Abstract
Cell motility plays a critical role in many physiological and pathological settings, ranging from wound healing to cancer metastasis. While cell migration on 2-dimensional (2-D) substrates has been studied for decades, the physical challenges cells face when moving in 3-D environments are only now emerging. In particular, the cell nucleus, which occupies a large fraction of the cell volume and is normally substantially stiffer than the surrounding cytoplasm, may impose a major obstacle when cells encounter narrow constrictions in the interstitial space, the extracellular matrix, or small capillaries. Using novel microfluidic devices that allow observation of cells moving through precisely defined geometries at high spatial and temporal resolution, we determined nuclear deformability as a critical factor in the cells’ ability to pass through constrictions smaller than the size of the nucleus. Furthermore, we found that cells with reduced levels of the nuclear envelope proteins lamins A/C, which are the main determinants of nuclear stiffness, passed significantly faster through narrow constrictions during active migration and passive perfusion. Given recent reports that many human cancers have altered lamin expression, our findings suggest a novel biophysical mechanism by which changes in nuclear structure and composition may promote cancer cell invasion and metastasis.
Keywords: Lamin, nucleus, mechanics, cancer, microfluidics, microstructures, metastasis, invasion, nuclear envelope
Introduction
The ability of cells to move within their environment is crucial for numerous physiological and pathological processes. During development, cell migration contributes to shaping the growing embryo and to forming nascent tissues; in mature organisms, immune cells are mobilized from the blood stream to enter sites of infection, and migration of epithelial cells and fibroblasts is vital for proper wound healing and tissue repair. In cancer metastasis, cell migration drives the invasion of tumor cells into the surrounding tissue and the dissemination to other organs. The fact that cancer metastasis, rather than the primary tumor, is responsible for up to 90% of all cancer deaths1 has been one of the primary motivators to study cell migration.
Traditionally, research on cell migration has involved observing cells moving on flat, two-dimensional (2-D) substrates, owing primarily to the convenience in sample preparation and the ability to image cells at high spatial and temporal resolution. These studies have resulted in tremendous insights into the intricate processes occurring during cell migration, particularly at focal adhesions and the actin cytoskeleton. Nonetheless, research carried out over the past decade indicates that cell migration in 3-D environments, as is the case in most physiological processes inside the human body, can substantially deviate from migration on 2-D substrates.2-6 Much of this work has focused on specific changes in cell morphology (e.g., spreading, cytoplasmic organization, etc.) or migration mode (e.g., amoeboid vs. mesenchymal migration) when comparing cells in 2-D vs. 3-D environments.4, 6 However, one additional and particularly important difference is that in 3-D migration, cells have to overcome the confinement of the surrounding extracellular matrix and other cells.2, 3, 7 In many cases, the openings in the cell’s 3-D environment can be as small as 2 to 30 μm in diameter,8, 9 i.e., substantially smaller than the cell diameter. Under these conditions, cells have two options: (1) to degrade/modify their environment to create sufficient space, for example, by secretion of matrix metalloproteinases (MMPs); or (2) to deform to fit through the available space. In the latter option, the deformability of the cell becomes an important factor in the ability of cells to pass through 3-D environments.
The cytoplasm is very flexible and can undergo large deformations; in addition, the cytoskeleton can actively remodel to take up the available space, allowing it to penetrate openings as small as 1 μm.10 The cell nucleus, on the other hand, is 2- to 10-times stiffer than the surrounding cytoplasm and occupies a large fraction of the cellular volume;11 with a typical diameter of 3–10 μm, the nucleus is larger than many of the pores encountered in the extracellular environment, so that cell movement through such constrictions requires substantial nuclear deformations.7, 10, 12, 13 Importantly, a recent study by Wolf and colleagues suggests that nuclear deformability constitutes a rate-limiting factor during non-proteolytic migration of cells through 3-D collagen matrices.10 As nuclear deformability is directly dependent on the expression of the nuclear envelope proteins lamins A and C,14, 15 we analyzed the effect of lamin expression on the ability of cells to transit constrictions smaller than the nuclear diameter during migration in 3-D environments or perfusion through microfluidic devices with precisely controlled constrictions. We found that mouse embryonic fibroblasts (MEFs) partially (Lmna+/−) or completely (Lmna−/−) lacking lamins A/C, for which we had previously demonstrated increased nuclear deformability,14, 15 passed significantly faster through the narrow constrictions than wild-type (Lmna+/+) controls.
Materials and Methods
Cell culture
MEFs from mice with homozygous (Lmna−/−) or heterozygous (Lmna+/−) deletion of the Lmna gene encoding lamins A/C, along with wild-type littermate controls (Lmna+/+), were generously provided by Dr. Colin Stewart.16 Human SV40 virus-transformed skin fibroblasts (GM00637J), originally obtained from Coriell Cell Repositories, were stably modified with a tetracyclin-regulated (Tet-off) GFP-lamin A construct17 provided by Dr. Tom Glover and maintained as described previously.18 NIH 3T3 fibroblasts were modified with mCherry–Histone-4 and GFP-LifeAct to allow visualization of nuclear position and F-actin distribution. All cells were maintained in Dulbecco Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), penicillin and streptomycin. Medium for live cell imaging was further supplemented with 25 mM HEPES buffer.
Device fabrication
The migration devices were fabricated at the Cornell NanoScale Science and Technology Facility (CNF) using standard lithography techniques. The perfusion devices were modeled on a design by the Fletcher laboratory19 and described previously.20, 21 In brief, the design consisted of 10 μm tall microfluidic channels that bifurcate multiple times and lead to 5 μm-wide constriction channels. A wide bypass channel is included to assure maintenance of uniform pressure drop across the constriction channels even if some channels are blocked.
The migration devices contain features of two different heights, produced from a mold composed of two layers: one 5 μm tall layer to form the constrictions channels, and a second, 250 μm tall layer to create cell collection areas directly before the constrictions (in which cells are seeded) and behind the constrictions (in which cells that have passed the constriction can gather). The constriction channels are made of a series of round pillars, with three narrow openings in a row, measuring 5, 3, and 2 μm in width, respectively (Fig. 1a). We created two variations of the migration devices; one of which consisted entirely of channels with narrow constrictions, the other additionally including regularly spaced 15 μm wide passages that were used to study cell migration in the 5 μm tall channels without substantial nuclear deformation. Large reservoirs on either side of the constrictions that can be filled with media were used to establish a chemotactic gradient across the device.
