Abstract
The BCR/ABL gene rearrangement is cytogenetically visualized in most chronic myeloid leukemia (CML) cases. About 5–10 % of CML patients lack its cytogenetic evidence, however, shows BCR/ABL fusion by molecular methods. We describe two CML patients with Philadelphia (Ph) negative (−ve) and BCR/ABL positive by fluorescence in situ hybridization (FISH). Both the cases were in chronic phase at diagnosis. Conventional cytogenetics and different FISH assays were adopted using BCR/ABL probes. Home-brew FISH assay using bacterial artificial clone (BAC) for BAC-CTA/bk 299D3 for chromosomal region 22q13.31-q13.32 was performed in case 1. Both the patients were Ph-ve. In first case, dual color dual fusion (DCDF)-FISH studies revealed 1 Red (R) 2 Green (G) 1 Fusion (F) signal pattern in 80 % of cells indicating BCR/ABL fusion signals on chromosomes 9 instead of Ph and 2G2F signal pattern in 20 % of cells indicating two BCR/ABL fusions on both chromosomes 9q34 on presentation. In second case, FISH studies revealed the 1R1G1F signal pattern indicating BCR/ABL fusion signals on chromosomes 9 instead of Ph in 100 % of cells at presentation. During follow-up, both the patients exhibited 2G2F signal pattern indicating two BCR/ABL fusions on both chromosomes 9q34, which indicated a clonal evolution in 100 % cells. Both the patients did not achieve therapeutic response. Relocation of BCR/ABL fusion sequence on sites other than 22q11 represents a rare type of variant Ph, the present study highlights the hot spots involved in CML pathogenesis and signifies their implications in Ph−ve BCR/ABL positive CML. This study demonstrated the genetic heterogeneity of this subgroup of CML and strongly emphasized the role of metaphase FISH, especially in Ph−ve CML cases, as it detects variations of the classical t(9;22).
Keywords: BCR/ABL, FISH, CML, Philadelphia chromosome
Introduction
The presence of Philadelphia (Ph) chromosome is characteristic chromosomal marker for more than 90 % of chronic myeloid leukemia (CML) cases. Cytogenetically, Ph chromosome is the result of the reciprocal translocation between chromosome 9 and 22 with 9q34 and 22q11 breakpoints. On molecular level, two hybrid genes are formed by this translocation: BCR/ABL, which is in the vast majority of the cases localized on the Ph chromosome. Most patients have breakpoints that result in a fusion mRNA in which either BCR exon b2 or b3 is joined to ABL exon a2. It is generally accepted that inception of BCR/ABL hybrid gene and its product plays one of the main role in pathogenesis of CML [1]. It is reported that around 2–4 % of patients with typical clinical features of CML do not posses Ph chromosome in bone marrow cells, however, are positive for BCR/ABL when analyzed by Southern blotting or reverse transcriptase-polymerase chain reaction (RT-PCR) [2]. Mostly, the rearranged gene localized on band 22q11 in these cases by in situ hybridization. Several investigators have reported localization of the fusion gene in region 9q34 [3–7], thereby indicating that it is not such a rare event [8]. Different mechanisms of origin of this rearrangement have been suggested. According to Hagemeijer et al. [3] it could be the result of a one step insertion. Viera et al. [9] hypothesized two translocations. Tanaka et al. [10] suggested segmental translocation which he defined as transposition.
To the best of our knowledge, only six cases with BCR/ABL positive CML with localization of fused gene on both the homologues of chromosome 9 in region 9q34 are reported among which the last case was reported in 2005 [11, 12]. Here, we report two more patients with Ph−ve, BCR/ABL positive CML with localization of fused gene on both the homologues of chromosome 9 in region 9q34 with the results of conventional cytogenetics and fluorescence in situ hybridization (FISH). We found these two cases out of 361 de novo CML cases studied. The presence of two copies of BCR/ABL fusion gene could be the sign of start of acceleration and/or blast crisis of CML. However, according to clinical findings, both patients were in chronic phase of CML at the first hematologic examination.
Materials and Methods
The study was approved by institutional ethics committee and scientific review committee.
