Abstract
Glucokinase is classified in bacteria based upon having ATP binding site and ‘repressor/open reading frames of unknown function/sugar kinases’ motif, the sequence of glucokinase gene (JN645812) of Staphylococcus aureus ATCC12600 showed presence of ATP binding site and ‘repressor/open reading frames of unknown function/sugar kinases’ motif. We have earlier observed glucokinase of S. aureus has higher affinity towards the substrate compared to other bacterial glucokinase and under anaerobic condition with increased glucose concentration S. aureus exhibited higher rate of biofilm formation. To establish this, 3D structure of glucokinase was built using homology modeling method, the PROCHECK and ProSA-Web analysis indicated this built glucokinase structure was close to the crystal structure. This structure was superimposed with different bacterial glucokinase structures and from the root-mean-square deviation values, it is concluded that S. aureus glucokinase exhibited very close homology with Enterococcus faecalis and Clostridium difficle while with other bacteria it showed high degree of variations both in domain and nondomain regions. Glucose docking results indicated -12.3697 kcal/mol for S. aureus glucokinase compared with other bacterial glucokinase suggesting higher affinity of glucose which correlates with enzyme kinetics and higher rate of biofilm formation.
Keywords: Biofilm, glucokinase, molecular docking, RMSD, ROK
Staphylococcus aureus (S. aureus) is a causative agent of many superficial deep-skin and soft tissue infections to life threatening diseases like endocarditis and a variety of toxin-mediated diseases including gastroenteritis, staphylococcal scalded-skin syndrome, and toxic shock syndrome[1,2]. It is known that one of the most common ways for bacteria to live is, by adherence to surfaces in the form of biofilms, where they are embedded in an extracellular polymeric matrix structures like exopolysaccharides and polysaccharide intercellular adhesin (PIA), whose synthesis requires high amount of glucose-6-phosphate (G-6-P)[3,4,5,6]. In most bacteria, glucose is transported by the phosphoenolpyruvate: sugar phosphotransferase system (PTS)[7], however, in S. aureus it can also be transported via PTS-independent systems and these systems functions according to the concentrations of external glucose. PTS-independent system functions through glucose permease (glk U) and glucokinase (glk A) genes when high external glucose concentration is present[8].
Glycolysis is a major pathway in S. aureus; 85%-90% of the glucose is catabolised through Embden-Meyerhof glycolytic pathway (EMP)[9,10]. The very first step of glycolysis is the formation of G-6-P, this is the ubiquitous anabolic intermediate that has essential role in the pathogen from energy generation in the catabolic reactions and upregulation of polysaccharide intracellular adhesion synthesis, virulence factors, cell wall synthesis and formation of small colony variants (SCV) are the key characteristic features of drug resistant strains like multi drug resistant (MDR) and vancomycin resistant S. aureus (VRSA). PIA is a polymer of N-acetyl glucosamine involved in the formation of adhesive exopolysaccharide matrix. This provides structural stability to biofilms, enhanced adhesion to surfaces made protection from host defenses and antibiotics[11,12]. Majority of G-6-P formation is catalysed by cytoplasmic glk A, this glk A in both Gram-positive and Gram-negative bacteria comprises of 315-321 residues with a monomeric mass of 33-35 kDa, Km values of glucokinase varied from 0.3-0.8 mM for glucose[13]. Glucokinases of bacteria are divided into 2 groups (1) glk A that belongs to ‘Repressor/open reading frames of unknown function/sugar kinases’ (ROK) family which is characterized by the presence of CXCGX(2)GCXE motifs, and (2) glk A without ROK motifs. However, in Archaea two types of ATP-dependent glk A are found one with ROK motif and the other without ROK motifs[14,15,16,17,18]. S. aureus glk A belongs to the Archaea group having both ATP binding site and ROK motifs.