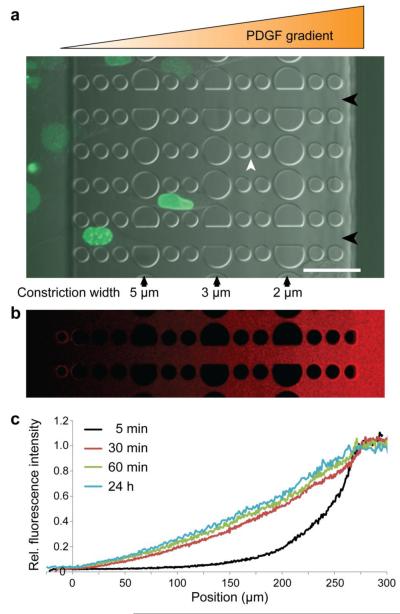
Figure 1. Overview of the microfluidic migration device.
a) Cells are seeded in a 250 μm tall chamber of the device (far left), which is separated from the other chamber (far right) by 5 μm tall constriction channels consisting of PDMS pillars. Larger pillars create a series of 5 μm, 3 μm, and 2 μm wide constrictions (black arrows). In the direction perpendicular to the gradient, 2 wide spacings between pillars (white arrowhead) allow the gradient to form uniformly even when cells are inside the constriction channels. Channels with wider spacing (15 μm, black arrowheads) are provided to assess the cell migration speed without nuclear confinement. Shown here are human skin fibroblasts expressing GFP-lamin A migrating through a device towards a PDGF chemotactic gradient. Scale bar: 50 μm. b) Gradient formation after 24 hours of fluorescently labeled (Texas Red) 70 kD dextran. c) Fluorescence intensity across the migration device during gradient formation with 70 kD fluorescent dextran, demonstrating that the gradient forms quickly (within 30 minutes) and is stable over 24 hours.
Molds of the devices were fabricated from SU-8 photoresist. Briefly, a silicon wafer was baked at 200°C for half an hour, then SU-8 was spun onto the wafer to obtain a 5 μm thick layer. To study the migration of cells along a chemotactic gradient in less confined conditions, we also created a variation of the migration device in which the channels had a height of 15 μm, but were otherwise identical. Wafers were baked at 60°C for 10 minutes, cooled, and UV exposed using the first mask. Wafers were then baked briefly at 95°C, cooled, and the unexposed SU-8 was dissolved in SU-8 developer. After development, the wafers were baked again at 200°C for half an hour, then SU-8 was spun on the wafers to obtain the 250 μm thick layer. The wafers were then baked at 50°C for 24 hours, exposed using the second mask, and baked at 95°C for 10 minutes. After cooling down, the wafers were developed, the heights of the structures were measured on a profilometer and the wafers were coated with (1H,1H,2H,2H-perfluorooctyl)trichlorosilane to facilitate detachment during replica production. Replicas of the SU-8 molds were produced using polydimethylsiloxane (PDMS). The two components of Sylgard 184 (Dow Corning) were mixed in the 1:10 ratio recommended by the manufacturer, the mixture was degassed to eliminate bubbles and poured over the wafer in a plastic petri dish. This assembly was then baked at 65°C for two hours. The resulting PDMS replica was carefully peeled away from the SU-8 structures and individual migration devices were cut out and placed on packing tape to prevent dust from accumulating on the clean surface. Inlets for cells were punched using 1.2 mm biopsy punches, and reservoirs for medium were created using 5 mm biopsy punches (Harris).
Microfluidic devices were assembled by bonding the PDMS replicas to glass slides. Glass slides were incubated in 0.2 M HCl overnight, rinsed with deionized water, isopropanol and water again, then dried off with filtered air. Both the glass slides and the PDMS replicas were exposed to an oxygen plasma for 5 minutes and the PDMS was then placed on the glass slides, which sealed instantly. The newly formed microfluidic devices were baked at 95°C on a hot plate for 5 minutes, doused and perfused with 70% ethanol to sterilize, followed by sterile water.
Validation of gradient formation in migration devices
Migration devices were passivated with 30 mg/mL BSA in PBS at 4°C overnight to reduce absorption of fluorescently labeled dextran to the PDMS surface. Prior to imaging, the BSA solution was replaced with PBS on the side the cells are loaded on and Texas Red–labeled 70-kDa dextran (Molecular Probes) dissolved at 10 mg/ml in PBS (Invitrogen) on the other side, mimicking the conditions for the migration experiments with a PDGF gradient. The devices were then covered with a glass coverslip to minimize evaporation, placed on the heated microscope stage or in a temperature controlled incubator and imaged at defined time-points for up to 24 hours. Line profiles of the fluorescence intensity were obtained using Zen light software (Zeiss) and the average profiles from at least 3 different measurements at each time point were used to determine the time course and stability of the gradient formation reported in Figure 1. As the PDGF used for the migration experiments has an even lower weight (~25 kDa) than the fluorescently labeled dextran, it is expected that the PDGF gradient forms even faster than the data presented in Figure 1.
Cell perfusion experiments
Experiments to measure passive nuclear deformability were performed as described previously.21 In brief, prior to the experiments, the microfluidic channels were treated with BSA (20 mg/mL in PBS) for 30 min to reduce cell adhesion. Cells suspended in PBS with 20 mg/mL bovine serum albumin (BSA) at a final concentration of 3 × 106 cells/mL were passed through a cell strainer and transferred into Exmire Microsyringes mounted on a microdialysis pump (CMA Microdialysis). A second syringe loaded with PBS + 20 mg/mL BSA was used to prime and occasionally flush the devices. Cells were perfused through the microfluidic device at a rate of 2.0 μL/min and imaged at 20× magnification with a MegaPlus 310T CCD camera (Roper Scientific) at 30 frames per second. Perfusion transit times through the 5 μm constrictions were determined from recorded 60-90 second long videos as described previously.19 Separate aliquots of the cell suspensions were imaged at 10× magnification with a Roper CoolSNAP HQ and cell sizes were determined using ImageJ software. All cells used for the perfusion experiments had a similar size distribution.
Micropipette aspiration
Experiments to measure nuclear stiffness were carried out following previously established protocols.22, 23 Micropipettes were pulled from glass capillary tubing (Sutter Instrument; 1 mm outer diameter; 0.5 mm inner diameter) with a Flaming/Brown Micropipette Puller (Model P-97, Sutter Instrument). Micropipettes were then cut to have an opening with an inner diameter of 4.0 to 5.5 μm. Subsequently, micropipettes were fire-polished and shaped using a Micro Forge (MF-900, Narishige). Micropipettes were immersed in PBS with 20 mg/mL BSA for 1 hour to minimize cell adhesion and backfilled with PBS. Cells incubated with Hoechst 33342 for 10 minutes at 37°C were suspended in DMEM + 10% FBS and placed in a microscope mounted glass-bottom culture dish. Individual cells in suspension were carefully positioned at the mouth of the micropipette using a negative aspiration pressure of ΔP = 10 mm H2O. Subsequently, the aspiration pressure was increased to ΔP = 75.5 mm H2O with a valve system developed in house, resulting in partial aspiration of the cell nucleus. Aspiration continued until an equilibrium position was reached and the nucleus stopped further advancing into the micropipette (typically less than 15 s). For a given aspiration pressure, the nuclear elasticity inversely scales with the ratio of the aspirated nuclear length, LP, and the micropipette diameter, D (Fig. 3).22 Cells in which the nucleus was positioned away from the micropipette were excluded from the analysis. Cell viability during the experiments was monitored using propidium iodide in the medium.
