Abstract
This study aimed to assess the prognostic influences of Wilms tumor 1 (WT1) gene mutations in cytogenetically normal acute myeloid leukemia (CN-AML) among Egyptian patients. Exon 7 of WT1 was screened for mutations in samples from 82 CN-AML patients out of 203 newly diagnosed AML patients, using a high-resolution capillary electrophoresis. Seven out of 82 AML patients (8.3 %) harbored WT1 mutations. There was no significant difference between the mutant WT1 and wild type AML patients as regard age, sex, French–American–British subtypes and the prevalence of success of induction remission therapy (P < 0.5). AML patients with mutant WT1 had shorter overall survival as compared to those patients with wild WT1 (HR = 1.38; 95 % CI 4.79–6.86; P = 0.004). In conclusion, CN-AML patients with WT1 gene mutation have poor clinical outcome. We recommend testing the WT1 mutations as part of molecularly based risk assessment and risk-adapted treatment stratification of patients with CN-AML.
Keywords: Wilms tumor 1, AML, Prognosis
Introduction
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous disease that accounts for 20 and 70 % of acute leukemia in children and adults respectively. The cytogenetic finding was considered as the cardinal marker for AML risk stratification. The cytogenetically normal acute myeloid leukemia (CN-AML) is a large cytogenetic subgroup of AML, representing ~45 % of adult patients with AML who are younger than 60 years. During the last decade, the prognostic stratification of CN-AML was based on several molecular marker including the nucleophosmin 1 (NPM1) gene, the fms-related tyrosine kinase 3 (FLT3) gene, the CCAAT/enhancer binding protein alpha (CEBPA) gene, the myeloid–lymphoid or mixed-lineage leukemia (MLL) gene, the neuroblastoma RAS viral oncogene homolog (NRAS) gene, the runt-related transcription factor 1 (RUNX1) gene and Wilms’ tumor gene (WT1) [1, 2].
The Wilms’ tumor gene (WT1) was identified as a tumor suppressor gene, which is located at chromosome 11p and encodes a transcription factor with an N-terminal transcriptional regulatory domain (exons 1–6) and a C-terminal 4-Cys2His2 zinc finger domain (exons 7–10) [3, 4].
WT1 expression occurs primarily in cells of the developing genitourinary and hematopoietic systems and is inversely correlated with the degree of differentiation in both systems. In hematopoiesis, expression occurs in CD34 progenitor cells but is absent in mature leukocytes. The precise role of WT1 in normal and malignant hematopoiesis remains controversial [5, 6]. It has been implicated in regulation of cell survival, proliferation and differentiation, and may function as an oncogene. However, this diversity may reflect both tissue specificity of downstream targets, expression of different isoforms, given that alternatively spliced isoforms, and post-transcriptional modifications are thought to control the cellular and functional properties of the protein [7, 8].
WT1 mutations have been found in about 10–15 % of cases of AML, 20 % of biphenotypic leukemia, but mutations are rare in acute lymphoblastic leukemia (ALL) [2]. In AML, the WT1 mutations are cluster mainly to exon 7 [9, 10].
The aim of this study was to evaluate the incidence and clinical impact of WT1 mutation in adult patients with CN-AML.
Subjects and Methods
Subjects and Treatment Protocols
The present study was carried out on 203 adult patients (21–74 years), from hematology unit, Mansoura Cancer Institute between June 2009 and January 2011, after signing written consent. The diagnosed of AML was based on the presence of blast cell ≥20 % in bone marrow (BM) smear. One hundred ninety-eight patients had de novo AML (14 M1, 59 M2, 12 M3, 65 M4, 39 M5, 7 M6 and 2 M7) and five patients had secondary AML. The diagnosis and FAB subtypes were confirmed by immunophenotyping using (Coulter Epics XL Flowcytometer PN 42372238 B, Coulter Corporation, Miami, Florida 33196, USA) to confirm diagnosis (Cyt. MPO, CD13, CD33, CD117) as a primary panel for myeloid lineage, (CD14, CD36, CD11b) for M4 and M5, (CD61, glycophorin A) for M6 and (CD41, CD42) for M7. The patients were observed for 12 months or until death. History taking and clinical examination for organomegaly were done for all patients.
