With worldwide transport of plants, exotic species spread disease to native relatives, but they can also provide disease resistance via hybrid breeding programs. American chestnut was nearly eliminated from North America by introduced chestnut blight, but resistant hybrids of American and Chinese chestnuts are being created. We studied the ecology and genetics of an unmanaged population of Chinese chestnut in North America. The forest dynamics of pure Chinese chestnut indicate that hybrids between the Chinese and American species could be successful colonizers of forests in North America. We challenge the paradigm that exotic tree species are wholly detrimental to native biodiversity.
Keywords: Dendrochronology, distribution, ecology, hybrid, invasive, root DNA barcoding.
Abstract
With the transport of plants around the globe, exotic species can readily spread disease to their native relatives; however, they can also provide genetic resistance to those relatives through hybrid breeding programmes. American chestnut (Castanea dentata) was an abundant tree species in North America until its decimation by introduced chestnut blight. To restore chestnut in North America, efforts are ongoing to test putative blight-resistant hybrids of Castanea dentata and Chinese chestnut (Castanea mollissima), but little is known about the ecology of C. mollissima. In a forest in northeastern USA in which C. mollissima has become established, we explored questions of stand dynamics, health and genetic relationships of C. mollissima offspring to an adjacent parent orchard. We found that C. mollissima was adapted and randomly distributed among native species in this relatively young forest. The genetics of the C. mollissima population compared with its parents indicated little effect of selection pressure as each of the parent trees contributed at least one offspring. The ease with which this exotic species proliferated calls to question why C. mollissima is rare elsewhere in forests of North America. It is likely that a time window of low animal predation allowed seedlings to establish, and the shallow soil at this site limited the maximum forest canopy height, permitting the characteristically short-statured C. mollissima to avoid suppression. Our results indicate that because C. mollissima exhibited pioneer species characteristics, hybrids between C. mollissima and C. dentata have the potential to be successful pioneer species of future forests in North America, and we challenge the paradigm that exotic tree species are wholly detrimental to native biodiversity. We contend that exotic tree species should be assessed not only by their level of threat to native species, but also by their potential positive impacts on ecosystems via hybrid breeding programmes.
Introduction
Anthropogenic activities increasingly influence natural systems worldwide; an example is the movement of plant species and their diseases around the globe. Land managers have a decreasing ability to keep ‘native’ and ‘exotic’ plants separate, especially when dealing with species of economic value, and as a consequence diseases have jumped from co-evolutionary plant hosts to their more susceptible relatives in other parts of the world. Although exotic species may harbour diseases, they also possess the genetic potential to resist those diseases, and tree breeders in the USA strive to capture those resistance genes to restore threatened native species (Michler et al. 2006; Merkle et al. 2007; Jacobs et al. 2013).
Chestnuts (genus Castanea, family Fagaceae) occur in temperate climates worldwide, and have ecological, economic and cultural importance in their native ranges of Europe, the Caucasus Mountains and eastern Asia (Conedera et al. 2004; Davis 2006). Historically, Castanea dentata (American chestnut) was also of great importance in eastern North America, until it was nearly extirpated from its native range in the early 20th century by chestnut blight disease, caused by the introduced fungus Cryphonectria parasitica (Anagnostakis 2012). The fungus was accidentally introduced from eastern Asia with early importation of Castanea crenata (Japanese chestnut) for commercial purposes (Powell 1898; Shear and Stevens 1916). Imported slightly later, Castanea mollissima (Chinese chestnut) has now been grown in cultivation in the USA for over a century. Its broad adaptability to soil and climatic conditions in eastern North America has made it commercially more valuable for nut production than the Japanese species (Hunt et al. 2009; Fei et al. 2012); however, there is still a desire to recapture the economical and ecological value of C. dentata as a forest tree and timber species. Current chestnut breeding programmes aimed at re-establishing American chestnut via hybridization of C. dentata and C. mollissima (and other Castanea), which are being carried out by organizations such as The American Chestnut Foundation and the Connecticut Agricultural Experiment Station (Burnham et al. 1986; Hebard 2001; Anagnostakis 2012). These breeding programmes aim to transfer resistance to chestnut blight from the genome of Asian Castanea species into Asian × American chestnut hybrids (Jacobs 2007; Anagnostakis 2012). One concern with hybrids is that although C. mollissima is a canopy co-dominant in forests of northern China (Steiner et al. 2009), the genetic stock available to breeding programmes in the USA seems to be limited in height, possibly due to its selection for nut production over timber form (Fei et al. 2012).
Ideally, hybrid Castanea will be able to establish and reproduce in habitats formerly occupied by C. dentata without becoming overly aggressive. There is some concern that breeding programmes will result in Castanea hybrids that are weedy and invasive and will displace native hardwoods (Jacobs 2007; Jacobs et al. 2013). Unlike Asian woody plants such as Lonicera spp., Ailanthis altissima and Elaeagnus spp., however, non-native Castanea, including C. mollissima, have not shown strong invasive tendencies in North America (Sakai et al. 2001; Zheng et al. 2004). There are few cases of C. mollissima escaping its orchard bounds to become established in the forest among native North American species. Test plantings of C. mollissima, C. dentata and Castanea hybrids made by the United States Department of Agriculture (USDA) in 1947–55 (Diller et al. 1964) were relocated and evaluated (Berry 1980; Schlarbaum et al. 1994), and only a few have persisted to date.
If Castanea hybrids are to survive and proliferate in North American forests, they must be adapted to North American forest conditions; however, little is known about the ecology, silvics and genetics of C. mollissima. There is only one case of naturalized C. mollissima in the USA reported in the scientific literature (Jaynes 1965; Anagnostakis 1992): a forest plot of ∼1 ha on a private property near Dayville, CT, USA. We conducted studies in this forest and in an adjacent orchard grove of ∼85-year-old C. mollissima trees, the ‘parent orchard.’ Castanea mollissima in this disjunct USA stand seem to be well integrated into a natural forest among native North American species.
The objectives of this study were to understand the factors that permitted C. mollissima to establish in a second-growth mixed forest in CT and, more generally, to understand if C. mollissima has the potential to persist in North American forests. The overarching goal of our research is to surmise, through observations and synthesis of the literature, the potential for Castanea hybrids to survive and compete effectively in future North American forests. This instance of C. mollissima infiltrating unmanaged land in CT is the only documented case of that species co-dominating a North American forest; therefore, we gained rare insight into its performance in an unmanaged setting. We examined forest composition, spatial distributions of woody stems, horizontal and vertical distributions of woody roots, stand dynamics and vigour of C. mollissima, and genetic relationships among all C. mollissima individuals. These analyses allowed us to explore spatial partitioning and competition above and below ground, disturbance history of the forest, associations of site quality and health of C. mollissima and the possibility of a genetic bottleneck between C. mollissima orchard parents and forest offspring.
Methods
Field site
The Dayville woodlot is a forest of ∼1 ha near Dayville, CT, USA (41–50′47″N, 071–53′15″W). The woodlot and a C. mollissima orchard immediately adjacent were visited in May 2012. The parent orchard sits on level ground atop a hill, and the woodlot is on a naturally terraced hillside with the slope ranging from 0 to ∼25° (0–47 %). Jaynes (1965) characterized the soil of the Dayville woodlot as sandy loam, with pH ranging from 4.4 to 4.8. At the time the parent orchard was planted in 1926, the woodlot was a grazed agricultural field that was subsequently abandoned and left to naturally regenerate into forest. The parent orchard, originally planted with 67 trees, was assessed for survival in 2012, and 28 trees were found, probably corresponding to the 28 found by Anagnostakis in 1992. Each parent tree was given a unique identification number preceded by a ‘P’ (e.g. P1).
The seeds from which the orchard trees grew were brought to the USA by the USDA in 1925 under the label of Plant Introduction (PI) 58602 (Anagnostakis 1992). While the seeds were officially imported from Nanking, in southern China, seeds were requested from other parts of the country, and especially the north, so that they might be more climatically adapted to the northern USA. Plant Introduction 58602 included seed collections from many small lots in China, though no records were kept of their exact origins (Anagnostakis 1992). It was subsequently dispersed widely across the USA (i.e. 7826 seedlings were sent out to 85 locations). Therefore, to determine if the survival and growth of C. mollissima in the Dayville woodlot was the result of a common genetic background, we assessed genetic relatedness within the orchard parent population, within the forest offspring population and between these populations.
