Abstract
Corynebacterium pseudotuberculosis (Cp) is a pathogenic bacterium that causes caseous lymphadenitis (CLA), ulcerative lymphangitis, mastitis, and edematous to a broad spectrum of hosts, including ruminants, thereby threatening economic and dairy industries worldwide. Currently there is no effective drug or vaccine available against Cp. To identify new targets, we adopted a novel integrative strategy, which began with the prediction of the modelome (tridimensional protein structures for the proteome of an organism, generated through comparative modeling) for 15 previously sequenced C. pseudotuberculosis strains. This pan-modelomics approach identified a set of 331 conserved proteins having 95-100% intra-species sequence similarity. Next, we combined subtractive proteomics and modelomics to reveal a set of 10 Cp proteins, which may be essential for the bacteria. Of these, 4 proteins (tcsR, mtrA, nrdI, and ispH) were essential and non-host homologs (considering man, horse, cow and sheep as hosts) and satisfied all criteria of being putative targets. Additionally, we subjected these 4 proteins to virtual screening of a drug-like compound library. In all cases, molecules predicted to form favorable interactions and which showed high complementarity to the target were found among the top ranking compounds. The remaining 6 essential proteins (adk, gapA, glyA, fumC, gnd, and aspA) have homologs in the host proteomes. Their active site cavities were compared to the respective cavities in host proteins. We propose that some of these proteins can be selectively targeted using structure-based drug design approaches (SBDD). Our results facilitate the selection of C. pseudotuberculosis putative proteins for developing broad-spectrum novel drugs and vaccines. A few of the targets identified here have been validated in other microorganisms, suggesting that our modelome strategy is effective and can also be applicable to other pathogens.
Background
Antimicrobial resistance involving a rapid loss of effectiveness in antibiotic treatment and the increasing number of multi-resistant microbial strains pose global challenges and threats. Thereby, efforts to find new drug and/or vaccine targets to control them are becoming indispensible. Corynebacterium pseudotuberculosis (Cp) is a pathogen of great veterinary and economic importance, since it affects animal livestock, mainly sheep and goats, worldwide, and its presence is reported in other mammals in several Arabic, Asiatic, East and West African and North and South American countries, as well as in Australia [1]. C. pseudotuberculosis is a Gram-positive, facultative intracellular, and pleomorphic organism; it is non-motile, although presenting fimbriae [2]. Based on rpoB gene (a β subunit of RNA polymerase), it shows a close phylogenetic relationship with other type strains of CMNR (Corynebacterium, Mycobacterium, Nocardia and Rhodococcus), a group that comprises genera of great medical, veterinary and biotechnological importance [1,3]. A recent study showed that phylogenetic analysis for the identification of Corynebacterium and other CMNR species based on rpoB gene sequences are more accurate than analyses based on 16S rRNA [4]. Its pathogenicity and biological impact have already led to the sequencing of various strains of this pathogen from a wide range of hosts [3]. The pathogen causes several infectious diseases in goat and sheep population (biovar ovis), including caseous lymphadenitis (CLA), a chronic contagious disease characterized by abscess formation in superficial lymph nodes and in subcutaneous tissues. In severe cases, biovar equi infects the lungs, kidneys, liver and spleen, thereby threatening the herd life of the infected animals [2,5]. The disease has been rarely reported in humans, as a result of occupational exposure, with symptoms similar to lymphadenitis abscesses [6-8]. The bacteria can survive for several weeks in soil in adverse conditions, what seems to contribute to its resistance and disease transmission [9,10]. Direct contact to infectious secretions or contaminated materials are the primary sources of pathogen transmission between animals, but most frequently the infection occurs through exposed skin lacerations [5]. Given the medical importance of Cp and a lack of efficient medicines, in this study we applied a computational strategy to search for new molecular targets from this bacterium.
Recently, computational approaches such as reverse vaccinology, differential genome analyses [11], subtractive and comparative microbial genomics have become popular for rapid identification of novel targets in the post genomic era [12], [13]. These approaches were used to identify targets in various human pathogens, like Mycobacterium tuberculosis [14], Helicobacter pylori [15], Burkholderia pseudomalleii [16], Neisseria gonorrhea [17], Pseudomonas aeruginosa [18] and Salmonella typhi [19]. In general, such approaches follow the principle that genes/proteins must be essential to the pathogen and preferably have no homology to the host proteins [20]. Nevertheless, essential targets that are homologous to their corresponding host proteins may also be molecular targets for structure-based selective inhibitors development. In this case, the targets must show significant differences in the active sites or in other druggable pockets, when pathogenic and host proteins are compared [21-23].
Once a molecular target is chosen, the conventional experimental methods for drug discovery consist of testing many synthetic molecules or natural products to identify lead compounds. Such practices are laborious, time consuming and require high investments [24,25]. On the other hand, computational methods for structure-based rational drug design can expedite the process of ligand identification and molecular understanding of interactions between receptor and ligand [26]. Such approaches are dependent on the availability of the structural information about the target protein. Considering the availability of experimental structures in PDB (Protein Data Bank) only for a low percentage of the known protein sequences, comparative modeling is frequently the method of choice for obtaining 3D coordinates for proteins of interest [27] for the development of specific drugs and docking analyses [28,29].
In this work, we used a modelomic approach for the predicted proteome of C. pseudotuberculosis species. This served to bridge the gap between raw genomic information and the identification of good therapeutic targets based on the three dimensional structures. The novelty of this strategy relies in using the structural information from high-throughput comparative modeling for large-scale proteomics data for inhibitor identification, potentially leading to the discovery of compounds able to prevent bacterial growth. The predicted proteomes of 15 C. pseudotuberculosis strains were modeled (pan-modelome) using the MHOLline workflow. Intra-species conserved proteome (core-modelome) with adequate 3D models was further filtered for their essential nature for the bacteria, using the database of essential genes (DEG). This led to the identification of 4 essential bacterial proteins without homologs in the host proteomes, which were employed in virtual screening of compound libraries. Furthermore, we investigated a set of 6 essential host homologs proteins. We observed residues of the predicted bacterial protein cavities that are completely different from the ones found in the homologous domains, and therefore could be specifically targeted. By applying this computational strategy we provide a final list of predicted putative targets in C. pseudotuberculosis, in biovar ovis and equi. They could provide an insight into designing of peptide vaccines, and identification of lead, natural and drug-like compounds that bind to these proteins.
Materials and methods
Genomes selection
Proteomes predicted based on the genomes of fifteen C. pseudotuberculosis strains, including both biovar equi and biovar ovis (Table 1) were used in this study. Most of these genomes were sequenced by our group and are available at NCBI. We downloaded the genome sequences in gbk format from the NCBI server (ftp://ftp.ncbi.nih.gov/genomes/Bacteria) and the corresponding protein sequences (curated CDSs) were exported using Artemis Annotation Tool [30] for further analyses.
Table 1.
Strains of C. pseudotuberculosis employed in the pan-modelome study, and their respective information regarding genomes statistics, disease prevalence and broad-spectrum hosts.
| Strains | GPID | NCBI Accession | Genome Size (Mb) | Number of Proteins | G+C% | Hosts/ Location |
Nitrate's Reduction/ Biovar |
Clinical Manifestation | Sequencing Technology |
|---|---|---|---|---|---|---|---|---|---|
| Cp1/06-A | 73235 | NC_017308.1 | 2.28 | 1,963 | 52.2 | Horse/USA | Positive/equi | Abscess | Illumina |
| Cp31 | 73223 | NC_017730.1 | 2.3 | 2,063 | 52.2 | Buffalo/Egypt | Positive/equi | Abscess | Ion Torrent, SOLiD v3 |
| Cp258 | 157069 | NC_017945.1 | 2.31 | 2,088 | 52.1 | Horse/Belgium | Positive/equi | Ulcerative lymphangitis | SOLiD v3 |
| Cp316 | 71591 | NC_016932.1 | 2.31 | 2,106 | 52.1 | Horse/USA | Positive/equi | Abscess | Ion Torrent |
| CpCIP52.97 | 61117 | NC_017307.1 | 2.32 | 2,057 | 52.1 | Horse/Kenya | Positive/equi | Ulcerative Lymphangitis | SOLiD v2 |
| Cp162 | 89445 | NC_018019.1 | 2.29 | 2,002 | 52.2 | Camel/UK | Positive/equi | Neck Abscess | SOLiD v3 |
| CpP54B96 | 77871 | NC_017031.1 | 2.34 | 2,084 | 52.2 | Antelope/S. Africa | Negative/ovis | CLA Abscess | Ion Torrent, SOLiD v3 |
| Cp267 | 73515 | NC_017462.1 | 2.34 | 2,148 | 52.2 | Lhama/USA | Negative/ovis | CLA Abscess | SOLiD v3 |
| Cp1002 | 40687 | NC_017300.1 | 2.34 | 2,097 | 52.2 | Goat/Brazil | Negative/ovis | CLA Abscess | 454, Sanger |
| Cp42/02-A | 73233 | NC_017306.1 | 2.34 | 2,051 | 52.2 | Sheep/Australia | Negative/ovis | CLA Abscess | Illumina |
| CpC231 | 40875 | NC_017301.1 | 2.33 | 2,095 | 52.2 | Sheep/Australia | Negative/ovis | CLA Abscess | 454, Sanger |
| CpI19 | 52845 | NC_017303.1 | 2.34 | 2,099 | 52.2 | Bovine/Israel | Negative/ovis | Bovine Mastitis Abscess | SOLiD v2 |
| Cp3/99-5 | 73231 | NC_016781.1 | 2.34 | 2,142 | 52.2 | Sheep/Scotland | Negative/ovis | CLA | Illumina |
| CpPAT10 | 61115 | NC_017305.1 | 2.34 | 2,089 | 52.2 | Sheep/Argentina | Negative/ovis | Lung Abscess | SOLiD v2 |
| CpFRC41 | 48979 | NC_014329.1 | 2.34 | 2,104 | 52.2 | Human/France | Negative/ovis | Necrotizing lymphadenitis | SOLiD v3 |
Pan-modelome construction
A high throughput biological workflow, MHOLline (http://www.mholline.lncc.br), was used to predict the modelome (complete set of protein 3D models for the whole proteome) for each Cp strain. MHOLline uses the program MODELLER [31] for protein 3D structure prediction through comparative modeling. Furthermore, the workflow includes BLASTp (Basic Local Alignment Search Tool for Protein) [32], HMMTOP (Prediction of transmembrane helices and topology of proteins) [33], BATS (Blast Automatic Targeting for Structures), FILTERS, ECNGet (Get Enzyme Commission Number), MODELLER and PROCHECK [34] programs. The protocol used here was modified accordingly from the original work by Capriles et al., 2010 [35]. Briefly, the input files of protein sequences were used in FASTA format for all strains because the MHOLline accepts only .faa format files for the whole process. Firstly, MHOLline selected the template structures available at the Protein data Bank (PDB) via BLASTp (version 2.2.18), using the default parameters (e-value ≤ 10e-5). Secondly, the program BATS refined the BLASTp search for template sequence identification into different groups namely G0, G1, G2 and G3. Only the protein sequences in the group G2, which are characterized by an e-value ≤ 10e-5, Identity ≥ 0.25 and LVI ≤ 0.7 (where LVI is a length variation index of the BATS program for sequence coverage, the lower the LVI value, the higher the sequence coverage and vice versa) were selected. Among the MHOLline output files, the group G2 contained the largest number of protein sequences (≥ 50% for each input file). Subsequently, the "Filter" tool classified the group G2 sequences into seven distinct quality models groups, from "Very High" to "Very Low" depending on the quality of the template structure for a given query protein sequence. The program MODELLER then modeled all these groups in an automated manner. The number of sequences in the group G2 varies for each C. pseudotuberculosis strain. Only the first four distinct quality model groups of G2 were taken into consideration in this study, these were: 1- Very High quality model sequences (identity ≥ 75%) (LVI ≤ 0.1), 2- High quality model sequences (identity ≥ 50%) and < 75%) (LVI ≤ 0.1), 3- Good quality model sequences (identity ≥ 50%) (LVI > 0.1 and ≤ 0.3) and 4- Medium to Good quality models (identity ≥ 35% and < 50%) (LVI ≤ 0.3) (http://www.mholline.lncc.br). The percentage of identity represents identity between query and template sequences, a LVI ≤ 0.1 is equivalent to coverage of more than 90%, while LVI ≤ 0.3 corresponds to coverage of more than 70%. Therefore, all protein 3D models considered in this study were built from sequences for which there existed a template with identity ≥ 35% and LVI coverage over 70%. Later on, the ECNGet tool assigned an Enzyme Commission (EC) number to each sequence in G2, according to the best PDB template. The MODELLER (v9v5) program performed the automated global alignment and 3D protein model construction. Finally, the program PROCHECK (v3.5.4) evaluated the constructed models based on their stereo-chemical quality. Additionally, transmembrane regions in the input protein sequences were predicted by HMMTOP, for putative vaccine and drug targets identification.
