Abstract
In stem cell biology, the dynamic addition and removal of 5-methylcytosines (5mCs) are necessary for lineage differentiation, nuclear reprogramming and embryonic development. Recent investigations have sought to understand the mechanisms of how 5mCs are added, and in particular how 5mCs are removed from DNA during embryogenesis.
In the last three years two compelling hypotheses (active and replication-dependent cytosine demethylation) have emerged to explain how 5mCs are dynamically removed from a mammalian genome. Active demethylation is a mechanism by which 5mCs are removed without the requirement for DNA replication. Replication-dependent demethylation occurs during semi-conservative DNA replication where 5mC on the parental strand is not copied onto the newly synthesized daughter strand during S-phase. As a result, at the completion of S-phase replication-dependent sensitive site/s are hemi-methylated on the double stranded DNA of sister chromatids. The cell-based models which have best shaped the two hypotheses have involved analysis of mouse gametes and newly formed mouse embryos including the one cell embryo called a zygote, and the blastomeres of cleavage stage embryos, particularly at the two and four cell stage. This is because in the zygtote, maternal and the paternal DNA are physically separate in their own pronuclei, and analysis of DNA replication can easily be incorporated into the experimental design by evaluating sister chromatids or using labeled nucleotides (Shen et al., 2014; Guo et al., 2014; Inoue et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011; Wossidlo et al., 2011). Identifying the mechanisms that promote active cytosine demethylation have specifically focused on the paternal pronucleus, which undergoes a remarkable global oxidation event converting 5mC to 5-hydroxymethylcytosine (5hmC) through the activity of Tet methylcytosine dioxigenase 3 (Tet3)(Gu et al., 2011; Wossidlo et al., 2011). In contrast, the maternal pronucleus does not acquire significant amounts of 5hmC. This has led to the general view that the paternal pronucleus is actively demethylated, whereas the maternal pronucleus is not. Although, biochemically 5hmC is still a methylated cytosine (albeit an oxidized one), and further processing is required to gain an unmethylated cytosine in its place.
Three recent papers, one in this issue (Shen et al., 2014), significantly change the way we think about the dynamic removal of 5mC around the time of fertilization (Guo et al., 2014; Shen et al., 2014; Wang et al., 2014). These new insights were made possible by the adaption of sequencing approaches to incredibly small sample sizes, and in some cases the use of different mouse strains as gamete donors. Combined, these papers now reveal the elegant yet complex dynamics by which 5mC is removed from the mouse genome during the first few days of life. Notably, there are differences in interpretation between the three papers with regard to the relative contribution of active verses replication-dependent demethylation, and solving this issue will require hypothesis driven exploration combined with genome-wide sequencing approaches. Critically, one of the major candidates for active removal of methylated cytosines from DNA, an enzyme called thymidine DNA glycosylase (TDG), has now been ruled out as having a role in zygotic DNA demethylation (Guo et al., 2014). Combined, this work should energize the field to sort out the relative contributions of active and replication-dependent genome-wide DNA demethylation, with or without 5hmC. Furthermore, this should trigger a new race towards finding novel strategies by which mammalian cells actively remove 5mC from the genome.
Prior to (Wang et al., 2014), a widely held view was that the maternal genome was not subject to active DNA demethylation. However Wang, Guo and now Shen and colleagues have overturned this assumption. Although, Wang and colleagues postulate that a significant fraction of the maternal genome is undergoing active demethylation before the 2-cell stage. Functionally, Guo and Shen independently prove that most of the methylated cytosines in the maternal and surprisingly the paternal pronucleus, as assayed by reduced representation bisulfite sequencing (RRBS), are removed in a replication-dependent manner in the zygote. Another surprise was the apparent lack of any role for TDG in actively replacing the oxidized products of 5mC with an unmodified base through base excision repair. TDG-dependent active demethylation represented an attractive potential mechanism to remove oxidized 5mC because further iterative oxidations of 5hmC to 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) create a modified base that is recognized and excised by TDG (He et al., 2011). Furthermore, deamination of 5hmC to 5-hydroxymethyluracil (5hmU) also creates a base that is recognized and excised by TDG (Cortellino et al., 2011). However, using an oocyte-specific TDG conditional knockout, there is now good reason to cast doubt on any major role of maternally transmitted TDG in the active removal of methylated cytosines from the zygotic genome (Guo et al., 2014).