In our earlier paper we discussed about the cloning, expression and characterization of S. aureus glucokinase suggests that the catalytic function of glk A shows considerable variation among bacteria and with human glucokinase (GCK)[19,20]. In the present study the sequence analysis showed presence of ROK motif indicating this enzyme is highly regulated in S. aureus, which is absent in other bacterial glk A, till now there is no crystal structure of S. aureus glk A gene reported, therefore; in the present study glk A structure was built using homology modeling method and comparative molecular docking of glucose into the substrate binding sites of S. aureus glk A, other bacterial glk A structures will reveal the binding mode variations and the strength of interactions with the substrate.
MATERIALS AND METHODS
Homology modeling of S. aureus glk A:
S. aureus glk A gene sequence (JN645812) was used in the construction of its structure which is not available in the protein data bank (PDB)[21] therefore; we have constructed its three-dimensional (3D) model by homology modeling method using Modeller 9v8 tool[22]. The S. aureus glk A protein sequence was subjected to basic local alignment search tool for proteins (BLASTp)[23] against PDB and the crystal structure of a putative glucokinase from Enterococcus faecalis (PDB ID:2QM1) showing the maximum identity of 40%[21,24] was chosen as template. A sequence alignment file was generated in PIR format for query and template sequences using ClustalX tool[25]. Python script was written and 20 best models were generated and the model showing lowest DOPE (discrete optimized protein energy) score was selected for further analysis.
The stereo chemical quality of the model was validated by PROCHECK and ProSAweb servers read the atomic co-ordinates of the 3D model and judge the quality of the structure. Ramachandran plot has been generated from PROCHECK[26] validation server was used to access the quality of the model by looking into the allowed and disallowed regions of the plot. Z-score values were generated from ProSAweb server that can determine the overall quality of the model and identity nearest to native NMR/X-ray crystal structure.
Structural comparison of S. aureus glk A with other bacterial glk A:
The comparative structural prediction studies to ensure the identity and variability of S. aureus glk A structure with other bacterial glk A structures were carried out using MATRAS (MArkovian TRAnsition of Structure) program which define the structural similarity score as the log-odds of two probabilities using a scheme similar to Dayhoff's amino acid substitution score. An alignment of superimposed structures and similarities were predicted scores and RMSD values for the following structures: the validated 3D model of S. aureus glk A was super imposed with other bacterial glucokinase such as Escherichia coli (1Q18), Enterococcus faecalis (2QM1), Clostridium difficile (Built structure), Streptococcus pneumonia (Built structure), Bacillus anthracis (Built structure), Renibacterium salmoninarum (Built structure) were recorded[27]. Wherever the PDB structures were not available the 3D structures for different bacterial glk A were built the following method mentioned earlier.
Molecular docking:
Molecular docking was carried out using Molecular Operating Environment MOE docking software tool (MOE 2011.10). The 3D structure of glucose was retrieved from PubChem[28] and its geometry was optimized in MOE working environment. The S. aureus glk A and other bacterial glk A structures were loaded individually into MOE software removing water molecules, hetero atoms and polar hydrogens were added. The structures were protonated at temperature of 300K, pH 7 and a salt concentration of 0.1. Generalized born implicit solvating environment was enabled with a dielectric constant of 1 and Van der Waals forces were enabled at a cut off value of 10 Å. Energy minimization was carried out in OPLS force field at a gradient of 0.05 to calculate the atomic coordinates of the protein that are local minima of molecular energy function and to determine low energy conformations; molecular dynamics simulations were carried out in the same force field. NVT statistical ensemble was used for the temperature held fixed to generate the trajectories. The most accurate Nose-Poincare-Anderson algorithm was enabled to solve the equation of motion during simulations. The initial temperature was set at 30 K and increased to a run time temperature of 300 K and the simulations were carried out for a total period of 10 nanoseconds and these stabilized conformations generated at the end of the simulations were used for molecular docking process.