All patients gave informed consent for both treatment and genetic analysis. All patients received intensive induction therapy (Cytarabine 100 mg/m2/day for 7 days i.v. continuous infusion and Daunorubicin 45 mg/m2/day for 3 days i.v.) consolidation therapy (Cytarabine 3 gm/m2/12 h for 3 days repeated for 3–6 cycle).
Methods
Cytogenetic and Molecular Genetic Analysis
Pretreatment blood samples from all patients were studied by chromosome banding analysis. The specimens were also analyzed by fluorescence in situ hybridization for the presence of t(8;21)(q22;q22) for M2, t(15;17)(q22;q12) for M3 inv(16)(p13q22) for M4e or 11q23 for M5.
Determination of Mutation Status in CN-AML
WT1 exons 7 were amplified using the polymerase chain reaction (PCR) with ~50 ng of genomic DNA, 1X QIAGEN Multiplex PCR Master Mix and 10 pmol of primers designed to flanking intronic regions: 6′ FAM end-labeled primer 7F (5′-GACCTACGTGAATGTTCACATG-3′), 7R (5′-ACCAACACCTGGATCAGACCT-3′). The annealing temperature was 60 °C, and 35 cycles of amplification were performed. Then the PCR products were analyzed by fragment analysis using high-resolution capillary electrophoresis (CE) on a ABI ® 310 Genetic Analyzer (PE Applied Biosystems, USA). The wild-type amplicon was 348 base pairs (bp). For samples with additional peaks representing mutant amplicons, the relative mutant level was calculated from the area under the curve and expressed as a percentage of total WT1 alleles (mutant/[wild type + mutant(s)] × 100).
Statistical Analysis
The statistical analysis of data were done by using excel program and SPSS version 16 (statistical package for social science). Qualitative data were described in the form of numbers and percentages. Quantitative data were described in the form of mean (±) standard deviation (SD). Statistical analysis were done by comparison between groups using Chi square test regarding qualitative data, while quantitative nonparametric data comparison were performed using one way ANOVA and paired sample t test. The probability of being by chance (P value) was calculated for all parameters (P is significant if ≤0.05 at confidence interval 95 %).
Result
Prevalence of WT1 Mutation in Exon 7
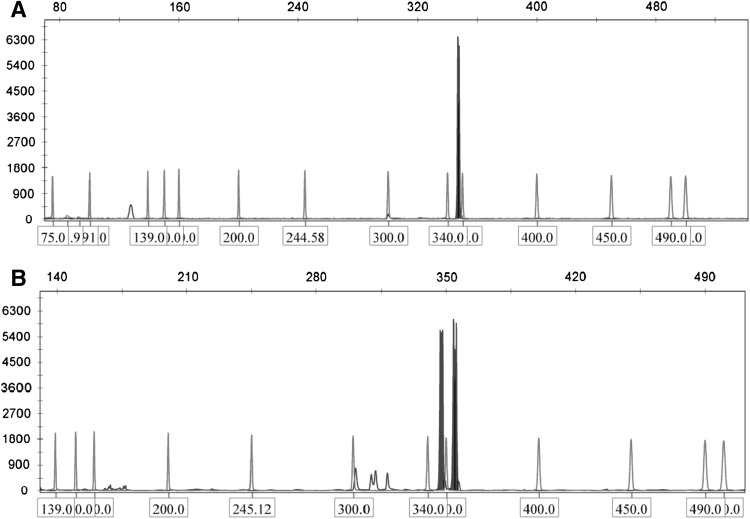
Cytogenetic studies revealed that 82 out of 203 AML patients were free from cytogenetic abnormalities (CN-AML). Mutant WT1 was detected in 7 out of 82 AML patients (8.3 %). The mutations levels range from 6 to 51 % in the form of additional peak at 355, 360, 373 and 390 base pair (Fig. 1).