Identification of intra-species conserved genes/proteins
The words genes and proteins are interchangeably used here but they refer to the same protein target of the pathogen. For the identification of highly conserved proteins with 3D models in all Cp strains (≥ 95% sequence identity), the standalone release of NCBI BLASTp+ (v2.2.26) was acquired from the NCBI ftp site (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/), installed on a local machine and a search was performed for all strains using Cp1002 as a reference genome. The highly conserved proteins were selected using a comparative genomics/proteomics approach using an all-against-all BLASTp analysis with cut off values of E = 0.0001 [12,17,20,36].
Analyses of essential and non-host homologous (ENH) proteins
To select conserved targets that were essential to the bacteria, a subtractive genomics approach was followed [20]. Briefly, the set of core-modelome proteins from C. pseudotuberculosis were subjected to the Database of Essential Genes (DEG) for homology analyses. DEG contains experimentally validated essential genes from 20 bacteria [37]. The BLASTp cutoff values used were: E-value = 0.0001, bit score ≥100, identity ≥ 35% [20].
Furthermore, the pool of essential genes was subjected to NCBI-BLASTp (E-value = 0.0001, bit score ≥100, identity ≥ 35%) against (human, equine, bovine and ovine proteomes) to identify essential non-host homologs targets [12]. The set of essential non-host homologous proteins were further crosschecked with the NCBI-BLASTp PDB database using default parameters to find any structural similarity with the available host homologs protein structures, keeping cutoff level to ≤ 15% for query coverage. These proteins were checked for their biochemical pathway using KEGG (Kyoto Encyclopedia of Genes and Genomes) [38], virulence using PAIDB (Pathogenicity island database) [39], functionality using UniProt (Universal Protein Resource) [40], and cellular localization using CELLO (subCELlular LOcalization predictor) [41]. The final list of targets was based on 12 criteria as described previously [20].
Analyses of essential and host homologous (EH) proteins
We have extrapolated our analyses and also considered protein targets that were predicted as essential to bacterial survival but showed homology to host proteins. This was based on the possibility to find differences between bacterial and host proteins to rationally design inhibitors. The pool of essential protein targets that showed cut off values equal or higher than those for essential non-host homologs through NCBI-BLASTp was treated as host homologous proteins. These were also analyzed for pathway involvement, virulence, functional annotation and cellular localization like essential non-host homologous proteins. To verify the presence of significant residue differences in druggable protein cavities, a structural comparison was performed for each pathogen and their corresponding host protein through the molecular visualization program PyMOL (v1.5, Schrodinger, LLC) (http://www.pymol.org). The related published data of each template structure for each host homolog was also crosschecked for information about these residues, based on the PDB code of each template structure as input in the PDBelite server [42]. Catalytic Site Atlas (CSA) was also consulted to get robust information of the active site residues for the druggable enzyme targets [43]. CSA is a database documenting enzyme active sites and catalytic residues in enzymes of 3D structure and has 2 types of entry, original hand-annotated entries with literature references and homologous entries, found by PSI-BLAST alignment to an individual original entry, using an e-value cut-off of 0.00005. CSA can be accessed via a 4-letter PDB code. The equivalent residue that aligns in the query sequence to the catalytic residue found in the original entry is documented. Though the DoGSiteScorer predicts the druggable protein cavities, the host homologous proteins were further subjected to CASTp (Computed Atlas of Surface Topography of Proteins) [44], Pocket-Finder and Q-SiteFinder [45] to get more reliable and robust results about the druggable cavities of the target proteins.
Prediction of druggable pockets
3D structure information and druggability analyses are important factors for prioritizing and validating putative pathogen targets [46,47]. As aforementioned, for druggability analyses, the final list of essential non-host and host homologous protein targets in PDB format, were subjected to DoGSiteScorer [48], an automated pocket detection and analysis tool for calculating the druggability of protein cavities. For each cavity detected the program returns the residues present in the pocket and a druggable score ranging from 0 to 1. The closer to 1 the obtained values are, the more druggable the protein cavity is predicted to be, i.e. the cavities are predicted to be more likely to bind ligands with high affinity [48]. The DoGSiteScorer also calculates volume, surface area, lipophilic surface, depth and other related parameters for each predicted cavity.
Virtual screening and docking analyses
The ligand library was obtained from the ZINC database, containing 11,193 drug-like molecules, with Tanimoto cutoff level of 60% [49]. Proteins were inspected for structural errors such as missing atoms or erroneous bonds and protonation states in MVD (Molegro Virtual Docker) [50]. The cavities predicted with DogSiteScorer (druggability ≥ 0.80) for all protein targets, were compared with the cavities detected by MVD. The most druggable cavity, according to DogSiteScorer, was subjected to virtual screening. MVD includes three search algorithms for molecular docking namely MolDock Optimizer [50], MolDock Simplex Evolution (SE), and Iterated Simplex (IS). In this work the MolDock Optimizer search algorithm, which is based on a differential evolutionary algorithm, was employed. The default parameters used for the guided differential evolution algorithm are a) population size = 50, b) crossover rate = 0.9, and c) scaling factor = 0.5. The top ranked 200 compounds for each protein were analyzed in Chimera for shape complementarity and hydrogen bond interactions, leading to the selection of a final set of 10 compounds for each target protein.
Results and discussion
Modelome and common targets in C. pseudotuberculosis species
Here we report the identification of common putative targets among 15 strains of C. pseudotuberculosis species based on the construction of genome scale protein three-dimensional structural models. Structural information of target proteins can aid in drug and/or vaccine design and in the discovery of new lead compounds [51]. The approach employed here generated high-confidence structural models through the MHOLline workflow (Figure 1) from orthologous protein. To identify the common conserved proteins with a sequence similarity of 95-100%, a comparative genomics approach was performed where all the BATS classified G2 sequences from "Very High" to "Medium to Good" quality, from 14 Cp strains, were aligned to the G2 sequences of Cp1002, assumed as a reference genome for this study. In total, a set of 331 protein sequences was selected, being conserved in all strains. An overview of the different steps involved in this computational approach for genome scale modelome and prioritization of putative drug and vaccine targets is given in Figure 2a-b.
Figure 1.
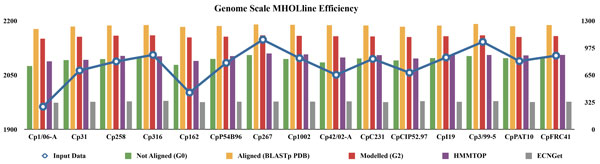
High-throughputness (efficiency) of the MHOLline biological workflow for genome-scale modelome (3D models) prediction. Predicted proteomes from the genomes of 15 C. pseudotuberculosis strains were fed to the MHOLline workflow in FASTA format. The blue line represents the number of input data, according to the left-hand side y-axis. The bars show the number in the form of MHOLline output data (according to the right-hand side y-axis) of: not aligned sequences (G0, green bars); sequences for which there is a template structure available at RCSB PDB (yellow bars); sequences with acceptable template structures that where modeled in the MHOLline workflow (G2, red bars); sequences with predicted transmembrane regions (HMMTOP, purple bars) and the number of sequences that were predicted as enzymes in each genome and were assigned an EC number (ECNGet, gray bars). The x-axis represents the C. pseudotuberculosis genomes used in this study.
Figure 2.
