Abstract
The era of genome sequencing has produced long lists of the molecular parts from which cellular machines are constructed. A fundamental goal in systems biology is to understand how cellular behavior emerges from the interaction in time and space of genetically encoded molecular parts, as well as non-genetically encoded small molecules. Networks provide a natural framework for the organization and quantitative representation of all the available data about molecular interactions. The structural and dynamic properties of molecular networks have been the subject of intense research. Despite major advances, bridging network structure to dynamics – and therefore to behavior – remains challenging. A key concept of modern engineering that recurs in the functional analysis of biological networks is modularity. Most approaches to molecular network analysis rely to some extent on the assumption that molecular networks are modular – that is, they are separable and can be studied to some degree in isolation. We describe recent advances in the analysis of modularity in biological networks, focusing on the increasing realization that a dynamic perspective is essential to grouping molecules into modules and determining their collective function.
Frameworks for Organizing Molecular Interactions
Because of the information available to date, much work has been done to define modules based on properties of network structure. A structural module can be viewed as a static representation of all the interactions that are possible between the elements of a module. In this case, the analysis of aggregated data sources provides enough information to define modules. However, knowledge of network structure is often not sufficient to infer function, and dynamic modularity can exist in the absence of structural modularity. A clear understanding of dynamic modularity emerges only when the time-dependent activity of molecular networks is monitored. As more dynamical data become available, it will be possible to complement our understanding of the structure of molecular networks with an understanding of their dynamics, and thereby to understand more about biological function, and how interactions between molecules generate cellular phenotypes.
Analysis of Network Structure
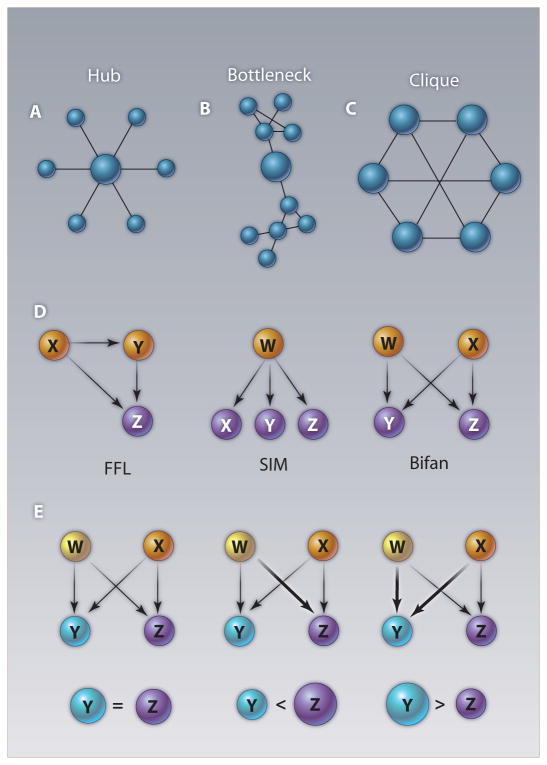
Generating data on molecular interactions at the genome scale is still a difficult enterprise. As of 2008, only about 20% of the predicted interactome in yeast has been experimentally characterized (1). Time-resolved molecular interaction data is even scarcer, so a substantial amount of research effort has been expended elucidating biological function from structural parameters of molecular interaction networks (2). The first step in finding modules from protein interaction data includes finding fully-connected subnetworks or cliques, or defective cliques which have lower connectivity (3, 4). Proteins on the shortest path between many pairs of nodes in the network, called bottlenecks, often represent interfaces between modules (5), whereas highly-connected nodes or hubs often lie in the center of modules (6).
Here we highlight examples of structural features of molecular networks for which the importance of dynamics is evident even in the absence of dynamical information. First, by mapping known three-dimensional structures of interacting proteins onto the yeast protein interaction network, two classes of hubs with distinct dynamical behavior were identified (7). Multi-interface hubs have several interaction surfaces and interact with multiple partners simultaneously, anchoring them into complexes that are stable across multiple conditions and time points. Single-interface hubs have one or sometimes two interaction surfaces and interact with partners one or two at a time. Multi- and single-interface hubs, defined by these biophysical properties, resemble party and date hubs, respectively, which were defined on the basis of their different signatures in gene expression data (8). Conclusively distinguishing between these two classes of hubs requires dynamic information.