Individual dockings were performed for S. aureus glk A and other bacterial glk A with glucose to find out the binding modes and affinity variations. These poses were generated by superposition of ligand atom triplets and triplets of receptor site points using alpha triangle docking placement methodology. The docked conformers are ranked by London dG scoring function to estimate the free energy of binding of the ligand from a given pose. The conformations thus were refined and rescored in the same force filed to remove the duplicate conformations. At the end of docking process the pose with least score was chosen from the total conformations and in each docking process the binding orientations of glucose was studied in the binding sites of S. aureus and other bacterial glucokinase[29,30,31].
RESULTS
Structural analysis of S. aureus glk A:
In our earlier studies we have cloned, sequenced (JN645812), expressed and characterized glk A of S. aureus. The catalytic properties of glk A and BLAST results of gene sequence revealed the presence of only one active enzyme in S. aureus, with a sequence having very low homology with both Gram-positive and Gram-negative bacteria (fig. 1). Further, the glk A sequence showed distinct ATP binding site and ROK motif which is indicated by three conserved C residues in the motif CNCGRSGCIE (fig. 2)[19].
Fig. 1.
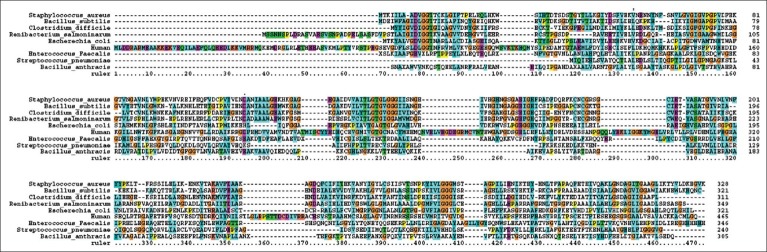
Multiple sequence alignment of 8 representative bacterial glk A.
S. aureus ATCC12600 (from the present study) Escherichia coli, Clostridium difficile, Enterococcus faecalis, Streptococcus pneumoniae, Bacillus anthracis, Bacillus subtilis and Renibacterium salmoninarum glk A. The homologous regions are shown in brown colour and variations are depicted in other colours.
Fig. 2.
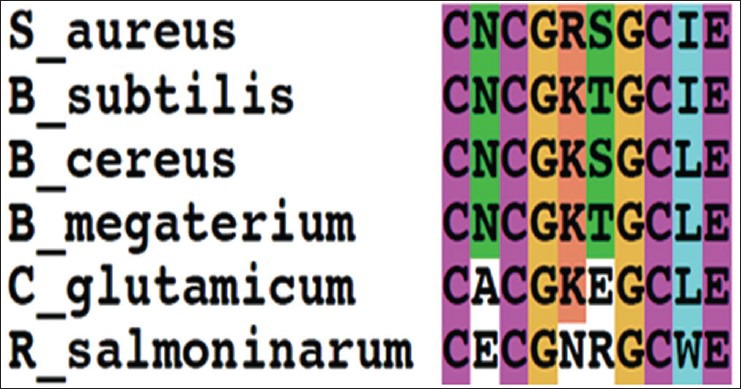
Multiple sequence alignment of S. aureus glk A with other bacterial glk A.
The multiple sequence alignment showed ROK (CXCGX(2)GCXE) motif.
The multiple sequence alignment results indicated differences with other bacterial glk A and so far crystal structure of S. aureus glk A was not available in the database therefore, the structure of S. aureus glk A was built using homology modeling method. In order to predict the structure of S. aureus glk A, 20 best models were generated for glk A using Modeler 9v8 tool for this the X-Ray crystallographic structure of glk A from Enterococcus faecalis (PDB ID: 2QM1) was used as template. The 19th model showed lowest DOPE score of -36384.98847 among 20 models and therefore this structure was considered for further study. The stereochemistry of the final model was verified by submitting to PROCHECK validation server and Ramachandran plot showed 91.7 % of the residues in most favorable region and no residues were found in the disallowed region. Most appropriately the plot was found with favorable comparison with X-Ray crystallographic data. The ProSA-Web evaluation of glk A 19 model revealed a compatible Z-Score value of -8.5 that falls in the range of native conformations of X-Ray crystal structure (fig. 3).
