Abstract
Lupus nephritis is a manifestation of SLE resulting from glomerular immune complex deposition and inflammation. Lupus nephritis demonstrates familial aggregation and accounts for significant morbidity and mortality. We completed a meta-analysis of three genome-wide association studies of SLE to identify lupus nephritis–predisposing loci. Through genotyping and imputation, >1.6 million markers were assessed in 2000 unrelated women of European descent with SLE (588 patients with lupus nephritis and 1412 patients with lupus without nephritis). Tests of association were computed using logistic regression adjusting for population substructure. The strongest evidence for association was observed outside the MHC and included markers localized to 4q11-q13 (PDGFRA, GSX2; P=4.5×10−7), 16p12 (SLC5A11; P=5.1×10−7), 6p22 (ID4; P=7.4×10−7), and 8q24.12 (HAS2, SNTB1; P=1.1×10−6). Both HLA-DR2 and HLA-DR3, two well established lupus susceptibility loci, showed evidence of association with lupus nephritis (P=0.06 and P=3.7×10−5, respectively). Within the class I region, rs9263871 (C6orf15-HCG22) had the strongest evidence of association with lupus nephritis independent of HLA-DR2 and HLA-DR3 (P=8.5×10−6). Consistent with a functional role in lupus nephritis, intra-renal mRNA levels of PDGFRA and associated pathway members showed significant enrichment in patients with lupus nephritis (n=32) compared with controls (n=15). Results from this large-scale genome-wide investigation of lupus nephritis provide evidence of multiple biologically relevant lupus nephritis susceptibility loci.
Keywords: lupus nephritis, genetic renal disease, epidemiology, outcomes, SLE
SLE is a prototypic autoimmune disease that disproportionately affects women (ratio of female to male patients, 9:1). It is characterized by the development of autoantibodies directed against nuclear and cellular components and the activation of inflammatory cascades, resulting in multisystem organ damage. Lupus nephritis (LN) is common, affecting 30%–40% of women of European descent with SLE and accounting for significant morbidity and mortality.1
CKDs are heavily influenced by both genetic and environmental factors.2 A genetic component to LN susceptibility is supported by an over-representation of LN among children of parents with SLE, familial aggregation of ESRD in African Americans with LN,3 linkage studies and candidate gene studies.4–7 Genome-wide association studies (GWAS) have been extremely successful in identifying susceptibility loci for many disease phenotypes, including SLE8–15 and related endophenotypes.16,17 Herein, we report the first large-scale GWAS to identify loci predisposing to LN among women with SLE.
Results
Table 1 summarizes the clinical characteristics of the 2000 unrelated women of European ancestry who meet the American College of Rheumatology (ACR) classification criteria for SLE and are included in this study. Of the 2000 patients with SLE, 588 had LN defined by ACR criteria and 1412 had SLE with no evidence of LN. Sets I and II are the largest contributors to the meta-analysis and had comparable proportions of patients with LN. Interestingly, the smallest sample, set III, had nearly double the proportion of LN and a slightly earlier median age of SLE onset. Set II had the highest proportion of patients with SLE from multiplex pedigrees (set I, 0.08; set II, 0.82; and set III, 0.43).
Table 1.
Clinical characteristics of women with SLE included in the meta-analysis of LN genetic susceptibility markers
| SLE Characteristic | Set I (n=1162) | Set II (n=690) | Set III (n=148) |
|---|---|---|---|
| Age at SLE diagnosis (yr) | 33 (25–43) | 33 (3–72) | 25 (8–62) |
| Mean ACR criteria (n)a | 5 (4–6) | 6 (5–7) | 6 (5–7) |
| ACR criteria for SLE | |||
| LN (proteinuria or cellular casts) | 310 (27) | 198 (29) | 80 (54) |
| Malar rash | 562 (48) | 419 (61) | 89 (60) |
| Discoid rash | 106 (9) | 82 (12) | 9 (6) |
| Photosensitivity | 832 (72) | 455 (66) | 73 (49) |
| Oral ulcers | 510 (44) | 293 (43) | 59 (40) |
| Arthritis | 918 (79) | 587 (85) | 117 (79) |
| Serositis (pericarditis or pleuritis) | 432 (37) | 325 (47) | 73 (49) |
| Neurologic (seizure or psychosis) | 106 (9) | 112 (16) | 27 (18) |
| Hematologic (leukopenia, lymphopenia, hemolytic anemia, or thrombocytopenia) | 706 (61) | 403 (59) | 91 (61) |
| Immunologic (anti-double-stranded DNA, anti-Smith, or antiphospholipid antibodies) | 772 (66) | 480 (70) | 115 (78) |
| Anti-nuclear antibody | 1102 (95) | 660 (96) | 144 (97) |
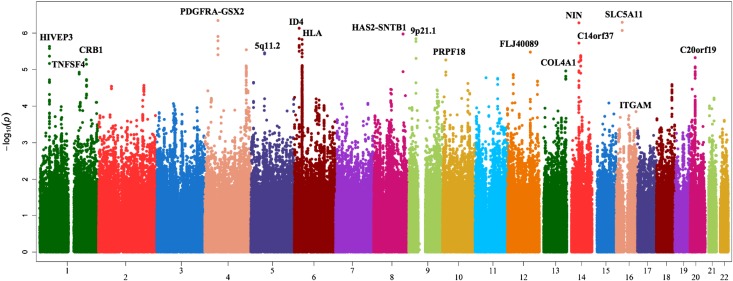
After applying quality control measures, 1,621,689 single nucleotide polymorphisms (SNPs) with genotype or imputed genotype data were available for these 2000 unrelated individuals. After adjusting for potential population substructure in each set via principal components, the meta-analysis inflation factor was 1.05; sets I–III had inflation factors of 1.16, 1.09, and 1.03, respectively. The inflation factor for the best P value (minimum of dominant, additive, and recessive) was 1.70. Supplemental Figure 1 shows the corresponding probability-probability plot. Given the exploratory nature of the study, we report the results considering all genetic models (Table 2) as well as only the additive model (Supplemental Table 1). P values were genomic control adjusted based on the best P value inflation factor. Supplemental Figure 2 presents the power analysis for the 588 patients with LN versus 1412 patients with SLE without nephritis. Figure 1 shows a Manhattan plot of the associations across the genome.
Table 2.
