Abstract
Coherent splicing networks arise from many discrete splicing decisions regulated in unison. Here, we examine the properties of robust, context-specific splicing networks. We propose that a subset of key splicing regulators, or “master splicing factors,” respond to environmental cues to establish and maintain tissue transcriptomes during development.
Design principles of robust splicing networks
The epigenetic landscape of differentiation defined by Conrad Waddington nearly 60 years ago proposed that homeostatic mechanisms maintained multiple irreversible cell states, providing an early suggestion that both stability and plasticity were critical components of biological networks (Waddington, 1957). While ensuing efforts focused on understanding the role of transcriptional programs in homeostasis or differentiation, the importance of the post-transcriptional contribution to these processes is now reaching the forefront. Several recent lines of evidence suggest that splicing networks are composed of highly interconnected events, conferring stability to the system while simultaneously maintaining responsiveness to external stimuli. This is the definition of a “robust” network (Kitano, 2004). Along with transcriptional and other post-transcriptional effects, splicing contributes a layer of regulation integral to the establishment of tissue transcriptomes.
While nearly all mammalian genes undergo alternative splicing (AS) in some context (Wang et al., 2008), only a minority of splicing events are conserved across evolution; nevertheless, this population has provided critical insights into the role of AS in gene networks (Barbosa-Morais et al., 2012; Merkin et al., 2012). Most conserved splicing events tend to maintain reading frame and encode alternative protein variants (Barbosa-Morais et al., 2012; Merkin et al., 2012). However, a substantial fraction of conserved splicing events introduce premature termination codons (PTC) and lead to downregulation of the transcript through the process of nonsense-mediated mRNA decay (NMD) (Baek and Green, 2005). Recent estimates from gene expression profiling suggest that in fact, as many as 10–30% of mammalian genes may be regulated by alternative splicing-coupled NMD (AS-NMD) in particular contexts (Lewis et al., 2003; Mendell et al., 2004; Weischenfeldt et al., 2012). The coordination of AS-NMD with alternative protein isoform production generates regulatory motifs in splicing networks. Understanding the behavior of these individual network motifs within the context of larger networks reveals insights into how splicing regulation impacts cell fate decisions.
Negative autoregulation maintains homeostasis
When compared to a simply regulated expression system (in which one gene product drives expression of another), a negatively autoregulated system (in which a gene product feeds back on its own production) is characterized by faster response times and reduced cell-to-cell variability in the concentration of a gene product. This consequently results in a single steady state of the gene product that is buffered against variations in transcriptional output or protein stability (Alon, 2007; Becskei and Serrano, 2000; Nevozhay et al., 2009; Rosenfeld et al., 2002). In Waddington’s metaphoric terms, this buffering contributes to the “canalization” that contains a cell within the steep valleys of the epigenetic landscape (Waddington, 1957). It is now understood that many RNA binding proteins (RBPs) undergo negative autoregulation through AS-NMD (Figure 1A). A key observation leading to this discovery was that AS-NMD events tend to be enriched in genes encoding splicing factors and are often ultraconserved, meaning they fall within regions of particularly high evolutionary conservation (Lareau et al., 2007; Ni et al., 2007; Wollerton et al., 2004). These studies specifically focused on AS-NMD regulation of SR proteins, a family of serine-arginine-rich RBPs with various roles in RNA processing and required for splicing (Long and Caceres, 2009), and hnRNP proteins, a diverse group of RBPs thought to function most commonly as splicing repressors (Martinez-Contreras et al., 2007). These proteins were shown to be able to bind their own transcripts, cause splicing of the NMD variant, and downregulate protein levels to maintain homeostatic protein expression (Lareau et al., 2007; Ni et al., 2007; Saltzman et al., 2008). Beyond AS-NMD regulation of SR and hnRNP proteins, negative autoregulation of RBPs can also occur via the production of nonfunctional or dominant negative protein isoforms (Damianov and Black, 2010). Additionally, proteins with enzymatic activity that lack the ability to bind RNA directly can nevertheless exert autoregulatory feedback by enzymatically modifying RBPs. One example of this type of feedback loop is the SR protein kinase Clk1, whose pre-mRNA undergoes increased productive splicing in response to small-molecule inhibition of its own kinase activity (Ninomiya et al., 2011).
Figure 1. Common motifs in splicing networks.
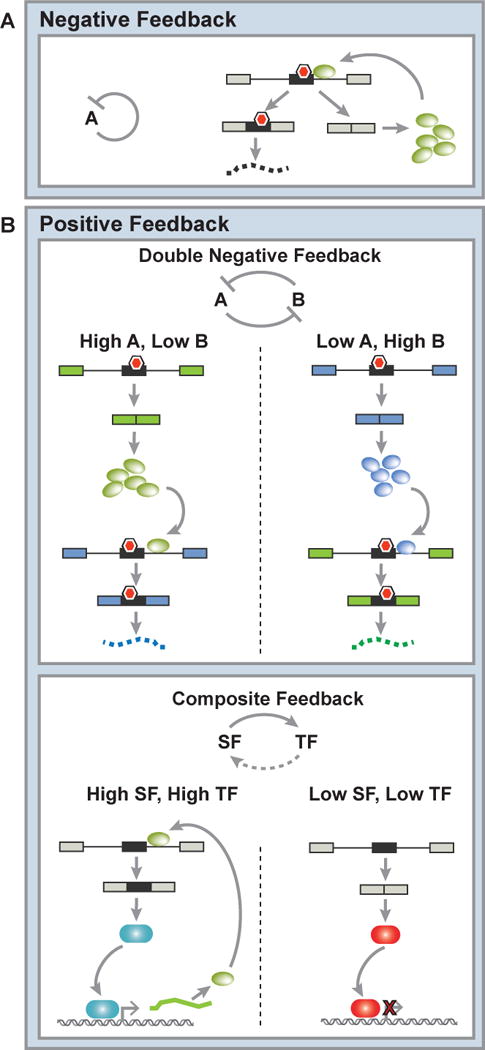
A) Negative feedback of RNA binding proteins (RBPs) occurs when a splicing factor regulates a nonsense-mediated decay-coupled splicing event within its own transcript to repress its own protein expression and maintain steady state expression levels. B) Top panel: Positive feedback in splicing networks can occur through a double negative feedback loop. Double negative feedback results from the cross-regulation of two RBPs, producing two steady states each characterized by the exclusive expression of one of the RBPs. Bottom panel: Positive feedback also arises through composite feedback with a splicing factor (SF) and one or more transcription factors (TF) that results in a double positive feedback loop and bistability. In this case, either the SF and TF are both expressed, or the SF and TF are both off. Red hexagons indicate premature termination codons.