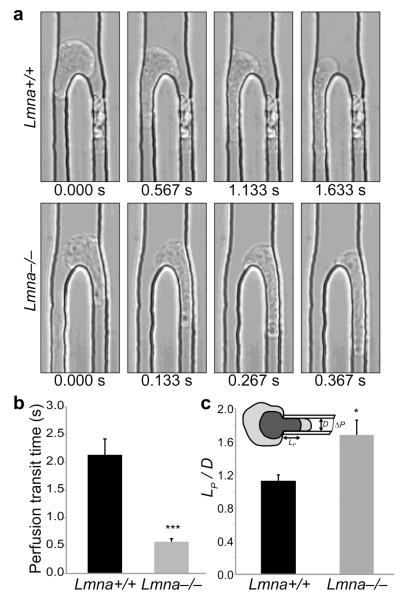
Figure 3. Lamin A/C-deficient cells have more deformable nuclei.
a) Representative images from high-speed time-lapse videos of cells being perfused through 5 μm wide microfluidic constriction channels, revealing that wild-type cells (Lmna+/+) take substantially longer to enter and move through the constriction channel than lamin A/C-deficient (Lmna−/−) cells. b) Perfusion transit times through 5 μm constriction channels in the perfusion experiments. N = 167 cells for Lmna+/+, 236 for Lmna−/−; ***, P < 0.0001. c) Micropipette aspiration measurements on wild-type and lamin A/C-deficient cells, demonstrating that lamin A/C-deficient cells have more deformable nuclei. The nuclear elasticity is inversely proportional to the ratio of the aspirated nuclear length, LP, and the micropipette diameter, D. N = 17 cells for Lmna+/+, 18 for Lmna−/−; *, P = 0.0105.
Cell migration through microfluidic constrictions
Cells for migration experiments were suspended in DMEM containing 10% FBS at a final concentration of 5 × 106 cells/mL. The migration devices were incubated with 0.2 mg/mL fibronectin (Millipore) in phosphate buffered saline (PBS) for at least 2 hours at 37°C. The fibronectin-coated devices were then filled with fresh medium and aliquots of 20,000 cells were added to each device (4 μl of 5 × 106 cells/mL). Live cell imaging experiments were carried out 24 hours after seeding cells into the devices. Immediately prior to imaging, the medium in both wells of the device was replaced with phenol red-free medium containing 25 mM HEPES (Gibco); for experiments using a chemotactic gradient, the well towards which the cells were migrating additionally contained 200 ng/mL PDGF as a chemoattractant. After medium replacement, glass coverslips were added to the top of the devices to limit evaporation, and the devices were placed in a temperature-controlled chamber on a Zeiss LSM 700 confocal microscope (AxioObserver) equipped with a CCD camera (CoolSNAP EZ, Photometrics) and a motorized stage (Zeiss). Time-lapse images were recorded every 2 or 10 minutes for 8 to 14 hours total. Image sequences used for the analysis of cell migration were acquired with a 20× objective in widefield/phase contrast mode. Fluorescent images were acquired in confocal mode with a 63× objective.
For the 48 hour migration studies, we quantified the number of cells successfully passing through the constriction channels using devices that did not contain the wider 15 μm channels. Cells were seeded in the devices; after 24 hours, the medium was replaced with imaging medium; the media in the reservoir on the other side of the constrictions was additionally supplemented with 200 ng/mL PDGF. Images were taken then and 24 hours later on an inverted microscope (Zeiss AxioObserver) with a 10× objective and a CCD camera (CoolSNAP EZ). The number of cells that had successfully passed through the constrictions was determined from these images. The proliferation rates of the different cell lines were measured by passaging the cells every 48 hours five-times in a row, counting the cells and each time re-seeding 1 × 106 cells in a T75 flask. The average fold-change over a 48 hour period was then determined from the cell counts and used to normalize the number of cells that had migrated through the constrictions.
Analysis of cell migration through microfluidic devices
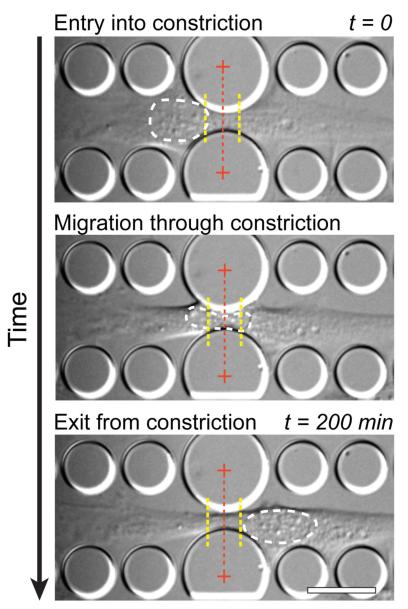
Time-lapse image sequences were collated into movies and corrected for drift in the x-y plane using a custom-written MATLAB algorithm. In most cases, the cells analyzed only went through one constriction within the time frame of the experiment and the data points presented here thus represent different cells. Movies of individual cells travelling through constrictions were cropped and the Manual Tracking plugin in ImageJ was used to track the position of the front and the back of the nucleus as it was passing through the constrictions. The migration transit time of a cell through a constriction was defined as the time between when the front of the cell crossed an imaginary line 5 μm in front of the constriction center and when the back of the nucleus passed a corresponding line 5 μm behind the constriction center (see dotted lines in Fig. 4). A similar analysis was performed on cells in the 15 μm wide channels to obtain values of the migration transit time of cells under non-constrained conditions. In addition, we measured the size and circularity of nuclei inside the 5 μm tall channels (but outside the narrow constrictions) using a custom-written MATLAB script.
Figure 4. Experimental approach to quantify the migration transit times of the nucleus through the narrow constrictions.
Time-lapse image sequences were used to measure the amount of time between when the cell nucleus enters and exits the constriction. In this example, a Lmna+/+ mouse embryonic fibroblast is shown. Entry into the constriction is defined as the time point at which the cell nucleus crosses a threshold (left yellow dotted line) located 5 μm in front of the constriction center; the nucleus is defined as having exited the constriction when the back of the nucleus crosses a threshold 5 μm past the center of the constriction (right yellow dotted line). The white dashed line indicates the outline of the nucleus for demonstration purposes. The center of the constriction is marked with a red dashed line. See Supplemental Data for representative time-lapse microscopy sequences of Lmna+/+, Lmna+/− and Lmna−/− cells. Scale bar: 20 μm.
Single cell migration experiments and analysis
Single cell migration assays were performed using small chambers built by punching 1 cm diameter holes into PDMS slabs approximately 5 mm thick and bonding these slabs onto glass slides to obtain multiple wells. The size of the glass slides was such that 6 wells could fit on one glass slide, allowing observation of the three cell types, with and without PDGF, during the same experiment. Before experiments, the wells were sterilized with 70% ethanol and incubated with 0.2 mg/mL fibronectin (Millipore) in PBS at 37°C for at least 2 hours. Cells were passaged, seeded at 100 cells/well, and allowed to adhere for at least 6 hours prior to the start of experiments. Immediately before imaging, the medium on the cells was replaced with phenol red-free medium containing 25 mM HEPES (Gibco). Half of the wells also contained 100 ng/mL PDGF; the concentration was chosen to be half-maximal concentration in the chemotactic gradient, thereby matching the average PDGF concentration inside the constriction channels. After replacing the medium, a glass slide was placed over the wells to limit evaporation; the PDMS assembly was then placed in a temperature-controlled chamber on a Zeiss LSM 700 confocal microscope (AxioObserver) equipped with a CCD camera (CoolSNAP EZ, Photometrics) and a motorized stage (Zeiss). Time-lapse images were recorded every 10 minutes for 8 to 14 hours total. Image sequences used for the analysis of cell migration were acquired with a 20× objective in widefield/phase contrast mode. Following the experiment, the position of the cells was tracked using the Manual Tracking plugin in ImageJ, and the average velocities were obtained. Cells that divided or were in contact with other cells were discarded from the analysis.