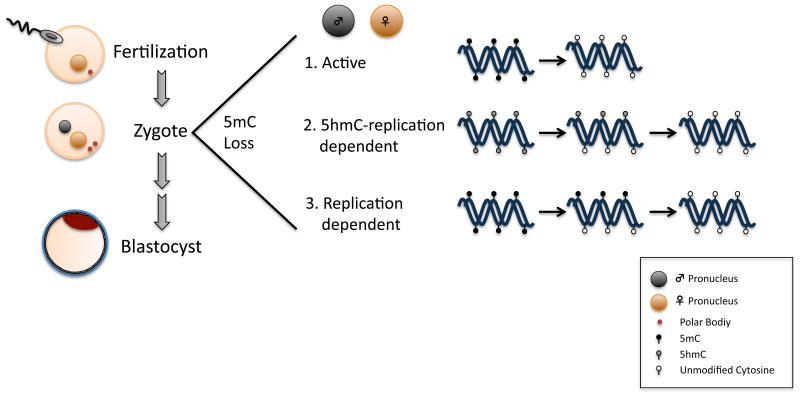
The replication-dependent hypothesis is perhaps less conceptually electrifying than enzymes en mass removing methylated cytosines from DNA, but it is incredibly effective. This mechanism can function downstream of Tet oxidation (5hmC-replication dependent), or in the absence of oxidation so long as the replication-coupled machinery is disabled or repressed (replication dependent). Failure to account for DNA replication or incorrect assumptions as to the number of times DNA has been replicated, will lead to erroneous over-estimations for the contribution of active DNA demethylation to removal of 5mC from the genome. In the work of Guo and colleagues RRBS was used to estimate that around 75% of demethylated loci in the paternal pronucleus and around 87% of demethylated loci in the female pronucleus do so by a replication-dependent mechanism with or without the activity of Tet3 (Guo et al., 2014). As further proof, hairpin bisulfite Sanger sequencing was used to independently support this claim. Similarly, Shen and colleagues in this issue have now reinforced the importance of replication- dependent demethylation to remove methylated cytosines from both the maternal and paternal genomes with Tet3's activity highly associated with replication. Taken together, these three papers elegantly demonstrate that both the maternal and paternal pronuclei use three different routes to remove 5mC from the genome (Figure 1). This work also serves as a reminder that conversion of 5mC to 5hmC is not implicit to active DNA demethylation.
Figure 1.
The male and female pronucleus each use three different mechanisms to remove 5mC from the genome (1-3). The specific pathway/s required for active removal of 5mC and 5hmC from the genome remain to be determined. DNA demethylation downstream of 5hmC occurs predominantly in the paternal pronucleus mostly by a 5hmC-replication dependent pathway. The female pronucleus mostly uses replication-dependent demethylation with a small contribution through 5hmC.
In conclusion, the field now has a tremendous opportunity to uncover the mechanisms downstream of Tet3 as well as Tet3-independent pathways that are responsible for actively removing methylated cytosines from zygotic DNA (Figure 1). It also raises important questions as to what protects and maintains some 5mCs from demethylation in the zygote and early embryo. Although these studies are changing the paradigm for how genomes become demethylated, it is important to take into account the genome being sampled. For example, RRBS represents a minor fraction of CpGs in the genome and is particularly suited for assaying genomic regions with higher CpG content such as CpG islands. Whole genome sequencing approaches with deep coverage are necessary to determine whether lessons learnt in RRBS can be applied to the tens of millions of CpGs that are not represented in an RRBS data set. One thing is certain, there will undoubtedly be more surprises in store as this critical area of investigation moves forward.
Acknowledgments
ATC is supported by funds from the NIH/NICHD 2 R01 HD058047
References
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo F, Li XL, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Bu TP, Hu B, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. This Issue. 2014 doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011 doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. Genome-scale base resolution analysis reveals distinctive roles of Tet3 and DNA replication in zygotic DNA demethylation 2014 [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Li G, Ci W, Li W, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature communications. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]