Forest structure
We conducted several forest-wide analyses of the Dayville woodlot to detect characteristics that might explain the invasion and persistence of C. mollissima at that location. To assess the sizes and distributions of living woody stems, all trees and shrubs >5 cm diameter-at-breast-height (DBH) in the forest were mapped by polar coordinates using a range finder and sighting compass, and the coordinates were used to create a ‘master map.’ All C. mollissima in the forest (hereafter ‘offspring’) were systematically given a unique identification number (1–72). For all trees, we recorded species identification and DBH, grouping them into 5-cm size classes. Anecdotal observations such as presence of woody vines, understorey composition and evidence of seedling regeneration were noted to aid the interpretation of our results.
Root DNA
Because forest establishment and individual tree growth are heavily influenced by soil qualities, root dynamics and below-ground competition, we analysed the distributions of woody roots at the study site. As with the above-ground forest structure, we were looking for below-ground explanations for the success of C. mollissima in the forest. Soil cores were taken from 13 locations at regularly spaced intervals throughout the forest (Fig. 1) using a 6.25-cm diameter soil probe. At each location, soil samples were taken at 0–15, 15–30, 30–45 and 45–60 cm, or until reaching bedrock. The most common depth to bedrock was 30 cm. In total, 25 soil cores were taken. Fine roots were removed from the soil by hand and dried in a desiccator using a Dri-Rite desiccant (Chicago, IL). Twelve fine root fragments were selected at random from each core for DNA extraction. Roots were placed in a wash solution of 0.01 % Triton X-100 in Tris–HCl and tumbled in a rotisserie at room temperature for 1 day to remove as much dirt and other contaminants as possible from the root surface. Roots were rinsed in nanopure water, and DNA extraction proceeded as described under relatedness and parentage (below).
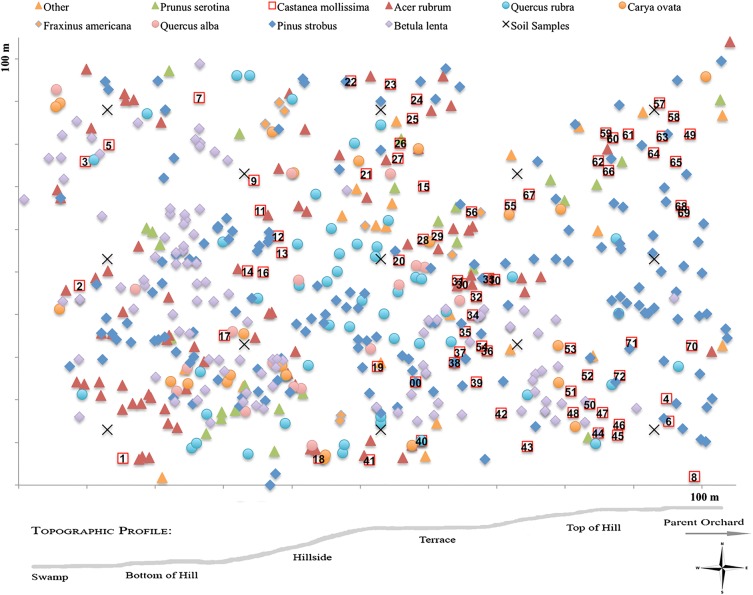
Figure 1.
Map of tree species in the naturalized stand of C. mollissima in CT, USA. Chestnuts are individually labelled by the unique identification number. Forest is 100 × 100 m. Topographic profile reveals microsites identified in the study.
To determine the species associated with root samples removed from soil cores, the rbcL genes from DNA of pooled root samples were sequenced using Illumina MiSeq (Illumina, Inc., San Diego, CA) run with rbcL PCR primers from Kress et al. (2009) and Illumina step-out PCR primers from the Illumina Customer Sequence Letter (2012). All DNA sequences were grouped by soil core (depth and location). Raw sequences were processed into FastA files to determine the number of reads and the number of bases per read. SAMtools (flagstat and idxstats) software was used to translate reads into BAM files for further processing and to confirm the percentage of properly paired and singleton reads and the percentage of mapped and unmapped reads (Li et al. 2009). Fastx_clipper was used to remove adapters and poor quality bases on both the 5′ and 3′ ends. Small reads (<30 bases) were discarded. Root rbcL sequences were cross-referenced against the GenBank® database to determine the best set of reference sequences for woody species present in the Dayville woodlot. Resolution to genus was achieved with 642 reference sequences. Read-to-reference mapping was performed using Bowtie2. Both global and local alignments were made, but because of their higher sensitivity and accuracy, global alignments were chosen for further analysis.
Tree health and vigour
We examined individual C. mollissima offspring trees to account for characteristics, history and environmental relationships that might help to elucidate the species' success in this forest. For each offspring we recorded canopy position (dominant, co-dominant, intermediate or suppressed), a rank of chestnut blight canker severity (using a system of 0–3 capturing the range of severity within that population based on the amount, size and level of healing of cankers) and the presence of epicormic branches, which indicate a decline in tree health. We distinguished six distinct microsites in the forest characterized by their slope position and proximity to the forest edge. The microsite of each offspring tree was recorded.
Dendrochronology
Twenty-five offspring of C. mollissima were chosen at random from among the population of 72, and two increment cores were taken at breast height from each. Cores were taken parallel to the slope of the hill to avoid tension wood. Cores were dried and sanded, and raw ring widths were measured using the Velmex measuring stage and Measure J2X tree ring software. Skeleton plots were used to align dates of the cores, and dated measurement files were checked for accuracy with the program COFECHA (Grissino-Mayer 2001).
Relatedness and parentage
We estimated genetic relatedness and parentage of the offspring to determine if the parent trees (in the orchard) contributed unequally to the offspring cohort. Leaves were collected for DNA extraction from each of the 72 offspring and the 28 parent trees in the orchard. A slingshot was used to sample leaves because of the canopy height (∼18 m) of the forest and orchard trees. The leaves were placed in 50 mL conical tubes and kept in a cooler with ice and subsequently in a freezer at −80°C. They were taken immediately from the freezer and freeze-dried for long-term storage. DNA was extracted in a buffer of cetyltrimethyl ammonium bromide and 2-mercaptoethanol and purified by phenol–chloroform extraction (Doyle and Doyle 1987; Zhao and Woeste 2011). Sodium chloride and cold isopropanol were used to precipitate the DNA, which was then dried and re-suspended in TE buffer. A nanodrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE) was used to quantify the DNA, which was diluted to ∼50 ng µL−1 with nanopure water. Eleven simple sequence repeat (SSR) markers (Table 2) were used for genotyping (Inoue et al. 2009; Kubisiak et al. 2013). To increase flexibility in post-PCR multiplexing, an M13 sequence (5′-AGTAAAACGACGGCCAGT-3′) was added to the 5′ end of each forward primer (Schuelke 2000), and three-primer PCR was performed including a dye (HEX or FAM) labelled with the same M13 sequence. The PCR thermal profile was a 4-min denaturation at 95 °C, 20 cycles of 30 s at 94 °C, 30 s at 65 °C and 1 min at 72 °C, with the annealing temperature stepping down 0.5 °C every cycle (from 65 °C to 55 °C), followed by 20 cycles of the same, with a constant annealing temperature of 55 °C. The final extension was 72 °C for 5 min. Genemapper (Applied BioSystems) was used to score the genotypes.
Table 2.