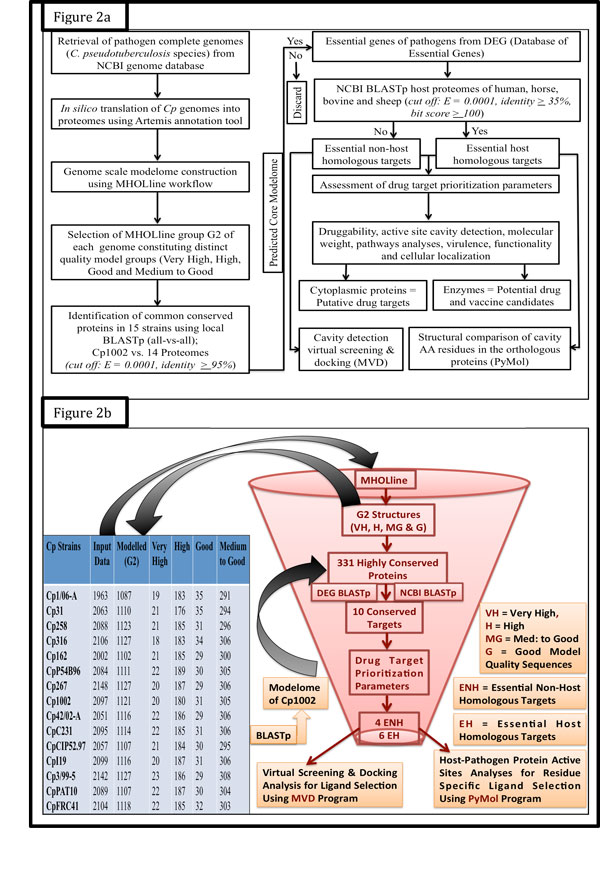
Overview of different computational steps employed in the identification of putative essential targets (non-host homologous and host homologous) for drugs and vaccines from the core-proteome of 15 C. pseudotuberculosis strains. Figure 2b. Intra-species subtractive modelomics workflow for conserved targets identification in C. pseudo tuberculosis species. The table (from left to right) represents the total number of protein sequences as an input data in fasta format fed to the MHOLline workflow (upper forward arrow). The remaining columns show the output data of group G2 (upper backward arrow), first by BATS and then by Filter tools of the MHOLline workflow respectively. Columns 4th-7th constitute the number of protein sequences of different qualities of all 15 Cp strains, where the sequences of 14 Cp strains were compared using BLASTp, to the sequences of Cp1002 strain as reference, for the identification of conserved protein targets (core-modelome). The funnel shows how this workflow processes and filters a large quantity of genomic data for putative drug and vaccine targets identification of a pathogen.
Identification of ENH and EH proteins as putative drug and/or vaccine targets
To identify essential proteins as putative therapeutic targets in C. pseudotuberculosis, from the set of core-modelome, these were compared to the Database of Essential Genes (DEG). Based on this filter, the number of selected targets was reduced drastically to a final set of only 10 targets. These were compared to the aforementioned corresponding host proteomes, leading to the identification of 4 essential non-host homologous proteins (ENH, Table 2) and 6 essential host homologous proteins (EH, Table 3).
Table 2.
Drug and/or vaccine targets prioritization parameters and functional annotation of the four essential non-host homologous putative targets.
| Gene and protein codes | Official full name | Number of cavities with Drug Scorea > 0.80 |
Number of cavities with Drug Scorea > 0.60 and < 0.80 |
Mol. Wt (KDa)b |
Functionsc | Cellular componentd | Pathwayse | Virulencef |
|---|---|---|---|---|---|---|---|---|
| Cp1002_0515 MtrA |
DNA-binding response regulator mtrA | 1 | 2 | 25.97 | MF: DNA binding, two-component response regulator activity. BP: Intracellular signal transduction, regulation of transcription, DNA-dependent | Intracellular/ Cytoplasm |
Two-component signaling systems | Yes |
| Cp1002_0742 IspH |
4-hydroxy-3-methylbut-2-enyl diphosphatereductase | 1 | 4 | 36.59 |
MF: Metal ion binding, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase activity, 3 iron, 4 sulfur cluster binding EC: 1.17.1.2 BP: Dimethylallyldiphosphate biosynthetic process, isopentenyldiphosphate biosynthetic process, mevalonate-independent pathway |
Cytoplasm | Inositol phosphate metabolism/ Pentose phosphate pathway/Terpene metabolism | Yes |
| Cp1002_1648 TcsR |
Two-component system transcriptional regulatory protein | 3 | 2 | 21.93 |
MF: Sequence-specific DNA binding, two-component response regulator activity, sequence-specific DNA binding transcription factor activity BP: Intracellular signal transduction, transcription, DNA-dependent |
Intracellular/ Cytoplasm |
Two-component system | Yes |
| Cp1002_1676 Nrdl |
Ribonucleoside-diphosphatereductase alpha chain | 1 | 1 | 88.02 |
MF: ATP binding, ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor BP: DNA replication |
Cytoplasm | Pyrimidine metabolism/ Purine metabolism | Yes |
aDruggability predicted with DoGSiteScorer software. A druggability score above 0.60 is considered to be good, but a score above 0.80 is favored [48].
bMolecular weight was determined using ProtParam tool (http://web.expasy.org/protparam/).
cMolecular function (MF) and biological process (BP) for each target protein was determined using UniProt.
dCellular localization of pathogen targets was performed using CELLO.
eKEGG was used to find the role of these targets in different cellular pathways.
fPAIDB was used to check if the putative targets are involved in pathogen's virulence.
Table 3.
Drug and/or vaccine targets prioritization parameters and functional annotation of the six essential host homologous putative targets.
| Gene and protein codes | Official full name | Number of cavities with Drug Scorea > 0.80 |
Number of cavities with Drug Scorea > 0.60 and < 0.80 |
Mol. Wt (KDa)b |
Functionsc | Cellular componentd | Pathwayse | Virulencef |
|---|---|---|---|---|---|---|---|---|
| Cp1002_0385 Adk |
Adenylate kinase | 1 | 0 | 24.120 |
MF: Kinase, Transferase, ATP binding BP: Nucleotide biosynthesis EC 2.7.4.3 |
Cytoplasm | Purine metabolism; AMP biosynthesis via salvage pathway | Yes |
| Cp1002_0692 GapA |
Glyceraldehyde-3-phosphate dehydrogenase A | 2 | 1 | 51.918 |
MF: Oxidoreductase, NAD binding, NADP binding, BP: glucose metabolic process EC 1.2.1.13 |
Cytoplasm | Glycolysis/Gluconeogenesis | Yes |
| Cp1002_0728 GlyA |
Serine hydroxymethyltransferase | 2 | 1 | 46.187 |
MF: Methyltransferase, Transferase BP: Amino-acid biosynthesis One-carbon metabolism EC 2.1.2.1 |
Cytoplasm | Amino-acid biosynthesis; glycine biosynthesis; One-carbon metabolism; tetrahydrofolate interconversion. | Yes |
| Cp1002_0738 FumC |
Fumaratehydratase class II | 2 | 0 | 49.767 |
MF: Lyase BP: Tricarboxylic acid cycle EC 4.2.1.2 |
Cytoplasm | Carbohydrate metabolism; tricarboxylic acid cycle; (S)-malate from fumarate |
Yes |
| Cp1002_1005 Gnd |
6-phosphogluconate dehydrogenase | 3 | 5 | 53.669 |
MF: Oxidoreductase BP: Pentose shunt EC 1.1.1.44 |
Cytoplasm | Carbohydrate degradation; pentose phosphate pathway; | No |
| Cp1002_1042 AspA |
Aspartate ammonia-lyase | 2 | 4 | 52.277 |
MF: Lyase EC 4.3.1.1 |
Cytoplasm | Alanine, aspartate and glutamate metabolism, Nitrogen metabolism | Yes |
aDruggability predicted with DoGSiteScorer software. A druggability score above 0.60 is usually considered, but a score above 0.80 is favored [48].
bMolecular weight was determined using ProtParam tool (http://web.expasy.org/protparam/).
cMolecular function (MF) and biological process (BP) for each target protein was determined using UniProt.
dCellular localization of pathogen targets was performed using CELLO.
eKEGG was used to find the role of these targets in different cellular pathways.
fPAIDB was used to check if the putative targets are involved in pathogen's virulence.
Among the ENH proteins, two targets were selected from a bacterial unique pathway, the two component signaling system. These targets are tcsR (two-component response regulator) and mtrA (two component sensory transduction transcriptional regulatory protein). While the tcsR is a novel protein target, as it is has not been described so far as a target in any organism, mtrA has been already reported as a target in Mycobacterium [52] and provides multidrug resistance to Mycobacterium avium [53]. Therefore, targeting mtrA in C. pseudotuberculosis may also be effective in controlling the infection of CLA. The remaining ENH protein targets, nrdI and ispH, also participate in biochemical pathways. NrdI (ribonucleoside-diphosphate reductase alpha chain) is a flavodoxin which contains a diferric-tyrosyl radical cofactor and it is involved in nucleotide metabolism in E. coli [54]. It has been reported as a putative target in several pathogens including C. pseudotuberculosis, Corynebacterium diphtheriae and Mycobacterium tuberculosis [20]. The target ispH (4-hydroxy-3-methylbut-2-enyl diphosphate reductase; EC 1.17.1.2) is an essential cytoplasmic enzyme in Escherichia coli [55]. This iron-sulfur protein plays a crucial role in terpene metabolism of various pathogenic bacteria [56,57] and it is a predicted target in Salmonella tyhpimurium [58] and Plasmodium falciparum [59]. It should be noted that according to the cut off threshold for NCBI-BLASTp that we have followed, ispH shows homology only to the human host. So, if human is not considered as a possible host, ispH can also be considered as a common putative target. The roles of these proteins in different metabolic pathways was confirmed from KEGG [38] and METACYC [60] databases.