The analysis of network motifs, pioneered by Uri Alon (9), provides information about structural modularity on the scale of a few interacting molecules. Network motifs can be recognized from network structure alone by finding groups of molecules connected in the network more often than would be expected at random. The feed-forward loop (FFL) and single-input module (SIM) are classic examples where just knowing network structure can provide some insight into dynamics and function (Fig. 1). For example, FFLs often play a role in noise filtering, turning on gene transcription only in response to sustained input signals (10). SIMs are often present in terminal differentiation cascades, where they lead to expression of a battery of functionally related genes in response to an activation signal (11). However, for another common network motif, the bifan, it is difficult to infer anything about its function from structure alone: Depending on the strength and sign of the interactions between nodes in the bifan motif, completely different dynamical behavior can result (12). A systematic analysis of network motif dynamics showed that several classes of real biological networks are enriched with network motifs that are robust to changes in dynamical parameters, like the FFL and SIM. The same networks were found to be depleted in network motifs that exhibit widely varying behavior in response to perturbation of their dynamical parameters, like the bifan (13). This work suggests that natural selection acts on network dynamics and that network structure and dynamics are coupled.
Fig. 1.
Hubs, bottlenecks, cliques, and network motifs are features of the topological structure of molecular networks that have been well-studied and can sometimes be used to infer function. (A) Hubs are nodes in the network with more interaction partners than average; they tend to be essential. (B) Bottlenecks are nodes that lie on the shortest path between many pairs of nodes in the network; they often lie at the interface between modules and also tend to be essential. (C) Cliques in protein interaction networks are indicative of protein complexes that function together. (D) The feed-forward loop (FFL) and single-input module (SIM) are network motifs whose output behavior varies little when dynamical parameters are changed, whereas (E) the bifan motif can generate widely different dynamics from different dynamical parameter combinations (12, 13).
Examination of condition-dependent expression of network motifs in the yeast regulatory network found some conditions enriched in FFLs and other conditions enriched in SIMs (11). In conditions that monitor the cell’s internal state (endogenous conditions), FFLs buffer multiple layers of internal signal processing, whereas in conditions that involve external stimuli (exogenous conditions), SIMs generate rapid responses to external conditions by expressing batteries of output genes appropriate to the condition. This work took advantage of the functional information inherent in the structure of FFLs and SIMs to predict dynamical behavior in different situations. It also revealed that hubs in some conditions are not hubs in others, showing that important features of network structure are transient and condition-specific (11). Another dynamic extension of the concept of network motifs is the activity motif. There, activity is defined using some quantitative functional genomics dataset overlaid onto the network structure. The network is then searched for series of nodes with quantitative patterns that are present more often than would be expected at random (14). For example, a chain of genes with decreasing transcription factor binding affinity might represent a just-in-time transcription program: The first gene with strongest affinity gets activated first, and so on, in a linear cascade.
Dynamic Modularity
These glimpses of dynamic signatures embedded in structural modules have led researchers toward the realization that network dynamics must be studied more systematically. The dynamics of molecular interactions can be studied at the genome scale by integrating time series of high-throughput datasets or datasets generated under various conditions. Even in the data-rich environment of functional genomics, however, the problem of relating network structure to dynamics remains very challenging. The problem is largely underdetermined because there are always more network parameters than data points available to constrain them. Accordingly, the necessary first step in such methods is to group molecules into modules and study the dynamic interactions between modules (15). Great care in designing the experiments helps to maximize the amount of information provided by these methods. Modular response analysis (MRA) is a classic framework for designing experiments to determine molecular network dynamics (15). On the basis of prior knowledge, a biological system is partitioned into functional modules such that each module can send and receive just one input from each other module. A series of experiments perturb each module separately and the framework of MRA is applied to the experimental results, thus enabling the strength of interaction between all the modules to be quantified, generating a dynamic modular network.
Although modularization reduces the size of the network and makes it less underdetermined, it pre-supposes that molecular networks can, in fact, be treated as separable modules (16, 17). An alternative view is that molecular interactions in the cell cannot be broken down into structural modules, that there is a large amount of crosstalk between molecules with unrelated functions. These two possibilities might reflect different evolutionary conditions. Nonmodular networks are more likely to evolve in stable environments, whereas modular networks are likely to evolve in organisms challenged by multiple environments (18, 19). The concepts of retroactivity and kinetic insulation provide a framework for understanding dynamic modularity.