Positive feedback generates bistability
In contrast to negative feedback, positive autoregulation is characterized by slower response times and increased noise in protein expression (Maeda and Sano, 2006). Biologically, the effect of positive feedback is to convert a graded input, such as a growth factor signal, into a binary output, such as proliferation vs. differentiation (Becskei et al., 2001). This is known as ultrasensitivity. If positive feedback is strong, it can produce two distinct steady states, or bistability, with stochastic fluctuations in gene expression driving the switch from one steady state to another (Alon, 2007; Blake et al., 2003). Unlike negative feedback, which generally returns a system to its original state after a perturbation, positive feedback can induce memory of a transient signal by reinforcing a new cell state even after the initiating signal has disappeared (Alon, 2007).
Splicing factors do not usually positively regulate the productive splicing of their own pre-mRNA. However, interaction between two different splicing factors can generate positive feedback and bistability. Double-negative feedback loops, in which two factors repress one another, can also generate two steady states, each with only one of the two factors expressed (Alon, 2007). As negative autoregulation of RBPs occurs so frequently, double-negative feedback is likely to be a common motif in splicing regulatory networks (Figure 1B). RBPs can also feature in positive feedback loops alongside transcription factors, for example during the regulation of developmental decisions (Figure 1B). A recent study investigated the role of positive feedback between OCT4, SRSF2, and the methyl-CpG-binding protein MBD2 in determining the switch between pluripotency and differentiation of embryonic stem cells (ESCs). Here, the ESC master transcription factor OCT4 drives expression of the splicing factor SRSF2, which generates an isoform of MBD2 that promotes OCT4 expression. If SRSF2 expression is sufficiently decreased, however, MBD2 is spliced to produce an isoform that represses expression of core ESC transcription factors. This composite positive feedback loop therefore maintains pluripotency in the first instance and helps drive differentiation in the second, based on the dosage of the components within the network (Lu et al., 2014). Similarly, the neuronal splicing regulator nSR100 participates in a positive feedback loop with REST, a transcriptional repressor of neurogenesis. In the presence of nSR100, REST is spliced to produce an isoform with greatly diminished repressive activity. As nSR100 is itself repressed by REST, a differentiated cell state characterized by high nSR100 activity and low REST activity is reinforced through positive feedback (Raj et al., 2011).
Crossregulation tunes steady states
The steady state expression level of a gene undergoing negative autoregulation tends toward the value of its repression threshold, which is the concentration of the gene product needed to repress its production by 50% (Alon, 2006). We might consequently predict that splicing factors that are negatively autoregulated would maintain similar steady state expression levels across tissues, regardless of transcriptional activity. Conversely, RBP expression patterns do indeed show cell type-specificity (de la Grange et al., 2010; Grosso et al., 2008), implying that additional post-transcriptional regulatory inputs must influence steady state. On an evolutionary timescale, the repression threshold and consequently the steady state level can be altered by mutation, for example by strengthening or weakening the binding site of a transcription factor at its own promoter. Tissue-to-tissue variation, however, cannot be explained by mutation. Moreover, as AS-NMD events within RBPs tend to be deeply conserved (Lareau et al., 2007; Ni et al., 2007), variations in RBP gene expression among species are unlikely to have arisen from differences in the strength of RBP binding to its transcript. Instead, extensive crossregulation between RBPs may be one mechanism that alters steady state RBP expression across cell types and across species. For example, splicing of an autoregulated poison cassette within one RBP can be competitively inhibited by the binding of a second RBP. This raises the effective concentration of the first RBP necessary for autoregulation and results in its increased steady state expression in the presence of the second RBP. Negative autoregulation thus likely cooperates with cross-regulation to ensure robust and tunable RBP expression patterns.
Data from a number of recent studies support the model that crossregulation of RBP negative feedback modulates splicing networks while maintaining their stability. We recently identified thousands of in vivo Rbfox2 binding sites and hundreds of Rbfox2-regulated splicing events across the mouse ESC transcriptome using iCLIP and RNAseq. From these parallel approaches, we identified a large class of Rbfox2-regulated splicing events that are rendered silent by NMD. A significant fraction of these NMD splicing events fell in the previously-described category of conserved autoregulatory splicing events within RBPs (Lareau et al., 2007; Ni et al., 2007). Cross-regulation of this autoregulatory splicing by Rbfox2 set the efficacy of autoregulation, and thus the steady-state expression of the gene (Figure 2A). This suggested that Rbfox2 tunes the autoregulation and gene expression of a network of RBPs, and that changes in Rbfox2 expression activate direct Rbfox2-dependent splicing changes as well as a cascade of secondary splicing changes (Jangi et al., 2014). These events, together, define the complete Rbfox2 splicing regulatory network.
Figure 2. RNA binding protein cross-regulation in splicing networks.
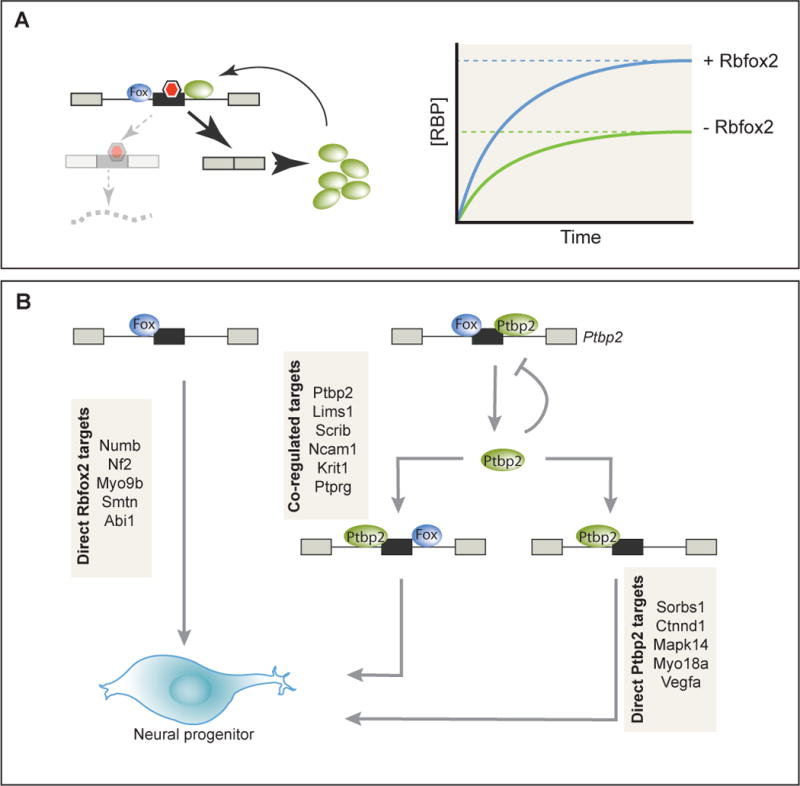
A) Model for the cross-regulation of an autoregulatory poison exon by Rbfox2. Binding of Rbfox2 inhibits inclusion of the poison exon (red hexagon indicates premature termination codon), leading to a higher steady-state expression level of the autoregulated protein in the presence of Rbfox2. B) Rbfox2 and Ptbp2 function in a splicing cascade during neuronal differentiation. Rbfox2 directly regulates a subset of targets involved in cytoskeletal organization. In addition, inhibition of autoregulatory splicing of Ptbp2 by Rbfox2 leads to Ptbp2 upregulation. Ptbp2, either in conjunction with Rbfox2 or independently, regulates the splicing of several genes involved in adherens junction formation. Together, these primary and secondary targets of Rbfox2 contribute to the specification of neural progenitor fate (Jangi et al., 2014; Licatalosi et al., 2012).