Prioritization parameters of drug and/or vaccine targets
Previous studies have shown several factors that can aid in determining the suitability of therapeutic targets [46]. The availability of 3D structural information, the main approach of our study, is very helpful in drug development. Other important factors for drug targets include preferred low MW and high druggability. On the other hand, for vaccine targets the information about subcellular localization is important and proteins that contain transmembrane motifs are preferred [36,46,61,62]. We have determined most of these prioritizing properties for the 10 essential proteins (Table 2 &3). Interestingly, according to the target-prioritizing criterion, all targets have a low MW, and are predicted to be localized in the cytoplasmic compartment of the Cp. Druggability evaluation with DoGSiteScorer [48] for all conserved targets allowed the prediction of numerous druggable cavities with at least one druggable cavity for each Cp target. For the 4 ENH proteins tcsR, mtrA, nrdI, and ispH, 3, 5, 5 and 2 cavities with score ≥ 0.80 were observed respectively. For each protein, the cavity that exhibited the highest druggability score was selected for docking analyses. For 6 EH targets, adk, gapA, glyA, fumC, gnd, and aspA, 1, 3, 3, 2, 8 and 6 cavities were observed respectively according to the aforementioned druggability score criteria (Table 2 &3). Here, in each case, the most druggable predicted cavity was structurally compared with the cavities in respective host proteins.
Virtual screening and molecular docking analyses of ENH targets
For each ENH target protein (mtrA, ispH, tcsR and nrdl), the top 200 drug-like molecules from virtual screening were visually inspected to select 10 molecules that showed favorable interactions with the target. The biological importance of each target and an analysis of the predicted protein-ligand interaction are described below. ZINC codes and MolDock scores of selected ligands, the number of hydrogen bonds as well as protein residues involved in these interactions, are shown in a table for each target protein (Tables 4, 5, 6, 7. Figures showing the predicted binding mode for one of the 10 selected ligands are also shown for each target (Additional files 1, 2, 3, 4, 5).
Table 4.
ZINC codes, MolDock scores and predicted hydrogen bonds for the ten compounds selected among the top ranking 200 molecules against Cp1002_0515 (MtrA, DNA-binding response regulator).
| ZINC IDs | MolDock score | Number of H-bonds/ residues interacting with the compound |
|---|---|---|
| 75109074 | -130.402 | 3 Thr73, Asp48, Arg116 |
| 12117405 | -115.838 | 3 Arg119, Arg118, Ala115 |
| 02546720 | -113.761 | 3 Thr73, Arg119 |
| 40266587 | -116.119 | 2 Asp48, Leu117 |
| 71405274 | -113.264 | 2 Arg116, Asp97 |
| 05687366 | -111.376 | 2 Arg119, Asp48 |
| 04730243 | -109.609 | 2 Arg119, Asp157 |
| 19720976 | -109.061 | 2 Arg119 |
| 72342680 | -108.299 | 2 Arg119, Asp157 |
Table 5.
ZINC codes, MolDock scores and predicted hydrogen bonds for the ten compounds selected among the top ranking 200 molecules against Cp1002_0742 (IspH, 4-hydroxy-3-methyl but-2-enyl diphosphate reductase).
| ZINC IDs | MolDock score | Number of H-bonds/ residues interacting with the compound |
|---|---|---|
| 00510419 | -151.376 | 7 Cys39, His68, Thr225, Ser250, Asn252 |
| 00529019 | -129.348 | 5 His68, Ser250, Asn252, Thr193, Thr193 |
| 04344036 | -135.156 | 8 Thr193, His151, His68, Ser251, Asn252, Ser250, Asn252 |
| 04632419 | -136.984 | 6 Cys39, Gly41, Ala100, Cys222, Thr193, Asn252 |
| 04730243 | -129.414 | 10 Cys222, Thr193, Asn252, His151, Ser250, His68, Ser250 |
| 05479451 | -129.963 | 9 Asn252, Ser250, Ser251, His68, Cys123, Cys39, Gly41 |
| 05775454 | -161.806 | 3 Asn252, Thr193, His68 |
| 16941408 | -126.163 | 6 Thr193, Asn252, Asn252, Ser250 |
| 04622741 | -127.816 | 12 Cys39, Cys123, His68, Cys222, Ser250, Ser251, His151, Thr193 |
| 14017317 | -129.664 | 8 Cys39, Glu153, His68, Asn252, Ser251, Asn252, His151, Thr193 |
Table 6.
ZINC codes, MolDock scores and predicted hydrogen bonds for the ten compounds selected among the top ranking 200 molecules against Cp1002_1648 (TcsR, Two component transcriptional regulator).
| ZINC IDs | MolDock score | Number of H-bonds/ residues interacting with the compound |
|---|---|---|
| 00510419 | -167.633 | 3 Val76, Gln185, Asn193 |
| 01617096 | -146.178 | 3 Ala74, Gln185, Arg191 |
| 32911447 | -148.424 | 3 Gln185, Ala70, Arg193 |
| 00091802 | -143.287 | 3 Val76, Ala51 |
| 67847806 | -156.655 | 4 Thr75, Thr75, Pro48, Val76 |
| 19399766 | -160.743 | 3 Val76, Val76, Ala51 |
| 16980834 | -147.631 | 4 Ala66, Val76 |
| 06269029 | -145.277 | 4 Thr75, Pro48, Val76 |
| 05934077 | -145.785 | 3 Arg191, Gln185 |
| 01647971 | -167.152 | 3 Thr75, Val76, Ala51 |
Table 7.
ZINC codes, MolDock scores and predicted hydrogen bonds for the ten compounds selected among the top ranking 200 molecules against Cp1002_1676 (NrdI).
| ZINC IDs | MolDock score | Number of H-bonds/ residues interacting with the compound |
|---|---|---|
| 01585114 | -151.406 | 6 Ser8, Ser7, Thr13, Asn12 |
| 04721321 | -144.134 | 7 Ser8, Ser7, Thr13, Leu116 |
| 17023683 | -140.718 | 6 Ser7, Ser8, Thr13, Thr13, Thr13 |
| 00510419 | -154.064 | 4 Thr10, Thr13, Ser8 |
| 01417445 | -138.997 | 4 Thr13, Tyr49, Ser8, Ser7 |
| 00042420 | -135.363 | 6 Tyr49, Ser7, Ser8, Thr13, Thr13, Thr13 |
| 00408361 | -133.535 | 6 Thr13, Ser7, Ser8, Tyr49, Thr48 |
| 15830653 | -153.83 | 4 Ser7, Thr13, Tyr49 |
| 00032839 | -139.327 | 6 Ser8, Ser7, Thr13, Thr13, Thr13 |
| 48212336 | -137.675 | 6 Ser7, Ser8, Ser8, Thr13, Ser54 |
Cp1002_0515 (MtrA, DNA-binding response regulator) is part of the two-component signal transduction system consisting of the sensor kinase (Histidine protein kinases, HKs) and the response regulator, MtrB and MtrA respectively. This system is highly conserved in Corynebacteria and Mycobacteria and it is essential for their survival to adapt to environmental changes. Homologs of MtrA and MtrB are present in many species of the genera Corynebacterium, Mycobacterium, Nocardia, Rhodococcus (CMNR), and others like Thermomonospora, Leifsonia, Streptomyces, Propionibacterium, and Bifidobacterium [63]. MtrA represents the fourth family member of the OmpR/PhoB family of response regulators. Like other family members, MtrA has been reported to be essential in M. tuberculosis [64]. It possesses an N-terminal regulatory domain and a C-terminal helix-turn-helix DNA-binding domain, already indicating that this response regulator functions as a transcriptional regulator, with phosphorylation of the regulatory domain modulating the activity of the protein [65]. Based on a comparison with a crystallographic structure of the MtrA template (2GWR, MtrA from M. tuberculosis), the active site residues involved in H-bond interactions with the crystallographic ligand are Val145, Gln151, Ile152 and Leu154. Although none of these residues is predicted to form hydrogen bonds with the ten selected docked ligands, these molecules were predicted to interact with other residues in the pocket. Table 4 shows the 10 selected ligands according to their minimum energy values and number of hydrogen bond interactions. ZINC75109074 (N-benzyl-N-[[2-(2-thienyl)-1H-imidazol-4-yl] methyl] prop-2-en-1-amine) is shown here as the top scoring ligand (Additional file 1).
Cp1002_0742 (IspH, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase) is an iron-sulfur oxidoreductase enzyme that plays a key role in the metabolism of terpenes in several pathogens. Terpenes constitute a large class of natural compounds. Their biosynthesis initiates with the building blocks isopentenyl-diphosphate (IPP) and dimethylallyldiphosphate (DMAPP), and differs in bacteria and mammals [57]. In bacteria and other pathogenic microorganisms the enzyme IspH catalyzes the last step in the production of IPP and DMAPP. The three structural units of the enzyme harbor a cubic iron-sulfur cluster at their center, enabling the enzyme to accomplish a challenging reaction by converting an allyl alcohol to two isoprene components. The iron-sulfur proteins normally participate in electron transfers. The IspH enzyme, thereby, in a similar fashion, binds the substrate directly to the iron-sulfur cluster [57]. In the template crystal structure of IspH (PDB 3KE8), it has been shown that His41, His74, His124, Thr167, Ser225, Ser226, Asn227 and Ser269 are the active site residues that are involved in hydrogen bond interactions with the ligand 4-hydroxy-3-methylbutyldiphosphate (EIP). Also, Cys12, Cys96, Cys197 and EIP have been shown to make metal interaction with the Fe4S4 (Iron/Sulfur Cluster). Although the ten selected drug-like compounds (Table 5) did not show any interaction with the aforementioned IspH residues, they are predicted to make very good hydrogen bond interactions with other surrounding residues of the predicted cavity. The predicted binding mode of the best scoring compound, ZINC00510419 is shown in Additional file 2. Good shape complementarity and 6 hydrogen bond interactions are observed in this complex.
Cp1002_1648 (TcsR, Two component transcriptional regulator) is a novel target without host homologs proteins. Differently from MtrA and IspH, in this case the template structure from Escherichia coli for TcsR did not contain any ligand (PDB 1A04), and no reported information was found about the ligand-residues interactions in their cavities. Therefore, among the cavities identified by MVD, the best cavity for virtual screening analysis was simply chosen based on the highest druggability score by the DogSiteScorer. Compound ZINC00510419 (Additional file 3) was the top-ranking compound, forming a network of 3 hydrogen bonds with Val76, Gln185 and Asn193. Table 6 lists the 10 compounds selected for this target.