Retroactivity
The concept of retroactivity emerged in the field of electric engineering. When electronic modules are connected in series, the dynamical properties of a downstream element may retroactively affect the functioning of an upstream element, for example by draining electrons too fast. An electrical module with low retroactivity can be mixed and matched with others to build more complex circuits in a modular way. Like electrical circuit elements, dynamic biological modules with low retroactivity may be mixed and matched in various combinations without altering the internal dynamics of each module. It is possible that molecular networks evolved to exhibit low retroactivity and improve their dynamic modularity. New methods to analyze retroactivity in molecular networks are emerging: A theoretical framework for such analysis has been proposed (20), and an algorithm was developed to partition molecular networks such that retroactivity between modules is minimized (21). Certainly functional modules designed for synthetic biology must exhibit low retroactivity, but it is an open question whether natural molecular networks have evolved the same characteristic.
Although MRA can aid in the generation of dynamic modular networks, the amount of retroactivity between modules cannot be determined with MRA, since it does not provide a mechanism to determine whether feedback from one module to another alters the internal dynamics of the receiving module. The only way to determine retroactivity between modules is to generate more detailed data about the dynamical parameters of the nodes inside the modules. As more such dynamical data becomes available, it will be possible to answer the question whether or not Nature evolves dynamical modules that are dynamically isolated by minimizing retroactivity. If the answer is no, that some functional modules are not dynamically isolated from each other, then kinetic insulation provides a way to reduce crosstalk by separating the timescales of dynamic signals that pass through the module from different inputs (22).
Kinetic Insulation
Both theoretical and experimental analyses indicate that a single molecular network can generate distinct outputs in response to input signals with different dynamical profiles. This phenomenon has been termed “kinetic insulation” (22), emphasizing the importance of dynamics to signaling specificity. For example, a progenitor cell in a tissue differentiation cascade can turn on different batteries of genes in response to input signals with different dynamic signatures. A classic example is the case where a mitogen-activated protein kinase (MAPK) cascade responds transiently to epidermal growth factor (EGF) but exhibits a sustained response to neural growth factor (NGF) (23). The PC12 cancer cell line is derived from adrenal gland tissue and can be thought of as an imperfect model of adrenal gland progenitor cells, which have the ability to differentiate into either sympathetic neurons or adrenal chromaffin cells that synthesize adrenaline. NGF generates sustained activation of MAPK in PC12 cells, leading to a specific gene expression program that causes the cells to differentiate into neuronal cells. In contrast, EGF activates the same MAPK only transiently, triggering a gene expression program that causes the cells to proliferate, presumably allowing them to differentiate into adrenaline-producing cells in response to a later cue. This tissue specification program is only possible because of the expression in the progenitor cell of different batteries of genes in response to different input signals. The MAPK cascade utilizes kinetic insulation to process different dynamic signals through the same set of molecules to generate different responses (23). The nuclear factor κB (NF-κB) regulatory network that is important in the mammalian inflammation response also exhibits kinetic insulation, producing transient or sustained activation of the NF-κB transcription factor in response to tumor necrosis factor alpha (TNF-α) or lipopolysaccharide (LPS) input signals, respectively (24).
Determining Dynamical Parameters of Cellular Networks
Dynamical models of molecular networks rely on knowledge of biochemical parameters like enzymatic rate constants, binding affinities, and protein expression levels determined over many decades of biochemical and molecular biological experiments. The assumption that a set of well-studied molecules can be treated as an isolated system is equivalent to the contention that they represent a functional module, and that the module dynamics will not be strongly affected by crosstalk with other molecules that have not been studied in the same functional context. There are a growing number of dynamical models of well-characterized molecular networks (25), which may reveal biological insight, keeping in mind the caveat that the assumption of modularity and lack of crosstalk may not hold in the face of future data.