Fig. 1.
Show the capillary electrophoresis of WT1 exon 7 a show the wild type peak at 348 bp b shows a wildtype peak at 348 bp and mutant peaks at 355
Patient Characteristics in Mutant Versus Wild WT1 Gene
There was no significant difference between the two groups regarding age, sex, WBCs, hemoglobin concentration levels, platelet counts and French–American–British subtypes (Table 1).
Table 1.
Clinical and demographic characteristics of the 82 CN-AML patients studied
| WT1 wild type | WT1 mutant type | P value | |||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| De novo | 73 | 91.3 | 7 | 8.7 | |
| Secondary | 2 | 100 | 0 | 0 | |
| FAB type | |||||
| M1 | 4 | 100 | 0 | 0 | 0.344 |
| M2 | 14 | 100 | 0 | 0 | |
| M4 | 27 | 84.4 | 5 | 15.6 | |
| M5 | 22 | 91.7 | 2 | 8.3 | |
| M6 | 6 | 100 | 0 | 0 | |
| Age | |||||
| 20–29 | 16 | 84.3 | 3 | 15.7 | 0.252 |
| 30–39 | 14 | 100 | 0 | 0 | |
| 40–49 | 9 | 81.8 | 2 | 18.2 | |
| 50–59 | 17 | 89.4 | 2 | 10.6 | |
| >60 | 19 | 100 | 0 | 0 | |
| Sex | |||||
| Female | 45 | 95.7 | 2 | 4.3 | 0.108 |
| Male | 30 | 85.7 | 5 | 14.3 | |
| WBC × 109/L | |||||
| <10 | 10 | 100 | 0 | 0 | 0.434 |
| 10–49.9 | 34 | 94.5 | 2 | 5.5 | |
| 50–99.9 | 17 | 85 | 3 | 15 | |
| >100 | 14 | 87.7 | 2 | 12.5 | |
| % BM blast | |||||
| <30 | 6 | 100 | 0 | 0 | 0.038 |
| 30–49 | 6 | 100 | 0 | 0 | |
| 50–79 | 32 | 82 | 7 | 18 | |
| >80 | 31 | 100 | 0 | 0 | |
| Organomegalya | |||||
| Spleenomegaly | 39 | 88.6 | 5 | 11.4 | 0.324 |
| Hepatomegaly | 42 | 87.5 | 6 | 12.5 | 0.127 |
| Hemoglobin | |||||
| Mean ± SD | 7.78 ± 1.55 | 8.5 ± 0.16 | 0.23 | ||
| Platelets | |||||
| Mean ± SD | 52.3 ± 40.5 | 43.8 ± 22.7 | 0.587 | ||
aSpleenomegaly (44 patient out of 75) & hepatomegaly (48 patient out of 75)
Response to Therapy and Clinical Outcome in WT1 Mutant-Positive Patients
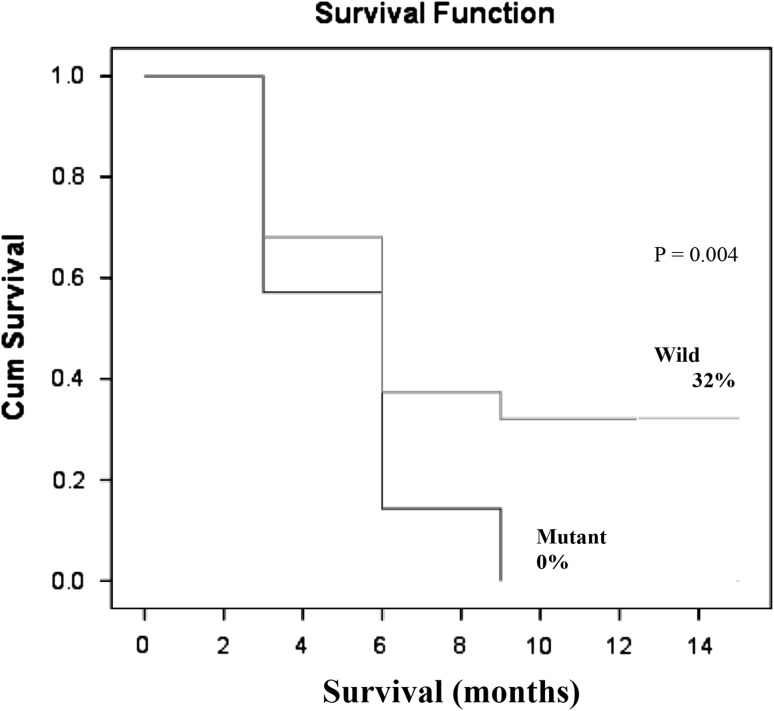
In univariable analysis, there was no significant difference in the induction remission rate (CR) between patients with mutant WT1 (28.6 %) and wild WT1 (29.3 %) (Table 2). On the other hand, the patients with mutant WT1 had shorter overall survival (OS) as compared to patients with wild type (HR = 1.38; 95 % CI 4.79–6.86; P = 0.004). The percentage of living patients with wild type WT1 and mutant one was 32.0 and 0.0 % respectively at 12 months follow up (Fig. 2).
Table 2.
Response to therapy in WT1 mutant and wild type patients
| Remission | Mutant type | Wild type | Total | P value | Odd ratio | |
|---|---|---|---|---|---|---|
| Yes | No | 2 | 22 | 24 | 0.966 | 1.038 |
| % | 28.6 | 29.3 | 29.2 | |||
| No | No | 5 | 53 | 58 | ||
| % | 71.4 | 70.7 | 70.8 | |||
| Total | No | 7 | 75 | 82 | ||
Fig. 2.
Overall survival (OS) in CN-AML patients according to the mutational status of WT1