Crossregulation of autoregulatory splicing has emerged as a common motif in several other splicing networks. Two members of the hnRNP family of splicing regulators, hnRNP L and hnRNP L-like, have been shown to bind their own and each other’s transcripts to regulate gene expression through AS-NMD (Rossbach et al., 2009). A genome-wide analysis of hnRNP binding and activity in human 293T cells revealed a high frequency of crossregulation between many hnRNP family members, including the regulation of several events previously shown to be associated with NMD (Huelga et al., 2012). More generally, hnRNP-bound transcripts were significantly enriched for RNA processing genes; 70% of all RBP transcripts were bound by at least one hnRNP, with greater than 25% of these showing evidence of gene expression or splicing regulation (Huelga et al., 2012). Similarly, the core spliceosomal protein SmB/B’ autoregulates its expression through AS-NMD (Saltzman et al., 2008). Depletion of SmB/B’ in human tissue culture cells led to widespread changes in splicing and expression of RBPs in a manner attributable to AS-NMD (Saltzman et al., 2011). CLIP signal enrichments of the RBP FUS and the SR protein Srsf3 in highly-conserved introns of other RBPs suggest that this is a general feature of splicing networks (Anko et al., 2012; Nakaya et al., 2013). Such interconnected networks may serve to stabilize splicing patterns within a particular cellular condition.
Signal amplification in splicing cascades
Crossregulated splicing events, like transcription factors and signaling components, can be integrated into cascades to achieve multiple responses to different inputs. CLIP and transcriptome analysis following changes in RBP activity that identified directly-bound splicing targets also identified many secondary or indirect targets that likely arise from changes in expression of the primary RBP targets (Huelga et al., 2012; Jangi et al., 2014; Saltzman et al., 2011). One biological function of this effect is to amplify the original signal, in this case the change in expression or activity of the RBP at the top of the cascade, while maintaining robustness against stochastic fluctuations in gene expression. Such a mechanism may determine splicing changes during the establishment of neuronal precursors in the developing brain. Here, the increase in Rbfox2 expression likely contributes to the increased expression of Ptbp2 through suppression of its autoregulatory AS-NMD. Several direct targets of Ptbp2 in mouse neocortex are also indirect targets of Rbfox2 in embryonic stem cells (Jangi et al., 2014; Licatalosi et al., 2012). This suggests that Rbfox2-dependent upregulation of Ptbp2 expands and reinforces the Rbfox2 splicing network that plays an integral role in the specification of neuronal lineages (Gehman et al., 2012; Jangi et al., 2014; Licatalosi et al., 2012) (Figure 2B). In this manner, a “master splicing regulator” – positioned at the top of a splicing cascade – could play a critical role in the determination of cell fate.
Master splicing regulators and cell identity
Studies in the definition of cell identity have revealed that the expression of a relatively small number of transcription factors, termed master transcription factors, can initiate commitment along a diverse array of developmental lineages (Young, 2011). Characterization of splicing changes across tissues (Merkin et al., 2012) as well as RNA-protein interaction maps (Witten and Ule, 2011) have raised the intriguing possibility that an analogous subset of master splicing regulators exists among splicing factors.
We propose that a putative master splicing regulator plays one or both of two critical roles in the implementation of a genetic program. First, the factor may be required for the proper differentiation or specification of a cell type. These factors are likely to be expressed in a tissue-specific manner. In the absence of the master splicing regulator, differentiation would be incomplete due to the misregulation of its splicing network. Conversely, ectopic expression of a master splicing regulator in a cell type where it is not normally expressed may drive the cell toward the phenotype of a cell in which it is normally expressed. Such master splicing regulators involved in lineage specification may generate switch-like behavior by directing splicing networks that contain positive feedback loops. Second, once a cell is committed to a lineage, a master splicing regulator may be required for maintaining homeostasis. This role probably requires secondary splicing regulators acting downstream of the master regulator to be negatively autoregulated, thus buffering against variations in the cellular environment. While master splicing regulators involved in maintaining cell identity may again be lineage-specific, the secondary splicing factors that they regulate, such as SR proteins and hnRNPs, may be ubiquitously expressed. Direct and indirect targets of homeostatic master splicing factors are likely to be functionally related, allowing for the regulation of specific cell functions.
Potential master splicing factors are likely to be highly regulated, such that their splicing networks are activated only in the correct settings. In order to perturb a stable splicing network sufficiently to transition to a new state, initiating signals would have to arise outside of the network itself. These signals are thus likely to be transcriptional, translational, post-translational, or otherwise splicing-independent. The various modes of splicing factor activation may have distinct effects on the kinetics and reversibility of the corresponding change in splicing network. Moreover, a master splicing factor may have dynamic expression patterns that vary over a range that is not limited by negative feedback. For example, transcriptional activation of a master splicing factor gene would steadily increase its expression and activity until reaching the threshold of autoregulation, with its splicing network maximally activated at this threshold.
Lineage-specifying splicing cascades
Several examples of RBPs functioning as master regulatory hubs driving broad splicing networks during development can be found in the literature. In some of these cases, autoregulation, crossregulation, and feedback play critical roles in initiating robust and dynamic splicing cascades with distinct outcomes. Although candidate master RBPs frequently function downstream of larger transcriptional programs, several of these systems also rely on AS regulation of transcription factors. The evidence presented below suggests that through this interplay of transcriptional and post-transcriptional effects, the activation of key RBPs may be necessary and even sufficient to establish cell identity. Here, we discuss instances and properties of splicing networks driven by master splicing regulators in the context of differentiation.