The behavior of a dynamical model might be insensitive to changes in individual parameters but remain sensitive to changes in combinations of those parameters. For example, the ratio of activity of a kinase and phosphatase acting on the same target might be constrained while allowing the expression levels and rates of the two enzymes to vary in concert over a wide range (26). Sensitivity analysis is a mathematical tool for identifying “stiff” dynamical parameters, which are combinations of biochemical parameters that when changed strongly affect the dynamical behavior of the model. Interestingly, most dynamical models in systems biology are “sloppy”, meaning that only a few combinations of parameters are stiff, whereas the other parameters can vary over an extremely wide range without altering output behavior (25). A dynamical model of the MAPK cascade that controls PC12 cell differentiation exhibits this characteristic (23). The stiffest dynamical parameter in the model is composed of biochemical parameters affecting the interaction between the Ras and Raf proteins, meaning that altering that interaction slightly has a strong effect on dynamical behavior.
Currently, most systems biology models are parameterized by reference to the extensive biochemical literature that has developed over many decades from studies of one or a few gene products in any given experiment. Knowing many parameter values accurately provides enormous predictive power, as recently demonstrated by a model of cytokinesis in yeast that relied on characterization of the biochemical parameters of interactions between actin, myosin, and associated proteins (27). It might seem natural to extend this practice to genome scale by developing high-throughput methods to measure individual biochemical parameters one at a time. However, the sloppy character of dynamical network models suggests that not all parameters, even if accurately measured, will provide the same amount of constraint on the model. In this case, using sensitivity analysis on the model and fitting all of the parameters at once to global time series data may identify which combinations of parameters are most important to measure.
Robustness and Evolvability
The uneven stiffness of dynamical parameters has interesting implications for the robustness and evolvability of molecular networks. The set of non-stiff dynamical parameters represents a “neutral space” where network phenotype is robust to parameter changes generated by molecular evolutionary processes (28). Changes in stiff dynamical parameters should be more strongly constrained by selection.
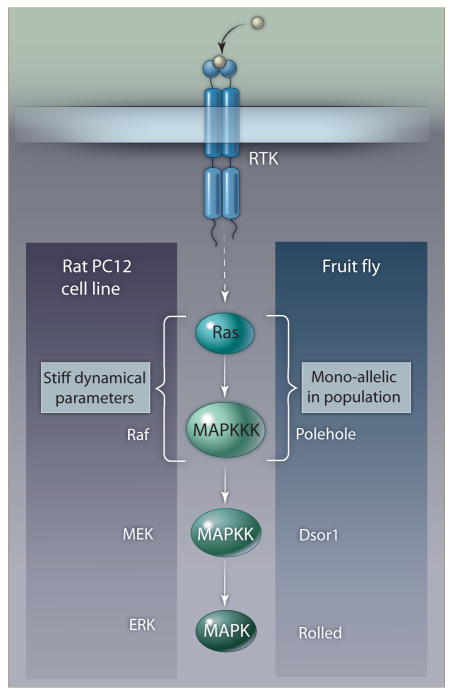
Reconstructing evolutionarily related families of molecular networks might reveal which combinations of parameters are sensitive and which are insensitive based on the extent of variation at the population level. An interesting example is again the network representing the response of rat adrenal gland progenitor cells to neural and epithelial growth factors. Analysis of a model of EGF versus NGF activation of the Ras-Raf-MEK-ERK MAPK pathway showed that the stiffest dynamical parameter was associated with Ras and Raf, the upstream elements of the pathway (23). A population genetic study of a homologous MAPK cascade in Drosophila showed that the fly Ras and Raf homologs exhibited the least polymorphism across the world-wide population of fruit flies, whereas other components of the pathway were more polymorphic, that is less constrained (29). Thus, a model of a MAPK cascade from rat made predictions about which proteins were dynamically constrained, and these predictions were confirmed at the population level in the homologous pathway in fruit flies (Fig. 2).
Fig. 2.
Dynamical constraints on a signaling pathway involved in tissue differentiation in rat correlate with allelic variation at the population level in the same pathway in fruit flies, revealing a link between dynamics and evolution. The “stiffest” combination of parameters was associated with the Ras and Raf proteins at the top of the pathway, and in fruit flies Ras and Raf exhibit the least allelic variation of all of the components of the pathway, consistent with the concept that stiff parameter combinations are constrained during evolution. The dotted box indicates that some accessory components of the two cascades are excluded here for clarity of presentation.
The proliferation of data about variability in human populations will be a useful resource for further studies of this type (30). Most studies of human population variability focus on variations that affect phenotype, usually associations with disease, but it should be remembered that even data about genotypic variability without phenotypic effect is useful, because it reveals which genes in a network are robust to variation in parameters (31, 32).