Summary of LN susceptibility markers with the strongest and most consistent evidence for association
| dbSNP Identifier | Genetic Model of Inheritance | Location | Genes within Interval (r2>0.20) | RA | Meta-Analysisa | Set I | Set II | Set III | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Genomic Control P Valueb | OR (95% CI) | Genomic Control P Value | OR (95% CI) | Genomic Control P Value | OR (95% CI) | Genomic Control P Value | |||||
| rs1364989 | R | 4q12 | PDGFRA, GSX2 | T | 3.41 (2.10 to 5.54) | 4.52E-07 | 4.57 (2.48 to 8.42) | 1.30E-06 | 2.32 (1.06 to 5.08) | 3.91E-02 | ||
| rs274068 | R | 16p12.1 | SLC5A11 | C | 2.85 (1.93 to 4.22) | 5.08E-07 | 2.26 (1.38 to 3.70) | 1.40E-03 | 3.87 (2.06 to 7.29) | 2.91E-05 | ||
| rs8012283 | A | 14q22.1 | NIN, SAV1, SPG3A, MAP4K5 | G | 1.64 (1.35 to 1.98) | 5.26E-07 | 1.68 (1.31 to 2.14) | 3.84E-05 | 1.59 (1.17 to 2.14) | 2.94E-03 | ||
| rs7773456 | D | 6p22.3 | None | G | 0.57 (0.46 to 0.70) | 7.36E-07 | 0.63 (0.48 to 0.83) | 1.02E-03 | 0.49 (0.35 to 0.7) | 7.97E-05 | ||
| rs7834765 | R | 8q24.12 | None | T | 3.15 (1.97 to 5.03) | 1.06E-06 | 3.79 (2.13 to 6.76) | 7.39E-06 | 2.48 (1.15 to 5.35) | 2.29E-02 | ||
| rs601162 | R | 9p21.1 | None | A | 3.11 (1.99 to 4.88) | 1.42E-06 | 2.85 (1.58 to 5.16) | 5.93E-04 | 3.49 (1.75 to 6.94) | 4.02E-04 | ||
| rs9267972 | D | 6p21.32 | C6orf10, NOTCh4 | A | 1.85 (1.46 to 2.33) | 1.49E-06 | 1.52 (1.15 to 2.02) | 4.25E-03 | 1.96 (1.33 to 2.89) | 7.32E-04 | 2.78 (1.28 to 6.04) | 1.03E-02 |
| rs4901847 | A | 14q23.1 | C14orf37 | T | 1.48 (1.28 to 1.71) | 1.71E-06 | 1.23 (1.02 to 1.48) | 3.92E-02 | 1.66 (1.32 to 2.10) | 2.23E-05 | 1.91 (1.22 to 3.00) | 5.16E-03 |
| rs752010 | D | 1p34.2 | HIVEP3 | C | 0.57 (0.45 to 0.71) | 2.31E-06 | 0.59 (0.44 to 0.79) | 4.15E-04 | 0.61 (0.42 to 0.89) | 1.02E-02 | 0.43 (0.2 to 0.93) | 3.31E-02 |
| rs6538678 | A | 12q23.1 | FLJ40089 | A | 1.57 (1.30 to 1.88) | 3.33E-06 | 1.52 (1.21 to 1.91) | 3.54E-04 | 1.63 (1.20 to 2.21) | 2.04E-03 | ||
| rs10041935 | R | 5q11.2 | None | C | 2.50 (1.71 to 3.66) | 3.46E-06 | 2.40 (1.49 to 3.88) | 3.86E-04 | 2.64 (1.44 to 4.85) | 1.94E-03 | ||
| rs2236178 | A | 20p11.23 | C20orf19, XRN2, NKX2-4, NKX2-2 | T | 0.68 (0.58 to 0.79) | 4.70E-06 | 0.70 (0.58 to 0.85) | 4.87E-04 | 0.78 (0.61 to 1.00) | 5.63E-02 | 0.45 (0.28 to 0.74) | 1.66E-03 |
| rs2786111 | R | 1q31.3 | CRB1, ZBTB41, ASPM, F13B, CFHR5 | C | 2.26 (1.6 to 3.19) | 5.31E-06 | 2.44 (1.56 to 3.82) | 1.10E-04 | 2.04 (1.19 to 3.52) | 1.10E-02 | ||
| rs648705 | R | 13q34 | COL4A1 | A | 0.48 (0.34 to 0.66) | 1.09E-05 | 0.42 (0.28 to 0.64) | 5.86E-05 | 0.57 (0.34 to 0.95) | 3.49E-02 | ||
| rs2399985 | D | 10p13 | PRPF18, FRMD4A | T | 1.65 (1.33 to 2.06) | 1.16E-05 | 1.58 (1.19 to 2.08) | 1.64E-03 | 1.76 (1.25 to 2.48) | 1.42E-03 | ||
The mode of inheritance is identical for each of the discovery, replication, and combined populations. Common variants listed in this table include only those with a level of statistical significance P≤0.05 across all three populations, a level of statistical significance P≤10−6 in the meta-analysis, and consistent effects across all populations (i.e., protective or at-risk effects). For multiple common variants in a single gene or gene region meeting these criteria, we list the single common variant with the lowest P value. Chromosome positions are based on build 36.1. dbSNP, Single Nucleotide Polymorphism Database; RA, reference allele; R, recessive; A, additive; D, dominant.
The numbers of participants were as follows: meta-analysis, 588 patients with LN versus 1412 patients without LN; set I, 310 patients with LN versus 852 patients without LN; set II, 198 patients with LN versus 492 patients without LN; and set III, 80 patients with LN versus 68 patients without LN.
Genomic control adjustment to the P value for the minimum of the dominant, additive, and recessive genetic models.
Figure 1.
Manhattan plot showing patients with LN in contrast with patients with lupus without LN.
LN Associations within the MHC
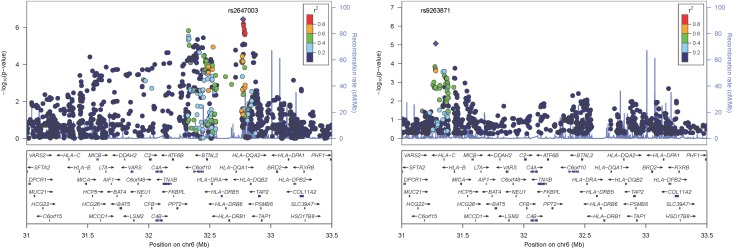
The MHC (chromosome 6: 25–32 Mb) is the most important region for SLE susceptibility, but its role in LN susceptibility is not established. In this study, the SNP most strongly associated with LN within the MHC was rs9267972 (odds ratio [OR], 1.85; 95% confidence interval [95% CI], 1.46 to 2.33; P=1.5×10−6; Figure 2A, Supplemental Table 2), which is located between NOTCH4 and C6orf10.
Figure 2.
HLA region associations with patients with LN in contrast with patients with SLE without LN. (A) Associations adjusting for principal components. (B) Associations adjusting for principal components and HLA SLE risk tagging SNPs rs9271366 (HLA-DR2) and rs2187668 (HLA-DR3).
Because HLA-DRB1*1501 (HLA-DR2) and HLA-DRB1*0301 (HLA-DR3) are well established SLE susceptibility loci, these two loci were examined using their tagSNPs rs9271366 (HLA-DR2) and rs2187668 (HLA-DR3).18 As assessed by these proxies, both HLA-DR2 (OR, 1.37; 95% CI, 1.09 to 1.71; P=0.06) and HLA-DR3 (OR, 1.55; 95% CI, 1.25 to 1.92; P=3.7×10−5) showed evidence of association with LN. Supplemental Table 2 summarizes the MHC associations.
To test whether the MHC associations observed with LN were influenced by linkage disequilibrium with HLA-DR2 and HLA-DR3, each SNP in the MHC region was tested for association while including the HLA-DR2 and HLA-DR3 tagSNPs as covariates in the logistic regression model (Figure 2B, Supplemental Table 3). In these analyses, the previously most significantly associated MHC SNP, rs9267972, was weakly associated with LN (OR, 1.35; 95% CI, 1.00 to 1.84; Padjusted=0.06). The most significantly associated MHC locus independent of HLA-DR2 and HLA-DR3 was rs9263871, located within HCG27 (OR, 1.7; 95% CI, 1.35 to 2.13; Padjusted= 8.5×10−6; Figure 2B, Supplemental Table 3) in the MHC class I region. No other MHC SNPs were associated with LN at Padjusted<1.0×10−5.