Drosophila sex determination
The up- or downregulation of a critical RBP, most likely one that is expressed in a tissue-restricted manner, could initiate a splicing network and downstream gene expression changes that lead to a stable change in cell identity. Perhaps the best understood and most remarkable example of a bistable splicing regulatory cascade dependent on autoregulation and crossregulation is the Drosophila melanogaster somatic sex determination pathway (Figure 3). Here, sex-specific expression of the splicing repressor Sex Lethal (Sxl) specifies female development through activation of the RBP Transformer (Tra) and repression of Male-Specific Lethal 2 (Msl2) (Schutt and Nothiger, 2000). Sxl, the master splicing regulator at the top of the sex-determination pathway, is unproductively spliced in developing male flies (Bell et al., 1991). During early embryogenesis, a transcriptional signal determined by the ratio of X chromosomes to autosomes (X:A) transiently activates an upstream Sxl promoter in female flies. The resulting transcript encodes active Sxl protein (Keyes et al., 1992). Active Sxl reinforces productive splicing from the constitutive downstream promoter in a positive feedback loop in females, while in males, the lack of active Sxl leads to continual production of nonfunctional Sxl. A key target of Sxl is the SR protein Tra, which is transcribed in both sexes but is only productively spliced in females in an Sxl-dependent manner (Sosnowski et al., 1989). Tra in turn regulates the splicing of two transcription factors, Fruitless (Fru) and Doublesex (Dsx), resulting in default male-specific isoforms in the absence of Tra and female-specific isoforms in its presence (Hertel et al., 1996; Lynch and Maniatis, 1996; Tian and Maniatis, 1993). Tra has also been suggested to positively feed back on Sxl splicing, further stabilizing the female differentiation program (Siera and Cline, 2008). In parallel, Sxl targets the dosage compensation regulator Msl2 to cause intron retention and a block in translation in females, while a functional protein is produced in males (Bashaw and Baker, 1995; Zhou et al., 1995). The downstream transcriptional regulators Msl2, Fru, and Dsx establish much of the sexual dimorphism in somatic tissues (Coschigano and Wensink, 1993). Although this Drosophila pathway is not conserved among insects, splicing-dependent positive feedback is a recurring theme in insect sex determination, suggesting that the use of a splicing cascade confers an evolutionary advantage in this bistable system (Salz, 2011).
Figure 3. Bistability in Drosophila sex determination.
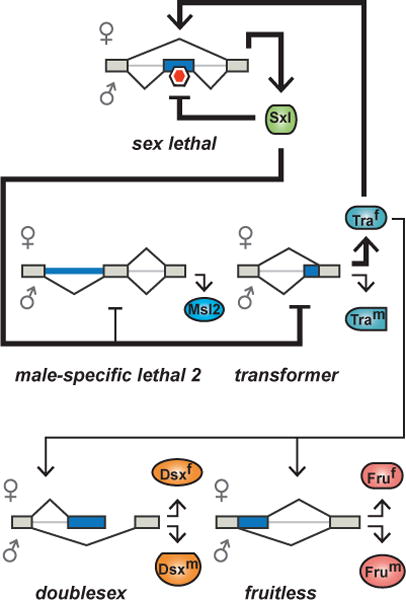
The Drosophila sex determination pathway initiates a cascade of splicing events within RNA binding proteins and transcription factors that regulates sexual dimorphism. The master regulator Sxl drives the female-specific isoform of Tra, which generates female-specific isoforms of the transcription factors Dsx and Fru. Positive feedback loops are indicated as bold arrows. Red hexagon indicates premature termination codon.
MBNL1 and cellular reprogramming
Analogous splicing cascades with such distinct phenotypic consequences have yet to be characterized in mammalian systems. Yet several recent reports have demonstrated that altering the expression of individual splicing factors can prime cells toward a different cell state. Splicing changes occurring during mesodermal differentiation correlate with the activity of the MBNL family of splicing regulators (Venables et al., 2013). More causally, in human and mouse fibroblasts, knockdown of MBNL1 increases the efficiency of reprogramming to iPSCs, while overexpression in human ESCs results in mesodermal splicing patterns (Han et al., 2013). The effect on reprogramming has been primarily attributed to MBNL1-dependent splicing of the transcription factor FOXP1, of which the ES-specific isoform (FOXP1-ES) represses differentiation-specific genes and activates key pluripotency genes due to an altered DNA binding domain (Gabut et al., 2011). Importantly, the increased reprogramming efficiency caused by MBNL1 knockdown is not phenocopied by overexpression of FOXP1-ES, implying that additional targets of MBNL1 are involved in this process (Gabut et al., 2011; Han et al., 2013). During cardiomyocyte maturation, MBNL1-dependent splicing changes are enriched for spliceosomal genes, and gene expression changes across this transition are enriched for components of the splicing machinery (Giudice et al., 2014). We can speculate that although a subset of splicing changes during these developmental transitions are likely direct targets of MBNL1, many of the events attributed to MBNL1 regulation may in fact arise from MBNL1-dependent modulations in the expression of other splicing components. These splicing cascades not only amplify the signal initiated by MBNL1 but may also increase robustness via the modulation of autoregulated RBPs.
T cell activation
Changes in the levels of a master splicing regulator can also trigger an irreversible transition between steady states along a single lineage. During T lymphocyte activation, the transmembrane receptor CD45 undergoes a splicing switch that has been suggested to initiate signaling through antigen receptors (Holmes, 2008). The activity of hnRNPLL is necessary and sufficient for the CD45 splicing switch, as well as for several other splicing events associated with hematopoietic cell differentiation and activation (Oberdoerffer et al., 2008; Topp et al., 2008). As a result, hnRNPLL has been hypothesized to be a master regulator of T cell activation (Holmes, 2008). hnRNPLL is rapidly upregulated upon stimulation of naïve T cells in a manner consistent with transcriptional activation (Oberdoerffer et al., 2008). In accordance with the cross-regulation observed in other splicing regulatory networks, several SR and hnRNP proteins containing AS-NMD cassettes undergo upregulation in two temporally distinct waves (Ip et al., 2011). Although hnRNPLL upregulation does not initiate a switch-like response as with Drosophila sex determination, it is likely that a cascade of splicing and transcriptional changes are sustained to produce a new, stable T cell state after a transient initiating transcriptional signal.
Splicing switches by opposing RBPs
A more common scenario in mammalian systems may be the toggling of splicing networks by RBPs that are expressed in a mutually exclusive manner between two conditions. The MBNL1 expression network that becomes activated during cardiac differentiation, for example, is likely opposed by an embryonic CELF1 network (Kalsotra et al., 2008). In the context of epithelial-mesenchymal transitions, the expression of Epithelial Splicing Regulatory Proteins 1 and 2 is quite tightly restricted to differentiated epithelial cell types. In contrast, the splicing patterns of less differentiated basal or mesenchymal cell types bear a signature of RBFOX2. Depletion of RBFOX2 or exogenous expression of ESRP1 in human mesenchymal breast cancer cells causes a partial reversion to an epithelial phenotype, suggesting that these factors and their targets play critical opposing roles in defining epithelial and mesenchymal cell states (Shapiro et al., 2011).