Case History
Patient: 1
A 27-year old CML male diagnosed outside in September 2002. At diagnosis, (September 2002, outside) Bone marrow aspiration revealed hypercellular marrow consisting of; blast 02 %, promyelocytes 04 %, myelocytes 19 %, metamyelocytes 25 %, band cells 30 %, polymorphs 05 %, lymphocytes 05 %, monocytes 02 %, eosinophils 02 %, basophils 02 %, late normoblast 04 %. Myeloid to erythroid ratio was 25:1 and megakaryocytes were seen, and findings were suggestive of CML-chronic phase (CP). Major-BCR region was detected in molecular studies qualitative RT-PCR. Patient was treated initially with hydroxyurea (HU).
On presentation at our institute, The Gujarat Cancer and Research Institute (GCRI), Ahmedabad (February, 2005), blood examination revealed; hemoglobin (Hb) level of 127 g/L, white blood cell (WBC) count of 20.1 × 109/L, platelet count 330 × 109/L. From September 2005, he was treated with imatinib mesylate (IM) (400 mg/day, Gleevec; Novartis, East Hanover, NJ, USA). During the treatment, follow-up bone marrow aspiration in April 2009 revealed controlled CML activity. On last follow-up on October 2010, blood examination revealed; hemoglobin (Hb) level of 125 g/L, WBC count of 14 × 109/L, platelet count 401 × 109/L. Patient did not achieve hematologic or cytogenetic response.
Patient: 2
A 30-year old male was referred to GCRI in September 2003 with complains of early fatigue and pain in abdomen. At presentation, laboratory data for blood examination revealed; Hb level of 86 g/L, WBC count of 302 × 109/L, platelet count 140 × 109/L. Bone marrow aspiration revealed hypercellular marrow; consisting of blast 04 %, promyelocytes 05 %, myelocytes 29 %, metamyelocytes 08 %, band cells 06 %, polymorphs 25 %, eosinophils 02 %, basophils 04 %, lymphocytes 03 %, early normoblasts 02 %, inter normoblast 05 %, late normoblast 06 % and reticulum cells 01 %. Myeloid to erythroid ratio was increased and megakaryocytes were seen, and findings were suggestive of CML-CP. Patient was initially treated with HU and from February 2007, he was treated with IM (400 mg/day, Gleevec; Novartis, East Hanover, NJ, USA). On the last follow-up on October 2011, blood examination revealed; Hb level of 101 g/L, WBC count of 21.9 × 109/L, platelet count 496 × 109/L. Patient did not achieve hematologic or cytogenetic response.
Conventional Cytogenetics
Short-term culture of bone marrow cells harvesting and GTG banding was performed according to standard procedures following karyotyping according to ISCN 2009 guidelines [13, 14].
FISH
Different FISH strategies were adopted; locus specific identifier (LSI) probes for BCR/ABL gene rearrangement, dual color dual fusion (DCDF), BCR/ABL-ES (extra signal) were performed in both the patients (Abbott Molecular-Vysis, Des Plaines, IL, USA) according to manufacturer’s protocol. In case 1, BCR/ABL + 9q34 Tricolor, Dual Fusion Translocation Probe (Abbott Molecular-Vysis, Des Plaines, IL, USA) was also performed.
Home-Brew FISH Probe
Bacterial artificial chromosomes (BACs) (a kind gift from Sanger center, Wellcome trust, UK) CTA/bk299D3 on 22q13.31-q13.32 tagged with spectrum green was used to know the mechanism behind localization of BCR/ABL fusion on 9q34 in case 1. Plasmid DNA was isolated by using Qiagen mini prep kit (USA). Nick translation and probe precipitation were done according to manufacturer’s instructions for labeling of BAC DNA (Abbott Molecular-Vysis, Des Plaines, IL, USA).
Results
Conventional Cytogenetics
The first cytogenetic examination revealed normal karyotype 46,XY in all metaphases in both the patients. At different time intervals, the follow-up cytogenetic examination was carried out in both the patients. In patient 1, cytogenetic studies were carried for five times, the karyotype description are as follows; in February 2005, the karyotype was 46,XY[14], in July 2005, 46,XY[10], in April 2009, 46,XY[10] and in April 2010, 46,XY[18], respectively. In patient 2, cytogenetic studies were carried for five times, the karyotype description are as follows; in September 2003, 46,XY[15], in August 2004, 46,XY[15], in September 2006, 46,XY[15], in August 2007, 46,XY[9] and in July 200946,XY[10], respectively. In both the patients the karyotype results were normal every time (Table 1).