LN Associations outside the MHC
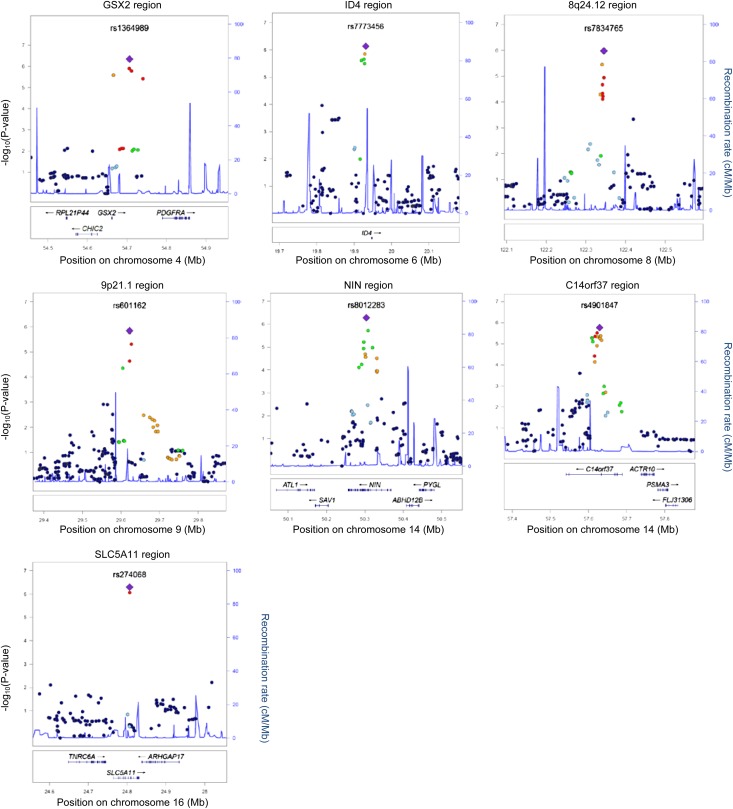
Five loci outside the MHC region showed comparable or stronger evidence of association than those within the MHC (approximate P=1.0×10−6; Figure 3, Table 2). The strongest statistical evidence of association with LN was with rs1364989 (OR, 3.41; 95% CI, 2.10 to 5.54; P=4.5×10−7) on 4q11-q13. This association localized to an intergenic region 83.5 kb upstream of PDGF receptor-α (PDGFRA). The next strongest statistical evidence of association with LN was with rs274068, located within an intronic region of sodium-dependent glucose cotransporter SCL5A11 at 16p12.1 (OR, 2.85; 95% CI, 1.93 to 4.22; P=5.1×10−7; Table 2). A potentially intriguing region of association was within 8q24.12 (rs7834765; OR, 3.15; 95% CI, 1.97 to 5.03; P=1.1×10−6). Although the associated SNPs in this region are intergenic and are not in linkage disequilibrium with any specific gene, the region contains a DNase I hypersensitivity cluster and HAS2 (see the Discussion). The remaining two regions with an approximate P=1×10−6 were in intergenic regions on 6p22 near ID4 and 9p21 (Table 2). Supplemental Tables 1 and 4 provide the summary statistics for the top non-MHC associations for the additive and all genetic models, respectively.
Figure 3.
Regional plots of LN loci. Genotyped and imputed SNPs are plotted with their meta-analysis P values (as -log10 values) as a function of genomic position (Human Genome Build 18) within a 500-kb region surrounding the most significant SNP. Recombination rates from the HapMap phase II CEU (Utah residents with ancestry from northern and western Europe) are plotted in blue to reflect the regional LD structure. In each region, the index SNP is represented by a purple diamond, and the color of all other SNPs (circles) indicates LD with the index SNP based on pairwise r2 values from HapMap CEU (red, r2>0.80; orange, r2=0.60–0.80; green, r2=0.40–0.60; light blue, r2=0.20–0.40; dark blue, r2<0.20). Known human genes in the UCSC Genome Browser (University of California, Santa Cruz, CA) are below each plot.
SLE Susceptibility Loci
Given the frequency of LN among patients with SLE and the manner in which SLE loci have been discovered (i.e., SLE cases versus unaffected controls), some previously discovered SLE susceptibility loci may be LN loci. This hypothesis was tested by taking 31 previously identified SLE susceptibility loci and testing for association with LN under the above case-only design (i.e., patients with LN versus patients with SLE without nephritis; Table 3). Three loci met the Bonferroni corrected significance level (α=0.05/31=1.61×10−3). The most strongly associated SLE susceptibility locus was rs2187668 within the HLA region (OR, 1.55; 95% CI, 1.25 to 1.92; P=3.67×10−5). This SNP tags the well known HLA-DR3 lupus-risk haplotype.18 However, the magnitude of the OR for LN was significantly lower than for SLE. The second most strongly associated SNP was rs2205960 within the 1q25.1 region near TNFSF4 (OR, 2.41; 95% CI, 1.59 to 3.64; P=7.82×10−5). The other SNP that showed association was rs9888739 within ITGAM, located on chromosome 16p11.2 (OR, 1.42; 95% CI, 1.17 to 1.72; P=1.34×10−3), which had an OR for LN that was comparable with SLE.10,19
Table 3.