A particularly well-characterized example of mutual exclusivity in splicing factor expression is found in the PTB/PTBP2 switch during neuronal differentiation. The neuronal splicing regulator PTBP2 is lowly expressed in most tissues and is upregulated at the protein level in neurons. The PTBP2 transcript contains an alternative exon that triggers NMD when skipped; inclusion of the exon requires positive transacting factors such as nSR100 in neurons, while repression is mediated by PTBP1 in undifferentiated cells (Calarco et al., 2009; Wollerton et al., 2004). Upon miRNA-mediated downregulation of PTBP1 during neuronal differentiation, the negative regulation of PTBP2 is relieved, leading to a switch from a PTB-driven to a PTBP2-driven AS network (Boutz et al., 2007; Makeyev et al., 2007; Spellman et al., 2007; Wollerton et al., 2004). Interestingly, depletion of PTBP1 in fibroblasts is sufficient for PTBP2 induction and neuronal trans-differentiation, arguing that PTBP1 is a master regulator of the undifferentiated cell state (Xue et al., 2013). It remains to be seen whether PTBP2 also regulates the AS-NMD event present in the PTBP1 transcript (Wollerton et al., 2004), as this would result in a double-negative feedback loop specifying neuronal versus non-neuronal lineages. Such dynamic regulation of otherwise stable splicing networks by opposing factors may be a common feature of diverging developmental programs.
Homeostatic splicing networks
Another function of a master splicing factor is to reinforce cell identity, for example by regulating a specific cell function or by controlling homeostatic splicing events. These factors may not be required to specify the cell’s lineage, but may be required to function within and respond to its environment. Master splicing regulators acting in this context are likely to have targets that work together to form a coherent cellular response. Homeostatic RBPs may also be important on short timescales. As intron removal can function as a rate-limiting step in gene expression (Bhatt et al., 2012), an RBP’s direct targets could be rapidly spliced upon its activation. Such temporal properties may be critical in mediating the transient and homeostatic responses discussed below.
Nova1/2 neural splicing networks
One of the best-characterized tissue-restricted splicing networks is regulated by the Nova proteins, Nova-1 and Nova-2, exclusively in neurons. Nova-1 and Nova-2 exhibit largely non-overlapping patterns of expression in different regions of the brain (Yang et al., 1998). Nova-1 null mice are born phenotypically normal but die soon after birth due to apoptosis of neurons of the hindbrain and ventral spinal cord, implying a survival role within differentiated neurons (Jensen et al., 2000). As with many splicing regulators, Nova proteins recognize a short sequence motif (in this case, YCAY) to regulate either exon inclusion or skipping, depending on the location of binding with respect to the regulated exon (Ule et al., 2006). Integrating this RNA map with splicing changes observed in Nova knockout mouse brains identified ~700 Nova-dependent splicing targets (Zhang et al., 2010). Many of these targets are involved in regulating synapse activity and axonal guidance (Ule et al., 2005). A number of Nova targets have also been proposed to interact with one another, suggesting that they may comprise a coherent network mediating synaptic plasticity (Ule et al., 2005). In addition, the involvement of Nova-2 targets in long-term potentiation has been suggested to fine-tune the kinetics and magnitude of neuronal excitability (Huang et al., 2005). These studies implicate the Nova proteins in the maintenance of a critical splicing network essential for neuronal survival and for controlling synapse activity. The regulation of functionally-related genes by a single family of RBPs to reinforce and refine cell identity may be a hallmark of master splicing factors.
Transient activation of Rbfox splicing networks
Post-translational modification of RBPs appears to play a critical role in establishing short-lived or reversible splicing networks. Although at least one Rbfox protein is expressed in a large number of tissues, differential activity of each factor may allow the Rbfox network to be regulated by unique signals in each tissue. In the brain, Rbfox1 and Rbfox2 show non-redundant splicing activity despite being expressed in many of the same neuronal cell types (Gehman et al., 2012). This is consistent with different modes of regulation for each Rbfox protein. For example, Erk1/2 activation by growth factor stimulation results in rapid phosphorylation of Rbfox2 in many cell types (M. Jangi and P. Sharp, unpublished observations; Carlson et al., 2011). If such post-translational modification results in differential localization or association with the splicing machinery, it is possible that the Rbfox2 splicing network could be toggled by growth factor signaling. In contrast, Rbfox1 localization and consequent splicing activity are transiently altered in response to neuronal depolarization. This change is self-limiting, as the localization change is reversed by an autoregulatory splicing event that antagonizes the effect of depolarization (Lee et al., 2009). No analogous shift in localization has been demonstrated for Rbfox2. Thus, since each Rbfox protein may respond differently to extracellular, splicing-independent signals and may also have unique binding partners, they could potentially regulate non-overlapping, reversible splicing networks in distinct contexts. This dynamic may be common to splicing factors that exist in paralogous isoforms, such as MBNL, CELF, and NOVA proteins.
Splicing-mediated stress responses
Homeostatic splicing networks have been implicated in coordinating rapid responses to a variety of stresses. A number of groups have reported the existence of a subset of introns with delayed splicing that seem to play a key part in this response. These introns are removed post-transcriptionally, after all other introns in the transcript have been removed, prior to export into the cytoplasm. While the initial studies focused on one or two specific introns, more recent work suggests that intron “detention” is a regulatory motif within larger gene networks (Bhatt et al., 2012; Denis et al., 2005; Hao and Baltimore, 2013; Ninomiya et al., 2011). In this manner, a reserve of otherwise completely spliced and polyadenylated messages that still contain one or two introns serves as a buffer, allowing the coordinated splicing and export of a large number of genes with precise kinetics upon receipt of a signal. This mechanism likely serves as a splicing-regulated positive counterpoint to the negative feedback enabled by AS-NMD, as activating splicing in response to signals could increase the amount of gene product available. The emerging evidence discussed below indicates that detained introns are involved in rapidly correcting deviations from homeostasis caused by cellular stressors.