SSR markers used in relatedness analysis. *From Inoue et al. (2009). †From Kubisiak et al. (2013).
| Locus | Primer sequence (5′–3′) | Size (bp) | Repeat | No. of alleles |
|---|---|---|---|---|
| CM004* | F:ACCACAGAGAGAGCCACACC | 189–270 | CT | 7 |
| R:TTTCATGAGCACGAAAGCTG | ||||
| CM005* | F:AAATAAAACCCCTCATCAACACA | 156–182 | CT | 8 |
| R:GAACTCAAAACCTCAAAACCTCA | ||||
| CM007* | F:TAGTCACGCCTCTCCGTCTT | 217–299 | AG | 5 |
| R:GCCATTTGAGGACTGAGGTT | ||||
| CM008* | F:CCCAAAATCAAAGTCTGAGCA | 143–159 | AC | 8 |
| R:CGTCGACTCTTCTCTATCTCCAA | ||||
| CM010* | F:GTTGGAGAGGTCGTCTCACG | 231–255 | (CT)(GT) | 6 |
| R:ATTGCGAGGAAAAGGAAACA | ||||
| CM018* | F:ACAACGATCCCAGACCAAAG | 172–224 | CT | 10 |
| R:CTAGGCGATCGGAGAGAGAC | ||||
| CM396† | F:AACTCCCACCACTCACATCC | 181–207 | CACACC | 6 |
| R:GTTTCTTTTTCGGACCATCCAGAACTC | ||||
| CM467† | F:CCCATGCCTACTACATTACAAA | 189–199 | TCG | 3 |
| R:GTTTCTTGTGGCCGATGGTGTAGATTT | ||||
| CM800† | F:TTATGGCAACCCTCCTGTTT | 140–154 | TC | 7 |
| R:GTTTCTTCTGAAATGATCGATGCTGCT | ||||
| CM883† | F:CAGCATCAGCACTCGTTCA | 204–218 | AGC | 5 |
| R:GTTTCTTGGGATTGAGAGGATGAAGCA | ||||
| CM945† | F:AGTGTGAGTGGGGAAGATGG | 215–249 | GAG | 11 |
| R:GTTTCTTTTtGGCTTCACTGCCAAAC |
Data analyses
Forest structure data were used to determine the number of stems per species, to create graphs of size-class distributions and to determine the percent basal area (BA) of each species. The polar location coordinates were translated into Euclidean coordinates, which were then used for Ripley's K analysis of spatial distribution in the statistical platform R to determine the spatial pattern of all trees in the forest. Ripley's Cross K was used to establish baseline forest characteristics, determining if there was spatial clustering or repulsion between C. mollissima and all other trees and between shade-tolerant and shade-intolerant tree species. Because field observations suggested that C. mollissima and Pinus strobus were clustered together, we used Ripley's Cross K to test those spatial relationships. Ripley's K and Cross-K statistics are indices to examine whether spatial point patterns are clustered, random or uniform. They combine methods of nearest neighbour counts and quadrat surveys into a second-order statistic that looks at the variances of distances to neighbours in two-dimensional space using concentric circles with increasing radii from each sample point (Haase 1995). For both analyses, an isotopic edge correction was used to eliminate the false empty space outside the edge of plot boundaries. Ripley's K values were converted to L-values [L = (K/π)1/2] for clear graphic representation. Chi-squared tests of association (α = 0.05) were used to evaluate the significance of associations between microsites, canker severity, canopy position and the presence of epicormic branches for C. mollissima.
The number of root sequence reads per genus was used as a proxy for root abundance in each soil core/location. Inverse modelling was used to fit functions for the distribution of roots relative to the distribution of above-ground stems, much like the techniques used to model seed dispersal from seed trap data (Nathan and Muller-Landau 2000; Jones et al. 2011). Contour-style maps were generated from the model showing the varying abundance of roots per genus across the forest. Because of orders of magnitude of difference in numbers of reads/genus/location, a log transformation of read number was applied for further analysis. Root abundance per genus was pooled across each sampling depth (15-cm increment) to show changes in abundance of all genera from the surface to 60 cm. Root abundance of C. mollissima within each microsite was compared with BA of the C. mollissima stems per microsite to determine if site conditions, root abundance and stem size were related.
Ring widths from the two cores from each tree were counted to determine the approximate ages/number of cohorts and averaged to analyse the growth history of each cohort. Tree-ring measurement files were processed with programmes FMT, YUX, CASE and finally JOLTS (http://web.utk.edu/~grissino/). JOLTS detects suppression and release events by calculating a moving average for a specified time window and specified sensitivity. For release events, a 10-year time window was used, based on the strategy of Nowacki and Abrams (1997), and the data were run using three sensitivity levels (minimum release factors): 10, 50 and 100 % growth change of raw ring widths. For suppression events, a 10-year time window was used with a minimum suppression factor of 100 %. Additionally, raw ring-width measurements were detrended for ontogenetic growth decline and compiled into a Master chronology using the program CRONOL (http://web.utk.edu/~grissino/). The National Oceanic and Atmospheric Administration's National Climate Data Center was used to gather precipitation data and cooling degree days in CT spanning the same years as the Master chronology (http://www.ncdc.noaa.gov/cag/time-series). Cooling degree days, calculated by summing each day's average temperature minus 18 °C, are proxies for heat and light and were used to compare relative amounts of heat and light throughout the years. Simple linear regression was used to test for correlations between stand-wide increment growth and each year's annual precipitation, each year's growing season precipitation, the previous autumn's precipitation and the previous 18 months of precipitation. Multiple linear regression was used to determine correlations between growth and precipitation and heat/light intensity per year.
Genetic diversity and relatedness were calculated using GenAlEx (Peakall and Smouse 2012). Pairwise relatedness, heterozygosity, Fst, Fis, Fit, private alleles and a genetic distance matrix were calculated between the two populations (parent and offspring), and tests for Hardy–Weinberg equilibrium were conducted for each locus. For pairwise relatedness, the Lynch and Ritland (1999) method was used. The genetic distance matrix was used to run a principal component analysis (PCA) based on correlations and SSR frequencies. For parentage analysis, the software CERVUS (Kalinowski et al. 2007) was used. Simulations were run for parent pairs of 10 000 simulated offspring, parent sexes unknown, with a 92 % genotype success rate. The proportion of candidate parents sampled was set at 0.6 because Jaynes found 35 reproducing orchard trees in 1965, yet for this study only 28 were remaining. A relaxed confidence (80 %) was used to assign parents. For offspring to which no parent pair could be confidently assigned, non-excluded parents were recorded, meaning those putative parents that had no loci mismatching. Randomness of the parent contribution to the offspring population was tested by bootstrapping with 95 % confidence.
Results
Forest structure
Common woody species (more than eight individuals) in the Dayville woodlot included P. strobus, C. mollissima, Betula lenta, Acer rubrum, Quercus rubra, Quercus alba, Carya ovata, Prunus serotina and Fraxinus americana (Figs 1 and 2, Table 1). Other species (less than eight individuals) included Populus grandidentata, Juniperus virginiana, Ulmus americana, Malus × domestica, Cornus alternifolia, Cornus florida, Euonymus alatus, Lonicera maackii and Sassafras albidum. Stem abundance and BA were determined for the most common species (Table 1). Pinus strobus was the most abundant species with 168 stems that accounted for 50 % of BA. Castanea mollissima was the fourth most abundant species with 72 stems, but it ranked second in BA, accounting for 18 %. Betula lenta was the second most abundant (108 stems), but its smaller stems accounted for only 8 % of the BA. Similarly, A. rubrum was the third most abundant species (80 stems) but accounted for only 5 % of BA. The other noteworthy species was Q. rubra, which comprised 50 stems and 9 % of BA.
Figure 2.
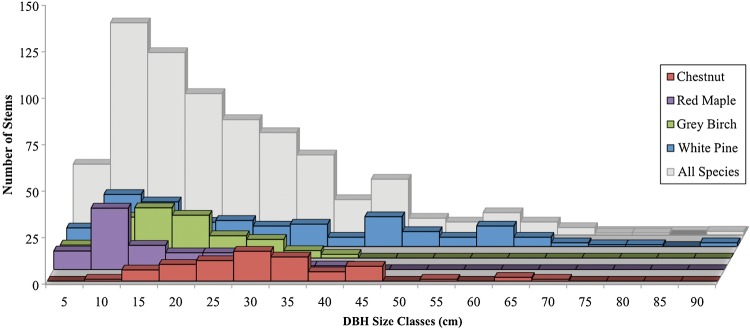
Size-class distributions of all species in the Dayville woodlot (grey) with the four most numerous species shown individually.
Table 1.
Species, stem densities and BA in the Dayville woodlot. *P. grandidentata, J. virginiana, U. americana, Malus × domestica, C. alternifolia, C. florida, E. alatus, L. maackii, S. albidum.
| Species | Stems (ha−1) | BA (m2 ha−1) | BA (%) |
|---|---|---|---|
| P. strobus | 168 | 15.07 | 50 |
| C. mollissima | 72 | 5.51 | 18 |
| Q. rubra | 50 | 2.84 | 9 |
| B. lenta | 108 | 2.55 | 8 |
| A. rubrum | 80 | 1.39 | 5 |
| P. serotina | 27 | 0.96 | 3 |
| C. ovate | 24 | 0.33 | 1 |
| F. americana | 8 | 0.32 | 1 |
| Q. alba | 18 | 0.28 | 1 |
| Others* | 23 | 1.12 | 4 |
Size-class distributions (Fig. 2) of the various species showed that P. strobus occupied the entire range of size classes, monotonically decreasing from 10 to 90 cm DBH. Castanea mollissima had a bell-shaped size distribution with peak abundance at 30 cm DBH. Pinus strobus and C. mollissima were the only species to have stems >50 cm DBH. Acer rubrum and B. lenta had bell-shaped distributions with peaks at 10 and 15 cm, respectively. Quercus rubra had fairly even size distributions from 10 to 40 cm.