Fig. 3.
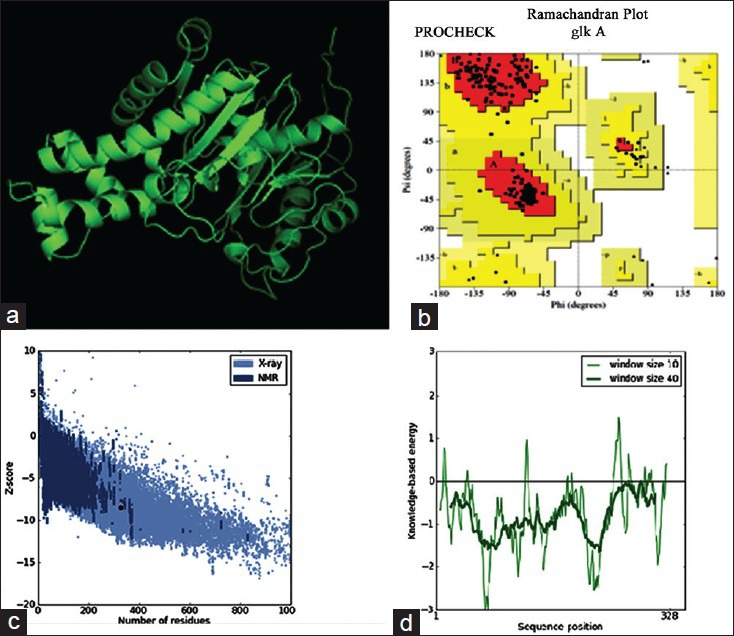
Molecular modeling of S. aureus glk A.
(a) The 3D structure of S. aureus glk A with lowest DOPE score built from modeller 9v8. (b) Ramachandran plot of S. aureus glk A structure. (c) The Z-score plot of glk A model. (d) Energy plot of glk A model.
Structural comparison with other bacterial glk A:
The S. aureus glk A structure was compared with other Gram-negative and -positive bacterial glk A. The S. aureus glk A structure showed very close homology with Enterococcus faecalis and Clostridium difficle, the RMSD values being 0.176 Å and 0.279 Å, respectively while with other bacteria Escherichia coli (8.732 Å), Streptococcus pneumonia (15.854 Å), Bacillus anthracis (15.317 Å) and Renibacterium salmoninarum (2.616 Å), the structures showed extensive variations both in the domain and nondomain regions. The identical regions are random throughout the alignment especially the structural superimposition of substrate binding regions showed much extent of variation and completely showing different conformations as indicated from the RMSD values (fig. 4).
Fig. 4.
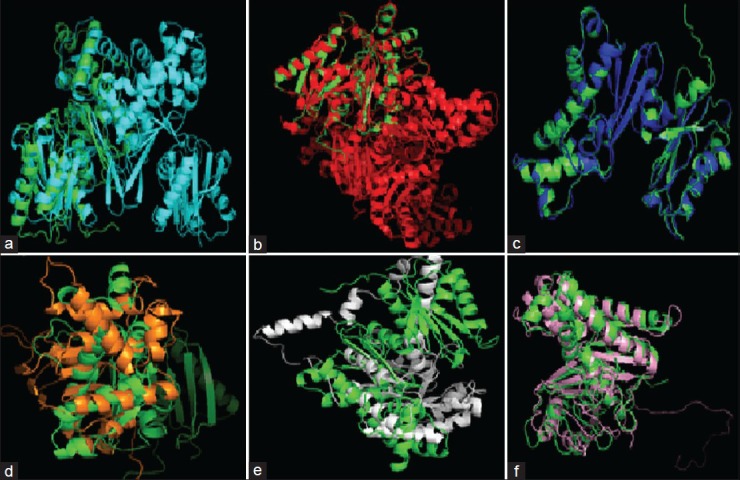
Super imposed structures of S. aureus glk A with other bacterial glk A.