During the splicing cycle, SR proteins undergo sequential phosphorylation and dephosphorylation mediated by the SRPK and Clk kinases and protein phosphatases (Mermoud et al., 1994; Roscigno and Garcia-Blanco, 1995). Through a mechanism yet to be defined, hyperosmotic stress and heat shock inhibit the activity of Clk1/4, leading to SR protein dephosphorylation and rapid splicing of SR protein-bound detained introns. One of the transcripts undergoing stress-dependent maturation is that encoding Clk1 itself. Detained introns surrounding a frame-preserving cassette exon in the Clk1 transcript are rapidly removed, thus reestablishing homeostatic levels of Clk kinase activity (Ninomiya et al., 2011). Beyond this autoregulatory loop, direct inhibition of Clk1/4 leads to expression changes of several splicing factors, presumably due to increased splicing of AS-NMD events within RBPs crossregulated by SR proteins, and activation of the p53 stress response (P. Sharp, unpublished results). These observations suggest that a large post-transcriptional regulatory network may be responsible for mediating a rapid and self-limiting cascade of transcriptome changes in response to stress signals. Although not an RBP, Clk1 can be classified as the master splicing regulator in this context. Interestingly, a significant fraction of genes that are differentially spliced upon UV-induced DNA damage encode the NMD isoforms of RBPs (Ip et al., 2011). It is possible that intron detention is playing a similar role in AS during DNA damage as during heat shock or hyperosmotic stress. Determining whether splicing-dependent responses are a common theme within stress responses will consolidate the role of splicing in the maintenance of homeostasis.
Master splicing regulators in disease
Evidence for RBPs playing a critical part in determining cell state comes from pathology caused by changes in RBP activity. While several genetic diseases have been attributed to mutations in cis that disrupt splicing factor binding sites within critical disease genes, many arise from the altered function of trans-acting splicing factors. Such disorders tend to have pleiotropic effects due to the dysregulation of entire post-transcriptional networks, thus shedding light on the targets regulated by the affected splicing factors in normal physiology. Disorders resulting in RBP dysfunction can be broadly categorized into RBP loss-of-function or toxic RNA gain-of-function and can be inherited, arise from autoreactivity, or develop in the context of cancer (Cooper et al., 2009). For example, the genetic disorder spinal muscular atrophy is the most frequent genetic cause of infant mortality and occurs due to loss of the SMN1 gene, encoding a protein involved in snRNP assembly. Although SMN1 is ubiquitously expressed, pathology is confined mainly to motor neurons and muscle, indicative of a cell-type-specific sensitivity to an altered splicing regulatory network (Crawford and Pardo, 1996). Myotonic dystrophy type I is caused by an expanded CUG repeat in the first exon of the gene DMPK. This results in nuclear sequestration of MBNL proteins, which bind CUG repeat hairpins, and subsequent disruption of the MBNL splicing network across several tissues (Wang et al., 2012). In parallel, CUG repeat expansion leads to activation of PKC signaling and hyperphosphorylation and stabilization of CELF proteins, further contributing to splicing dysregulation (Kuyumcu-Martinez et al., 2007). Recently, the role of apparently constitutive splicing regulators in myelodysplastic syndromes and acute myelocytic leukemia has come to the forefront. Recurrent mutations in Sf3b1, U2af35, and other splicing factors in these syndromes have suggested that the regulation of a specific splicing network influences cell survival and proliferation in hematopoietic lineages (Yoshida et al., 2011). Such dysregulation or loss of specific splicing regulators in particular contexts argues for the establishment and regulation of broad biological networks by a single upstream splicing factor.
Identifying novel master splicing regulators
To expand upon the studies discussed here, how can we identify the master splicing regulators of a particular cell state? The input signals that activate splicing networks may provide key insights into which factors are contributing most strongly to the network. As proposed here, these inputs may frequently be post-translational or transcriptional. While transient RBP-dependent changes in cell state are crucial for normal physiology, it has proven to be difficult to identify RBP targets of signaling pathways (Heyd and Lynch, 2011). Here, we will focus on identifying the transcriptional inputs into splicing networks, in particular those within irreversible developmental programs.
The emerging wealth of data in transcriptional networks affords a tractable approach to identify master splicing regulators in diverse cell systems. It was recently shown that master transcription factors bind and regulate a subset of ~250 large transcriptional enhancer regions, termed superenhancers, within most cell types (Hnisz et al., 2013). The genes proximal to these superenhancers are enriched for key cell state determinants, including lineage-specific transcription factors (Hnisz et al., 2013; Whyte et al., 2013). Beyond transcription factors, however, many additional genes driven by superenhancers are likely to initiate and maintain critical processes during differentiation. The subset of tissue-specific splicing factors activated by superenhancers may fit these criteria. Specifically, we propose that superenhancer-activated splicing factors could directly and rapidly impact the cellular transcriptome as master splicing regulators. In the cases where RBPs are among the primary targets of master transcription factors, it is possible that the ectopic activation of such master splicing regulators is necessary and sufficient to substantially alter cell identity.
Transcriptional superenhancers at RBP genes
We predict that as with master transcription factors, 1–4 master splicing factors may be transcriptionally controlled in any particular cell type. Indeed, across a panel of 86 human tissues and cell lines, hierarchical clustering by the presence of a superenhancer near an RBP gene separates samples largely by tissue type, with ~4–5 RBP genes associated with a superenhancer in each cell type (M. Jangi and P. Sharp, unpublished observations; Hnisz et al., 2013). Conversely, individual RBPs may be activated by superenhancers across multiple tissues. For example, consistent with a suggested role in initiating and maintaining irreversible splicing networks during differentiation, the splicing factor MBNL1 is marked with a superenhancer across several differentiated cell types, including brain, adipose, and adult but not fetal gastrointestinal tract (M. Jangi and P. Sharp, unpublished observations; Hnisz et al., 2013).
In contrast to master splicing regulators, we posit that secondary splicing regulators would contain AS-NMD splicing events cross-regulated by master RBPs and would be less strongly transcriptionally modulated. Several previously-identified AS-NMD targets of Rbfox2, including Tia1, Tra2a, and Snrnp70, lack transcriptional superenhancers across all tissues examined but have varying expression across many of these cell types (Grosso et al., 2008; Hnisz et al., 2013; Jangi et al., 2014). These observations support the notion that an initiating transcriptional signal, indicated by a superenhancer, drives expression of a master splicing factor that further regulates a cascade of additional RBPs to establish sustained condition-specific splicing networks. Examples of potential master splicing factors activated by tissue-specific superenhancers are outlined in Table 1.
Table 1.