Statistical analysis
Experiments were performed at least three independent times. Statistical analysis was performed with PRISM 3.0 software (GraphPad) and SAS v 9.13 (SAS Institute 2003). Data representing two groups were compared by unpaired Student’s t-test with Welch’s correction to allow for different variances. For samples that did not follow a Gaussian distribution (perfusion transit times), data were compared with the non-parametric Mann-Whitney test. Data representing three groups (migration and perfusion transit times and cell number studies; Figs. 4, 5 and 7) were compared using an omnibus F-test and Tukey-Kramer multiple-means-analysis. Statistical correlation between constriction size and migration transit time for each cell line was analyzed using the Pearson test (Fig. 5). The data in the cell number study (Fig. 7) were transformed using a log(x+1) transformation prior to analysis to better meet the assumptions of variance of the ANOVA. Data are expressed as arithmetic mean ± S.E.M. For all experiments, a two-tailed P-value of less than 0.05 was considered significant.
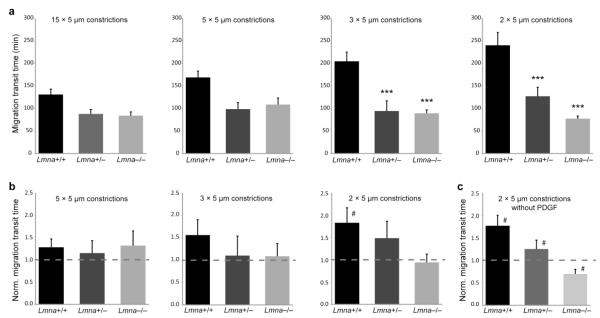
Figure 5. Reduced lamin A/C levels facilitate migration through narrow constrictions.
a) Migration transit times for Lmna+/+, Lmna+/− and Lmna−/− cells migrating through constrictions 15, 5, 3, or 2 μm wide and 5 μm tall. Wild-type (Lmna+/+) cells take significantly longer to pass through the 2 × 5 μm2 and 3 × 5 μm2 constrictions than cells with reduced lamin A/C levels. For wild-type cells, but not Lmna+/− and Lmna−/− cells, migration transit times were negatively correlated with constriction size (P < 0.01). N = 24 to 41 cells per genotype, constriction; ***, P < 0.001 compared to Lmna+/+ cells. b) Migration transit times through narrow constrictions normalized to the corresponding migration transit times through 15 μm-wide channels, which are larger than the nucleus. Wild-type cells have impaired migration efficiency through the smallest constrictions, indicated by increased normalized migration transit times, while cells with reduced levels of lamins A/C maintain comparable migration transit times for all constriction sizes. N = 24 to 41 cells per genotype, constriction; #, P < 0.05 compared to migration transit times through 15 μm channels. c) Migration transit times through 2 × 5 μm2 constrictions normalized to the 15 μm channels without the presence of a chemoattractant gradient; #, P < 0.05 compared to migration transit times through 15 μm channels.
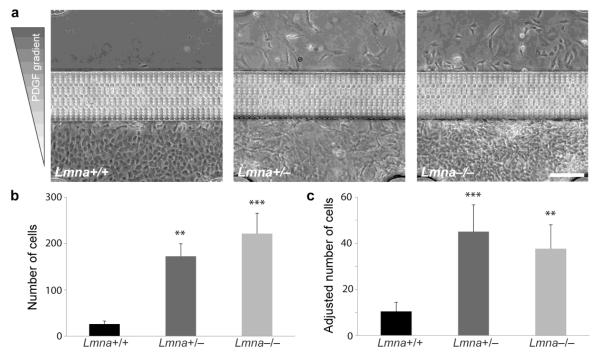
Figure 7. Cells with reduced levels of lamin A/C migrate through narrow constrictions more efficiently.
a) Representative images of migration devices after 48 hours of migration, revealing increased numbers of Lmna+/− and Lmna−/− cells that have passed through the constrictions. b) Quantitative analysis of cell numbers that have passed through the channels after 48 hours. Cells with a reduced expression of lamins A/C had significantly larger numbers than wild-type cells. c) Cell numbers were adjusted for the different proliferation rates between cell lines. The adjusted data confirmed that cells with reduced levels of lamins A/C were more efficient at migrating through the narrow constrictions. N = 8 (from three independent experiments); **, P < 0.01, ***, P < 0.001, compared to Lmna+/+ cells. Scale bar: 200 μm.
Results
Nuclear deformation during migration in confined 3-D environments
It is now well established that cell motility in 3-D environments can substantially deviate from cell migration on flat substrates.5, 6 Recent reports further suggest that the deformability of the cell, and particularly the nucleus, may present a rate-limiting factor in 3-D migration.7, 10, 24 However, most current strategies to study cell migration in 3-D environments are limited in their ability to quantitatively assess the effect of the physical environment on single cells: collagen matrices used for 3-D cell migration contain randomly distributed pores of variable size10 and the dense collagen network can make high resolution imaging challenging; transwell migration assays and Boyden chambers, on the other hand, have precisely defined pore sizes but do not allow real-time imaging of the cells travelling through the narrow constrictions at high resolution. We have therefore designed a novel microfluidic device in which cells migrate along a chemotactic gradient through well-defined constrictions (Fig. 1). The device is composed of an array of 5 μm tall channels containing a series of increasingly narrower constrictions measuring 5, 3 and 2 μm in width, respectively. Small lateral openings (Fig. 1a, white arrowhead) between the channels assure a uniform chemotactic gradient across the constrictions and nutrient supply to the cells, while intermittent 15 μm wide channels (Fig. 1a, black arrowhead) serve as controls to account for inherent differences in migration speed. By imaging the fluorescence intensity distribution of TexasRed-labeled dextran, we confirmed that gradients formed rapidly (<30 min) within the device and remained stable over the time-course of the experiments (Fig. 1b, c). Using this device, we obtained high resolution time-lapse sequences of fibroblasts moving along a PDGF gradient through the constrictions, revealing substantial nuclear deformations as the cells squeezed through the constrictions (Fig. 2a-d), including deformation of the nuclear lamina (Fig. 2c). In some cases, what appeared like buckling of the nuclear lamina was observed in cells expressing GFP-lamin A (Fig. 2c), possibly due to the presumably more rigid nuclear lamina in these cells. Importantly, the cells took up the entire height of the channels and adhered to both the top and the bottom of the fibronectin-coated channels (Fig. 2b), indicating that the device presents a true 3-D environment to the cell. Furthermore, the height of the channels (5 μm) was sufficient to confine the nucleus, as evidenced by the reduction in nuclear height as cells entered the channels (Fig. 2d).