Reported SLE susceptibility loci and risk of LN among loci included in the LN meta-analysis
| Gene | dbSNP Identifier | Genetic Model of Inheritance | Location | Samples (n) | RA | OR (95% CI) | Genomic Control P Value | Directiona | References | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Set I | Set II | Set III | |||||||||
| IL12RB2 | rs1874791 | D | 1p31.2 | 1987 | A | 0.85 (0.68 to 1.06) | 5.88E-01 | − | + | − | 41 |
| PTPN22 | rs2476601 | A | 1p13.2 | 690 | A | 0.83 (0.22 to 3.1) | 9.84E-01 | ? | + | ? | 10, 41–43 |
| FCGR2A | rs1801274 | D | 1q23.3 | 1786 | A | 1.12 (0.89 to 1.41) | 5.42E-01 | + | + | ? | 10, 43, 44 |
| TNFSF4 | rs2205960 | R | 1q25.1 | 1770 | T | 2.41 (1.59 to 3.64) | 7.82E-05 | + | + | ? | 9, 41 |
| NMNAT2 | rs2022013 | D | 1q25.3 | 1969 | C | 1.07 (0.86 to 1.32) | 8.23E-01 | − | − | − | 9, 10, 41 |
| IL10 | rs3024505 | A | 1q32.1 | 690 | A | 0.89 (0.66 to 1.21) | 7.67E-01 | ? | − | ? | 9, 41 |
| LYST | rs9782955 | D | 1q42.3 | 832 | T | 0.86 (0.63 to 1.18) | 6.05E-01 | ? | − | − | 41 |
| IFIH1 | rs1990760 | R | 2q24.2 | 1779 | C | 0.80 (0.57 to 1.08) | 2.96E-01 | + | + | ? | 41 |
| STAT4 | rs7574865 | D | 2q32.2 | 1137 | T | 1.38 (1.06 to 1.81) | 2.84E-02 | − | ? | ? | 9, 10, 41, 43, 45 |
| PXK | rs6445975 | D | 3q14.3 | 1988 | G | 0.98 (0.8 to 1.21) | 1.00E+00 | + | + | − | 10, 43 |
| BANK1 | rs10516487 | A | 4q24 | 1834 | A | 0.84 (0.72 to 0.99) | 4.10E-02 | − | − | ? | 9–12, 41, 43 |
| PTTG1 | rs2431697 | D | 5q33.3 | 1781 | C | 0.86 (0.69 to 1.07) | 2.60E-01 | + | + | ? | 10, 41, 43 |
| HLA-DR2 | rs9271366 | D | 6p21.32 | 1798 | G | 1.37 (1.09 to 1.71) | 5.73E-02 | + | + | + | |
| HLA-DR3 | rs2187668 | D | 6p21.32 | 1955 | T | 1.55 (1.25 to 1.92) | 3.67E-05 | + | + | + | 11, 41, 43 |
| UHRF1BP1 | rs11755393 | D | 6p21.31 | 1844 | G | 1.08 (0.87 to 1.34) | 8.04E-01 | − | − | ? | 41 |
| ATG5 | rs6568431 | A | 6q21 | 1931 | A | 1.11 (0.96 to 1.29) | 1.68E-01 | + | + | − | 10, 41, 43 |
| TNFAIP3 | rs6920220 | D | 6q23.3 | 1965 | A | 0.92 (0.75 to 1.14) | 5.51E-01 | − | − | + | 41 |
| ICA1 | rs10156091 | A | 7p21.3 | 689 | T | 0.93 (0.66 to 1.32) | 1.00E+00 | ? | − | ? | 10, 41, 43 |
| JAZF1 | rs849142 | R | 7p15.1 | 1993 | C | 1.05 (0.83 to 1.35) | 1.00E+00 | − | − | + | 41 |
| IRF5 | rs12537284 | A | 7q32.1 | 1800 | A | 1.02 (0.84 to 1.23) | 1.00E+00 | − | + | ? | 10, 41 |
| BLK | rs2248932 | R | 8p23.1 | 1836 | A | 1.32 (0.97 to 1.78) | 9.30E-02 | + | + | ? | 9, 10 |
| LYN | rs7829816 | R | 8q12.1 | 1119 | G | 1.30 (0.72 to 2.36) | 6.65E-01 | − | ? | ? | 10 |
| PHRF1-IRF7 | rs4963128 | R | 11p15.5 | 680 | T | 0.62 (0.34 to 1.13) | 2.02E-01 | ? | − | ? | 10, 41, 43 |
| PDHX-CD44 | rs2732552 | A | 11p13 | 1966 | T | 0.82 (0.7 to 0.94) | 1.15E-02 | − | − | − | 46 |
| DDX6 | rs503425 | R | 11q23.3 | 1130 | C | 2.02 (1.15 to 3.53) | 2.30E-02 | − | ? | ? | 9, 41 |
| SLC15A4 | rs6486730 | R | 12q24.32 | 1073 | G | 1.11 (0.79 to 1.56) | 9.11E-01 | − | ? | ? | 41 |
| ITGAM | rs9888739 | A | 16p11.2 | 1841 | T | 1.42 (1.17 to 1.72) | 1.34E-03 | + | + | ? | 10, 41, 43 |
| IRF8 | rs12444486 | R | 16q24.1 | 1148 | T | 0.96 (0.72 to 1.29) | 1.00E+00 | − | ? | ? | 41 |
| CD226 | rs727088 | D | 18q22.2 | 1842 | A | 1.13 (0.88 to 1.44) | 5.95E-01 | + | + | ? | 47 |
| UBE2L3 | rs5754217 | D | 22q11.21 | 1974 | T | 0.98 (0.79 to 1.21) | 1.00E+00 | − | + | + | 10, 41, 43 |
| SCUBE1 | rs2071725 | D | 22q13.2 | 1794 | A | 0.89 (0.69 to 1.14) | 6.68E-01 | − | − | ? | 10 |
dbSNP, Single Nucleotide Polymorphism Database; RA, reference allele; D, dominant; A, additive; R, recessive.
Symbols represent the direction of the risk effect in sets I, II, and III, respectively: ?, not in meta-analysis; −, OR <1; +, OR >1.
The number of SLE risk polymorphisms that an individual carries (i.e., genetic load) may influence risk and may be a strong predictor of LN. However, after adjusting for the above three SLE loci, the risk of LN as a function of the sum of the number of risk alleles across the remaining 28 loci was not statistically significant.
Gene Expression in Renal Biopsies
Renal LN gene expression profiles were examined for the associated regions identified in the LN GWAS meta-analysis (Tables 2 and 3); clinical characteristics of the samples are described in Supplemental Table 5. Thirteen transcripts (Table 4) were detectable in the LN susceptibility regions displaying the strongest GWAS association (Table 2). Most of these 13 transcripts were regulated in the glomerular and tubulointerstitial compartments of patients with LN compared with living donor controls (Table 4). In both compartments, PDGFRA and COL4A1 had the greatest mRNA expression increase, whereas ID4 was underexpressed (Table 4). Similarly, several transcripts from other previously reported SLE GWAS were also regulated in both renal compartments of patients with LN versus living donors (Table 4). Among the established and confirmed SLE susceptibility genes, ITGAM exhibited the highest and most significant change within the glomerular compartment (Table 4). To verify the relevance of those genes in LN disease, an equivalent number of unassociated loci were randomly chosen. The mRNA level of each transcript located around each of the random SNPs showed a significant lower percentage of significantly regulated transcripts (Supplemental Tables 6 and 7).
Table 4.