Superenhancer-driven candidate master RNA binding proteins (RBPs)
| RBPs | Tissue | Targets | References |
|---|---|---|---|
| RBM24 | Muscle myoblasts | Myogenin | (Jin et al., 2010; Miyamoto et al., 2009) |
| RBM38 | Skeletal muscle | p21 | (Jin et al., 2010; Li et al., 2010; Miyamoto et al., 2009) |
| RBM20 | adult cardiac muscle, Skeletal muscle | Titin | (Linke and Bucker, 2012) |
| RBFOX1 | Skeletal muscle | F1γ, α-actinin | (Jin et al., 2003) |
| RBFOX2 | Epithelial cells | Fgfr2, Enah, Ctnnd1, Fn1 | (Shapiro et al., 2011; Venables et al., 2009) |
| HNRNPA0 | Hematopoietic stem cells | Egr1/2, Gfi1 | (Young et al., 2014) |
| MBNL1 | Brain, adipose, adult gastrointestinal tract | FOXP1 | (Gabut et al., 2011) |
| ZNF638/NP220 | Brain | Unknown | n/a |
RBM24 and RBM38: Conserved drivers of myogenesis
Identification of cell type-specific transcriptional superenhancers proximal to RBPs may lead to the discovery of novel master splicing regulators. Skeletal muscle myoblasts, for example, possess three splicing factors that are marked with proximal superenhancers. One of these is CELF2, a well-studied splicing regulator that has been previously implicated in determining muscle-specific splicing patterns (Kalsotra et al., 2008). The remaining two are less well-characterized members of the RRM-containing family of RBPs – RBMS3 and RBM24 – and also show no evidence of containing AS-NMD splicing events. Little is known about the effect of RBMS3 on RNA metabolism, although its primarily cytoplasmic localization implies a role in regulating RNA stability as opposed to splicing (Jayasena and Bronner, 2012). Interestingly, both Rbm24 and its close paralog Rbm38, which is marked with a superenhancer in mature skeletal muscle, have been found to regulate myogenesis in part through post-transcriptional regulation of myogenin and p21 (Jin et al., 2010; Miyamoto et al., 2009) (Figure 4). Rbm24 is a target of the muscle-specific master transcription factor MyoD and may thus establish a post-transcriptional regulatory network early in muscle differentiation (Li et al., 2010). Indeed, overexpression of either Rbm24 or Rbm38 promotes murine myogenesis, while loss of Rbm24 inhibits myogenesis in both murine and Xenopus models (Jin et al., 2010; Li et al., 2010; Miyamoto et al., 2009). The evolutionary conservation of this pathway indicates that post-transcriptional regulation affords particular advantageous characteristics. In addition, in support of the combinatorial control and co-regulation that characterizes RBP networks, it has also been speculated that Rbm38 works cooperatively to activate Rbfox-dependent splicing specifically in muscle (Heinicke et al., 2013; Zhang et al., 2008). Rbm24 moreover interacts with Rbfox1 by yeast two-hybrid (Lim et al., 2006). These observations may explain the increased use of Rbfox-dependent alternative exons in muscle in spite of similar Rbfox expression patterns in muscle compared to several other tissues lacking Rbfox splicing signatures (Figure 4). Master RBPs such as Rbm24 and Rbm38 could thus have a significant contribution to, and may in some cases be sufficient for, the cell type-specific gene expression networks thought to be driven primarily by master transcription factors. Taken together, RBPs identified by proximity to transcriptional superenhancers are likely to function as master splicing regulators upstream of broad cell type-specific splicing networks.
Figure 4. Master splicing factors in myogenesis.
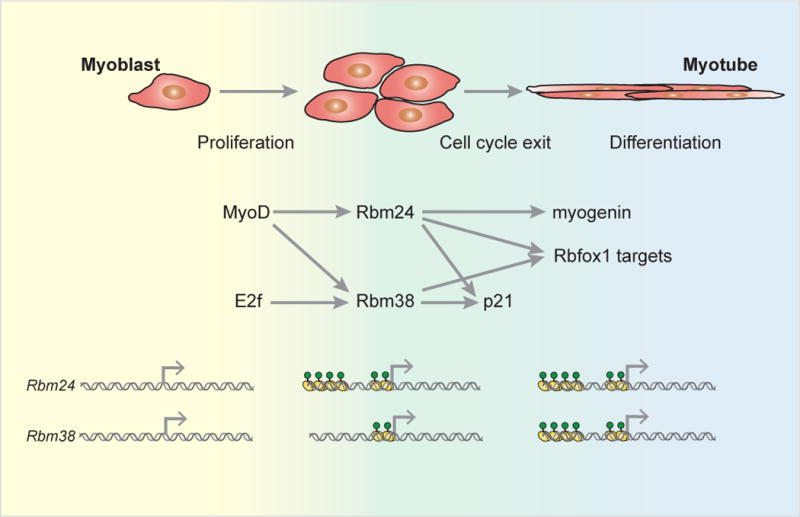
Rbm24 and Rbm28 are master RNA binding proteins in skeletal muscle differentiation and maintenance. The master transcription factor MyoD activates Rbm24 and Rbm38 in skeletal muscle myoblasts, concurrent with E2f-dependent activation of cell proliferation and Rbm38 activation. In a negative feedback loop that counters E2f activity, Rbm24 and Rbm38 initiate exit from the cell cycle by stabilization of p21. Rbm24 drives differentiation through the stabilization of the critical myogenesis factor myogenin. Both Rbm24 and Rbm38 control muscle-dependent splicing in differentiated myotubes through the co-regulation of Rbfox1 target splicing events (Jin et al., 2010; Li et al., 2010; Miyamoto et al., 2009; Zhang et al., 2008). Bottom panel: Rbm24 acquires an active transcriptional superenhancer in skeletal muscle myoblasts (yellow and green circles) and retains the mark in differentiated myotubes, in which the Rbm38 superenhancer also becomes active (Hnisz et al., 2013).
Future Perspectives
Over the past decade, high-throughput RNA sequencing technologies have advanced the study of alternative splicing from a single-regulator, single-target approach to a more global analysis of splicing networks. The difficulty now lies in synthesizing the complex information generated in these studies within the conceptual framework of a biological program. The general systems biology principles presented in this review will aid in the parsing of new data into homeostatic, developmental, or as-yet undefined regulatory networks.
Identifying the master and secondary splicing regulators driving context-specific splicing patterns is being furthered by bioinformatics approaches. Several groups have undertaken computational analyses that integrate RBP consensus motifs and other RNA features with transcriptomics and RNA-protein interaction maps to identify primary RBP contributors and tissue-specific splicing parameters (Barash et al., 2010; Lim et al., 2011; Zhang et al., 2010). These “splicing codes” have made apparent that different subsets of genomically-encoded cis elements are active in different tissues, suggesting that the balance of splicing factors expressed in a given cell type is a primary determinant of its splicing patterns. Moving forward, it will be critical to integrate the splicing cascades outlined here into these splicing codes to achieve a more comprehensive understanding of the regulatory layers that function in different physiological contexts. Such studies will also allow for a more quantitative definition of master splicing regulators, for example by requiring a factor to regulate, directly and indirectly, a minimum fraction of splicing events in a particular condition to qualify as a master regulator.