References
- 1.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu XW, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 3.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:27. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu HY, Paccanaro A, Trifonov V, Gerstein M. Predicting interactions in protein networks by completing defective cliques. Bioinformatics. 2006;22:823–829. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- 5.Yu HY, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:713–720. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 7.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 8.Han JDJ, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJM, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 9.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 10.Milo R, Itzkovitz S, Kashtan N, Levitt R, Shen-Orr S, Ayzenshtat I, Sheffer M, Alon U. Superfamilies of evolved and designed networks. Science. 2004;303:1538–1542. doi: 10.1126/science.1089167. [DOI] [PubMed] [Google Scholar]
- 11.Luscombe NM, Babu MM, Yu HY, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 12.Ingram PJ, Stumpf MPH, Stark J. Network motifs: structure does not determine function. BMC Genomics. 2006;7:108–120. doi: 10.1186/1471-2164-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prill RJ, Iglesias PA, Levchenko A. Dynamic properties of network motifs contribute to biological network organization. PLoS Biol. 2005;3:1881–1892. doi: 10.1371/journal.pbio.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chechik G, Oh E, Rando O, Weissman J, Regev A, Koller D. Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat Biotechnol. 2008;26:1251–1259. doi: 10.1038/nbt.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kholodenko BN, Kiyatkin A, Bruggeman FJ, Sontag E, Westerhoff HV, Hoek JB. Untangling the wires: A strategy to trace functional interactions in signaling and gene networks. Proc Natl Acad Sci USA. 2002;99:12841–12846. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, Longabaugh W, Vuthoori M, Whitehead K, Madar A, Suzuki L, Mori T, Chang DE, DiRuggiero J, Johnson CH, Hood L, Baliga NS. A Predictive Model for Transcriptional Control of Physiology in a Free Living Cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 17.Reiss DJ, Baliga NS, Bonneau R. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics. 2006;7:22. doi: 10.1186/1471-2105-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci USA. 2005;102:13773–13778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parter M, Kashtan N, Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol. 2007;7:8. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Vecchio D, Ninfa AJ, Sontag ED. Modular cell biology: retroactivity and insulation. Mol Syst Biol. 2008;4:16. doi: 10.1038/msb4100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saez-Rodriguez J, Gayer S, Ginkel M, Gilles ED. Automatic decomposition of kinetic models of signaling networks minimizing the retroactivity among modules. Bioinformatics. 2008;24:i213–219. doi: 10.1093/bioinformatics/btn289. [DOI] [PubMed] [Google Scholar]
- 22.Behar M, Dohlman HG, Elston TC. Kinetic insulation as an effective mechanism for achieving pathway specificity in intracellular signaling networks. Proc Natl Acad Sci USA. 2007;104:16146–16151. doi: 10.1073/pnas.0703894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown KS, Hill CC, Calero GA, Myers CR, Lee KH, Sethna JP, Cerione RA. The statistical mechanics of complex signaling networks: nerve growth factor signaling. Phys Biol. 2004;1:184–195. doi: 10.1088/1478-3967/1/3/006. [DOI] [PubMed] [Google Scholar]
- 24.R. Cheong, A. Hoffmann, A. Levchenko, Understanding NF-kB signaling via mathematical modeling. Mol. Syst. Biol. 4 (2008).
- 25.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldbeter A, Koshland DE. An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 28.Wagner A. Robustness and evolvability: a paradox resolved. Proc R Soc B-Biol Sci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riley RM, Jin W, Gibson G. Contrasting selection pressures on components of the Ras-mediated signal transduction pathway in Drosophila. Mol Ecol. 2003;12:1315–1323. doi: 10.1046/j.1365-294x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 30.Pennisi E. Breakthrough of the year - Human genetic variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- 31.Chen YQ, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang CS, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim PM, Korbel JO, Gerstein MB. Positive selection at the protein network periphery: Evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci USA. 2007;104:20274–20279. doi: 10.1073/pnas.0710183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.RA would like to thank Marcelo Behar, Kevin Brown, Ryan Gutenkunst and Alex Hoffmann for stimulating discussions. R.P.A. was support by NIH grant T32-HG003198–04. T.E. was supported by an Alfred P. Sloan Foundation Fellowship.