(a) S. aureus glk A structure (green) with Escherichia coli (cyan), (b) Clostridium difficile (blue), (c) Enterococcus faecalis (red), (d) Streptococcus pneumoniae (orange), (e) Bacillus anthracis (white) and (f) Renibacterium salmoninarum (wheat) were generated by using MATRAS programme.
Molecular docking:
The docking of glucose into the substrate binding sites of S. aureus glk A and other bacterial glk A structures revealed variable binding orientations along with different docking scores of -12.3697 for S. aureus glk A, -9.2766 for Clastridium difficlae glk A, -6.3493 for Bacillus anthrasis glk A, -10.7487 for Bacillus subtillis glk A, -8.3749 for Ranibacterium salmoninarum glk A, -6.9410 for Streptococcus pneumonia glk A, -5.5898 for E. coli glk A and -9.1023 for Enterococcus faecalis glk A, respectively. Lowest score indicates greater stability of ligand enzyme complex and the docking scores indicate that S. aureus glk A forms more stable complex with glucose than with other bacterial glk A. Glucose is found to be interacting with V77, N114, S161 and G160 in S. aureus glk A forming a total of eight hydrogen bonds (Table 1) and (fig. 5). These results indicate higher affinity glucose towards glk A compared with other bacterial glk A correlating with the kinetic data and rate of biofilm formation in increased glucose concentration. Thus, rate of G-6-P formation in S. aureus probably playing key role in the biofilm formation.
TABLE 1.
MOLECULAR DOCKING INTERACTION OF GLUCOSE INTO S. AUREUS AND OTHER BACTERIAL GLK A
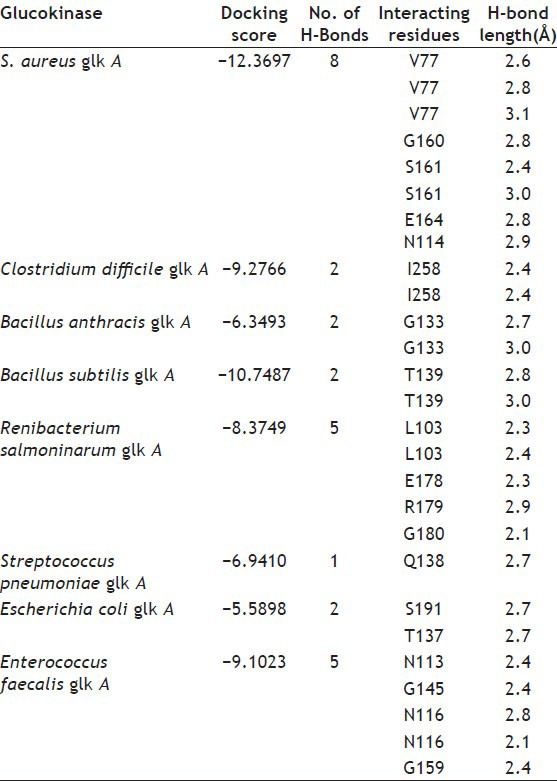
Fig. 5.