While a great deal of evidence reinforces the notion that modulation of a single RBP can initiate cell state transitions, we posit that a large supporting cast of splicing factors also plays crucial roles in establishing and maintaining cell identity. Understanding the co-regulation of direct and indirect splicing changes, as well as gene expression changes arising from transcriptional regulation, will be essential in determining the roles of both master and secondary splicing factors in the establishment of tissue transcriptomes. Splicing-independent roles of RBPs, such as the regulation of mRNA localization and stability, will also need to be addressed to define the contribution of various post-transcriptional regulatory mechanisms to cell state maintenance. Once identified within larger regulatory networks in specific cellular contexts, modulation of master splicing factor levels could provide a more precise alternative to transcription factor-based cellular reprogramming and manipulation. Future genetic, genomic, and biochemical analyses will elucidate the critical and nuanced role of the RBP repertoire in defining gene regulatory networks across mammalian development.
Acknowledgments
We thank all members of the Sharp lab, in particular Paul Boutz, Sara Dubbury, and Hiroshi Suzuki for helpful discussion and critical comments on the manuscript. We also thank Jeff Gore and Nicole Vega for insightful discussion. This work was supported by United States Public Health Service grants RO1-GM34277, R01-CA133404, U54-CA112967 to P.A.S. and partially by the Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute.
References
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, Florida: CRC Press; 2006. [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nature reviews Genetics. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome biology. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Baker BS. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development. 1995;121:3245–3258. doi: 10.1242/dev.121.10.3245. [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. The EMBO journal. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, M KA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & development. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Carlson SM, Chouinard CR, Labadorf A, Lam CJ, Schmelzle K, Fraenkel E, White FM. Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Science signaling. 2011;4:rs11. doi: 10.1126/scisignal.2002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes & development. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiology of disease. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. Rna. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic acids research. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M, Jr, Otis TS, Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes & development. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice J, Xia Z, Wang ET, Scavuzzo MA, Ward AJ, Kalsotra A, Wang W, Wehrens XH, Burge CB, Li W, et al. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nature communications. 2014;5:3603. doi: 10.1038/ncomms4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic acids research. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Baltimore D. RNA splicing regulates the temporal order of TNF-induced gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11934–11939. doi: 10.1073/pnas.1309990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinicke LA, Nabet B, Shen S, Jiang P, van Zalen S, Cieply B, Russell JE, Xing Y, Carstens RP. The RNA binding protein RBM38 (RNPC1) regulates splicing during late erythroid differentiation. PloS one. 2013;8:e78031. doi: 10.1371/journal.pone.0078031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Lynch KW, Hsiao EC, Liu EH, Maniatis T. Structural and functional conservation of the Drosophila doublesex splicing enhancer repeat elements. Rna. 1996;2:969–981. [PMC free article] [PubMed] [Google Scholar]
- Heyd F, Lynch KW. Degrade, move, regroup: signaling control of splicing proteins. Trends in biochemical sciences. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes N. Immunology. A splicing switch for T cells. Science. 2008;321:646–647. doi: 10.1126/science.1162294. [DOI] [PubMed] [Google Scholar]
- Huang CS, Shi SH, Ule J, Ruggiu M, Barker LA, Darnell RB, Jan YN, Jan LY. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123:105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell reports. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome research. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes & development. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena CS, Bronner ME. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-beta signaling. The Journal of cell biology. 2012;199:453–466. doi: 10.1083/jcb.201204138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jin D, Hidaka K, Shirai M, Morisaki T. RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes to cells : devoted to molecular & cellular mechanisms. 2010;15:1158–1167. doi: 10.1111/j.1365-2443.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. The EMBO journal. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nature reviews Genetics. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Molecular cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes & development. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Bourdelas A, Carron C, Shi DL. The RNA-binding protein Seb4/RBM24 is a direct target of MyoD and is required for myogenesis during Xenopus early development. Mechanisms of development. 2010;127:281–291. doi: 10.1016/j.mod.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes & development. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA, Bucker S. King of hearts: a splicing factor rules cardiac proteins. Nature medicine. 2012;18:660–661. doi: 10.1038/nm.2762. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. The Biochemical journal. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, et al. Alternative Splicing of MBD2 Supports Self-Renewal in Human Pluripotent Stem Cells. Cell stem cell. 2014 doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes & development. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- Maeda YT, Sano M. Regulatory dynamics of synthetic gene networks with positive feedback. Journal of molecular biology. 2006;359:1107–1124. doi: 10.1016/j.jmb.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Molecular cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Advances in experimental medicine and biology. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AI. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. The EMBO journal. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Hidaka K, Jin D, Morisaki T. RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes to cells : devoted to molecular & cellular mechanisms. 2009;14:1241–1252. doi: 10.1111/j.1365-2443.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- Nakaya T, Alexiou P, Maragkakis M, Chang A, Mourelatos Z. FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns. Rna. 2013;19:498–509. doi: 10.1261/rna.037804.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & development. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya K, Kataoka N, Hagiwara M. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. The Journal of cell biology. 2011;195:27–40. doi: 10.1083/jcb.201107093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, O’Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Molecular cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. Rna. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. Journal of molecular biology. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Molecular and cellular biology. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Molecular and cellular biology. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes & development. 2011;25:373–384. doi: 10.1101/gad.2004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK. Sex determination in insects: a binary decision based on alternative splicing. Current opinion in genetics & development. 2011;21:395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt C, Nothiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 2000;127:667–677. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS genetics. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siera SG, Cline TW. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene sex-lethal by its target transformer. Genetics. 2008;180:1963–1981. doi: 10.1534/genetics.108.093898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski BA, Belote JM, McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989;58:449–459. doi: 10.1016/0092-8674(89)90426-1. [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Molecular cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. Rna. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. Nova regulates brain-specific splicing to shape the synapse. Nature genetics. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. Cancer-associated regulation of alternative splicing. Nature structural & molecular biology. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, et al. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nature communications. 2013;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- Waddington CH. The Strategy of the Genes. London: George Allen and Unwin; 1957. [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome biology. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends in genetics : TIG. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Molecular cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Young DJ, Stoddart A, Nakitandwe J, Chen SC, Qian Z, Downing JR, Le Beau MM. Knockdown of Hnrnpa0, a del(5q) gene, alters myeloid cell fate in murine cells through regulation of AU-rich transcripts. Haematologica. 2014;99:1032–1040. doi: 10.3324/haematol.2013.098657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes & development. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yang Y, Scott MJ, Pannuti A, Fehr KC, Eisen A, Koonin EV, Fouts DL, Wrightsman R, Manning JE, et al. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. The EMBO journal. 1995;14:2884–2895. doi: 10.1002/j.1460-2075.1995.tb07288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