Spatial distribution
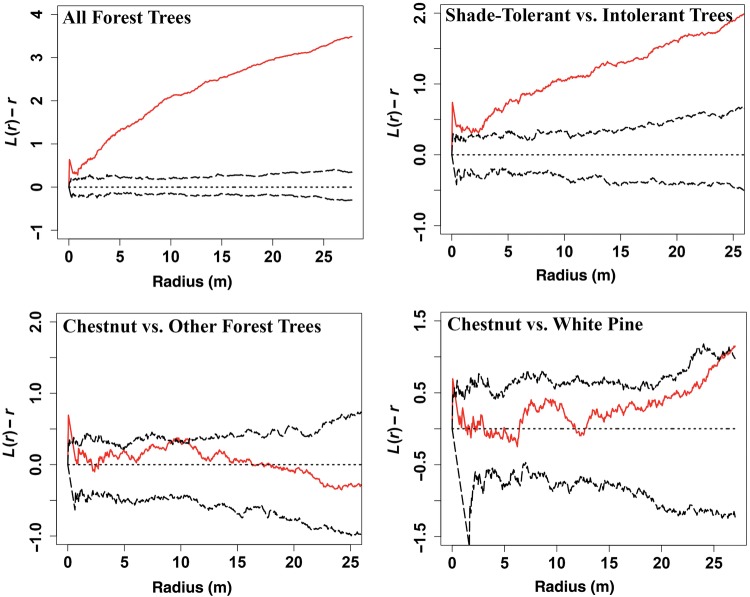
Ripley's K analysis (Fig. 3) indicated that all forest trees were clustered. Ripley's Cross-K analysis showed that shade-tolerant trees clustered around shade-intolerant trees, which is consistent with expectations of a naturally generated, unmanaged forest. Ripley's Cross-K analysis of C. mollissima showed that it was randomly distributed among all other forest trees and randomly distributed with respect to P. strobus, though the two species showed a trend towards clustering with each other. These two species were the largest in the forest, and the most likely pioneer colonizers due to the proximity of seed sources, their biological affinity for full sun, the soil types and the conditions that likely prevailed in the abandoned pasture that became the Dayville woodlot.
Figure 3.
Ripley's K and Cross-K (converted to L-values) indicating patterns of spatial distribution. All forest trees are clustered. Shade-tolerant trees cluster around shade-intolerant trees. Chestnut is randomly distributed among all forest trees. Chestnut does not significantly cluster with white pine.
Root dynamics
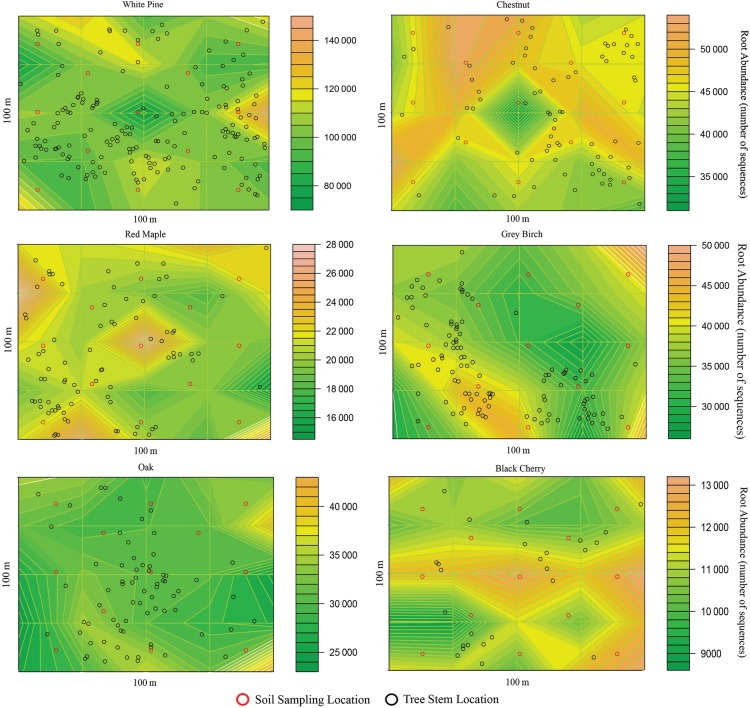
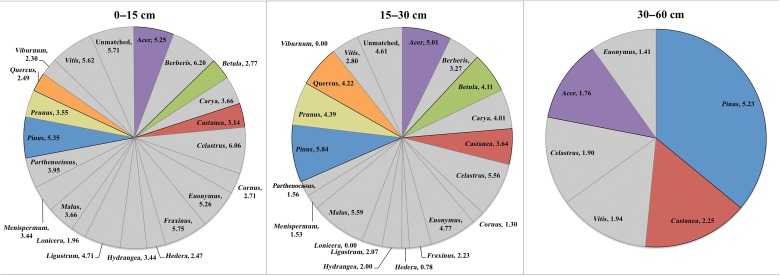
Analysis of DNA sequences and inverse modelling showed that the roots of common woody genera were distributed across the forest, and their abundance was variable across the woodlot and at different depths, but the abundance and distribution of roots did not necessarily reflect the abundance and distribution of stems (Fig. 4). Roots of Castanea were found throughout the forest but were not associated with microsite or BA (data not shown). Roots of shrub genera were concentrated in the upper 15 cm of the soil, with the exception of Euonymus, which was found down to the 30–60 cm range. Trees generally comprised a smaller proportion of the root biomass in the upper 15 cm of soil and a larger proportion below 15 cm (Fig. 5). Pinus, Castanea and Acer made up the majority of roots found at the 30–60 cm depth range. Vine roots from Celastrus and Vitis occurred in large numbers throughout the depth profile. Large stems of these vines were observed throughout the forest.
Figure 4.
Roots of all genera are distributed throughout the Dayville woodlot, but abundance varies spatially per genus. Warm colours indicate higher root abundance.
Figure 5.
Abundance of roots of all woody genera in the Dayville woodlot at three different depths. Values are the log transformation of the number of sequence reads. Species of interest to this study are highlighted.
Tree health and vigour
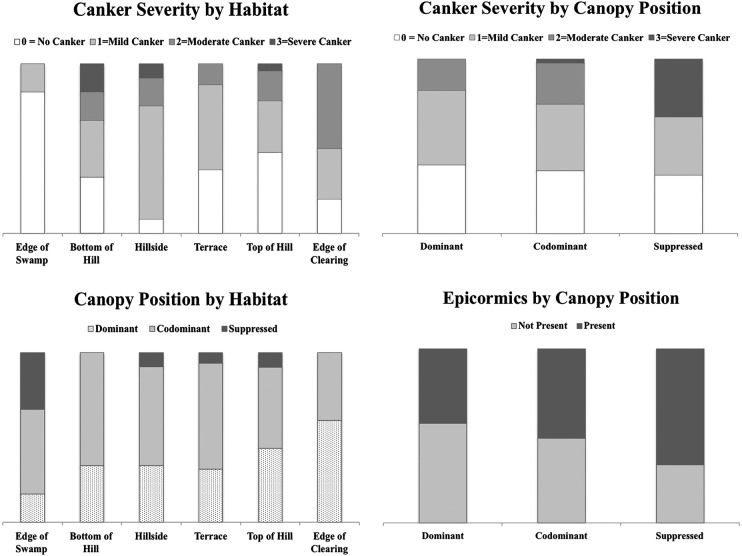
The spatial distribution of chestnut size classes, canopy positions, disease severity and microsites revealed no evident groupings of these attributes. There were no significant associations of attributes except that severe canker was significantly associated with suppressed canopy position (χ2 = 17.271, df = 9, P = 0.0446; Fig. 6). Canker severity was not affected by habitat (χ2 = 17.462, df = 10, P = 0.0647). There were no significant associations between canopy position and habitat (χ2 = 15.613, df = 15, P = 0.482), nor between canopy position and the presence/absence of epicormic branches (χ2 = 3.515, df = 3, P = 0.3188). The power of these tests was limited by small sample sizes: canopy position (dominant = 29, co-dominant = 37, intermediate = 0, suppressed = 6), habitat (edge of swamp = 6, bottom of hill = 6, hillside = 12, terrace = 15, top of hill = 23, edge of clearing = 10), canker severity (no canker = 27, small-healed canker = 28, large-healed canker = 14, severe canker = 3) and epicormic branches (absent = 37, present = 35).