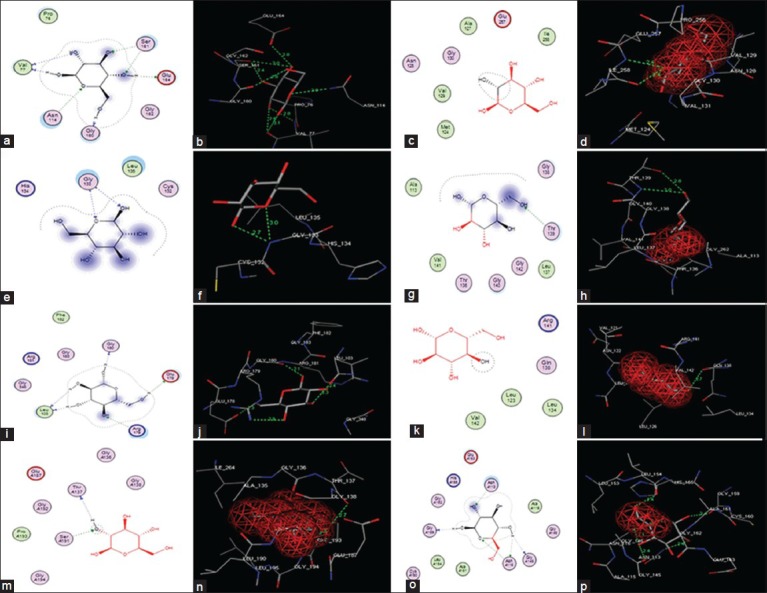
Molecular docking of glucose in to S. aureus glk A and other bacterial glk A active sites
(a) Two-dimensional (2D) linear representation of glucose interaction in the S. aureus glk A active site. (b) Three-dimensional (3D) graphical representation of glucose interaction in the active site of S. aureus glk A. (c, d) Clostridium difficile glk A. (e, f) Bacillus anthracis glk A. (g, h) Bacillus subtilis glk A. (i, j) Renibacterium salmoninarum glk A. (k, l) Streptococcus pneumonia glk A. (m, n) Escherichia coli glk A and (o, p) Enterococcus faecalis glk A.
DISCUSSION
S. aureus is a notorious pathogen and its infections ranging from minor skin infections to life threatening diseases is compounded with the occurrence of multi drug resistant strains of S. aureus[1,2]. One of the main factors in the pathogenesis of S. aureus is through biofilm formation. The biofilm formation contributes in developing this pathogen to become increasingly resistant to various classes of antibiotics[12]. Also, this increased rate of G-6-P formation in the pathogen probably elevates pentose phosphate pathway resulting in rapid nucleotide synthesis and followed by bacteremia in the human host[32,33,34].
In our earlier studies we have cloned, sequenced and characterized glk A gene which showed considerable variations in the sequence and functions with other bacterial glk A[11,19,20,35]. Our earlier findings have shown presence of ROK motif in the S. aureus glk A sequence and absent in other bacterial glucokinases which prompted us to perform structural comparison studies between S. aureus glk A and other bacterial glk A structures to predict their role in the host pathogen interactions. So far, glk A crystal structure is not present in PDB therefore, we have used homology modeling method to built the 3D structure of glk A[24]. This glk A structure was built using Modeller 9V8 and was validated through PROCHECK which showed 91.7% residues were in the allowed region and no residues were found in disallowed region. The ProSA-Web analysis indicated the built S. aureus glk A structure was close to the crystal structure with Z score -8.5 (fig. 3) therefore for all our structural comparison studies this built structure was used interestingly the glk A structure showed considerable identity with Enterococcus faecalis and Clostridium difficlae, while with other bacteria it showed high degree of variations as indicated from RMSD values.
The docking of glucose with S. aureus glk A structure showed very low docking score compared with other bacterial glk A structures indicating highly stable complex was generated with S. aureus glk A, this is probably due to the difference in the binding orientations and strength of these interactions suggest that the rate of catalysis is probably high with S. aureus glk A compared with other bacterial glk A[27,29,30] (Table 1 and fig. 5). These results aptly corroborated with our earlier kinetic results of S. aureus glk A[19,20]. In an earlier study we have observed S. aureus grown under anaerobic condition with elevated glucose concentration showed higher rate of biofilm formation[36]. In increased glucose concentration G-6-P formation is catalysed through glk A therefore; having very high affinity with glucose compared with other bacterial glk A through light in the survival, colonization and pathogenesis of S. aureus especially in relapsed episodes of infection.