Discussion
The WT1 mutations were identified in 7 out of 82 CN-AML patients (8.5 %). This percentage of cases is slightly lower than that reported by Virappane et al. [8] (10.0 %) and Paschka et al. [11] (10.7 %). The difference could be explained on the basis that in the present study we analyzed WT1 mutation in exons 7 only while the other two studies analyzed mutations in exons 7 and 9.
In the current study, there was no significant difference between the WT1 mutant and wild type as regard age, sex, WBCs count, hemoglobin concentration, platelet count and French–American–British subtypes. Similar findings were reported by Virappane et al. [8]. On the other hand Paschka et al. [11] and Gaidzik et al. [2], demonstrated that there is significant difference between the WT1 mutant and wild type as regard WBC count.
The BM blast cell counts were significantly higher in AML subgroup with mutant WT1 as compared to those with wild type AML. This finding is in agreement with that of Virappane et al. [8], Paschka et al. [11] and Gaidzik et al. [2]. They stated that the WT1 mutation might induce high proliferation capacity of blast cells.
Our study revealed that there was a non-significant difference between the WT1 mutant and wild type in rate of induction remission (IR). This was in agreement with the finding of Paschka et al. [11], and Gaidzik et al. [2], who did not found a relation between WT1 mutations and achievement of IR. On the other hand, Virappane et al. [8] found that the patients with WT1 mutations had an inferior response to induction chemotherapy, with a low CR rate and the induction death rate was higher.
In the present study CN-AML patients with mutant WT1 had shorter overall survival (2.7 months) as compared to those patients with wild type (5.8 months); the difference was statistically significant (P = 0.004). This result is in agreement with that of Paschka et al. [11] and Virappane et al. [8] who found that mutations in the WT1 gene are independent predictors for worse DFS and OS in CN-AML. The effect of WT1 mutation on patient’s survival could be explained on the basis that patients with mutated WT1 have high expressers of ERG and BAALC more frequently than patients with unmutated WT1 [12]. Over expression of both the ERG and the BAALC genes has been associated with an adverse prognosis. Moreover, WT1 mutations would be expected to abolish, impair, or change the DNA binding ability of the WT1 protein to its target genes, including to those that encode proteins involved in the regulation of normal hematopoiesis (RARA, CSF1), apoptosis (BCL2, BCL2A1, BAK1), cell cycle (CCNE1, CDKN1A), gene transcription (MYC, PAX2, MYB, EGR1), and cell proliferation (TGFB1, PDGFA) [11, 13].