Renal expression of the genes having a SNP closely localized as assessed by HGU133A arrays from European Renal cDNA Bank Consortium patients with LN compared with controls
| SNP | Genes in the SNP Region | Glomerular Compartment | Tubulointerstitial Compartment | |||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | Gene Symbol | Fold Change | q | P Value | Fold Change | q | P Value | |
| SNPs from Table 2 | ||||||||
| rs1364989 | 5156 | PDGFRAa | 2.57 | <1.1E-04 | 2.86E-05 | 2.40 | <1.3E-04 | 6.03E-06 |
| rs7773456 | 3400 | ID4a | 0.76 | 6.1E-04 | 6.20E-03 | 1.04 | 1.8E-01 | 5.98E-01 |
| rs7834765 | 3037 | HAS2a | 0.95 | 4.3E-02 | 6.22E-02 | 0.92 | 4.2E-03 | 2.53E-02 |
| 6641 | SNTB1a | 1.16 | 2.2E-03 | 2.94E-04 | 1.02 | 2.0E-01 | 5.97E-01 | |
| rs8015968 | 60485 | SAV1a | 1.06 | 2.4E-01 | 6.52E-01 | 1.27 | <1.3E-04 | 1.71E-03 |
| 11183 | MAP4K5a | 1.24 | 2.2E-03 | 5.64E-04 | 1.08 | 7.0E-03 | 5.14E-04 | |
| rs752010 | 59269 | HIVEP3a | 0.99 | 1.0E+00 | 7.05E-01 | 0.93 | 1.1E-02 | 2.82E-02 |
| rs2236178 | 55857 | C20orf19a | 0.95 | 1.3E-01 | 4.15E-01 | 1.20 | 1.5E-03 | 7.83E-03 |
| rs2786111 | 23418 | CRB1a | 0.98 | 1.4E-01 | 2.91E-01 | 0.93 | 1.9E-02 | 6.15E-02 |
| 259266 | ASPM | 1.11 | 1.6E-01 | 1.44E-01 | 1.01 | 2.0E-01 | 5.16E-01 | |
| rs648705 | 1282 | COL4A1a | 2.61 | <1.1E-04 | 7.77E-10 | 1.89 | <1.3E-04 | 7.72E-06 |
| rs2399985 | 8559 | PRPF18a | 1.58 | <1.1E-04 | 3.57E-08 | 1.19 | 1.7E-02 | 5.11E-02 |
| 55691 | FRMD4Aa | 1.12 | 7.3E-02 | 4.04E-02 | 0.99 | 1.0E+00 | 6.84E-01 | |
| SNPs from Table 3 | ||||||||
| rs2205960 | 7292 | TNFSF4a | 0.91 | 2.0E-02 | 2.65E-02 | 0.95 | 2.5E-02 | 7.33E-02 |
| rs2022013 | 23057 | NMNAT2a | 0.96 | 1.2E-01 | 3.69E-01 | 0.94 | 2.1E-02 | 3.21E-02 |
| rs7601754 | 6775 | STAT4a | 1.68 | 1.0E-03 | 3.13E-04 | 0.95 | 8.0E-02 | 3.27E-01 |
| rs10516487 | 55024 | BANK1a | 1.06 | 1.4E-01 | 4.03E-01 | 1.11 | 2.3E-02 | 9.82E-03 |
| rs2431697 | 9232 | PTTG1a | 1.58 | <1.1E-04 | 1.10E-05 | 1.20 | 8.4E-03 | 4.07E-03 |
| rs2187668 | 3123 | HLA-DR3 (HLA-DRB1, allele 0301)a | 1.88 | <1.1E-04 | 1.24E-06 | 2.22 | <1.3E-04 | 6.96E-07 |
| rs6568431 | 9474 | ATG5a | 1.11 | 4.3E-02 | 1.71E-02 | 1.22 | <1.3E-04 | 8.58E-05 |
| rs6920220 | 7128 | TNFAIP3a | 1.19 | 4.7E-02 | 1.00E-01 | 0.85 | 8.4E-03 | 7.14E-02 |
| rs4917014 | 10320 | IKZF1a | 1.01 | 1.0E+00 | 7.93E-01 | 0.93 | 4.3E-03 | 1.44E-02 |
| rs10239340 | 3663 | IRF5a | 1.01 | 1.0E+00 | 8.67E-01 | 0.92 | 1.2E-02 | 3.98E-02 |
| rs2248932 | 640 | BLKa | 1.12 | 1.7E-01 | 1.67E-01 | 0.95 | 3.5E-02 | 1.06E-01 |
| rs9937837 | 3684 | ITGAMa | 3.21 | <1.1E-04 | 9.83E-08 | 1.08 | 7.6E-02 | 1.29E-01 |
| rs5754217 | 7332 | UBEL2L3a | 0.96 | 1.1E-01 | 2.87E-01 | 1.12 | 6.6E-03 | 1.31E-02 |
dbSNP, Single Nucleotide Polymorphism Database.
Genes significantly regulated in patients with LN compared with controls (q<0.05).
Because LN is primarily characterized by intraglomerular histopathology, we focused our analysis on the entire glomerular transcript data set (961 genes regulated in patients with LN versus living donors with a q value <0.05, and a fold change ≥1.5 and ≤0.7 for the upregulated and downregulated genes, respectively). Using Ingenuity Pathway Analysis, the PDGF signaling pathway containing PDGFRA showed a significant enrichment of LN regulated transcripts with 12 of 79 molecules being differentially expressed (P=0.01 compared with a random gene set), corroborating the role of PDGFRA in LN. Independent of predefined canonical pathways, a co-citation literature-based transcriptional network built from the 961 LN genes using GePS software identified PDGFRA as one of the main nodes.
Discussion
This study presents the first GWAS of LN among individuals with SLE. This study is particularly notable because it incorporates genetic data across the three largest GWAS of SLE in individuals of European descent, focuses on unrelated women, and assesses the biologic relevance of genes in proximity of the genetic associations. Given that the frequency of LN among individuals diagnosed with SLE is high, a GWAS of patients with LN versus healthy controls will detect SLE-predisposing loci that do not influence the risk of nephritis. Thus, the study design reported contrasts LN patients with lupus patients without nephritis.
The most significant evidence of an association with LN was observed with rs1364989, located 83 kb from PDGFRA, a gene that encodes the α polypeptide of the PDGF receptor. PDGF and its associated pathways act to promote local cell proliferation, synthesis of extracellular matrix, chemotaxis, and cytokine production, and play a regulatory role in inflammation.20 Both PDGFRA and PDGFRB are expressed in renal glomeruli and in the interstitium. Signaling through the PDGFRA pathway can activate interstitial and mesangial cells, which can lead to renal fibrosis and tubulointerstitial scarring.20 Overproduction of PDGF, the ligand for PDGFRA and PDGFRB, has been implicated in the cellular proliferation and extracellular matrix production that accompanies experimental GN in animal models of SLE (NZB/W and MRL-lpr mice) and spontaneous LN in humans PDGF and PDGFR gene expression is known to be increased in kidney tissue from patients with proliferative forms of GN, including LN, IgA nephropathy, and Henoch–Schönlein purpura.21,22 Administration of the tyrosine kinase inhibitor imatinib to NZB/W mice with established lupus immune complex mediated GN improved survival and reduced proteinuria, tubulointerstitial fibrosis, and renal invasion of macrophages and monocytes.23,24 Similar effects were observed in MRL-lpr mice.25 Mesangial cell proliferation is effectively inhibited by anti-PDGF antibodies in animal models of GN with resultant reductions in mesangial matrix deposition and glomerulosclerosis.26,27 The expression data presented in this report provide evidence that increased PDGFRA mRNA is found in both the renal glomeruli and the tubulointerstitium of patients with LN, suggesting that this gene may mediate the pathogenesis of LN. Integrative analysis of gene expression data, pathway mapping, and coregulation transcriptional networks provided multiple lines of evidence for a key role of PDGFRA in LN. Together, these data suggest that common variation associated with PDGFRA may influence the development of LN in humans. Further analyses are needed to define the causal variation and specific mechanisms impacting the PDGFRA function in LN.
The second strongest signal of association was observed with rs274068, an intronic SNP located within SLC5A11 on chromosome 16p12.1. This sodium-dependent glucose cotransporter is responsible for active cellular uptake of glucose and other nutrients. SLC5A11 was initially proposed as a candidate gene for SLE because of evidence of linkage and interaction between the pericentromeric region of chromosome 16 and chromosome 1q23.28 A candidate gene association analysis performed in 95 Korean patients with SLE implicated SLC5A11 polymorphisms in susceptibility to SLE and select disease manifestations.29 These authors identified gene–gene interactions between SLC5A11 and immune-function genes (TNF, LTA, and C4) and the T and B cell costimulatory receptor gene (PDCD1), a confirmed LN susceptibility locus among European descendants.30
Hyaluronan synthase 2 (HAS2) located on 8q24 leads to the production of the extracellular matrix component hyaluronan. Hyaluronan accumulates in the renal cortex in the scarring phase of immune-mediated kidney disease, producing pathologic renal fibrosis. For example, hyaluronan deposition appears to be prominent in MRL-Fas(lpr) mice with renal disease, an effect mediated in part by the local synthesis of HAS2. In a previous study, large amounts of hyaluronan were deposited in the cortical interstitium of MRL-Fas(lpr) mice with autoimmune renal injury, but not in congenic MRL-++ mice.31 Steady state mRNA levels of HAS2 correlated with the activity of kidney disease. Enhanced synthesis of hyaluronan may be mediated by the effects of proinflammatory cytokines because TNF-α, IFN-γ, and their combination markedly enhanced hyaluronan synthesis and HAS2 gene expression in kidney tubule cell lines in this study. Our findings support a role for HAS2 in LN pathogenesis via the inflammatory activation, which is observed in LN in humans.