Table 1.
Details of karyotype results and DCDF-FISH-BCR/ABL results at presentation and on follow-up
| No | Date | Karyotype | FISH LSI-DCDF-BCR/ABL | RT-PCR | ||
|---|---|---|---|---|---|---|
| Signal pattern | Interpretation-Interphase | Interpretation-Metaphase | ||||
| Patient 1 | 3-Sep-02 | – | – | Major BCR | ||
| 10-Feb-05 | 46,XY[14] | – | – | |||
| 7-Jul-05 | 46,XY[10] | 1R2G1F (80 %) | Positive-deletion of ABL on der(9) | Fusion for BCR/ABL on der(9) | ||
| 2G2F (20 %) | Positive-Variant signal pattern | Fusion for BCR/ABL on both der(9) | ||||
| 7-Sep-06 | – | 2R2G (73 %) | Negative | – | ||
| 1R1G1F (24 %) | Positive-deletion of ABL/BCR on der(9) | |||||
| 2G2F (3 %) | Positive-Variant signal pattern | |||||
| 23-Apr-09 | 46,XY[10] | 2G2F (50 %) | Positive-Variant signal pattern | Fusion for BCR/ABL on both der(9) | ||
| 1R2G1F (28 %) | Positive-deletion of ABL on der(9) | Fusion for BCR/ABL on der(9) | ||||
| 2R2G (22 %) | Negative | Negative | ||||
| 15-Apr-10 | 46,XY[18] | 2G2F (100 %) | Positive-Variant signal pattern | Fusion for BCR/ABL on both der(9) | ||
| Patient 2 | 2-Sep-03 | 46,XY[15] | – | – | ||
| 24-Aug-04 | 46,XY[15] | |||||
| 1-Sep-06 | 46,XY[15] | 1R1G1F (100 %) | Fusion for BCR/ABL on der(9) | |||
| 24-Aug-07 | 46,XY[9] | – | ||||
| 31-Jul-09 | 46,XY[10] | 2G2F (100 %) | Fusion for BCR/ABL on both der(9) | |||
FISH
BCR/ABL-DCDF
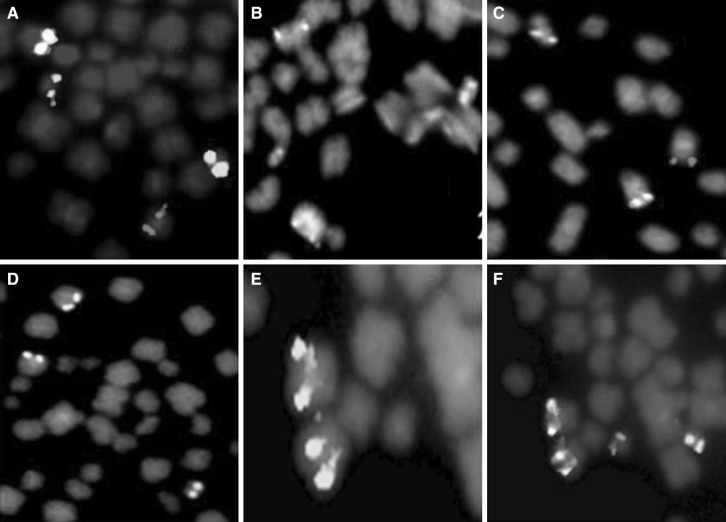
The details of DCDF-FISH results at diagnosis and on follow-up are described in Table 1 for both patients (Figs. 1a, b, 2a1, a2).
Fig. 1.
a, b LSI-BCR/ABL-DCDF. c, d LSI-BCR/ABL-ES. e, f LSI-BCR/ABL-Tri-color
Fig. 2.