In conclusion; the S. aureus glk A structure generated in the present study was highly validated as it was found close to the crystal structure with no residues found in the disallowed region. This structure showed close homology with glk A structures of Clostridium difficle and Enterococcus faecalis while with other bacteria it showed extensive variations both in the domain and non-domain regions. In the present study we have observed distinct differences in structures of S. aureus glk A and other bacterial glk A, also S. aureus glk A showed very high affinity towards glucose compared to other bacterial glk A as evidenced from molecular docking studies and kinetic studies of S. aureus glk A indicating the rate of G-6-P formation is high in S. aureus thus, leading to increased rate of biofilm formation.
Footnotes
Kumar, et al.: Structural and Functional Analysis of Staphylococcus aureus Glucokinase
REFERENCES
- 1.Fidalgo S, Vázquez F, Mendoza MC, Pérez F, Méndez FJ. Bacteremia due to Staphylococcus epidermidis: Microbiologic, epidemiologic, clinical, and prognostic features. Rev Infect Dis. 1990;12:520–8. doi: 10.1093/clinids/12.3.520. [DOI] [PubMed] [Google Scholar]
- 2.Frederiksen MS, Espersen F, Frimodt-Møller N, Jensen AG, Larsen AR, Pallesen LV, et al. Changing epidemiology of pediatric Staphylococcus aureus bacteremia in Denmark from 1971 through 2000. Pediatr Infect Dis J. 2007;26:398–405. doi: 10.1097/01.inf.0000261112.53035.4c. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent Infections. Science. 1999;284:1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Joyce CM, Grindley ND. Method for determining whether a gene of Escherichia coli is essential: Application to the polA gene. J Bacteriol. 1984;158:636–43. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasa I. Towards the identification of the common features of bacterial biofilm development. Int Microbiol. 2006;9:21–8. [PubMed] [Google Scholar]
- 6.Xia M, Lunsford RD, McDevitt D, Iordanescu S. Rapid method for the identification of essential genes in Staphylococcus aureus. Plasmid. 1999;42:144–9. doi: 10.1006/plas.1999.1422. [DOI] [PubMed] [Google Scholar]
- 7.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: Carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–94. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Götz F, Bannerman T, Schleifer K. The Genera Staphylococcus and Macrococcus. Prokaryotes. 2006;4:5–75. [Google Scholar]
- 9.Blumenthal HJ, Huettner CF, Montiel FA. Comparative aspects of glucose catabolism in Staphylococcus aureus and S. epidermidis. Ann N Y Acad Sci. 1974;236:105–14. doi: 10.1111/j.1749-6632.1974.tb41485.x. [DOI] [PubMed] [Google Scholar]
- 10.Strasters KC, Winkler KC. Carbohydrate metabolism of Staphylococcus aureus. J Gen Microbiol. 1963;33:213–29. doi: 10.1099/00221287-33-2-213. [DOI] [PubMed] [Google Scholar]
- 11.Gould IM. Treatment of bacteraemia: Meticillin-resistant Staphylococcus aureus (MRSA) to vancomycin-resistant S. aureus (VRSA) Int J Antimicrob Agents. 2013;42:17–21. doi: 10.1016/j.ijantimicag.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Xiong YQ, Sadykov MR, Fey PD, Lei MG, Lee CY, et al. Tricarboxylic Acid Cycle-Dependent Attenuation of Staphylococcus aureus: In Vivo Virulence by Selective Inhibition of Amino Acid Transport. Infect Immun. 2009;77:4256–64. doi: 10.1128/IAI.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strasters KC, Winkler KC. Carbohydrate metabolism of Staphylococcus aureus. J Gen Microbiol. 1963;33:213–29. doi: 10.1099/00221287-33-2-213. [DOI] [PubMed] [Google Scholar]
- 14.Kengen SW, de Bok FA, van Loo ND, Dijkema C, Stams AJ, De Vos WM. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269:17537–41. [PubMed] [Google Scholar]
- 15.Kengen SW, Tuininga JE, de Bok FA, Stams AJ, de Vos WM. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:30453–7. doi: 10.1074/jbc.270.51.30453. [DOI] [PubMed] [Google Scholar]