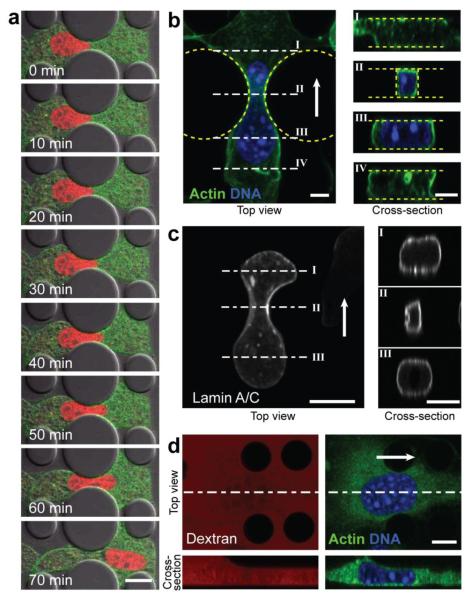
Figure 2. Nuclear deformation during migration through narrow constrictions.
a) Time-lapse image sequence of NIH 3T3 cells expressing fluorescently tagged Histone-4 (red) and actin (green) migrating through a 3 μm wide constriction, revealing substantial nuclear deformation. Scale bar: 20 μm. b) Top-view and cross-sections of a confocal 3-D reconstruction of a cell migrating through a 3 μm-wide constriction, stained for DNA (blue) and F-actin (green). The cross-sections (right) demonstrate that the cell takes up the entire height of the device and that the nucleus fills out the constriction. Scale bar: 5 μm. White arrow denotes direction of migration. c) Confocal 3-D reconstruction of a cell expressing GFP-lamin A migrating through a 2 μm-wide constriction. The cross-section through the constriction (II) suggests compression and buckling of the nuclear lamina inside the constriction. Scale bar: 5 μm. d) Confocal top- and side-view of a single cell entering the 5 μm tall constriction channel inside the migration device. The channels were filled with fluorescently labeled dextran (red; left images) and cells were stained for DNA (blue) and expressed fluorescent actin (green). As the cell enters the 5 μm tall channel, the cell and nuclear height adjusts to the available height of the channel (compare bottom left and right images). Scale bar: 10 μm.
Cells with lower lamin A/C expression have more deformable nuclei
Given the large nuclear deformations required to pass through the narrow constrictions in our microfluidic channels and observed during non-proteolytic cell migration in dense collagen matrices,10 we hypothesized that cells with more deformable nuclei may be able to transit faster through constrictions that are smaller than the nuclear diameter. The deformability of the nucleus is primarily governed by the chromatin and the mechanical properties of the nuclear envelope.11 In particular, the nuclear envelope proteins lamins A/C are one of the major determinants of nuclear deformability, and MEFs from lamin A/C-deficient mice (Lmna−/−) have substantially softer nuclei than cells from wild-type littermates (Lmna+/+), with heterozygous cells (Lmna+/−) displaying an intermediate phenotype.14 As the Lmna+/+, Lmna+/− and Lmna−/− MEFs have been well characterized in terms of their nuclear mechanics14, 15 and their migration on 2-D substrates,25-28 we focused our subsequent studies on these cells. To confirm the increased nuclear deformability of the lamin A/C-deficient cells during passage through narrow constrictions, we perfused cells through microfluidic channels with constrictions smaller than the size of the nucleus (Fig. 3a). The lamin A/C-deficient cells passed through the constrictions significantly faster than wild-type controls (Fig. 3a-b), indicating that they had softer nuclei that required less time (and energy) to deform. We further confirmed the increased nuclear deformability of the lamin A/C-deficient cells using nuclear micropipette aspiration assays (Fig. 3c), consistent with previous results of cells with reduced levels of lamins A/C.29-31
Cells with lower lamin A/C levels migrate faster through narrow constrictions
To test whether reduced levels of lamins A/C and the associated increased nuclear deformability could improve the ability of cells to migrate through confined environments, we measured the migration transit time (Fig. 4) of Lmna+/+, Lmna+/− and Lmna−/− MEFs cells migrating through precisely-defined constrictions of 5 μm height and 5 μm, 3 μm, or 2 μm width (see Fig. 1). The cells with reduced lamin A/C levels (Lmna+/− and Lmna−/−) had significantly shorter migration transit times through the 3 μm and 2 μm wide constrictions than the wild-type cells (Fig. 5a; see also Supplemental Data for representative videos); the difference between the Lmna+/− and Lmna−/− cells was not statistically significant. In the 5 μm wide constrictions, the cells with lower lamin A/C levels displayed a trend towards faster migration transit times, but the difference between the cell lines was not quite statistically significant (P = 0.057).
To account for inherent differences in the migration speeds between the three cell lines in unconfined conditions (Fig. 6a, b), we normalized migration transit times through the various constrictions by the corresponding migration transit times in the 15 μm-wide channels for each cell line. This normalization allows for a more general comparison of the effect of small constrictions on the migration behavior of the different cell lines. A normalized migration transit time larger than ‘1’ indicates that cells from a given cell line take longer to migrate through a given constriction than the same cells would take to pass through a large channel that does not require nuclear deformation. For the 5 μm-wide constrictions, all three cell lines had normalized migration transit times close to ‘1’, indicating that this constriction size had no significant effect on cell migration (Fig. 5b, left graph). However, when further reducing the constriction size, this resulted in progressively increasing normalized migration transit times for the wild-type cells, particularly in the 2 μm-wide constrictions (Fig. 5b, right graph). These data suggest that in wild-type cells, nuclear deformability becomes rate-limiting when passing through openings substantially smaller than the nucleus. In contrast, for cells with reduced levels of lamins A/C, restricting the constriction size down to as small as 2 × 5 μm2 had no effect on the migration transit time. In fact, for these cells the migration transit times through the narrow constrictions were indistinguishable from those for the 15 μm-wide channels, i.e., the normalized migration transit time remained close to ‘1’ (Fig. 5b, right graph), suggesting that the nuclei of these cells do not significantly resist deformation in the constriction sizes tested.
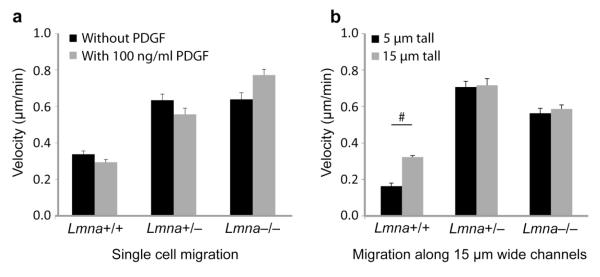
Figure 6. Cell migration velocity in unconfined conditions.
a) Single cell migration analysis of mouse embryonic fibroblasts migrating on 2-D fibronectin-coated glass substrates, indicating that wild-type (Lmna+/+) mouse embryonic fibroblasts migrate slower than Lmna+/− and Lmna−/− cells. PDGF stimulation had no significant effect on migration velocity. b) Migration velocity in the direction of a chemoattractant (PDGF) gradient in 15 μm wide channels with heights of 5 μm (3-D migration) or 15 μm (2.5-D migration). Consistent with the 2-D migration experiments, wild-type cells moved more slowly than the lamin A/C-deficient cells; unlike the lamin A/C-deficient cells, wild-type cells had impeded migration speeds in the 5 μm tall channels compared to the 15 μm tall channels, indicating a possible effect of partial nuclear compression (Fig. 2D).