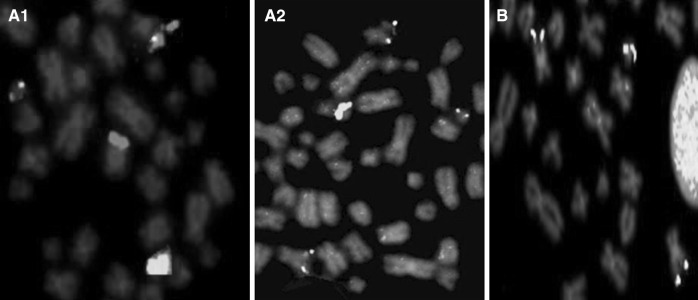
a1 LSI-BCR/ABL-DCDF at diagnosis, a2 LSI-BCR/ABL-DCDF on follow-up, b LSI-BCR/ABL-ES
Patient: 1
Total four times FISH studies were carried out; In July 2005, 80 % cells exhibited 1Red(R) 2Green(G)1F(fusion) signal pattern, metaphase FISH indicated BCR/ABL fusion on der(9), whereas 20 % cells exhibited 2G2F signal pattern, metaphase FISH indicated two BCR/ABL fusions on both der(9). In September 2006, 73 % cells were negative for BCR/ABL fusion, whereas 24 % cells exhibited 1R2G1F signal pattern and 3 % cells exhibited 2G2F signal pattern as earlier. In April 2009, 50 % cells exhibited 2G2F signal pattern, 28 % cells exhibited 1R2G1F signal pattern and 22 % cells were negative. On last follow-up in April 2010, 100 % cells were with 2G2F signal pattern, metaphase FISH indicated two BCR/ABL fusions on both der(9).
Patient: 2
DCDF-FISH was carried out twice; in September 2006, 100 % cells exhibited 1R2G1F signal pattern, metaphase FISH indicated BCR/ABL fusion on der(9). On last follow-up in July 2009, 100 % cells exhibited 2G2F signal pattern, metaphase FISH indicated two BCR/ABL fusions on both der(9).
Other BCR/ABL FISH strategies
In both the patients FISH with BCR/ABL-ES also showed unusual signal pattern 1R1G1F and 1G2F in both interphase and metaphase. The signal pattern 1R1G1F indicated, 1R on normal 9, 1 G on chromosome 22 and fusion on der(9) (Fig: 1c). The signal pattern 1G2F indicated, 1 G on chromosome 22 and 2 fusion on both the chromosome 9 [Fig: 1d]. FISH with BCR/ABL-Tri color showed no deletion of ABL/ASS observed [Fig. 1e–f].
Home-Brew FISH Probe
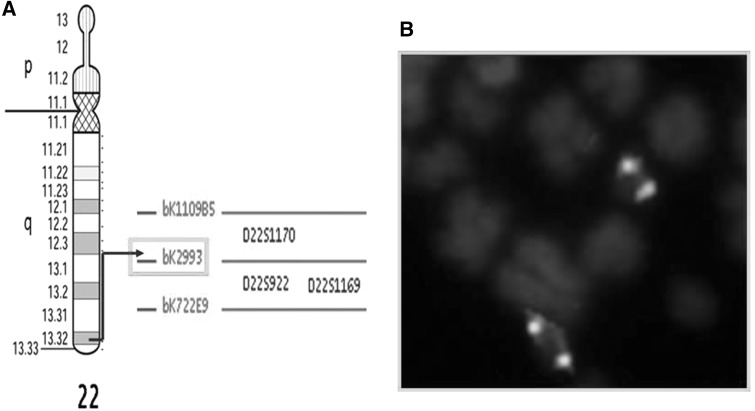
The patient1 was having BCR/ABL fusion on both the 9 chromosome instead of Ph. BAC-FISH was carried out using CTA/bk299D3 situated on terminal region of q arm of chromosome 22, below the BCR gene. It was tagged with green spectrum. Both of the green signals for BAC-CTA/bk299D3 were present on 22q13.32. It indicated that, only part of BCR gene was inserted into 9q34, whereas rest of the part was remained on der(22), hence provided a clue for insertion mechanism (Fig. 3a, b).