Cp1002_1676 (NrdI, protein) belongs to the nrdI protein family, a unique group of metalloenzymes that are essential for cell-proliferation [66]. It is classified as a ribonucleotide reductase (RNR), an iron-dependent enzyme that belongs to class Oxidoreductases (EC 1.17.4.1) acting on CH or CH2 groups with a disulfide as acceptor [67]. The class Ia enzyme supplies deoxynucleotides during normal aerobic growth. The class Ib RNR plays a similar role although its function in E. coli is not clear, but it is reported to be expressed under oxidative stress and iron-limited conditions [68]. Class I RNR enzymes have two homodimeric subunits, α2 (NrdE), where nucleotide reduction takes place, and β2 (NrdF) containing an unidentified metallocofactor for initiating nucleotide reduction in α2. Although the exact function of NrdI within RNR has not yet been fully characterized, it is found in the same operon as NrdE and NrdF, and encodes an unusual flavodoxin, a bacterial electron-transfer protein that includes a flavin mononucleotide that has been proposed to be involved in metallocofactor biosynthesis and/or maintenance. It has also been proposed that NrdI plays an important role in E. coli class Ib RNR cluster assembly. Recent in vitro studies have shown that a stable diferric-tyrosyl radical (FeIII2-Y·) and dimanganese (III)-Y· (MnIII2-Y·) cofactors are active in nucleotide reduction [69]. The first one can be formed by self-assembly from FeII and O2 while the later cofactor can be generated from MnII-2-NrdF, but only in the presence of O2 and NrdI protein [54,69]. RNR is responsible for the de novo conversion of ribonucleoside diphosphates into deoxyribonucleoside diphosphates and it is essential for DNA synthesis and repair [70]. The active site residues of RNR, in the template structure of NrdI protein (PDB 3N3A), include Ser8, Ser9, Ser11, Ser48, Asn13, Asn83, Thr14, Tyr49, Ala89 and Gly91, all of which are involved in a hydrogen bond network with the cofactor flavin mononucleotide isoalloxazine ring (FMN, PDB 3N3A) [71]. Interestingly, two of these residues, Ser8 and Tyr49, were predicted to make hydrogen bonds with all 10 selected ligands (Table 7). The interaction between the top scoring compound ZINC01585114 (5-nitro-3, 4-diphenyl-2-furamide) and the residues from the predicted target cavities are shown in Additional file 4.
Furthermore, the drug-like molecule ZINC00510419 (3,4-bis (5-methylisoxazole-3-carbonyl)-1,2,5-oxadiazole 2-oxide was among the top ten selected molecules for three of the pathogen target proteins, showing good H-bond interactions. It ranked first against the targets Cp1002_0742 (MolDock score = -151.376, no. of H-bonds = 7) and Cp1002_1648 (MolDock score = -167.633, no. of H-bonds = 3) and ranked fourth against the target Cp1002_1676 (MolDock score = -154.064, no. of H-bonds = 4).
Essential host homologous as putative targets
To compare the predicted EH protein targets to their host homologs, two approaches were taken. First, ClustalX (v2.1, http://www.clustal.org), a multiple sequence alignment program, was used to find different residues between bacterial and host proteins. As expected, a high percentage of residues was found to be conserved, but significant differences were also observed. Most percentage identities are between 35 and 50 (Table 8), except for fumarate hydratase, which shows 54% sequence identity to human and equine homologous proteins, but no hits in bovine and ovine proteomes.
Table 8.
Percentage of sequence identity between C. pseudotuberculosis and host homologous proteins.
| Protein Locus tag | Official full name | Percentage of Sequence Identity# | |||
|---|---|---|---|---|---|
| HS* | EC* | BT* | OA* | ||
| Cp1002_0385 Adk | Adenylate kinase | 38 | 36 | 35 | 35 |
| Cp1002_0692 GapA | Glyceraldehyde-3-phosphate dehydrogenase A | 39 | 40 | 41 | 41 |
| Cp1002_0728 GlyA | Serine hydroxymethyltransferase | 43 | 45 | 45 | 45 |
| Cp1002_0738 FumC | Fumaratehydratase class II | 54 | 54 | No Hits | No Hits |
| Cp1002_1005 Gnd | 6-phosphogluconate dehydrogenase | 48 | 48 | 48 | 48 |
| Cp1002_1042 AspA | Aspartate ammonia-lyase | 39 | 39 | 39 | 39 |
Next, to determine if the observed differences could be exploited in rational design of ligands selective to bacterial proteins, we focused on the predicted druggable cavities. A structural alignment to the host homologous proteins was performed and the cavities were compared in PyMol. In most cases, the DogSiteScorer predicted more than one cavity for each input Cp protein structure. The number of residues in the bacterial predicted cavity that differ from the residues in the cavity of the host protein, for all druggable pockets, varied from zero to seven (Table 9).
Table 9.
Comparison of the residues from druggable cavities in C. pseudotuberculosis proteins and the corresponding residues in structurally aligned host protein cavities.
| Protein Loci | Bacterial Residues for the Most Druggable Cavity Predicted by DGSS Server# | HS* | EC* | BT* | OA* |
|---|---|---|---|---|---|
|
Cp1002_0692 (Glyceralderayde 3-phosphate dehydrogenase) |
Lys157 | Asp35 | Asp33 | Asp33 | Asp33 |
| Val174 | Thr52 | Thr50 | Thr50 | Thr50 | |
| Arg229 | Thr103 | Thr101 | Thr101 | Thr101 | |
| Asn311 | Ala183 | Ala181 | Ala181 | Ala181 | |
|
Cp1002_0385 (Adenylate kinase) |
Phe35 | Leu50 | Leu52 | Leu52 | Leu43 |
| Ile53 | Met68 | Met70 | Met70 | Met61 | |
| Thr64 | Val79 | Val81 | Val81 | Val72 | |
|
Cp1002_0728 (Serine hydroxymethyltransferase) |
Cys70 | Ala88 | Thr86 | Thr86 | Thr86 |
| Ala99 | Ser121 | Ser119 | Ser119 | Ser119 | |
| Ala101 | Ser123 | Ser121 | Ser121 | Ser121 | |
| Trp177 | Thr204 | Thr202 | Thr202 | Thr202 | |
| Pro361 | Ala397 | Ala395 | Ala395 | Ala395 | |
|
Cp1002_1005 (6-phosphogluconate dehydrogenase) |
Ser55 | Thr35 | Thr161 | Thr35 | Thr35 |
| Met94 | Leu74 | Leu200 | Leu74 | Leu74 | |
| Gln96 | Lys76 | Lys202 | Lys76 | Lys76 | |
| Val104 | Phe84 | Phe210 | Phe84 | Phe84 | |
| Ile148 | Val128 | Val254 | Val128 | Val128 | |
| Gln268 | Lys248 | Lys374 | Lys248 | Lys248 | |
| Pro269 | His249 | Tyr375 | His249 | His249 | |
|
Cp1002_1042 (Aspartate ammonia-lyase) |
Gln193 | His235 | His257 | His235 | His235 |
| Ile428 | Lys470 | Lys492 | Lys470 | Lys470 | |
| His447 | Leu489 | Leu511 | Leu489 | Leu489 | |
#Drug score ≥ 0.80
*HS = Homo sapiens, EC = Equus caballus, BT = Bos taurus, OA = Ovis aries
For conserved host-homologous targets Cp1002_0385 (adk, Adenylate kinase), Cp1002_0692 (gapA, Glyceraldehyde 3-phosphate dehydrogenase), Cp1002_0728 (glyA, Serine hydroxymethyltransferase), Cp1002_0738 (fumC, Fumarate hydratase class II/fumarase), Cp1002_1005 (gnd, 6-Phosphogluconate dehydrogenase) and Cp1002_1042 (aspA, Aspartate ammonia-lyase/aspartase), three, four, five, zero, seven and three different residues were observed, respectively. Then, a more detailed analysis was performed for the predicted highest druggable cavity for each protein. The results are described below, together with information about the biological importance of each target protein.
Cp1002_0692 (GapA, Glyceraldehyde 3-phosphate dehydrogenase, GAPDH/G3PDH, EC 1.2.1.12) catalyzes the sixth step of glycolysis. In addition, GAPDH has recently been shown to be involved in several non-metabolic processes, including transcription activation, initiation of apoptosis [72] fast axonal or axoplasmic transport and endoplasmic reticulum to Golgi vesicle shuttling [73,74]. This enzyme has been reported as an anti-trypanosomatid and anti-leishmania drug target in structure-based drug design efforts [21-23]. Furthermore, it has been shown as an interesting putative drug and vaccine target in malaria pathogenesis [75]. Comparison of protein cavities reveals significant differences between bacterial and host proteins, with replacement of bacterial Lys157, Arg229 and Asn311 by Asp, Thr and Ala, respectively. Such differences result in a more basic cavity in bacteria, making it possible to rationally design selective ligands, especially negatively charged molecules, which interact with Lys157 and Arg229, or compounds able to form hydrogen bond to Asn311 (Additional file 5a).
Nucleoside monophosphate kinases vitally participate in sustaining the intracellular nucleotide pools in all living organisms. Cp1002_0385 (Adk, Adenylate kinase, EC 2.7.4.3) is a ubiquitous enzyme, which catalyzes the reversible Mg2+-dependent transfer of the terminal phosphate group from ATP to AMP, releasing two molecules of ADP [76]. Only one highly druggable cavity was predicted for adenylate kinase, with a druggability score = 0.81. Three residues in the bacteria cavity were different from the hosts: Leu, Met and Val in the hosts replaced Phe35, Ile53 and Thr64, respectively (Additional file 5b). These differences impact the cavity volume, since aromatic and bulky Phe is replaced by Leu, and the ability to make hydrogen bonds, through the replacement of a Thr by a Val. Therefore; the bacterial cavity is smaller and more hydrophilic, making it possible to envision rational design of selective ligands that interact with Thr64.