Effect of lamin expression on cell migration is independent of PDGF-gradient
To test whether the results depend on the specific chemotactic gradient used here (i.e., PDGF), we also performed experiments on cells migrating through the constriction channels without a chemotactic gradient. We found that the experimental observation remained the same: the Lmna+/+ cells took significantly longer to pass through the narrow (2 × 5 μm2) constrictions than the 15 μm-wide constrictions (P < 0.001), and the Lmna+/+ cells had significantly increased normalized migration transit times through the 2 × 5 μm2 constrictions compared to Lmna−/− cells (Fig. 5c). Just as in the experiments with the PDGF gradient, Lmna+/− cells displayed an intermediate phenotype between the Lmna+/+ and the Lmna−/− cells. Taken together, these results suggest that differences in lamin A/C expression and the associated changes in nuclear deformability, rather than differential responses to the chemotactic gradient, are responsible for the different abilities to pass through constrictions smaller than the nuclear diameter.
We conducted additional assays in which cells were induced to migrate through the constrictions in the absence or presence of a chemotactic gradient while being maintained in a cell culture incubator. For these experiments, we used devices without the 15 μm-wide channels to ensure that all cells had migrated through the narrow constriction channels. After 48 hours, significantly more Lmna+/− and Lmna−/− cells had passed through the constriction than wild-type cells (Fig. 7a b), confirming the results obtained in the time-lapse microscopy studies. However, we observed that lamin A/C-deficient cells proliferate faster than wild-type cells (doubling times were 0.68, 1.0, and 1.6 days for the Lmna−/−, Lmna+/−, and Lmna+/+ cells, respectively), which could affect the number of cells on both sides of the constriction after 48 hours despite identical initial seeding densities. We therefore also compared migration efficiency after normalizing the cell numbers to the proliferation rate for each cell line. Importantly, even after accounting for the different proliferation rates, cells with reduced levels of lamins A/C had significantly higher migration efficiencies through the narrow constrictions than wild-type cells, while differences between Lmna+/−, and Lmna−/− cells were not statistically significant (Fig. 7c). Experiments performed without a PDGF gradient produced similar findings (data not shown). Taken together, these data demonstrate that cells with reduced levels of lamins A/C, which have more deformable nuclei, are significantly more efficient at migrating through constrictions smaller than the size of the nucleus.
Discussion
Using novel microfluidic migration devices that enable imaging of nuclear deformation during cell passage through tight constrictions in 3-D environments, we found that reduced levels of lamins A/C result in enhanced transit efficiency during passive perfusion and active cell migration. These results provide further support for an important role of nuclear mechanics in 3-D cell migration and indicate that changes in nuclear envelope composition, which often affect nuclear deformability, can have important implications in the ability of cells to move through dense tissues or other confined spaces. Cells embedded in dense collagen matrices show severe nuclear deformations when MMP activity is inhibited, and reducing the pore size between collagen fibers to less than ≈6 μm2 (or about 1/10 of the cross-sectional area of the unconfined nucleus) causes stalled cell migration and nuclei forming long protrusions as they unsuccessfully attempt to pass through the constriction.10 Similarly, breast cancer cells moving through microfluidic PDMS channels have significantly decreased rates of migration when the channel size is reduced below 8 μm × 5 μm,12 and osteosarcoma cell migration is impaired when channel width is reduced below 20 μm.13
In our experiments, we did not observe permanent arrest of cell migration, even in the 2 × 5 μm2 constrictions; however, when using migration channels with similar design but a channel height of only 3 μm, migration efficiency was substantially lower than in the 5 μm tall constriction channels, particularly for the wild-type cells (data not shown). Importantly, even though the cells in our experiments had not reached the ‘nuclear deformation limit’ beyond which 3-D cell migration stalls,10 nuclear deformability had nonetheless a strong effect on migration efficiency through constrictions smaller than the unconfined nuclear diameter. In wild-type cells, migration transit times negatively correlated with a decrease in constriction size (P < 0.01), and migration through 2 × 5 μm2-sized constrictions was significantly slower than through the 15 × 5 μm2-sized reference channels. In contrast, cells with reduced levels of lamins A/C, which have more deformable nuclei,14, 15 needed less time to pass through the narrow constrictions than wild-type cells; furthermore, their migration transit times were independent of the size of the constriction, at least down to the 2 × 5 μm2-sized constrictions used in our experiments.
To further compare the migration behavior of cells in 2-D and 3-D environments, we also performed single cell migration experiments on 2-D glass substrates. In these experiments, wild-type cells were significantly slower than lamin-A/C deficient cells (Fig. 6), suggesting some inherent cell line-to-cell line variation and prompting us to use normalized migration transit times for further analysis to account for differences in migration speed in unconfined conditions. In addition, we also used migration devices with 15 μm tall channels to assess the migration speeds under the influence of a chemotactic gradient in 2.5-D, i.e., cells adhering to the side-walls and bottom or top, but not the top and bottom simultaneously. Similar to the results of the 2-D experiments, the wild-type cells migrated more slowly than the lamin A/C-deficient cells. Interestingly, while we observed similar migration speeds between 2.5-D and 3-D migration for the lamin A/C-deficient cells (Fig. 6b), the wild-type cells had lower migration velocities in the 5 μm tall channels (3-D migration) compared to the 15 μm tall channels (2.5-D migration), suggesting that wild-type cells already resist nuclear compression when reducing the available height down to 5 μm (see Fig. 2d), impairing their migration. Thus, normalizing the migration transit times with the (faster) transit times through the 15 μm wide sections in the 15 μm tall channels, rather than the (slower) times for the 5 μm tall channels as done in the current analysis, would result in even greater differences between the wild-type and lamin A/C-deficient cells.
As PDGF is a known effector of lamin A/C phosphorylation,32 and lamin A/C can conversely affect several signaling pathways involved in cell migration,33 we performed additional experiments to assess the effect of the PDGF chemoattractant on cell migration in wild-type and lamin A/C-deficient cells in more detail. Wild-type cells, but not lamin A/C-deficient cells, responded to the PDGF gradient by increasing their migration speed through the constriction channels compared to cells migrating without a PDGF gradient. However, this effect was independent of the constriction size (data not shown); hence, the PDGF gradient had little effect on the normalized migration transit times (compare Figs. 5b, c). Importantly, migration transit times for wild-type cells were always significantly longer than corresponding transit times for lamin A/C-deficient cells, regardless of the absence or presence of a PDGF gradient. Thus, the rate-limiting role of nuclear deformability in 3-D migration through narrow constrictions, governed by levels of lamins A/C, was independent of the chemoattractant used in the experiments.