In conclusion, CN-AML patients with WT1 gene mutation have poor clinical outcome. We recommend testing the WT1 mutations as part of molecularly based risk assessment and risk-adapted treatment stratification of patients with CN-AML.
Conflict of interest
The authors stated that there is no conflict of interest.
References
- 1.Brown P. Adding WT1 to childhood AML alphabet soup. Blood. 2009;113(23):5696–5697. doi: 10.1182/blood-2009-03-207282. [DOI] [PubMed] [Google Scholar]
- 2.Gaidzik V, Schlenk R, Moschny S, Becker A, Bullinger L, Corbacioglu A, Krauter J, Brigitte S, Ganser A, Hartmut D, Konstanze D. Prognostic impact of WT1 mutations in cytogenetically normal acute myeloid leukemia: a study of the German–Austrian AML study group. Blood. 2009;113(19):4505–4511. doi: 10.1182/blood-2008-10-183392. [DOI] [PubMed] [Google Scholar]
- 3.Scott R, Stiller C, Walker L. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet. 2006;43:705–715. doi: 10.1136/jmg.2006.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mrózek K, Marcucci G, Paschka P, Whitman S, Bloomfield C. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hohenstein P, Hastie N. The many facets of the Wilms’ tumour gene, WT1. Hum Mol Genet. 2006;15:196–201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Han Y, Suarez Saiz F, Minden M. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 7.Ariyaratana S, Loeb D. The role of the Wilms tumour gene (WT1) in normal and malignant haematopoiesis. Expert Rev Mol Med. 2007;9:1–17. doi: 10.1017/S1462399407000336. [DOI] [PubMed] [Google Scholar]
- 8.Virappane P, Gale G, Hills R, Kakkas I, Summers S, Stevens J, Allen C, Green C, Quentmeier H, Drexler H, Burnett A, Linch D, Bonnet D, Andrew Lister T, Fitzgibbon J. Mutation of the Wilms’ tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol. 2008;26(33):5429–5435. doi: 10.1200/JCO.2008.16.0333. [DOI] [PubMed] [Google Scholar]
- 9.Wagner K, Patek C, Cunningham A, Taylor A, Hooper M, Ansell J. C-terminal truncation of WT1 delays but does not abolish hematopoiesis in embryoid bodies. Blood Cells Mol Dis. 2002;28(3):428–435. doi: 10.1006/bcmd.2002.0529. [DOI] [PubMed] [Google Scholar]
- 10.Owen C, Fitzgibbon J, Paschka P. The clinical relevance of Wilms tumour 1 (WT1) gene mutations in acute leukaemia. Hematol Oncol. 2010;28(1):13–19. doi: 10.1002/hon.931. [DOI] [PubMed] [Google Scholar]
- 11.Paschka P, Marcucci G, Ruppert A, Whitman S, Mrózek K, Maharry K, Langer C, Baldus C, Zhao W, Powell B, Baer M, Carroll A, Caligiuri M, Kolitz J, Larson R, Bloomfield C. Wilms tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldus C, Thiede C, Soucek S, Bloomfield C, Thiel E, Ehninger G. BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: prognostic implications. J Clin Oncol. 2006;24:790–797. doi: 10.1200/JCO.2005.01.6253. [DOI] [PubMed] [Google Scholar]
- 13.Marcucci G, Maharry K, Whitman S, Vukosavljevic T, Paschka P, Langer C, Mrózek K, Baldus C, Carroll A, Powell B, Kolitz J, Larson R, Bloomfield C. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]