In contrast with the GWAS of SLE and other autoimmune diseases in individuals of European ancestry, the MHC does not dominate the list of most strongly LN-associated regions. In fact, the LN ORs for MHC loci were comparable with that of non-MHC loci in women of European descent. Although some studies suggested an association with HLA-DR2 and HLA-DR3, the two best established MHC susceptibility loci for SLE, not all studies are in agreement regarding the role of the MHC in LN.32,33 In our study that contrasts patients with LN with patients with lupus without nephritis, modest associations were observed with both of these MHC class II alleles (as assessed by proxies). Analyses conditioned on rs9271366 and rs2187668 indicate that loci outside of MHC class II genes may also contribute to LN risk, as seen in other studies of SLE.34 We posit that genetic variation in the MHC may strongly predispose individuals to develop SLE, but non-MHC factors may play a greater role in promoting the development of LN.
Evaluation of the association between other previously identified SLE susceptibility loci and LN indicates that only a small number of these loci seem to significantly influence the development of LN. In this analysis, SNPs in or near TNFSF4 and ITGAM were associated with LN, albeit not as strongly as the LN susceptibility loci listed in Table 2. We hypothesize that LN loci do not necessarily predispose to SLE but rather influence the response of the kidney to the immunologic challenges posed by SLE, as exemplified in the case of PDGFRA.
Although this is the first GWAS of LN among individuals with SLE, the meta-analysis has limitations. First, some SLE cases with no evidence of LN may develop LN in the future. Common to case-control studies, future conversion or misclassification reduces statistical power and results in conservative estimates of effect size. Second, the overall presence of LN was chosen as the case phenotype because more than one subclass of LN can be present at the same time (e.g., proliferative occurring with membranous lesions), and the type of subclass a patient has can change over the course of disease. Other indicators of renal disease (e.g., albuminuria or GFR) were not selected as the renal phenotype because they are influenced by prior and current medication use (e.g., immunosuppressants). Although using LN allowed us to identify genetic susceptibility toward renal involvement in SLE, more homogeneous phenotypes might increase power for specific risk loci. Third, although this study examined patients with SLE who were of European descent, LN is more prevalent and severe among patients with SLE who have other ethnic backgrounds. Finally, this study incorporates data from three large-scale GWAS of SLE in women of European descent and is well powered for intermediate effect sizes. Although it is the largest study to date, additional well powered studies are needed that include both male and female participants and other ethnicities and that attempt to replicate the findings in this study.
In summary, we report novel LN susceptibility loci, including loci near PDGRFA, HAS2, and SLC5A11, among women with SLE of European descent. Although an overlap between previously identified SLE and LN susceptibility loci (i.e., TNFSF4, ITGAM) exists, it is not strong and underscores the need to search for LN susceptibility loci independent of SLE loci. Delineating the genetic factors that lead to LN among patients with SLE may facilitate our understanding of the etiology of LN, provide improved SLE complication risk prediction, and ultimately motivate novel therapeutic targets.
Concise Methods
This study reports the results of a meta-analysis for LN based on three GWAS of unrelated women of European descent with SLE. The three GWAS used include the following: set I, samples genotyped using Illumina HumanHap550v1 as described by Hom et al.11; set II, samples genotyped using Illumina HumanHap300 as described by Harley et al.10; and set III, samples genotyped using Affymetrix 5.0 as described by Graham et al.8 The primary inference of this study is based on a female-only case–case (i.e., patients with SLE with nephritis versus patients with SLE without nephritis) analysis to minimize the potential influence of SLE susceptibility variants in our tests of association with LN. To account for potential population substructure, principal component analyses were computed using SNPs that passed quality control.35 All tests of association were computed and adjusted for five principal components in the set I samples and four principal components in the set II and set III samples. Within each GWAS set, autosomal SNPs that passed quality control filters and exhibited a minor allele frequency >0.01 were used to impute the approximately 2.5 million SNPs in HapMap2 (Build 35) using IMPUTE.36 The reference set for imputation was the Utah residents with ancestry from northern and western Europe available via the HapMap. Within each GWAS set, logistic regression models included the above-stated principal components and accounted for the imputation uncertainty via the posterior genotype probabilities.36 Dominant, additive, and recessive genetic models were computed assuming at least 10 and 20 individuals homozygous for the minor allele were observed. Because of the modest sample size, this requirement caused many SNPs to not have a test computed in set III. The evidence of association from each of the three GWAS sets was weighted by sample size and combined using the weighted inverse normal method as implemented in METAL.37 To account for any inflation, the genomic control-adjusted P value was reported.
Renal biopsies from 15 healthy pretransplant living donor controls and 32 patients with LN were collected from an international multicenter repository (European Renal cDNA Bank).38 Healthy controls had normal kidney function (i.e., GFR between 80 and 120 ml/min per 1.73 m2 estimated using the Modification of Diet in Renal Disease equation). Glomerular and tubulointerstitial compartments of kidney biopsies were microdissected.38–40 The mRNA extracted from each renal compartment was analyzed using Affymetrix Human Genome U133A Genechips (Affymetrix, Santa Clara, CA).40 mRNA expression levels generated from the biopsies of patients with LN and from healthy controls (no lupus, no nephritis) were compared using unpaired t tests. A q value (false discovery rate) <0.05 was considered significant.
Disclosures
T.W.B. and R.R.G. are full-time employees of Genentech, Inc.
Supplementary Material
Acknowledgments
Out-of-study controls for the set I and set II GWAS were provided by Peter K. Gregersen at the Robert S. Boas Center for Genomics and Human Genetics, Feinstein Institute for Medical Research (Manhasset, NY). The authors also acknowledge computing support from the Wake Forest School of Medicine Center of Public Health Genomics.
The authors gratefully acknowledge the following funding sources for this research. Genotyping funds for set I were provided by Genentech. The Alliance for Lupus Research funded the International Consortium for Systemic Lupus Erythematosus Genetics and the genotyping for set II. Set III was funded by grants from the National Institutes of Health (AI063274 and AR052125 to P.M.G. and AR043247 to K.L.M.; National Research Service Award 5F32-AR50927 to R.R.G.), the Lupus Foundation of Minnesota (to P.M.G. and K.L.M.-S.), and the Arthritis Foundation (to K.L.M.-S.). This study was also supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK079912 and P30-DK081943 to M.K.), the Alliance for Lupus Research (M.K., B.P.T., L.A.C., and M.K.), and the Swedish Research Council of Medicine (to M.E.A.-R.), as well as a research fellowship from the National Kidney Foundation (FLB1245 to C.C.B.), an Arthritis Research UK Special Strategic Award (19289 to T.J.V.), and a Kirkland Scholar Award (to L.A.C). Sample collections and additional financial support were also obtained from grants from the National Institutes of Health (KL2-TR000143 to S.A.C.; R01-AR033062, N01-AR062277, RC2-AR058951, and UL1-TR000165 to E.E.B. and R.P.K.; P60-AR053308, K24-AR02175, R01-AR052300, and UL1-TR000004 to L.A.C.; AR057028 to F.Y.D.; IMMA 9, 5R37AI024717-25, 5P01AI083194, 5P01AR049084, PR094002, and 3R37AI024717-23S1 to J.B.H.; R01-AR057172 to C.O.J.; HL092397 to M.I.K.; R01-AR046588, National Institute of Arthritis and Musculoskeletal and Skin Diseases [NIAMS] Progression of Cardiovascular Disease in Lupus K24-AR002213 to S.M.; AR43727 and UL1-RR025005 to M.P.; R01-AR043814 to B.P.T.; R01-AR057172 to C.O.J.; and R01-HL56266 and R01-DK070941 to B.I.F.).