Cp1002_0728 (GlyA, Serine hydroxymethyltransferase EC 2.1.2.1) is an enzyme that plays an important role in cellular one-carbon pathways by catalyzing the reversible, simultaneous conversions of L-serine to glycine (retro-aldol cleavage) and tetrahydrofolate to 5,10-methylenetetrahydrofolate [77]. In Plasmodium, serine hydroxymethyltransferase (SHMT) has been reported as an attractive drug target [78]. For this protein 3 residues were observed different between bacteria and host: Ala99 and Ala101 replaced two Ser residues while Trp177 replaced Thr (Additional file 5c). At first glance these changes could have a big impact in the active site, generating a considerably more hydrophilic pocket in the hosts. However, careful inspection of the pocket reveals that the side chains of these residues are not turned towards the pocket, in such a way that these differences probably would not allow rational design of selective ligands.
Cp1002_0738 (FumC, Fumaratehydratase class II/fumarase EC 4.2.1.2) catalyzes the reversible hydration/dehydration of fumarate to S-malate during the ubiquitous Krebs cycle, through the aci-carboxylate intermediate subsequent to olefin production [79]. There are two classes of fumarases; Class I fumarases, composed of heat-labile, iron-sulfur (4Fe-4S) homodimeric enzymes, only found in prokaryotes; and Class II fumarases, made of thermostable homotetrameric enzymes [80] found in both prokaryotic and eukaryotic mitochondria. Class II belongs to a superfamily that also includes aspartate-ammonia lyases, arginino-succinatases, d-crystallins and 3-carboxy-cis, cis-muconate lactonizing enzymes. All these enzymes release fumarate from different substrates, ranging from adenylosuccinate to malate [81-84]. FumC of Escherichia coli is the first member of class II fumarases family whose structure has been solved and provided most of the structural information [85]. Inhibition of fumarase in the tricarboxylic acid cycle (TCA) has been reported as a potential molecular target of bismuth drugs in Helicobacter pylori [86]. Comparison of the active site cavity of this protein, which is formed in the interface of three monomers, revealed no differences between bacteria and hosts (additional file 5d).
Cp1002_1005 (Gnd, 6-Phosphogluconate dehydrogenase EC 1.1.1.44) is an enzyme from the pentose phosphate pathway. It forms ribulose 5-phosphate from 6-phosphogluconate. The enzyme 6-phosphogluconate dehydrogenase is a potential drug target for the parasitic protozoan Trypanosoma brucei, the causative organism of human African trypanosomiasis [87]. Three druggable sites with score > 0.80 were detected in this protein. As opposed to the observation for other proteins, the most druggable predicted cavity (score = 0.88) was not the active site. Leu, Lys and Val residues in the hosts replace residues Met94, Gln96 and Ile148 in the bacterial cavity, respectively (Additional file 5e). The most significant of these differences is the replacement of Gln by Lys, which could make binding of negative molecules more favorable to the host proteins.
Cp1002_1042 (AspA, Aspartate ammonia-lyase/aspartase EC 4.3.1.1) catalyzes the deamination of aspartic acid to form fumarate and ammonia [88]. Recent progresses to prepare enantiopure l-aspartic acid derivatives, highly valuable tools for biological research and chiral building blocks for pharmaceuticals and food additives, make it a target of interest for industrial applications. On the other hand, the important role that it plays in microbial nitrogen metabolism makes it a putative drug target in overcoming bacterial pathogenesis [89]. Based on the sequence alignment for this protein, two significant differences in residues are observed in the most druggable pocket: bacterial His447 and Ile428 are replaced by Leu and Lys in host proteins. Such differences should allow rational ligand design. It is interesting to note that additional differences in the position of helices that contain these residues increase the difference between the active sites (Additional file 5f).
Based on the above-mentioned analyses, we conclude that it would be difficult to rationally design selective ligands for Cp1002_0738 (FumC, Fumaratehydratase class II), since no residue differences were observed in the most druggable cavity, and for Cp1002_0728 (GlyA, Serine hydroxymethyltransferase), where the side chains of differing residues are not turned toward the druggable pocket. On the other hand, for putative essential and homologous targets that include Cp1002_0692 (GapA, Glyceraldehyde 3-phosphate dehydrogenase), Cp1002_0385 (Adk, Adenylate kinase), Cp1002_1005 (Gnd, 6-Phosphogluconate dehydrogenase) and Cp1002_1042 (AspA, Aspartate ammonia-lyase), significant differences were observed in druggable pockets, suggesting that despite the existence of a host homologous protein they could be good targets for the design of ligands, selective only to the bacterial proteins.
Conclusion
Here, for the first time, the genomic information was used to determine the conserved predicted proteome of 15 strains of C. pseudotuberculosis, along with their three-dimensional structural information. Even though the structural information discussed is fully computationally predicted, and could therefore deviate from eventually solved experimental structures, we have been careful to concentrate on the analysis of protein models for which there were good templates which provided high quality models, minimizing this concern. The data presented here can effectively contribute in guiding further research for antibiotics and vaccines development. The final dataset can provide valuable information in designing molecular biology and immunization experiments in animal models for validating the targets of a pathogen, as well as in experimental structure determination protocols.
The criterion for target selection in C. pseudotuberculosis was stringent, resulting in a small set of prioritized putative drug and vaccine targets, of which four are essential and non-homologous and six are essential and host homologous proteins. For the latter, a detailed structural comparison between the residues of the predicted cavities of host and pathogen proteins has been performed, showing in most cases the potential for the development of selective ligands. Therefore, we suggest that the whole set can be considered for antimicrobial chemotherapy, especially the four essential non-host homologous targets.
The in silico approaches followed in this study might aid in the development of novel therapeutic drugs and vaccines in a broad-spectrum of hosts at intraspecies level against C. pseudotuberculosis. Furthermore, the strategy described here could also be applied to other pathogenic microorganisms.
Authors' contributions
Coordinated entire work: SSH RSF VA DB. Performed all in silico analyses: SSH RSF ST SBJ NBS FDP LCG. Cross-analyzed genome contents, pan-modelome construction, conserved pan-modelome, subtractive modelome approach, virtual screening & docking analyses and residue level structural comparison: SSH RSF ST FDP AI SCS SA DB AGT. Provided timely consultation and reviewed the manuscript: VA AI SCS SA DB NBS LCG AA AM AS VACA AGT. Read and approved the final manuscript: RSF SSH ST AI SCS SBJ SA DB NBS LCG AGTAA AM AS VA. Conceived and designed the work: SSH RSF VA DB. Analyzed the data: SSH RSF ST AI SCS SBJ SA DB NBS LCG AA AB LJ AGTAM AS VA. Wrote the paper: SSH RSF ST.
Conflict of interest
The authors declare that they have no competing interests.
Supplementary Material
Docking representation of the best drug-like compound ZINC75109074 in the most druggable protein cavity of Cp1002_0515 (MtrA, DNA-binding response regulator). Three hydrogen bonds were observed with Thr73, Asp48 and Arg116.
Docking representation of compound ZINC00510419 in the most druggable protein cavity of Cp1002_0742 (IspH, 4-hydroxy-3-methyl but-2-enyl diphosphate reductase). Residues Cys39, Thr225, Ser250, His68 and Asn252 are predicted to make seven hydrogen bonds to this ligand.
Docking representation of the best drug-like compound ZINC00510419 in the most druggable protein cavity of Cp1002_1648 (TcsR, Two component transcriptional regulator). Hydrogen bonds were observed with residues Val76, Gln185 and Asn193.
Docking representation of the best drug-like compound ZINC04721321 in the most druggable protein cavity of Cp1002_1676 (NrdI protein). Hydrogen bonds were observed with residues Ser8, Thr13 and Leu116.
Comparison among the most druggable cavities from essential bacterial and the respective host homologue proteins. Protein structures are shown as cartoon (green for the bacterial protein and gray for Ovis aries host protein). Other host proteins are not shown for simplicity, but the same substitutions were present in all host proteins analyzed. Residues that differ in the bacterial and host cavity are highlighted in sticks and labeled (bacterial labels in green and host labels in black). a) Cp1002_0692 (Glyceralderayde 3-phosphate dehydrogenase); b) Cp1002_0385 (adenylate kinase); c) Cp1002_0728 (serine hydroxymethyltransferase); d) Cp1002_0738 (fumarate hydratase class II) the site shown is formed by three monomers, which are represented in green, blue and orange. No residues are highlighted, since the active sites are identical between bacteria and host; e) Cp1002_1005 (6-phosphogluconate dehydrogenase); f) Cp1002_1042 (aspartate ammonia-lyase). Figures were prepared with the PyMol.
Contributor Information
Syed Shah Hassan, Email: hassan_chemist@yahoo.com.
Sandeep Tiwari, Email: sandip_sbtbi@yahoo.com.
Luís Carlos Guimarães, Email: luisguimaraes.bio@gmail.com.
Syed Babar Jamal, Email: syedbabar.jamal@gmail.com.
Neha Barve Sharma, Email: nehabarve2006@gmail.com.
Siomar de Castro Soares, Email: siomars@gmail.com.
Síntia Almeida, Email: sintiaalmeida@gmail.com.
Amjad Ali, Email: amjad_uni@yahoo.com.
Arshad Islam, Email: arshad.cgl@gmail.com.
Fabiana Dias Póvoa, Email: fabianapovoa@gmail.com.
Vinicius Augusto Carvalho de Abreu, Email: vini.abreu@gmail.com.
Neha Jain, Email: mymailtoneha@gmail.com.
Antaripa Bhattacharya, Email: antaripa1210@gmail.com.
Lucky Juneja, Email: ljpreet88@gmail.com.
Anderson Miyoshi, Email: miyoshi@icb.ufmg.br.
Artur Silva, Email: asilva@ufpa.br.
Debmalya Barh, Email: dr.barh@gmail.com.
Adrian Gustavo Turjanski, Email: adrian@qi.fcen.uba.ar.
Vasco Azevedo, Email: vasco@icb.ufmg.br.
Rafaela Salgado Ferreira, Email: rafaelasf@gmail.com.
Acknowledgements
We acknowledge financial support from the funding agencies CNPq, CAPES and FAPEMIG. Hassan S.S acknowledges the receipt of fellowship under "TWAS-CNPq Postgraduate Fellowship Program" for doctoral studies.