Furthermore, the improved migration efficiency of the lamin A/C-deficient cells during 3-D migration through narrow constrictions could not be explained by differences in nuclear size, as the cells with reduced lamin A/C levels had larger nuclei than the control cells, with nuclear cross-sectional areas of 330 ± 10 μm2, 320 ± 10 μm2, and 210 ± 10 μm2 for Lmna−/−, Lmna+/− and Lmna+/+ MEFs, respectively. In fact, the larger nuclear size of the Lmna−/− and Lmna+/− cells would be expected to impair cell migration through narrow constrictions, pointing to an even greater effect of lamin A/C levels on cell motility. Further support for the importance of nuclear lamins and nuclear deformability on 3-D cell migration comes from a recent study in which cells expressing progerin, a lamin A mutant that results in more rigid nuclei,34, 35 had difficulties passing between microposts positioned 6 μm apart, but showed no difference in unconfined migration.36 Similarly, ectopic overexpression of lamin A dramatically reduced the ability of neutrophil-like cells to pass through narrow constrictions during passive perfusion or active migration.20 Importantly, neutrophils normally have highly lobulated nuclei and downregulate lamins A/C and other nuclear envelope proteins during granulopoiesis while upregulating expression of lamin B receptor (LBR);37 it has long been speculated that this change in nuclear envelope composition and the associated changes in nuclear shape and stiffness enhance the ability of neutrophils to migrate through microscopic openings between endothelial cells during extravasation and promote mobility in tissues during infection.7, 20 Developmentally or environmentally regulated changes in lamin A/C expression have also been postulated to contribute to enhanced nuclear deformability in stem cells and may contribute to the correct trafficking of hematopoietic stem cells.29, 38
Interestingly, even though Lmna+/− cells have only 50% reduced levels of lamins A/C and their nuclear deformability falls between that of Lmna−/− and wild-type cells, the ability of Lmna+/− cells to migrate through narrow constrictions was virtually indistinguishable from that of Lmna−/− cells (Figs. 5, 7). One possible reason is that lamins A/C form an extended part of the LINC (Linker of nucleoskeleton and cytoskeleton) complex.39, 40 Consequently, complete loss of lamins A/C will not only increase nuclear deformability, which should enhance migration efficiency, but can also disrupt force transmission between the nucleus and the cytoskeleton, which could negatively affect cell migration.24 Consistent with this idea, recent studies found that complete loss of lamins A/C or LINC complex disruption impairs the physical coupling between the actin cytoskeleton and nucleus, resulting in impaired cell polarization and migration on 2-D substrates25-28, 41 and in 3-D collagen matrices.42 Cells with reduced (but not absent) levels of lamins A/C, such as the Lmna+/− MEFs, may thus benefit from the increased nuclear deformability caused by reduced lamin A/C expression while retaining near-normal nucleocytoskeletal coupling.
In conclusion, the role of lamins A/C in contributing to nuclear stiffness has important implications in determining the ability of cells to pass through narrow constrictions during 3-D migration or perfusion through narrow capillaries. These properties are not only relevant in various physiological processes, but are also attracting increasing interest in the study of cancer metastasis, as altered expression of lamins has been reported in a variety of human cancers (reviewed in 33). Breast cancer tumor cells, for example, often have reduced expression of lamins A/C,43, 44 and loss of lamins A/C negatively correlates with disease-free survival.43 The increased nuclear deformability associated with reduced lamin A/C levels may promote invasion of metastatic cancer cells into surrounding tissues and spreading through the vascular and lymphatic system, thereby constituting a substantial risk factor on top of gene-regulatory changes linked to altered lamin expression.33 Thus, an improved understanding of the multifaceted function of lamins has the potential to uncover novel therapeutic targets in cancer metastasis and diseases caused by mutations in nuclear envelope proteins.
Supplementary Material
Supplemental Video 1: Lmna+/+ cell migrating through a narrow constriction. Representative time-lapse video of a wild-type (Lmna+/+) mouse embryonic fibroblast migrating through a 3 × 5 μm2 constriction. To aid the viewer, the nucleus is outlined with a dashed white line at the beginning of the video. The vertical white lines indicate the beginning (left) and end (right) of the constriction, as defined for the migration transit time measurements (compare with Figure 4). Time measurements (indicated in the top left corner of the image) are based relative to the nucleus entering the constriction. In this video, the migration transit time was 3:20 h. The time interval between images was 10 minutes; scale bar: 25 μm.
Supplemental Video 2: Lmna+/− cell migrating through a narrow constriction. Representative time-lapse video of a Lmna+/− mouse embryonic fibroblast migrating through a 3 × 5 μm2 constriction. To aid the viewer, the nucleus is outlined with a dashed white line at the beginning of the video. The vertical white lines indicate the beginning (left) and end (right) of the constriction, as defined for the migration transit time measurements (compare with Figure 4). Time measurements (indicated in the top left corner of the image) are based relative to the nucleus entering the constriction. In this video, the migration transit time was 1:40 h. The time interval between images was 10 minutes; scale bar: 25 μm.
Supplemental Video 3: Lmna−/− cell migrating through a narrow constriction. Representative time-lapse video of a lamin A/C-deficient (Lmna−/−) mouse embryonic fibroblast migrating through a 3 × 5 μm2 constriction. To aid the viewer, the nucleus is outlined with a dashed white line at the beginning of the video. The vertical white lines indicate the beginning (left) and end (right) of the constriction, as defined for the migration transit time measurements (compare with Figure 4). Time measurements (indicated in the top left corner of the image) are based relative to the nucleus entering the constriction. In this video, the migration transit time was 1:40 h. The time interval between images was 10 minutes; scale bar: 25 μm.
Acknowledgements
The authors thank Philipp Isermann for providing plasmids (GFP-LifeAct, mCherry–Histone-4), advice, and some MATLAB image analysis scripts; Drs. Colin Stewart and Tom Glover for providing cells and reagents; Dr. Amy Rowat for design and fabrication of some of the perfusion devices; Dr. Sarah Jandricic for help with statistical analysis; Kathy Zhang and Ileana D’Aloisio for quantification of cell migration assays; and Rachel Gilbert for helpful discussions. This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECCS-0335765). This work was supported by National Institutes of Health awards [R01 NS059348 and R01 HL082792]; the Department of Defense Breast Cancer Idea Award [BC102152]; a National Science Foundation CAREER award to Lammerding J [CBET-1254846]; and a Pilot Project Award by the Cornell Center on the Microenvironment & Metastasis through Award Number U54CA143876 from the National Cancer Institute. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations, symbols, and terminology
- 2-D
two-dimensional
- 3-D
three-dimensional
- ANOVA
analysis of variance
- BSA
bovine serum albumin
- DMEM
Dulbecco Modified Eagle Medium
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- LINC
linker of nucleoskeleton and cytoskeleton
- MEF
mouse embryonic fibroblast
- MMP
matrix metalloproteinase
- PBS
phosphate buffered saline
- PDGF
platelet derived growth factor
- PDMS
polydimethylsiloxane
Bio
Dr. Jan Lammerding is an Assistant Professor in the Department of Biomedical Engineering and the Weill Institute for Cell and Molecular Biology at Cornell University. He received a Bachelor of Engineering degree from the Thayer School of Engineering at Dartmouth College, a Diplom Ingenieur degree in Mechanical Engineering from the University of Technology Aachen, Germany, and a Ph.D. in Biological Engineering from the Massachusetts Institute of Technology (MIT). Before joining Cornell University, Dr. Lammerding served as a faculty member at Harvard Medical School/Brigham and Women’s Hospital (BWH) while also teaching in the Department of Biological Engineering at MIT. At Cornell, the Lammerding laboratory is developing novel experimental techniques to investigate the interplay between cellular mechanics and function, with a particular emphasis on the cell nucleus and its response to mechanical forces. Dr. Lammerding has won several prestigious awards, including a National Science Foundation CAREER Award, an American Heart Association Scientist Development Grant, and the BWH Department of Medicine Young Investigator Award. Dr. Lammerding has published over 40 peer-reviewed articles, including in Nature and PNAS. His research is supported by grants from the National Institutes of Health, the National Science Foundation, the Department of Defense Breast Cancer Research Program, the American Heart Association, and the Progeria Research Foundation.