The following individuals were members of the European Renal cDNA Bank–Kroener-Fresenius Biopsy Bank at the time of this study: C.D. Cohen, M. Fischereder, H. Schmid, P.J. Nelson, M. Kretzler, D. Schloendorff, and W. Samtleben (Munich, Germany); J.D. Sraer and P. Ronco (Paris, France); M.P. Rastaldi and G. D’Amico (Milan, Italy); F. Mampaso (Madrid, Spain); P. Doran and H.R. Brady (Dublin, Ireland); D. Moenks (Gottingen, Germany); P. Mertens and J. Floege (Aachen, Germany); N. Braun and T. Risler (Tübingen, Germany), L. Gesualdo and F.P. Schena (Bari, Italy); J. Gerth and G. Wolf (Jena, Germany); R. Oberbauer and D. Kerjaschki (Vienna, Ausria); B. Banas and B.K. Kraemer (Regensburg, Germany); H. Peters and H.H. Neumayer (Berlin, Germany); K. Ivens and B. Grabensee (Dusseldorf, Germany); R.P. Wuethrich (Zurich, Switzerland); and V. Tesar (Prague, Czech Republic).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013050446/-/DCSupplemental.
References
- 1.Cervera R, Khamashta MA, Font J, Sebastiani GD, Gil A, Lavilla P, Aydintug AO, Jedryka-Góral A, de Ramón E, Fernández-Nebro A, Galeazzi M, Haga HJ, Mathieu A, Houssiau F, Ruiz-Irastorza G, Ingelmo M, Hughes GR, European Working Party on Systemic Lupus Erythematosus : Morbidity and mortality in systemic lupus erythematosus during a 5-year period. A multicenter prospective study of 1,000 patients. Medicine (Baltimore) 78: 167–175, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Freedman BI, Bowden DW, Rich SS: Genetic basis of kidney disease. In: Brenner & Rector's The Kidney, edited by Taal M, 9th Ed., Philadelphia, Elsevier Saunders, 2012, pp 1554–1569 [Google Scholar]
- 3.Freedman BI, Wilson CH, Spray BJ, Tuttle AB, Olorenshaw IM, Kammer GM: Familial clustering of end-stage renal disease in blacks with lupus nephritis. Am J Kidney Dis 29: 729–732, 1997 [DOI] [PubMed] [Google Scholar]
- 4.Quintero-Del-Rio AI, Kelly JA, Kilpatrick J, James JA, Harley JB: The genetics of systemic lupus erythematosus stratified by renal disease: Linkage at 10q22.3 (SLEN1), 2q34-35 (SLEN2), and 11p15.6 (SLEN3). Genes Immun 3[Suppl 1]: S57–S62, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Quintero-del-Rio AI, Kelly JA, Garriott CP, Hutchings DC, Frank SG, Aston CE, Harley JB: SLEN2 (2q34-35) and SLEN1 (10q22.3) replication in systemic lupus erythematosus stratified by nephritis. Am J Hum Genet 75: 346–348, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karassa FB, Trikalinos TA, Ioannidis JP, Fc gamma RIIIA-SLE meta-analysis investigators : The Fc gamma RIIIA-F158 allele is a risk factor for the development of lupus nephritis: a meta-analysis. Kidney Int 63: 1475–1482, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG: Association of programmed cell death 1 polymorphisms and systemic lupus erythematosus: A meta-analysis. Lupus 18: 9–15, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, Plenge RM, Behrens TW, Wither JE, Rioux JD, Fortin PR, Graham DC, Wong AK, Vyse TJ, Daly MJ, Altshuler D, Moser KL, Gaffney PM: Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet 40: 1059–1061, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Xu JH, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Li Y, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ: Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 41: 1234–1237, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA, International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) : Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40: 204–210, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, Ballinger DG, Kosoy R, Demirci FY, Kamboh MI, Kao AH, Tian C, Gunnarsson I, Bengtsson AA, Rantapää-Dahlqvist S, Petri M, Manzi S, Seldin MF, Rönnblom L, Syvänen AC, Criswell LA, Gregersen PK, Behrens TW: Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358: 900–909, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, Truedsson L, Pons-Estel BA, Witte T, D’Alfonso S, Barizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, González-Escribano MF, Martin J, Abderrahim H, Alarcón-Riquelme ME: Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet 40: 211–216, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Li R, Yang W, Zhang J, Hirankarn N, Pan HF, Mok CC, Chan TM, Wong RW, Mok MY, Lee KW, Wong SN, Leung AM, Li XP, Avihingsanon Y, Lee TL, Ho MH, Lee PP, Wong WH, Wong CM, Ng IO, Yang J, Li PH, Zhang Y, Zhang L, Li W, Baum L, Kwan P, Rianthavorn P, Deekajorndej T, Suphapeetiporn K, Shotelersuk V, Garcia-Barceló MM, Cherny SS, Tam PK, Sham PC, Lau CS, Shen N, Lau Y, Ye DQ: Association of CD247 with systemic lupus erythematosus in Asian populations. Lupus 21: 75–83, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Okada Y, Shimane K, Kochi Y, Tahira T, Suzuki A, Higasa K, Takahashi A, Horita T, Atsumi T, Ishii T, Okamoto A, Fujio K, Hirakata M, Amano H, Kondo Y, Ito S, Takada K, Mimori A, Saito K, Kamachi M, Kawaguchi Y, Ikari K, Mohammed OW, Matsuda K, Terao C, Ohmura K, Myouzen K, Hosono N, Tsunoda T, Nishimoto N, Mimori T, Matsuda F, Tanaka Y, Sumida T, Yamanaka H, Takasaki Y, Koike T, Horiuchi T, Hayashi K, Kubo M, Kamatani N, Yamada R, Nakamura Y, Yamamoto K: A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus erythematosus in Japanese. PLoS Genet 8: e1002455, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, Leung AM, Li XP, Avihingsanon Y, Wong CM, Lee TL, Ho MH, Lee PP, Chang YK, Li PH, Li RJ, Zhang L, Wong WH, Ng IO, Lau CS, Sham PC, Lau YL, Asian Lupus Genetics Consortium : Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet 6: e1000841, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kariuki SN, Franek BS, Kumar AA, Arrington J, Mikolaitis RA, Utset TO, Jolly M, Crow MK, Skol AD, Niewold TB: Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther 12: R151, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung SA, Taylor KE, Graham RR, Nititham J, Lee AT, Ortmann WA, Jacob CO, Alarcón-Riquelme ME, Tsao BP, Harley JB, Gaffney PM, Moser KL, Petri M, Demirci FY, Kamboh MI, Manzi S, Gregersen PK, Langefeld CD, Behrens TW, Criswell LA, SLEGEN : Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet 7: e1001323, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, Morrison J, Richardson A, Walsh EC, Gao X, Galver L, Hart J, Hafler DA, Pericak-Vance M, Todd JA, Daly MJ, Trowsdale J, Wijmenga C, Vyse TJ, Beck S, Murray SS, Carrington M, Gregory S, Deloukas P, Rioux JD: A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38: 1166–1172, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, Chen W, Zhu C, McEver RP, Kimberly RP, Alarcón-Riquelme ME, Vyse TJ, Li QZ, Wakeland EK, Merrill JT, James JA, Kaufman KM, Guthridge JM, Harley JB: A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet 40: 