This article has been published as part of BMC Genomics Volume 15 Supplement 7, 2014: Proceedings of the 9th International Conference of the Brazilian Association for Bioinformatics and Computational Biology (X-Meeting 2013). The full contents of the supplement are available online at http://www.biomedcentral.com/bmcgenomics/supplements/15/S7.
References
- Hassan SS, Schneider MP, Ramos RT, Carneiro AR, Ranieri A, Guimaraes LC, Ali A, Bakhtiar SM, Pereira Ude P, dos Santos AR. et al. Whole-genome sequence of Corynebacterium pseudotuberculosis strain Cp162, isolated from camel. Journal of bacteriology. 2012;194(20):5718–5719. doi: 10.1128/JB.01373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorella FA, Pacheco LG, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Veterinary research. 2006;37(2):201–218. doi: 10.1051/vetres:2005056. [DOI] [PubMed] [Google Scholar]
- Soares SC, Trost E, Ramos RT, Carneiro AR, Santos AR, Pinto AC, Barbosa E, Aburjaile F, Ali A, Diniz CA, Genome sequence of Corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. Journal of biotechnology. 2012. [DOI] [PubMed]
- Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. Journal of clinical microbiology. 2005;43(4):1934–1936. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LH. Caseous lymphadenitis in small ruminants. Vet Clin North Am Food Anim Pract. 2001;17(2):359–371. doi: 10.1016/s0749-0720(15)30033-5. vii. [DOI] [PubMed] [Google Scholar]
- Peel MM, Palmer GG, Stacpoole AM, Kerr TG. Human lymphadenitis due to Corynebacterium pseudotuberculosis: report of ten cases from Australia and review. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1997;24(2):185–191. doi: 10.1093/clinids/24.2.185. [DOI] [PubMed] [Google Scholar]
- Luis MA, Lunetta AC. [Alcohol and drugs: preliminary survey of Brazilian nursing research] Revista latino-americana de enfermagem. 2005;13 doi: 10.1590/s0104-11692005000800018. Spec No:1219-1230. [DOI] [PubMed] [Google Scholar]
- Mills AE, Mitchell RD, Lim EK. Corynebacterium pseudotuberculosis is a cause of human necrotising granulomatous lymphadenitis. Pathology. 1997;29(2):231–233. doi: 10.1080/00313029700169944. [DOI] [PubMed] [Google Scholar]
- Augustine JL, Renshaw HW. Survival of Corynebacterium pseudotuberculosis in axenic purulent exudate on common barnyard fomites. American journal of veterinary research. 1986;47(4):713–715. [PubMed] [Google Scholar]
- Yeruham I, Friedman S, Perl S, Elad D, Berkovich Y, Kalgard Y. A herd level analysis of a Corynebacterium pseudotuberculosis outbreak in a dairy cattle herd. Veterinary dermatology. 2004;15(5):315–320. doi: 10.1111/j.1365-3164.2004.00388.x. [DOI] [PubMed] [Google Scholar]
- Perumal D, Lim CS, Sakharkar KR, Sakharkar MK. Differential genome analyses of metabolic enzymes in Pseudomonas aeruginosa for drug target identification. In silico biology. 2007;7(4-5):453–465. [PubMed] [Google Scholar]
- Barh D, Gupta K, Jain N, Khatri G, Leon-Sicairos N, Canizalez-Roman A, Tiwari S, Verma A, Rahangdale S, Shah Hassan S, Conserved host-pathogen PPIs. Integrative biology : quantitative biosciences from nano to macro. 2013. [DOI] [PubMed]
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B. et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- Asif SM, Asad A, Faizan A, Anjali MS, Arvind A, Neelesh K, Hirdesh K, Sanjay K. Dataset of potential targets for Mycobacterium tuberculosis H37Rv through comparative genome analysis. Bioinformation. 2009;4(6):245–248. doi: 10.6026/97320630004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Singh SK, Ghosh P, Mukherjee R, Mitter S, Bandyopadhyay D. In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. In silico biology. 2006;6(1-2):43–47. [PubMed] [Google Scholar]
- Chong CE, Lim BS, Nathan S, Mohamed R. In silico analysis of Burkholderia pseudomallei genome sequence for potential drug targets. In silico biology. 2006;6(4):341–346. [PubMed] [Google Scholar]
- Barh D, Kumar A. In silico identification of candidate drug and vaccine targets from various pathways in Neisseria gonorrhoeae. In silico biology. 2009;9(4):225–231. [PubMed] [Google Scholar]
- Sakharkar KR, Sakharkar MK, Chow VT. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. In silico biology. 2004;4(3):355–360. [PubMed] [Google Scholar]
- Rathi B, Sarangi AN, Trivedi N. Genome subtraction for novel target definition in Salmonella typhi. Bioinformation. 2009;4(4):143–150. doi: 10.6026/97320630004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barh D, Jain N, Tiwari S, Parida BP, D'Afonseca V, Li L, Ali A, Santos AR, Guimaraes LC, de Castro Soares S. et al. A novel comparative genomics analysis for common drug and vaccine targets in Corynebacterium pseudotuberculosis and other CMN group of human pathogens. Chemical biology & drug design. 2011;78(1):73–84. doi: 10.1111/j.1747-0285.2011.01118.x. [DOI] [PubMed] [Google Scholar]
- Aronov AM, Verlinde CL, Hol WG, Gelb MH. Selective tight binding inhibitors of trypanosomal glyceraldehyde-3-phosphate dehydrogenase via structure-based drug design. Journal of medicinal chemistry. 1998;41(24):4790–4799. doi: 10.1021/jm9802620. [DOI] [PubMed] [Google Scholar]
- Singh S, Malik BK, Sharma DK. Molecular modeling and docking analysis of Entamoeba histolytica glyceraldehyde-3 phosphate dehydrogenase, a potential target enzyme for anti-protozoal drug development. Chemical biology & drug design. 2008;71(6):554–562. doi: 10.1111/j.1747-0285.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- Suresh S, Bressi JC, Kennedy KJ, Verlinde CL, Gelb MH, Hol WG. Conformational changes in Leishmania mexicana glyceraldehyde-3-phosphate dehydrogenase induced by designed inhibitors. Journal of molecular biology. 2001;309(2):423–435. doi: 10.1006/jmbi.2001.4588. [DOI] [PubMed] [Google Scholar]
- Adams CP, Brantner VV. Estimating the cost of new drug development: is it really 802 million dollars? Health affairs. 2006;25(2):420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nature reviews Drug discovery. 2004;3(8):711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Congreve M, Murray CW, Blundell TL. Structural biology and drug discovery. Drug discovery today. 2005;10(13):895–907. doi: 10.1016/S1359-6446(05)03484-7. [DOI] [PubMed] [Google Scholar]
- Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- Cavasotto CN, Phatak SS. Homology modeling in drug discovery: current trends and applications. Drug discovery today. 2009;14(13-14):676–683. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Behera DK, Behera PM, Acharya L, Dixit A, Padhi P. In silico biology of H1N1: molecular modelling of novel receptors and docking studies of inhibitors to reveal new insight in flu treatment. Journal of biomedicine & biotechnology. 2012;2012:714623. doi: 10.1155/2012/714623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural RJ. ARTEMIS: a tool for displaying and annotating DNA sequence. Briefings in bioinformatics. 2000;1(2):199–200. doi: 10.1093/bib/1.2.199. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Current protocols in protein science / editorial board, John E Coligan [et al] 2007. Chapter 2:Unit 2 9. [DOI] [PubMed]
- Mount DW. Using the Basic Local Alignment Search Tool (BLAST) CSH protocols. 2007. 2007:pdb top17. [DOI] [PubMed]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17(9):849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Capriles PV, Guimaraes AC, Otto TD, Miranda AB, Dardenne LE, Degrave WM. Structural modelling and comparative analysis of homologous, analogous and specific proteins from Trypanosoma cruzi versus Homo sapiens: putative drug targets for chagas' disease treatment. BMC genomics. 2010;11:610. doi: 10.1186/1471-2164-11-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadio AK, Kioshima ES, Teixeira MM, Martins NF, Maigret B, Felipe MS. Comparative genomics allowed the identification of drug targets against human fungal pathogens. BMC genomics. 2011;12:75. doi: 10.1186/1471-2164-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Ou HY, Zhang CT. DEG: a database of essential genes. Nucleic acids research. 2004;32(Database):D271–272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Park YK, Lee S, Choi D, Oh TK, Hur CG, Kim JF. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic acids research. 2007;35(Database):D395–400. doi: 10.1093/nar/gkl790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database : the journal of biological databases and curation. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein science : a publication of the Protein Society. 2004;13(5):1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, Gore SP. et al. PDBe: Protein Data Bank in Europe. Nucleic acids research. 2012;40(Database):D445–452. doi: 10.1093/nar/gkr998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic acids research. 2004;32(Database):D129–133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic acids research. 2006;34(Web Server):W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21(9):1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- Aguero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AW, Chen F. et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nature reviews Drug discovery. 2008;7(11):900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, Nasrullah I, Tahir S, Tong Y. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PloS one. 2012;7(8):e43080. doi: 10.1371/journal.pone.0043080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkamer A, Kuhn D, Rippmann F, Rarey M. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics. 2012;28(15):2074–2075. doi: 10.1093/bioinformatics/bts310. [DOI] [PubMed] [Google Scholar]
- Voigt JH, Bienfait B, Wang S, Nicklaus MC. Comparison of the NCI open database with seven large chemical structural databases. Journal of chemical information and computer sciences. 2001;41(3):702–712. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. Journal of medicinal chemistry. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nature reviews Drug discovery. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Li Y, Zeng J, He ZG. Characterization of a functional C-terminus of the Mycobacterium tuberculosis MtrA responsible for both DNA binding and interaction with its two-component partner protein, MtrB. Journal of biochemistry. 2010;148(5):549–556. doi: 10.1093/jb/mvq082. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Do JS, Freeman R, Bennett JG, Semret M, Behr MA. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrobial agents and chemotherapy. 2006;50(2):461–468. doi: 10.1128/AAC.50.2.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo JA, Stubbe J. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14383–14388. doi: 10.1073/pnas.0807348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer S, Coulson A, McLennan N, Masters M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. Journal of bacteriology. 2001;183(24):7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl M, Hintz M, Reichenberg A, Kollas AK, Wiesner J, Jomaa H. Microbial isoprenoid biosynthesis and human gammadelta T cell activation. FEBS letters. 2003;544(1-3):4–10. doi: 10.1016/s0014-5793(03)00483-6. [DOI] [PubMed] [Google Scholar]