Footnotes
Conflicts of Interest Patricia M. Davidson, Celine Denais, Maya C. Bakshi, and Jan Lammerding declare that they have no conflicts of interest.
Ethical Standards No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26(10):4045–56. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, Yang JT, Konstantopoulos K. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J Cell Biol. 2013;202(5):807–24. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188(1):11–9. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. J Cell Sci. 2012;125(Pt 24):5917–26. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedl P, Sahai E, Weiss S, Yamada KM. New dimensions in cell migration. Nat Rev Mol Cell Biol. 2012;13(11):743–7. doi: 10.1038/nrm3459. [DOI] [PubMed] [Google Scholar]
- 7.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23(1):55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoitzner P, Pfaller K, Stossel H, Romani N. A close-up view of migrating Langerhans cells in the skin. J Invest Dermatol. 2002;118(1):117–25. doi: 10.1046/j.0022-202x.2001.01631.x. [DOI] [PubMed] [Google Scholar]
- 9.Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. Principles of interface guidance and microvesicle dynamics. IntraVital. 2012;1(1):32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201(7):1069–84. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson PM, Lammerding J. Broken nuclei - lamins, nuclear mechanics, and disease. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Chin LK, Bourouina T, Liu AQ, VanDongen AM. Nuclear deformation during breast cancer cell transmigration. Lab Chip. 2012;12(19):3774–8. doi: 10.1039/c2lc40477j. [DOI] [PubMed] [Google Scholar]
- 13.Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS One. 2012;7(1):e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281(35):25768–80. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 15.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113(3):370–8. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147(5):913–20. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glynn MW, Glover TW. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum Mol Genet. 2005;14(20):2959–69. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 18.Verstraeten VL, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci U S A. 2011;108(12):4997–5002. doi: 10.1073/pnas.1019532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbluth MJ, Lam WA, Fletcher DA. Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab Chip. 2008;8(7):1062–70. doi: 10.1039/b802931h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem. 2013;288(12):8610–8. doi: 10.1074/jbc.M112.441535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isermann P, Davidson PM, Sliz JD, Lammerding J. Assays to measure nuclear mechanics in interphase cells. Curr Protoc Cell Biol. 2012 doi: 10.1002/0471143030.cb2216s56. Chapter 22. Unit22 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deguchi S, Maeda K, Ohashi T, Sato M. Flow-induced hardening of endothelial nucleus as an intracellular stress-bearing organelle. J Biomech. 2005;38(9):1751–9. doi: 10.1016/j.jbiomech.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Shao JY, Xu J. A modified micropipette aspiration technique and its application to tether formation from human neutrophils. J Biomech Eng. 2002;124(4):388–96. doi: 10.1115/1.1486469. [DOI] [PubMed] [Google Scholar]
- 24.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204(5):669–82. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95(11):5462–75. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93(7):2542–52. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci U S A. 2011;108(1):131–6. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329(5994):956–9. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F, Speicher DW, Discher DE. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341(6149):1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanovska I, Swift J, Harada T, Pajerowski JD, Discher DE. Physical plasticity of the nucleus and its manipulation. Methods Cell Biol. 2010;98:207–20. doi: 10.1016/S0091-679X(10)98009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104(40):15619–24. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fields AP, Tyler G, Kraft AS, May WS. Role of nuclear protein kinase C in the mitogenic response to platelet-derived growth factor. J Cell Sci. 1990;96(Pt 1):107–14. doi: 10.1242/jcs.96.1.107. [DOI] [PubMed] [Google Scholar]
- 33.Ho CY, Lammerding J. Lamins at a glance. J Cell Sci. 2012;125(Pt 9):2087–93. doi: 10.1242/jcs.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2006;103(27):10271–6. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verstraeten VL, Ji JY, Cummings KS, Lee RT, Lammerding J. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 2008;7(3):383–93. doi: 10.1111/j.1474-9726.2008.00382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Booth-Gauthier EA, Du V, Ghibaudo M, Rape AD, Dahl KN, Ladoux B. Hutchinson-Gilford progeria syndrome alters nuclear shape and reduces cell motility in three dimensional model substrates. Integr Biol (Camb) 2013;5(3):569–77. doi: 10.1039/c3ib20231c. [DOI] [PubMed] [Google Scholar]
- 37.Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, Karakesisoglou I, Hodzic D, Olins DE. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88(4):203–14. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin JW, Spinler KR, Swift J, Chasis JA, Mohandas N, Discher DE. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc Natl Acad Sci U S A. 2013;110(47):18892–7. doi: 10.1073/pnas.1304996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172(1):41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152(6):1376–89. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286(30):26743–53. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu PH, Marchand J, Celedon A, Hale CM, Sun SX, Hodzic D, Wirtz D. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wazir U, Ahmed MH, Bridger JM, Harvey A, Jiang WG, Sharma AK, Mokbel K. The clinicopathological significance of lamin A/C, lamin B1 and lamin B receptor mRNA expression in human breast cancer. Cell Mol Biol Lett. 2013;18(4):595–611. doi: 10.2478/s11658-013-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capo-chichi CD, Cai KQ, Smedberg J, Ganjei-Azar P, Godwin AK, Xu XX. Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin J Cancer. 2011;30(6):415–25. doi: 10.5732/cjc.010.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video 1: Lmna+/+ cell migrating through a narrow constriction. Representative time-lapse video of a wild-type (Lmna+/+) mouse embryonic fibroblast migrating through a 3 × 5 μm2 constriction. To aid the viewer, the nucleus is outlined with a dashed white line at the beginning of the video. The vertical white lines indicate the beginning (left) and end (right) of the constriction, as defined for the migration transit time measurements (compare with Figure 4). Time measurements (indicated in the top left corner of the image) are based relative to the nucleus entering the constriction. In this video, the migration transit time was 3:20 h. The time interval between images was 10 minutes; scale bar: 25 μm.
Supplemental Video 2: Lmna+/− cell migrating through a narrow constriction. Representative time-lapse video of a Lmna+/− mouse embryonic fibroblast migrating through a 3 × 5 μm2 constriction. To aid the viewer, the nucleus is outlined with a dashed white line at the beginning of the video. The vertical white lines indicate the beginning (left) and end (right) of the constriction, as defined for the migration transit time measurements (compare with Figure 4). Time measurements (indicated in the top left corner of the image) are based relative to the nucleus entering the constriction. In this video, the migration transit time was 1:40 h. The time interval between images was 10 minutes; scale bar: 25 μm.
Supplemental Video 3: Lmna−/− cell migrating through a narrow constriction. Representative time-lapse video of a lamin A/C-deficient (Lmna−/−) mouse embryonic fibroblast migrating through a 3 × 5 μm2 constriction. To aid the viewer, the nucleus is outlined with a dashed white line at the beginning of the video. The vertical white lines indicate the beginning (left) and end (right) of the constriction, as defined for the migration transit time measurements (compare with Figure 4). Time measurements (indicated in the top left corner of the image) are based relative to the nucleus entering the constriction. In this video, the migration transit time was 1:40 h. The time interval between images was 10 minutes; scale bar: 25 μm.