Fig. 3.
a Genomic address of BAC-CTA/bk299D3 used in present case. b Partial metaphase with green signals of BAC-CTA/bk299D3 on 22q13.32
Discussion
Cryptic BCR/ABL rearrangements can be found in cases with a normal karyotype [15] and in cases with complex karyotype in which the t(9;22) is not detected by conventional cytogenetic analysis. Incidence of such normal cases is reported to be around 1 % cases. These rearrangements can be revealed by FISH [16]. Complex rearrangements in some cases represent secondary changes arising likely from two consecutive translocations with a total of four breaks, or may arise from multiple simultaneous breaks in some patients. The first translocation results in t(9;22)(q34;q11). The second translocation occurs involving a break distal at band 9q34 and another break on a third chromosome [17]. The most frequent location of the BCR/ABL fusion gene in complex chromosomal rearrangement is 22q11.2 [7], but in rare variant cases BCR/ABL is translocated on other sites. At least 25 cases [18] described in the literature showed fusion gene located at 9q34. Despite the advances made in defining translocations related oncogenes in CML, the molecular mechanisms of leukemogenesis in the Ph−ve BCR/ABL positive CML cases remain to be defined.
Of special interest, the patients in present study had two BCR/ABL fusion gene in each cell located at 9q34. Six similar Ph−ve cases with double BCR/ABL fusion genes have been published previously [12]; four were in chronic phase, two were in myeloid blast crisis. In agreement with hypothesis of Hagemeijer et al. [3], we presume that the first molecular and cytogenetic event in bone marrow cells of both patients could be the insertion of the 5′ part of the BCR gene within or at the 5′ side of the ABL gene on chromosome 9. The second event was duplication of the rearranged chromosome 9 together with the loss of normal chromosome 9.
Previously reported cases with two Ph chromosomes or double BCR/ABL fusion signals have been found in the bone marrow cells of the patients in transformation to accelerate and/or in blast crisis of CML. During the transformation the additional chromosomal abnormalities to Ph chromosome could be seen in some patients and are considered as indicating unfavorable prognosis. The same applies for patients with BCR/ABL fusion gene on 9q34 [2]. Clinically, both the patients in present investing were in chronic phase at the time of diagnosis. Essential results were obtained by FISH with specific LSI BCR/ABL probe, which determined the location of fused gene on both chromosomes no. 9 and gave warning of poor prognosis of the patients. It was repeatedly hypothesized, based on the course of the disease of previously published cases that prognosis of patients with two Ph chromosomes and/or BCR/ABL gene located on one or both chromosome 9 is generally very poor [11]. Out of six cases reported in the literature, only one received imatinib treatment and achieved complete response [12]. Both the patients received imatinib treatment; it may be stated that the therapy with Glivec proved to be efficient from clinical point of view. The unusual location of BCR/ABL sequences on a chromosome 9 and the duplication of this chromosome apparently did not confer hematologic response. However for cytogenetic response, it needs further follow-up studies.
The current investigation documented that patient with normal karyotype showed BCR/ABL rearrangement. These patients may have atypical signal pattern of BCR/ABL rearrangement. In addition, we have prioritized observations from metaphase chromosomes, whereas most studies depended on interphase FISH analysis. Such analysis improves the knowledge of the genesis and the outcomes of CML, which remains a more heterogeneous disease than previously believed.
References
- 1.Heim S, Mitelman F. Cancer cytogenetics. 2. New York): Wiley; 1995. pp. 33–68. [Google Scholar]
- 2.Verma RS, Chandra P. Clinical significance of reverse BCR/ABL gene rearrangement in Ph negative chronic myelogenous leukemia. Leuk Res. 2000;24:631–635. doi: 10.1016/S0145-2126(00)00034-5. [DOI] [PubMed] [Google Scholar]
