression Omnibus, accession number GSE51052. Simulation results and programming code for simulation, sequencing and simulation data analysis, and reproduction of figures is publicly available at http://datadryad.org/, doi:10.5061/dryad.ch352.
References
- Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells. 2002;7:629–638. doi: 10.1046/j.1365-2443.2002.00549.x. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol. Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin N, Ehrenberg M. Mutations in 23 S Ribosomal RNA Perturb Transfer RNA Selection and can Lead to Streptomycin Dependence. J. Mol. Biol. 1994;235:813–824. doi: 10.1006/jmbi.1994.1041. [DOI] [PubMed] [Google Scholar]
- Bordbar A, Monk J, King Z, Palsson B. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15 doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis P. Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus. 2008 doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- Brewster RC, Weinert FM, Garcia HG, Song D, Rydenfelt M, Phillips R. The Transcription Factor Titration Effect Dictates Level of Gene Expression. Cell. 2014;156:1312–1323. doi: 10.1016/j.cell.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol. Microbiol. 2010;78:796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol. Microbiol. 2011;80:772–785. doi: 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA Abundance and Codon Usage in Escherichia coli at Different Growth Rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-Induced Protein Misfolding as a Dominant Constraint on Coding-Sequence Evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective Charging of tRNA Isoacceptors Explains Patterns of Codon Usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- Fender A, Sissler M, Florentz C, Giegé R. Functional idiosyncrasies of tRNA isoacceptors in cognate and noncognate aminoacylation systems. Biochimie. 2004;86:21–29. doi: 10.1016/j.biochi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Garza-Sánchez F, Gin JG, Hayes CS. Amino Acid Starvation and Colicin D Treatment Induce A-site mRNA Cleavage in Escherichia coli. J. Mol. Biol. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 2011;80:1204–1219. doi: 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 Rescues Ribosomes in 3′ Untranslated Regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CL, Marashi F. Two kinetically distinct tRNAile isoaccepton in Escherichia coli C6. Nucleic Acids Res. 1980;8:2023–2037. doi: 10.1093/nar/8.9.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome Rescue by tmRNA Requires Truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Ehrenberg M. tmRNA-induced Release of Messenger RNA from Stalled Ribosomes. J. Mol. Biol. 2005;350:897–905. doi: 10.1016/j.jmb.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Jacques N, Dreyfus M. Translation initiation in Escherichia coli: old and new questions. Mol. Microbiol. 1990;4:1063–1067. doi: 10.1111/j.1365-2958.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Feaga HA. Resolving nonstop translation complexes is a matter of life or death. J. Bacteriol. 2014:01490–14. doi: 10.1128/JB.01490-14. JB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a Peptide Tagging System in Degradation of Proteins Synthesized from Damaged Messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-Sequence Determinants of Gene Expression in Escherichia Coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying Absolute Protein Synthesis Rates Reveals Principles Underlying Allocation of Cellular Resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol. Microbiol. 2008;68:462–473. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian S-B. Cotranslational Response to Proteotoxic Stress by Elongation Pausing of Ribosomes. Mol. Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 2005;58:456–466. doi: 10.1111/j.1365-2958.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Myers G, Blank HU, Söll D. A Comparative Study of the Interactions of Escherichia coli Leucyl-, Seryl-, and Valyl-Transfer Ribonucleic Acid Synthetases with Their Cognate Transfer Ribonucleic Acids. J. Biol. Chem. 1971;246:4955–4964. [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. Selective Ribosome Profiling Reveals the Cotranslational Chaperone Action of Trigger Factor In Vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Ehrenberg M. Rate of Translation of Natural mRNAs in an Optimizedin VitroSystem. Arch. Biochem. Biophys. 1996;328:9–16. doi: 10.1006/abbi.1996.0136. [DOI] [PubMed] [Google Scholar]
- Perez F, Granger BE. IPython: A System for Interactive Scientific Computing. Comput. Sci. Eng. 2007;9:21–29. [Google Scholar]
- Qian W, Yang J-R, Pearson NM, Maclean C, Zhang J. Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. Identification of Endogenous SsrA-tagged Proteins Reveals Tagging at Positions Corresponding to Stop Codons. J. Biol. Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-Limiting Steps in Yeast Protein Translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. Widespread Regulation of Translation by Elongation Pausing in Heat Shock. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MA. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 2001;307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- Sørensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Björk GR, Ehrenberg M. Over Expression of a tRNALeu Isoacceptor Changes Charging Pattern of Leucine tRNAs and Reveals New Codon Reading. J. Mol. Biol. 2005;354:16–24. doi: 10.1016/j.jmb.2005.08.076. [DOI] [PubMed] [Google Scholar]
- Subramaniam AR, Pan T, Cluzel P. Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl. Acad. Sci. 2013a;110:2419–2424. doi: 10.1073/pnas.1211077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, DeLoughery A, Bradshaw N, Chen Y, O’Shea E, Losick R, Chai Y. A serine sensor for multicellularity in a bacterium. eLife. 2013b;2 doi: 10.7554/eLife.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process: Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Vind J, Sørensen MA, Rasmussen MD, Pedersen S. Synthesis of Proteins in Escherichia coli is Limited by the Concentration of Free Ribosomes: Expression from Reporter Genes does not always Reflect Functional mRNA Levels. J. Mol. Biol. 1993;231:678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- Wallace EWJ, Airoldi EM, Drummond DA. Estimating Selection on Synonymous Codon Usage from Noisy Experimental Data. Mol. Biol. Evol. 2013;30:1438–1453. doi: 10.1093/molbev/mst051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design Parameters to Control Synthetic Gene Expression in Escherichia coli. PLoS ONE. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004;32:733. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegian CD, Stent GS, Martin EM. Intracellular condition of Escherichia coli transfer RNA. Proc. Natl. Acad. Sci. U. S. A. 1966;55:839–846. doi: 10.1073/pnas.55.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. A Primary Role for Release Factor 3 in Quality Control during Translation Elongation in Escherichia coli. Cell. 2011;147:396–408. doi: 10.1016/j.cell.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Goldman E, Zubay G. Clustering of Low Usage Codons and Ribosome Movement. J. Theor. Biol. 1994;170:339–354. doi: 10.1006/jtbi.1994.1196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.