Figure 6.
Association of canker severity with habitat and canopy position and canopy position with habitat and presence of epicormic branches for chestnut. Canker severity by habitat: χ2 = 17.462, df = 10, P = 0.0647. Canopy position by habitat: χ2 = 15.613, df = 15, P = 0.482. Canker severity by canopy position: χ2 = 17.271, df = 9, P = 0.0446. Presence of epicormic branches by canopy position: χ2 = 3.515, df = 3, P = 0.3188.
Dendrochronology
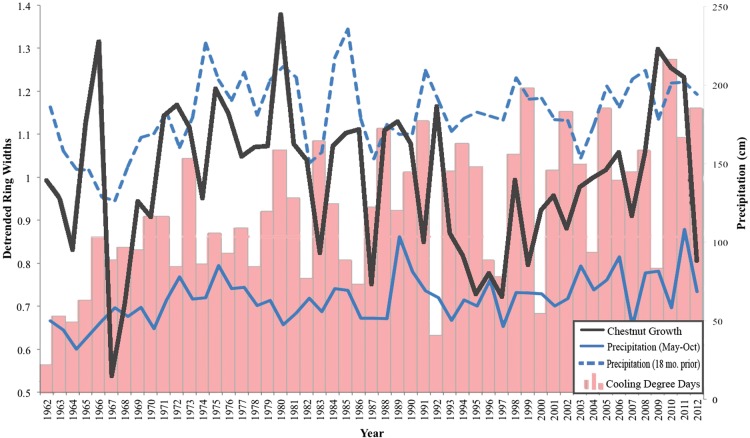
Growth rings indicated that although C. mollissima stems varied somewhat in size, they were similar in age; all were around 50 years old, and they comprised a single cohort. The growth curve indicated that the C. mollissima trees had grown freely without prolonged periods of suppression or release, and the JOLTS analysis confirms that there were no significant suppression or release events in any of the sampled trees, and therefore no indications of stand-wide disturbance in the forest, at any of the sensitivity levels.
A master chronology of growth increments for C. mollissima compared with precipitation and cooling degree days (an indication of growing-season heat) in CT revealed a mixture or significant and non-significant correlations between growth and precipitation windows (Fig. 7). Growth was correlated (at α = 0.05) with growing-season precipitation (P = 0.042, R2 = 0.081) and the previous 18 months' precipitation (P = 0.044, R2 = 0.079), but not with the previous autumn's precipitation (P = 0.895, R2 = 0.000), each year's annual precipitation (P = 0.325, R2 = 0.019) or with cooling degree days (P = 0.885, R2 = 0.000). Mixed linear models revealed that growing-season precipitation (B = 0.255, P = 0.125) and previous 18 months' precipitation (B = 0.16, P = 0.131) were the only combination of tested factors significantly correlated with growth (P = 0.041, R2 = 0.124).
Figure 7.
Master chronology of Chinese chestnut growth (detrended ring widths) overlaid on growing-season precipitation, previous 18 months' precipitation and cooling degree days (to show relative heat and light over the years). Multiple regression reveals that growing-season precipitation (B = 0.255, P = 0.125) and previous 18 months' precipitation (B = 0.16, P = 0.131) are significantly related to C. mollissima's increment growth (P = 0.041), but only account for a small part of the variation (R2 = 0.124).
Relatedness and parentage
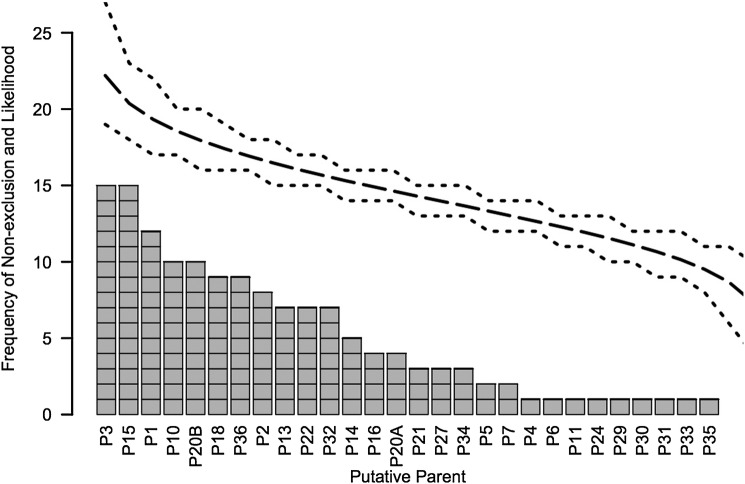
We produced a unique microsatellite genotype for each Castanea on the site based on 11 loci (Table 2). A PCA of genetic distance showed all trees in both parent and offspring populations to be effectively the same population with high pairwise relatedness among all trees but few fixed alleles in the population (Fst = 0.005, Fis = 0.072, Fit = 0.077). Molecular variance within individuals was 92 %, among individuals was 7 % and among populations (parents versus offspring) was 1 %. Parent and offspring populations shared many alleles, with the exception of three private alleles in the parent population and seven private alleles in the offspring population (Table 3). Most loci were in Hardy–Weinberg equilibrium except CM396, CM800 and CM004 in the parent population and CM396, CM467 and CM800 in the offspring population. The CERVUS analysis of parentage showed that each parent contributed at least one offspring, with some parents contributing to as many as 15 offspring. There was no genetic bottleneck evident, and parental contribution to the offspring population was random (Fig. 8).
Table 3.
Allele information, heterozygosity and tests for the Hardy–Weinberg equilibrium for each locus in each population.aNumber of alleles observed in the population. bObserved heterozygosity. cExpected heterozygosity. dNumber of private alleles per locus, with frequency in parentheses. eProportion of loci with missing data, averaged over individuals. fHardy–Weinburg test includes degrees of freedom (df), Chi-square value, P-value and significance: ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001.
| Population | Locus name | Naa | Hob | Hec | PAd (frequency) | Missinge | Tests for Hardy–Weinburg equilibriumf |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| df | χ2 | P-value | Significance | |||||||
| Orchard parents (N = 28) | CM005 | 8.000 | 0.929 | 0.783 | 0 | 0 | 28 | 34.677 | 0.179 | ns |
| CM008 | 7.000 | 0.893 | 0.798 | 0 | 0 | 21 | 19.617 | 0.546 | ns | |
| CM010 | 6.000 | 0.667 | 0.652 | 1 (0.019) | 0 | 15 | 8.031 | 0.923 | ns | |
| CM018 | 10.00 | 0.815 | 0.762 | 1 (0.019) | 0 | 45 | 33.842 | 0.888 | ns | |
| CM396 | 5.000 | 1.000 | 0.678 | 0 | 0 | 10 | 29.596 | 0.001 | *** | |
| CM467 | 3.000 | 0.852 | 0.647 | 0 | 0 | 3 | 7.078 | 0.069 | ns | |
| CM800 | 7.000 | 0.741 | 0.765 | 0 | 0 | 21 | 36.486 | 0.019 | * | |
| CM004 | 5.000 | 0.550 | 0.679 | 0 | 0.04 | 10 | 19.960 | 0.030 | * | |
| CM007 | 5.000 | 0.565 | 0.723 | 1 (0.022) | 0.07 | 10 | 10.517 | 0.396 | ns | |
| CM883 | 4.000 | 0.481 | 0.488 | 0 | 0.04 | 6 | 4.150 | 0.656 | ns | |
| CM945 | 9.000 | 0.818 | 0.801 | 0 | 0.07 | 36 | 44.043 | 0.168 | ns | |
| Forest offspring (N = 72) | CM005 | 8.000 | 0.881 | 0.790 | 0 | 0 | 28 | 31.201 | 0.308 | ns |
| CM008 | 8.000 | 0.819 | 0.776 | 1 (0.007) | 0 | 28 | 31.012 | 0.317 | ns | |
| CM010 | 5.000 | 0.754 | 0.683 | 0 | 0 | 10 | 13.208 | 0.212 | ns | |
| CM018 | 9.000 | 0.859 | 0.777 | 0 | 0 | 36 | 32.050 | 0.657 | ns | |
| CM396 | 6.000 | 0.871 | 0.635 | 1 (0.036) | 0 | 15 | 40.196 | 0.000 | *** | |
| CM467 | 3.000 | 0.886 | 0.666 | 0 | 0 | 3 | 15.943 | 0.001 | ** | |
| CM800 | 7.000 | 0.761 | 0.750 | 0 | 0 | 21 | 32.702 | 0.050 | * | |
| CM004 | 7.000 | 0.577 | 0.577 | 2 (0.024) | 0.04 | 21 | 19.632 | 0.545 | ns | |
| CM007 | 4.000 | 0.473 | 0.604 | 0 | 0.014 | 6 | 12.467 | 0.052 | ns | |
| CM883 | 5.000 | 0.455 | 0.520 | 1 (0.009) | 0.027 | 10 | 8.911 | 0.541 | ns | |
| CM945 | 11.00 | 0.892 | 0.819 | 2 (0.019) | 0.014 | 55 | 51.033 | 0.627 | ns | |
Figure 8.