- 3.Hagemeijer A, Buijs A, Smit E, Janssen B, Creemers G-J, Van der Plas D, Grosveld G. Translocation of BCR to chromosome 9: a new cytogenetic variant detected by FISH in two Ph-negative, BCR-positive patients with chronic myeloid leukemia. Genes Chromosom Cancer. 1993;8:237–245. doi: 10.1002/gcc.2870080406. [DOI] [PubMed] [Google Scholar]
- 4.Mohamed AN, Koppitch F, Varterasian M, Karanes C, Yao K-L, Sarkar FH. BCR/ABL fusion located on chromosome 9 in chronic myeloid leukemia with a masked Ph chromosome. Genes Chromosom Cancer. 1995;13:133–137. doi: 10.1002/gcc.2870130210. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi N, Miura I, Ohshima A, Utsumi S, Nimura T, Hashimoto K, Saito M, Miura A. Duplication of chromosome 9 carrying a BCR/ABL chimeric gene in Philadelphia chromosome negative chronic myeloid leukemia. Cancer Genet Cytogenet. 1996;89:166–169. doi: 10.1016/0165-4608(96)00039-8. [DOI] [PubMed] [Google Scholar]
- 6.Reddy KS, Grove B. A Philadelphia-negative chronic myeloid leukemia with a BCR/ABL fusion gene on chromosome 9. Cancer Genet Cytogenet. 1998;107:48–50. doi: 10.1016/S0165-4608(98)00064-8. [DOI] [PubMed] [Google Scholar]
- 7.Sessarego M, Fugazza G, Bruzzone R, Ballestrero A, Miglino M, Bacigalupo A. Complex chromosome rearrangements may locate the bcr/abl fusion gene sites other than 22q11. Hematologica. 2000;85:35–39. [PubMed] [Google Scholar]
- 8.Yehuda O, Abeliovich D, ben-Neriah S, Sverdlin I, Cohen R, Varadi G, Orr R, Ashkenazi YJ, Heyd J, Lugassy G, Yehuda DB. Clinical implications of fluorescence in situ hybridization analysis in 13 chronic myeloid leukemia cases: Ph negative and variant Ph positive. Cancer Genet Cytogenet. 1999;114:100–107. doi: 10.1016/S0165-4608(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 9.Vieira L, Alves AC, Marques B, Reis I, Jorge G, Ambrosio A, deSousa AB, Boavida MG. Insertion of the 5 part of BCR within the ABL gene at 9q34 in a Philadelphia negative chronic myeloid leukemia. Cancer Genet Cytogenet. 1999;114:17–21. doi: 10.1016/S0165-4608(99)00036-9. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, Arif M, Kyo T, Dohy H, Kamada N. Transposition of duplicated chromosomal segment involving fused BCR–ABL gene or ABL oncogene alone in chronic myeloid leukemia and Ph chromosome positive acute leukemia with complex karyotypes. Cancer Genet Cytogenet. 2000;119:8–14. doi: 10.1016/S0165-4608(99)00206-X. [DOI] [PubMed] [Google Scholar]
- 11.Michalová K, Zemanová Z, Bkezinová J, et al. Location of the BCR/ABL fusion genes on both chromosomes 9q34 in Ph negative chronic myeloid leukemia. Leuk Lymphoma. 2002;43(8):1695–1700. doi: 10.1080/1042819021000003063. [DOI] [PubMed] [Google Scholar]
- 12.Dufva IH, Karle H, Brondum-Nielsen K, et al. Chronic myeloid leukaemia with BCR–ABL fusion genes located to chromosomes 9, cyclic leukocytosis and nodal T-lymphoblastic transformation–durable complete remission following imatinib therapy. Leukemia. 2005;19(4):671–673. doi: 10.1038/sj.leu.2403662. [DOI] [PubMed] [Google Scholar]
- 13.Verma RS, Babu A. Human chromosomes. In: Verma RS, Babu A, editors. Manual of basic techniques. New York: Pergamon Press; 1995. [Google Scholar]
- 14.ISCN . In: International system of human cytogenetic nomenclature. Shaffer LG, Slovak ML, Campbell LJ, editors. Basel: S Karger AG; 2009. [Google Scholar]
- 15.Haigh S, Cuthbert G. Fluorescence in situ hybridization characterization of different cryptic BCR/ABL rearrangements in chronic myeloid leukemia. Cancer Genet Cytogenet. 2004;155:132–137. doi: 10.1016/j.cancergencyto.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Morel F, Herry A, Bris M, et al. Contribution of fluorescence in situ hybridization analyses to the characterization of masked and complex Philadelphia chromosome translocations in chronic myelocytic leukemia. Cancer Genet Cytogenet. 2003;147:115–120. doi: 10.1016/S0165-4608(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 17.Calabrese G, Stuppia L, Franhchi GP, et al. Complex Translocations of the Ph Chromosome and Ph negative CML Arise from similar mechanisms, as Evidenced by FISH analysis. Cancer Genet Cytogenet. 1994;78:153–159. doi: 10.1016/0165-4608(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 18.Brahmbhatt MM, Patel PS, Trivedi PJ, et al. Unusual location of BCR/ABL fusion gene in four CML patients with masked Philadelphia chromosome. Int J Med Med Sci. 2010;2(3):38–43. [Google Scholar]