- 16.Koga S, Yoshioka I, Sakuraba H, Takahashi M, Sakasegawa S, Shimizu S, et al. Biochemical characterization, cloning, and sequencing of ADP-dependent (AMP-forming) glucokinase from two hyperthermophilic archaea, Pyrococcus furiosus and Thermococcus litoralis. J Biochem. 2000;128:1079–85. doi: 10.1093/oxfordjournals.jbchem.a022836. [DOI] [PubMed] [Google Scholar]
- 17.Labes A, Schonheit P. ADP-dependent glucokinase from the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324. Arch Microbiol. 2003;180:69–75. doi: 10.1007/s00203-003-0563-2. [DOI] [PubMed] [Google Scholar]
- 18.Sakuraba H, Yoshioka I, Koga S, Takahashi M, Kitahama Y, Satomura T, et al. ADP-dependent glucokinase/phosphofructokinase, a novel bifunctional enzyme from the hyperthermophilic archaeon Methanococcus jannaschii. J Biol Chem. 2002;277:12495–8. doi: 10.1074/jbc.C200059200. [DOI] [PubMed] [Google Scholar]
- 19.Lakshmi HP, Yeswanth S, Prasad UV, Vasu D, Swarupa V, Kumar PS, et al. Cloning, expression and characterization of glucokinase gene involved in the glucose-6-phosphate formation in Staphylococcus aureus. Bioinformation. 2013;9:169–73. doi: 10.6026/97320630009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddi KV, Kumar PS, Prasad UV, Yeswanth S, Swarupa V, Vasu D, et al. Isolation, purification and characterization of glucokinase from Staphylococcus aureus. J Pharm Res. 2013;6:205–9. [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TH, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using modeller. Curr Protoc Bioinformatics. 2006;15:1–30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalton JA, Jackson RM. An evaluation of automated homology modelling methods at low target template sequence similarity. Bioinformatics. 2007;23:1901–8. doi: 10.1093/bioinformatics/btm262. [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereo chemical quality of protein structures. J Appl Cryst. 1993;26:283–91. [Google Scholar]
- 27.Kawabata T. MATRAS a program for protein 3D structure comparison. Nucleic Acids Res. 2003;31:3367–9. doi: 10.1093/nar/gkg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:623–33. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamata K, Mitsuya M, Nishimura T, Eiki J, Nagata Y. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 2004;12:429–38. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Kumar YN, Kumar PS, Sowjenya G, Rao VK, Yeswanth S, Prasad UV, et al. Comparison and correlation of binding mode of ATP in the kinase domains of Hexokinase Family. Bioinformation. 2012;8:543–7. doi: 10.6026/97320630008543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar PS, Kumar YN, Prasad UV, Yeswanth S, Swarupa V, Sowjenya G, et al. In silico designing and molecular docking of a potent analog against Staphylococcus aureus porphobilinogen synthase. J Pharm Bioallied Sci. 2014;6:158–66. doi: 10.4103/0975-7406.135246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;20:339:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 33.Severin A, Tabei K, Tenover F, Chung M, Clarke N, Tomasz A. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J Biol Chem. 2004;279:3398–407. doi: 10.1074/jbc.M309593200. [DOI] [PubMed] [Google Scholar]
- 34.Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, et al. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun. 2002;70:6373–82. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu ZL. Phosphatase-coupled universal kinase assay and kinetics for first-order-rate coupling reaction. PLoS One. 2011;6:e23172. doi: 10.1371/journal.pone.0023172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeswanth S, Kumar YN, Prasad UV, Swarupa V, Rao VK, Sarma PV. Cloning and characterization of L-lactate dehydrogenase gene of Staphylococcus aureus. Anaerobe. 2013;24:43–8. doi: 10.1016/j.anaerobe.2013.09.003. [DOI] [PubMed] [Google Scholar]