- Span I, Wang K, Wang W, Zhang Y, Bacher A, Eisenreich W, Li K, Schulz C, Oldfield E, Groll M. Discovery of acetylene hydratase activity of the iron-sulphur protein IspH. Nature communications. 2012;3:1042. doi: 10.1038/ncomms2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaimas K, Eils R, Konig R. Identifying essential genes in bacterial metabolic networks with machine learning methods. BMC systems biology. 2010;4:56. doi: 10.1186/1752-0509-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Sharma YD. Inhibition of Plasmodium falciparum ispH (lytB) gene expression by hammerhead ribozyme. Oligonucleotides. 2007;17(2):189–200. doi: 10.1089/oli.2007.0075. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic acids research. 2010;38(Database):D473–479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey CR, Rohwer A, Oellien F, Marhofer RJ, Braschi S, Oliveira G, McKerrow JH, Selzer PM. A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen, Schistosoma mansoni. PloS one. 2009;4(2):e4413. doi: 10.1371/journal.pone.0004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther GJ, Shanmugam D, Carmona SJ, Doyle MA, Hertz-Fowler C, Berriman M, Nwaka S, Ralph SA, Roos DS, Van Voorhis WC. et al. Identification of attractive drug targets in neglected-disease pathogens using an in silico approach. PLoS neglected tropical diseases. 2010;4(8):e804. doi: 10.1371/journal.pntd.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker M, Mack C, Bott M. Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. Journal of bacteriology. 2011;193(5):1237–1249. doi: 10.1128/JB.01032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrt TC, Deretic V. An essential two-component signal transduction system in Mycobacterium tuberculosis. Journal of bacteriology. 2000;182(13):3832–3838. doi: 10.1128/jb.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland N, Mack TR, Yu M, Hung LW, Terwilliger TC, Waldo GS, Stock AM. Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry. 2007;46(23):6733–6743. doi: 10.1021/bi602546q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Follmann H. The Ribonucleotide Reductases - a Unique Group of Metalloenzymes Essential for Cell-Proliferation. Struct Bond. 1983;54:27–91. [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annual review of biochemistry. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Monje-Casas F, Jurado J, Prieto-Alamo MJ, Holmgren A, Pueyo C. Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. The Journal of biological chemistry. 2001;276(21):18031–18037. doi: 10.1074/jbc.M011728200. [DOI] [PubMed] [Google Scholar]
- Cotruvo JA, Stubbe J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry. 2010;49(6):1297–1309. doi: 10.1021/bi902106n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends in biochemical sciences. 1992;17(3):119–123. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- Boal AK, Cotruvo JA, Stubbe J, Rosenzweig AC. Structural basis for activation of class Ib ribonucleotide reductase. Science. 2010;329(5998):1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, Larochette N, Zamzami N, Jan G, Kroemer G, Brenner C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene. 2007;26(18):2606–2620. doi: 10.1038/sj.onc.1210074. [DOI] [PubMed] [Google Scholar]
- Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelieres FP, Marco S, Saudou F. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013;152(3):479–491. doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- Bressi JC, Verlinde CL, Aronov AM, Shaw ML, Shin SS, Nguyen LN, Suresh S, Buckner FS, Van Voorhis WC, Kuntz ID. et al. Adenosine analogues as selective inhibitors of glyceraldehyde-3-phosphate dehydrogenase of Trypanosomatidae via structure-based drug design. Journal of medicinal chemistry. 2001;44(13):2080–2093. doi: 10.1021/jm000472o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhowmick I, Andersen J, Srinivasan P, Narum DL, Bosch J, Miller LH. Binding of aldolase and glyceraldehyde-3-phosphate dehydrogenase to the cytoplasmic tails of Plasmodium falciparum merozoite duffy binding-like and reticulocyte homology ligands. mBio. 2012;3(5) doi: 10.1128/mBio.00292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinzoni M, Haouz A, Grana M, Munier-Lehmann H, Shepard W, Alzari PM. The crystal structure of Mycobacterium tuberculosis adenylate kinase in complex with two molecules of ADP and Mg2+ supports an associative mechanism for phosphoryl transfer. Protein science : a publication of the Protein Society. 2006;15(6):1489–1493. doi: 10.1110/ps.062163406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appaji Rao N, Ambili M, Jala VR, Subramanya HS, Savithri HS. Structure-function relationship in serine hydroxymethyltransferase. Biochimica et biophysica acta. 2003;1647(1-2):24–29. doi: 10.1016/s1570-9639(03)00043-8. [DOI] [PubMed] [Google Scholar]
- Sopitthummakhun K, Thongpanchang C, Vilaivan T, Yuthavong Y, Chaiyen P, Leartsakulpanich U. Plasmodium serine hydroxymethyltransferase as a potential anti-malarial target: inhibition studies using improved methods for enzyme production and assay. Malaria journal. 2012;11:194. doi: 10.1186/1475-2875-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechaly AE, Haouz A, Miras I, Barilone N, Weber P, Shepard W, Alzari PM, Bellinzoni M. Conformational changes upon ligand binding in the essential class II fumarase Rv1098c from Mycobacterium tuberculosis. FEBS letters. 2012;586(11):1606–1611. doi: 10.1016/j.febslet.2012.04.034. [DOI] [PubMed] [Google Scholar]
- Woods SA, Schwartzbach SD, Guest JR. Two biochemically distinct classes of fumarase in Escherichia coli. Biochimica et biophysica acta. 1988;954(1):14–26. doi: 10.1016/0167-4838(88)90050-7. [DOI] [PubMed] [Google Scholar]
- Sampaleanu LM, Vallee F, Slingsby C, Howell PL. Structural studies of duck delta 1 and delta 2 crystallin suggest conformational changes occur during catalysis. Biochemistry. 2001;40(9):2732–2742. doi: 10.1021/bi002272k. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang Y, Woolridge EM, Arora V, Petsko GA, Kozarich JW, Ringe D. Crystal structure of 3-carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida, a fumarase class II type cycloisomerase: enzyme evolution in parallel pathways. Biochemistry. 2004;43(32):10424–10434. doi: 10.1021/bi036205c. [DOI] [PubMed] [Google Scholar]
- Toth EA, Yeates TO. The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway. Structure. 2000;8(2):163–174. doi: 10.1016/s0969-2126(00)00092-7. [DOI] [PubMed] [Google Scholar]
- Tsai M, Koo J, Yip P, Colman RF, Segall ML, Howell PL. Substrate and product complexes of Escherichia coli adenylosuccinate lyase provide new insights into the enzymatic mechanism. Journal of molecular biology. 2007;370(3):541–554. doi: 10.1016/j.jmb.2007.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver TM, Levitt DG, Donnelly MI, Stevens PP, Banaszak LJ. The multisubunit active site of fumarase C from Escherichia coli. Nature structural biology. 1995;2(8):654–662. doi: 10.1038/nsb0895-654. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhou Q, Ge R. Inhibition of fumarase by bismuth(III): implications for the tricarboxylic acid cycle as a potential target of bismuth drugs in Helicobacter pylori. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2012;25(1):95–102. doi: 10.1007/s10534-011-9485-7. [DOI] [PubMed] [Google Scholar]
- Ruda GF, Campbell G, Alibu VP, Barrett MP, Brenk R, Gilbert IH. Virtual fragment screening for novel inhibitors of 6-phosphogluconate dehydrogenase. Bioorganic & medicinal chemistry. 2010;18(14):5056–5062. doi: 10.1016/j.bmc.2010.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Dunbar J, Jayasekera MM, Viola RE, Farber GK. The structure of L-aspartate ammonia-lyase from Escherichia coli. Biochemistry. 1997;36(30):9136–9144. doi: 10.1021/bi9704515. [DOI] [PubMed] [Google Scholar]
- de Villiers M, Puthan Veetil V, Raj H, de Villiers J, Poelarends GJ. Catalytic mechanisms and biocatalytic applications of aspartate and methylaspartate ammonia lyases. ACS chemical biology. 2012;7(10):1618–1628. doi: 10.1021/cb3002792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Docking representation of the best drug-like compound ZINC75109074 in the most druggable protein cavity of Cp1002_0515 (MtrA, DNA-binding response regulator). Three hydrogen bonds were observed with Thr73, Asp48 and Arg116.
Docking representation of compound ZINC00510419 in the most druggable protein cavity of Cp1002_0742 (IspH, 4-hydroxy-3-methyl but-2-enyl diphosphate reductase). Residues Cys39, Thr225, Ser250, His68 and Asn252 are predicted to make seven hydrogen bonds to this ligand.
Docking representation of the best drug-like compound ZINC00510419 in the most druggable protein cavity of Cp1002_1648 (TcsR, Two component transcriptional regulator). Hydrogen bonds were observed with residues Val76, Gln185 and Asn193.
Docking representation of the best drug-like compound ZINC04721321 in the most druggable protein cavity of Cp1002_1676 (NrdI protein). Hydrogen bonds were observed with residues Ser8, Thr13 and Leu116.
Comparison among the most druggable cavities from essential bacterial and the respective host homologue proteins. Protein structures are shown as cartoon (green for the bacterial protein and gray for Ovis aries host protein). Other host proteins are not shown for simplicity, but the same substitutions were present in all host proteins analyzed. Residues that differ in the bacterial and host cavity are highlighted in sticks and labeled (bacterial labels in green and host labels in black). a) Cp1002_0692 (Glyceralderayde 3-phosphate dehydrogenase); b) Cp1002_0385 (adenylate kinase); c) Cp1002_0728 (serine hydroxymethyltransferase); d) Cp1002_0738 (fumarate hydratase class II) the site shown is formed by three monomers, which are represented in green, blue and orange. No residues are highlighted, since the active sites are identical between bacteria and host; e) Cp1002_1005 (6-phosphogluconate dehydrogenase); f) Cp1002_1042 (aspartate ammonia-lyase). Figures were prepared with the PyMol.