Frequency of chestnut parent contribution to the offspring population. Parents were assigned to offspring based on statistical likelihood or non-exclusion due to matching alleles. Long dashed line is the observed parent randomness, and dotted lines are 95 % confidence envelope for randomness.
Discussion
Castanea mollissima is not native to North America, and while it has been cultivated and grown commercially, it is rarely found integrated into North American forests (Schlarbaum et al. 1994). In the Dayville woodlot, however, 50-year-old C. mollissima trees were numerous, co-dominant in the canopy and randomly distributed alongside native North American species. No native C. dentata had been found in the area for at least 100 years. The species composition, size-class distributions and historical records of the land and forest suggested that it developed through natural colonization and succession and had not been directly managed by humans (Figs 1–3). The presence and age of the trees, along with the mix of significant and non-significant relationships between precipitation, temperature and growth, indicated that the climate and soil were sufficient for C. mollissima to establish and to maintain their canopy position as the forest structure developed, and the growth increment data showed little competition pressure or periods of adversity in the past (Fig. 7).
Ecologically, the forest-grown C. mollissima resemble C. dentata, with several analogues to the development of a disjunct stand of C. dentata growing outside of its range in West Salem, WI (Paillet and Rutter 1989). In WI, C. dentata was functionally an exotic species that colonized land with shallow, sandy soil of low pH. As at Dayville, the West Salem site had been released from grazing, and pioneer trees grew there under open conditions without competition from other large trees, becoming dominant or co-dominant in the canopy alongside Quercus, Carya, Betula spp. and others (Paillet and Rutter 1989). Despite the fact that the founding population in West Salem was small and closely related (like the C. mollissima in our study), the trees in WI did not show any detrimental effects of inbreeding and exhibited heterozygous advantage (Pierson et al. 2007). The West Salem C. dentata comprised ∼25 % of the BA in the forest, similar to the almost 20 % of C. mollissima found at Dayville; and like the C. mollissima at Dayville, the growth forms of the C. dentata in WI ranged from multi-stemmed and shrub-like to single-stem erect trees (Paillet and Rutter 1989). Tree-ring analyses of the C. dentata in WI also showed no direct relationship between tree growth and precipitation or heat (Paillet and Rutter 1989; McEwan et al. 2006). In contrast, however, the C. dentata population has been dying back and diminishing due to the introduction of chestnut blight (Paillet and Rutter 1989; McEwan et al. 2006; Gilland et al. 2012), while the C. mollissima in CT have continued to grow in the forest despite the presence of chestnut blight.
Chestnut blight
Chestnut blight cankers were found on almost all of the C. mollissima trees, but there was no evidence of mortality caused by blight, similar to previous observations in the parent orchard (Jaynes 1965). Blight severity did not appear to be correlated with differences in site characteristics or location (Fig. 6). There was no link between blight severity and parentage. Because of the long infestation of chestnut blight in the area, it seems that all surviving C. mollissima parent trees were capable of producing blight-tolerant offspring. Canopy suppression was the only negative impact observed on the most severely blighted offspring (Fig. 6). The suppressed C. mollissima individuals appeared to have become so in recent years, and in addition to canopy suppression they also suffered from stressors such as wind damage and vine competition. In contrast, other Castanea species such as C. dentata and C. sativa (European chestnut) either die or are at a severe competitive disadvantage when exposed to chestnut blight (Paillet 2002; Zlatanov et al. 2013). This means that C. mollissima can survive and compete in the forest without complete blight resistance, but trees in the lower end of the resistance spectrum are expected to grow more poorly. Based on the observations of wild C. mollissima in China (Steiner et al. 2009) and suppressed C. dentata stump sprouts in the northeastern USA (Paillet 1984), it appears that blight severity causes canopy suppression more than canopy suppression worsens blight symptoms. There appears to be genetic variation in blight resistance within pure C. mollissima populations (Steiner et al. 2009), indicating that not all genes for blight resistance are fixed in the species. The presence of blight-susceptible C. mollissima individuals requires an explanation as to why natural selection in China has not purged blight-susceptible alleles from the population. Current hypothetical models for the genetics of blight resistance (Burnham 1981; Burnham et al. 1986; Hebard 1994, 2006) do not account for the variability in blight resistance in C. mollissima over evolutionary time.
Genetic relatedness and heterozygosity
All C. mollissima trees in the Dayville woodlot, parents and offspring, were closely related and effectively a single breeding population. Mating and parentage appeared random, but because there were 67 orchard trees planted originally, and because all forest C. mollissima are the offspring of at least one of the 28 surviving orchard trees, it is likely that selection pressure has already worked against the non-viable parents and offspring, leaving no trace of the unfit and poorly adapted trees. Of the SSRs used for determining relatedness, CM396 had unusually high heterozygosity and was significantly out of Hardy–Weinberg equilibrium in both the parents and the offspring. CM800 and CM467 also had unusually high heterozygosity, especially in the forest offspring population (Table 3). These markers are located within the quantitative trait locus that confers resistance to the root pathogens in the genus Phytophthora (Tatiana Zhebentyayeva, Clemson University, personal communication), which are known to be detrimental and/or lethal to Castanea species, especially C. dentata (Rhoades et al. 2003). It is possible the deviations from allele frequency expectations that we observed at these loci are the consequence of selection favouring the heterozygous state.
Invasive ecology of C. mollissima and co-occurring species
The scarcity of other unmanaged populations of C. mollissima throughout eastern North America calls to question why the species has not been as successful elsewhere. It has been suggested that C. mollissima cannot compete with native North American forest species because of its short mature canopy height and spreading growth habit (Griffin 2000; Fei et al. 2012). Other factors potentially limiting the spread of C. mollissima into North American forests include predation of seeds and seedling herbivory by deer, as well as direct competition from invasive understorey plant species. At the Dayville site, shallow (average 30 cm deep) sandy soils on top of granite bedrock limited forest canopy height to ∼18–20 m, and C. mollissima trees were able to grow to that height and maintain a competitive position in the canopy. Additionally, it is likely that low predation pressure from a relatively small deer population in CT prior to 1965 allowed more seeds to be dispersed and hoarded by squirrels (Jaynes 1965). We found that C. mollissima were producing nuts in this forest, as evidenced by chestnut burs found on the ground under offspring trees, but there appeared to be no tree seedlings, other than several Fraxinus seedlings, under 5 cm DBH. The understorey was dominated by non-native shrubs such as Lonicera, Berberis, E. alatus and Ligustrum that could outcompete tree seedlings and shelter seed predators such as rabbits and small rodents. The root analysis revealed high abundance of, and therefore high below-ground competition from exotic invasive species such as E. alatus, Berberis spp. and Celastrus orbiculatus (Fig. 5). It follows that natural reproduction of any Castanea species, including blight-resistant hybrids, will face the same difficulties as C. mollissima and many North American forest natives (Richardson et al. 2011; Nuttle et al. 2013).