152–154, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Ostendorf T, Eitner F, Floege J: The PDGF family in renal fibrosis. Pediatr Nephrol 27: 1041–1050, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Gesualdo L, Di Paolo S, Milani S, Pinzani M, Grappone C, Ranieri E, Pannarale G, Schena FP: Expression of platelet-derived growth factor receptors in normal and diseased human kidney. An immunohistochemistry and in situ hybridization study. J Clin Invest 94: 50–58, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda M, Shikata K, Makino H, Sugimoto H, Ota K, Akiyama K, Hirata K, Ota Z: Gene expression of PDGF and PDGF receptor in various forms of glomerulonephritis. Am J Nephrol 17: 25–31, 1997 [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa T, Inoue H, Sasahara M: Platelet-derived growth factor and renal disease. Curr Opin Nephrol Hypertens 21: 80–85, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Zoja C, Corna D, Rottoli D, Zanchi C, Abbate M, Remuzzi G: Imatinib ameliorates renal disease and survival in murine lupus autoimmune disease. Kidney Int 70: 97–103, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Sadanaga A, Nakashima H, Masutani K, Miyake K, Shimizu S, Igawa T, Sugiyama N, Niiro H, Hirakata H, Harada M: Amelioration of autoimmune nephritis by imatinib in MRL/lpr mice. Arthritis Rheum 52: 3987–3996, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Javaid B, Quigg RJ: Treatment of glomerulonephritis: Will we ever have options other than steroids and cytotoxics? Kidney Int 67: 1692–1703, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Kurogi Y: Mesangial cell proliferation inhibitors for the treatment of proliferative glomerular disease. Med Res Rev 23: 15–31, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Tsao BP, Cantor RM, Grossman JM, Kim SK, Strong N, Lau CS, Chen CJ, Shen N, Ginzler EM, Goldstein R, Kalunian KC, Arnett FC, Wallace DJ, Hahn BH: Linkage and interaction of loci on 1q23 and 16q12 may contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum 46: 2928–2936, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Tsai LJ, Hsiao SH, Tsai LM, Lin CY, Tsai JJ, Liou DM, Lan JL: The sodium-dependent glucose cotransporter SLC5A11 as an autoimmune modifier gene in SLE. Tissue Antigens 71: 114–126, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Prokunina L, Gunnarsson I, Sturfelt G, Truedsson L, Seligman VA, Olson JL, Seldin MF, Criswell LA, Alarcón-Riquelme ME: The systemic lupus erythematosus-associated PDCD1 polymorphism PD1.3A in lupus nephritis. Arthritis Rheum 50: 327–328, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Feusi E, Sun L, Sibalic A, Beck-Schimmer B, Oertli B, Wüthrich RP: Enhanced hyaluronan synthesis in the MRL-Fas(lpr) kidney: Role of cytokines. Nephron 83: 66–73, 1999 [DOI] [PubMed] [Google Scholar]
- 32.Lindqvist AK, Alarcón-Riquelme ME: The genetics of systemic lupus erythematosus. Scand J Immunol 50: 562–571, 1999 [DOI] [PubMed] [Google Scholar]
- 33.Marchini M, Antonioli R, Lleò A, Barili M, Caronni M, Origgi L, Vanoli M, Scorza R: HLA class II antigens associated with lupus nephritis in Italian SLE patients. Hum Immunol 64: 462–468, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Barcellos LF, May SL, Ramsay PP, Quach HL, Lane JA, Nititham J, Noble JA, Taylor KE, Quach DL, Chung SA, Kelly JA, Moser KL, Behrens TW, Seldin MF, Thomson G, Harley JB, Gaffney PM, Criswell LA: High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet 5: e1000696, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D, Rotter JI, Torres E, Taylor KD, Goldstein DB, Reich D: Long-range LD can confound genome scans in admixed populations. Am J Hum Genet 83: 132–135, author reply 135–139, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J, Howie B, Myers S, McVean G, Donnelly P: A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39: 906–913, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen CD, Frach K, Schlöndorff D, Kretzler M: Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int 61: 133–140, 2002 [DOI] [PubMed] [Google Scholar]
- 39.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R: NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res 37[Database issue]: D885–D890, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M, European Renal cDNA Bank (ERCB) Consortium : Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 41.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Rantapaa-Dahlqvist S, Baechler EC, Brown EE, Alarcon GS, Edberg JC, Ramsey-Goldman R, McGwin G, Jr, Reveille JD, Vila LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Ronnblom L, Criswell LA, Syvanen AC, Behrens TW, Graham RR: A large-scale replication study identifies TNIP1 PRDM1 JAZF1 UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 41: 1228–1233, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW: Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 75: 504–507, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham RR, Hom G, Ortmann W, Behrens TW: Review of recent genome-wide association scans in lupus. J Intern Med 265: 680–688, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Karassa FB, Trikalinos TA, Ioannidis JP: Role of the Fcgamma receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum 46: 1563–1571, 2002 [DOI] [PubMed] [Google Scholar]
- 45.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, Li W, Masters SL, Booty MG, Carulli JP, Padyukov L, Alfredsson L, Klareskog L, Chen WV, Amos CI, Criswell LA, Seldin MF, Kastner DL, Gregersen PK: STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med 357: 977–986, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lessard CJ, Adrianto I, Kelly JA, Kaufman KM, Grundahl KM, Adler A, Williams AH, Gallant CJ, Anaya JM, Bae SC, Boackle SA, Brown EE, Chang DM, Criswell LA, Edberg JC, Freedman BI, Gregersen PK, Gilkeson GS, Jacob CO, James JA, Kamen DL, Kimberly RP, Martin J, Merrill JT, Niewold TB, Park SY, Petri MA, Pons-Estel BA, Ramsey-Goldman R, Reveille JD, Song YW, Stevens AM, Tsao BP, Vila LM, Vyse TJ, Yu CY, Guthridge JM, Bruner GR, Langefeld CD, Montgomery C, Harley JB, Scofield RH, Gaffney PM, Moser KL: Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am J Hum Genet 88: 83–91, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lofgren SE, Yin H, Delgado-Vega AM, Sanchez E, Lewen S, Pons-Estel BA, Witte T, D'Alfonso S, Ortego-Centeno N, Martin J, Alarcon-Riquelme ME, Kozyrev SV: Promoter insertion/deletion in the IRF5 gene is highly associated with susceptibility to systemic lupus erythematosus in distinct populations but exerts a modest effect on gene expression in peripheral blood mononuclear cells. J Rheumatol 37: 574–578, 2010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.