There are concerns that future C. mollissima × C. dentata hybrids could become invasive and displace native hardwoods (Jacobs 2007; Jacobs et al. 2013); however, evidence from the Dayville woodlot and the basic biology of Castanea spp. make this unlikely. Problematic invasive woody species in North America, such as Ailanthis altissima, Lonicera spp., Rhamnus cathartica, Pyrus calleryana and Elaeagnus umbellata, are aggressive partly because they have small seeds that are dispersed by wind or passed through the guts of animals, giving them a wide dispersal range, and they tend to be broadly adapted to varying soil and light conditions (Binggeli 1996; Rejmánek 1996; Guo et al. 2000; Zheng et al. 2004). Castanea seeds are not wind dispersed and are killed when consumed by animals, rather than passing through guts intact. Predation pressure impedes, not enables, their dispersal until the predator population is satiated. Additionally, all Castanea spp. are similar in terms of their site requirements and climatic limits (Fitzsimmons 2006; Hunt et al. 2009; Fei et al. 2012), which means that they are broadly adapted climatically, but narrowly adapted edaphically (McCament and McCarthy 2005; Mellano et al. 2012). As such, it is likely that Castanea seedlings only exhibit growth advantage over other hardwood genera such as Quercus and Prunus under optimal soil conditions. This probably explains the discrepancy in the studies of Paillet and Rutter (1989), Jacobs and Severeid (2004), McEwan et al. (2006) and Jacobs et al. (2009), where Castanea was found to have superior growth over other genera, and Clark et al. (2012), Griscom and Griscom (2012), and Gauthier et al. (2013) where Castanea did not outperform other genera. Finally, Castanea species are nutritious to wildlife and to humans (McCarthy and Meredith 1988; Senter et al. 1994; Paillet 2006; Burke 2013), provide complex habitat structure in forest communities (Zlatanov et al. 2013) and mast even under extreme weather conditions such as drought (Gilland et al. 2012); therefore, they could be beneficial to future natural and anthropogenic landscapes. Synthesis from the literature and Dayville C. mollissima observations predict that Castanea hybrids can proliferate in certain areas of North America and confer ecological benefits while not completely displacing other desirable genera such as Quercus and Prunus.
Conclusions
The seeming ease with which C. mollissima established in the Dayville woodlot begs for an explanation as to why similar stands of C. mollissima are rare elsewhere in North America. Likely explanations are that a rare time window of low seed and seedling predation allowed seedlings to establish, and the shallow soil depth at this particular site limited the growth of the native forest canopy to a height attainable by the characteristically short-statured C. mollissima. Hybrids may need the genetic potential to grow taller than C. mollissima on more productive sites if they are to persist. However, most of the C. mollissima germplasm in the USA was selected for nut production and so the full range of its growth potential in forest environments has yet to be explored. The formation of this stand of C. mollissima and the disjunct stand of C. dentata in WI indicates that Castanea species and their hybrids may play a valuable ecological role as pioneer species in the reforestation and reclamation of degraded lands such as fallow farm fields. Short-statured, C. mollissima-like hybrids may be desirable components of highly disturbed, anthropogenic environments, conferring more economic and ecological benefits than other exotics that have become invasive. These results imply that introduced tree species in North America could be assessed not only by their level of threat to native species (Parker et al. 1999) but also by their potential positive impacts on North American ecosystems and their usefulness in programmes to breed certain resilient traits into native North American species.
Sources of Funding
Our work was funded by a Fred M. van Eck scholarship from the Hardwood Tree Improvement and Regeneration Center at Purdue University, West Lafayette, IN, USA.
Contributions by the Authors
A.C.M. performed most of the field measurements, laboratory analyses, data analyses and table/figure creation, and wrote most of the manuscript. K.E.W. provided guidance on genetic analyses and interpretations and helped write the manuscript. S.L.A. provided access to the study site, background information for the study and helped write the manuscript. D.F.J. contributed to experimental design and logistics, and helped write the manuscript.
Conflicts of Interest Statement
None declared.
Acknowledgements
The authors thank Charles Michler and Harmony Dalgleish for their guidance on this project. Nick LaBonte, Kyle Earnshaw and Philip Crystal assisted with field measurements and laboratory analysis. Two anonymous reviewers improved this manuscript. Mention of a trademark, proprietary product or vendor does not constitute a guarantee or warranty of the product by the US Dept. of Agriculture and does not imply its approval to the exclusion of other products or vendors that may also be suitable.
Literature Cited
- Anagnostakis SL. Survival and naturalization of Chinese chestnut in Connecticut. Annual Report for the Northern Nut Growers Association. 1992;83:24–27. [Google Scholar]
- Anagnostakis SL. Connecticut chestnut research: breeding and biological control. New Haven, CT, USA: The Connecticut Agricultural Experiment Station; 2012. [Google Scholar]
- Berry FH. Evaluation of chestnut test plantings in the eastern United States. 1980. Research Paper NE-454. Broomall, PA, USA: Northeastern Forest Experiment Station, 5 p. [Google Scholar]
- Binggeli P. A taxonomic, biogeographical and ecological overview of invasive woody plants. Journal of Vegetation Science. 1996;7:121–124. [Google Scholar]
- Burke KL. Chestnuts and wildlife. The Journal of the American Chestnut Foundation. 2013;27:8–11. [Google Scholar]
- Burnham CR. Blight-resistant American chestnut: there's hope. Plant Disease. 1981;65:459–460. [Google Scholar]
- Burnham CR, Rutter PA, French DW. Breeding blight-resistant chestnuts. Plant Breeding Reviews. 1986;4:347–397. [Google Scholar]
- Clark S, McNab H, Loftis D, Zarnoch S. American chestnut growth and survival five years after planting in two silvicultural treatments in the southern Appalachians. Forests. 2012;3:1017–1033. [Google Scholar]
- Conedera M, Manetti MC, Giudici F, Amorini E. Distribution and economic potential of the sweet chestnut (Castanea sativa Mill.) in Europe. Ecologia Mediterranea. 2004;30:179–193. [Google Scholar]
- Davis DE. Historical significance of American chestnut to Appalachian culture and ecology. In: Steiner KC, Carlson JE, editors. Restoration of American chestnut to forest lands. 2006. Natural Resources Report NPS/NCR/CUE/NRR - 2006/001. Washington, DC, USA: National Park Service. [Google Scholar]
- Diller JD, Clapper RB, Jaynes RA. Cooperative test plots produce some promising Chinese and hybrid chestnut trees. 1964. Research Paper NE-25. Radnor, PA, USA: Northeastern Forest Experiment Station, 8 p. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Fei S, Liang L, Paillet F, Steiner K, Fang J, Shen Z, Wang Z, Hebard F. Modelling chestnut biogeography for American chestnut restoration. Diversity and Distributions. 2012;18:754–768. [Google Scholar]
- Fitzsimmons S. Planting and growing chestnut trees. The Pennsylvania Chapter of the American Chestnut Foundation Planting Manual. 2006:15 p. [Google Scholar]
- Gauthier MM, Zellers KE, Löf M, Jacobs DF. Inter-and intra-specific competitiveness of plantation-grown American chestnut (Castanea dentata) Forest Ecology and Management. 2013;291:289–299. [Google Scholar]
- Gilland KE, Keiffer CH, McCarthy BC. Seed production of mature forest-grown American chestnut (Castanea dentata (Marsh.) Borkh) The Journal of the Torrey Botanical Society. 2012;139:283–289. [Google Scholar]
- Griffin GJ. Blight control and restoration of the American chestnut. Journal of Forestry. 2000;98:22–27. [Google Scholar]
- Griscom HP, Griscom BW. Evaluating the ecological niche of American chestnut for optimal hybrid seedling reintroduction sites in the Appalachian ridge and valley province. New Forests. 2012;43:441–455. [Google Scholar]
- Grissino-Mayer HD. Evaluating crossdating accuracy: a manual and tutorial for the computer program COFECHA. Tree-Ring Research. 2001;57:205–221. [Google Scholar]
- Guo Q, Brown JH, Valone TJ, Kachman SD. Constraints of seed size on plant distribution and abundance. Ecology. 2000;81:2149–2155. [Google Scholar]
- Haase P. Spatial pattern analysis in ecology based on Ripley's K function: introduction and methods of edge correction. Journal of Vegetation Science. 1995;6:575–582. [Google Scholar]
- Hebard FV. The American Chestnut Foundation breeding plan: beginning and intermediate steps. Journal of the American Chestnut Foundation. 1994;8:21–28. [Google Scholar]
- Hebard FV. Backcross breeding program produces blight-resistant American chestnuts. Ecological Restoration. 2001;19:252–254. [Google Scholar]
- Hebard FV. The backcross breeding program of the American chestnut foundation. Journal of the American Chestnut Foundation. 2006;19:55–77. [Google Scholar]
- Hunt K, Gold MA, Reid W, Warmund MR. Growing Chinese chestnuts in Missouri. Extension publications (MU); 2009. [Google Scholar]
- Inoue E, Ning L, Hara H, Ruan S, Anzai H. Development of simple sequence repeat markers in Chinese chestnut and their characterization in diverse chestnut cultivars. Journal of the American Society for Horticultural Science. 2009;134:610–617. [Google Scholar]
- Jacobs DF. Toward development of silvical strategies for forest restoration of American chestnut (Castanea dentata) using blight-resistant hybrids. Biological Conservation. 2007;137:497–506. [Google Scholar]
- Jacobs DF, Severeid LR. Dominance of interplanted American chestnut (Castanea dentata) in southwestern Wisconsin, USA. Forest Ecology and Management. 2004;191:111–120. [Google Scholar]
- Jacobs DF, Selig MF, Severeid LR. Aboveground carbon biomass of plantation-grown American chestnut (Castanea dentata) in absence of blight. Forest Ecology and Management. 2009;258:288–294. [Google Scholar]
- Jacobs DF, Dalgleish HJ, Nelson CD. A conceptual framework for restoration of threatened plants: the effective model of American chestnut (Castanea dentata) reintroduction. New Phytologist. 2013;197:378–393. doi: 10.1111/nph.12020. [DOI] [PubMed] [Google Scholar]
- Jaynes RA. Natural regeneration from a 40-year-old Chinese chestnut planting. Journal of Forestry. 1965;65:29–31. [Google Scholar]
- Jones AF, Erickson DL, Bernall MA, Berminghang E, Kress WJ, Herrel EA, Muller-Landau HC, Turner BL. The roots of diversity: below ground species richness and rooting distributions in a tropical forest revealed by DNA barcodes and inverse modeling. PloS ONE. 2011;6:e9. doi: 10.1371/journal.pone.0024506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences of the USA. 2009;106:18621–18626. doi: 10.1073/pnas.0909820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisiak TL, Nelson CD, Staton ME, Zhebentyayeva T, Smith C, Olukolu BA, Fang GC, Hebard FV, Anagnostakis S, Wheeler N, Sisco PH, Abbott AG, Sederoff RR. A transcriptome-based genetic map of Chinese chestnut (Castanea mollissima) and identification of regions of segmental homology with peach (Prunus persica) Tree Genetics and Genomics. 2013;9:557–571. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCament CL, McCarthy BC. Two-year response of American chestnut (Castanea dentata) seedlings to shelterwood harvesting and fire in a mixed-oak forest ecosystem. Canadian Journal of Forest Research. 2005;35:740–749. [Google Scholar]
- McCarthy MA, Meredith FI. Nutrient data on chestnuts consumed in the United States. Economic Botany. 1988;42:29–36. [Google Scholar]
- McEwan RW, Keiffer CH, McCarthy BC. Dendroecology of American chestnut in a disjunct stand of oak chestnut forest. Canadian Journal of Forest Research. 2006;36:1–11. [Google Scholar]
- Mellano MG, Beccaro GL, Donno D, Torello Marinoni D, Boccacci P, Canterino S, Cerutti AK, Bounous G. Castanea spp. biodiversity conservation: Collection and characterization of the genetic diversity of an endangered species. Genetic Resources and Crop Evolution. 2012;59:1727–1741. [Google Scholar]
- Merkle SA, Andrade GM, Nairn CJ, Powell WA, Maynard CA. Restoration of threatened species: a noble cause for transgenic trees. Tree Genetics and Genomes. 2007;3:111–118. [Google Scholar]
- Michler CH, Pijut PM, Jacobs DF, Meilan R, Woeste KE, Ostry ME. Improving disease resistance of butternut (Juglans cinerea), a threatened fine hardwood: a case for single-tree selection through genetic improvement and deployment. Tree Physiology. 2006;26:121–128. doi: 10.1093/treephys/26.1.121. [DOI] [PubMed] [Google Scholar]
- Nathan R, Muller-Landau HC. Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Trends in Ecology and Evolution. 2000;15:278–285. doi: 10.1016/s0169-5347(00)01874-7. [DOI] [PubMed] [Google Scholar]
- Nowacki GJ, Abrams MD. Radial-growth averaging criteria for reconstructing disturbance histories from presettlement-origin oaks. Ecological Monographs. 1997;67:225–249. [Google Scholar]
- Nuttle T, Royo AA, Adams MB, Carson WP. Historic disturbance regimes promote tree diversity only under low browsing regimes in eastern deciduous forest. Ecological Monographs. 2013;83:3–17. [Google Scholar]
- Paillet FL. Growth-form and ecology of American chestnut sprout clones in northeastern Massachusetts. Bulletin of the Torrey Botanical Club. 1984;111:316–328. [Google Scholar]
- Paillet FL. Chestnut: history and ecology of a transformed species. Journal of Biogeography. 2002;29:1517–1530. [Google Scholar]
- Paillet FL. Chestnut and wildlife. In: Steiner KC, Carlson JE, editors. Restoration of American chestnut to forest lands. 2006. Natural Resources Report NPS/NCR/CUE/NRR - 2006/001. Washington, DC, USA: National Park Service. [Google Scholar]
- Paillet FL, Rutter PA. Replacement of native oak and hickory tree species by the introduced American chestnut (Castanea dentata) in southwestern Wisconsin. Canadian Journal of Botany. 1989;67:3457–3469. [Google Scholar]
- Parker IM, Simberloff D, Lonsdale WM, Goodell K, Wonham M, Kareiva PM, Williamson MH, Von Holle B, Moyle PB, Byers JE, Goldwasser L. Impact: toward a framework for understanding the ecological effects of invaders. Biological Invasions. 1999;1:3–19. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson SAM, Keiffer CH, McCarthy BC, Rogstad SH. Limited reintroduction does not always lead to rapid loss of genetic diversity: an example from the American Chestnut (Castanea dentata; Fagaceae) Restoration Ecology. 2007;15:420–429. [Google Scholar]
- Powell GH. The European and Japanese chestnuts in the eastern United States. Vol. 42. Delaware: Delaware College Agricultural Experiment Station; 1898. [Google Scholar]
- Rejmánek M. A theory of seed plant invasiveness: the first sketch. Biological Conservation. 1996;78:171–181. [Google Scholar]
- Rhoades CC, Brosi SL, Dattilo AJ, Vincelli P. Effects of soil compaction and moisture on incidence of Phytophthora root rot on American chestnut (Castanea dentata) seedlings. Forest Ecology and Management. 2003;184:47–54. [Google Scholar]
- Richardson DM, Carruthers J, Hui C, Impson FAC, Miller JT, Robertson MP, Rouget M, Le Roux JJ, Wilson JRU. Human-mediated introductions of Australian acacias—a global experiment in biogeography. Diversity and Distributions. 2011;17:771–787. [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O'Neil P, Parker IM, Thompson JN, Weller SG. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Schlarbaum SE, Anagnostakis SL, Morton MC. Evaluation of experimental chestnut plantings in Eastern North America. In: Double ML, MacDonald WL, editors. Proceedings of the International Chestnut Conference. 1994. pp. 52–66. Morgantown, WV, USA: West Virginia University Press. [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Senter SD, Payne JA, Miller G, Anagnostakis SL. Comparison of total lipids, fatty acids, sugars, and nonvolatile organic acids in nuts from four Castanea species. Journal of the Science of Food and Agriculture. 1994;65:223–227. [Google Scholar]
- Shear CL, Stevens NE. The discovery of the chestnut-blight parasite (Endothia parasitica) and other chestnut fungi in Japan. Science. 1916;43:173–176. doi: 10.1126/science.43.1101.173-a. [DOI] [PubMed] [Google Scholar]
- Steiner K, Paillet F, Hebard F, Fei S, Fitzsimmons S. The American Chestnut Foundation expedition to China. Journal of the American Chestnut Foundation. 2009;23:9–21. [Google Scholar]
- Zhao P, Woeste K. DNA markers identify hybrids between butternut (Juglans cinerea L.) and Japanese walnut (Juglans ailantifolia Carr.) Tree Genetics and Genomes. 2011;7:511–533. [Google Scholar]
- Zheng H, Wu Y, Ding J, Binion D, Fu W, Reardon R. Invasive plants of Asian origin established in the United States and their natural enemies. Vol. 1. West Virginia: USDA Forest Service FHTET; 2004. [Google Scholar]
- Zlatanov T, Schleppi P, Velichkov I, Hinkov G, Georgieva M, Eggertsson O, Zlatanova M, Vacik H. Structural diversity of abandoned chestnut (Castanea sativa Mill.) dominated forests: implications for forest management. Forest Ecology and Management. 2013;291:326–335. [Google Scholar]