Abstract
Cytologically, the species of Passiflora with known chromosome number can be divided into four groups: (1) 2n = 12, 24, 36; (2) 2n = 24; (3) 2n = 18, 72; and (4) 2n = 20. The base chromosome number proposed for the genus is x = 6, with x = 9, x = 10 and x = 12 being considered secondary base numbers. In the present study, variability of 5S and 45S rDNA sites was investigated in 20 species of these four groups to check the reliability of this hypothesis. In the group with x = 6, five diploid species (2n = 12) exhibit two 5S rDNA sites and two (P. capsularis, P. morifolia and P. rubra) or four (P. misera 2x and P. tricuspis) 45S rDNA sites. The hexaploid cytotype of P. misera had 12 45S rDNA sites and six 5S rDNA. A tetraploid species, P. suberosa, had ten 45S rDNA sites and four 5S rDNA sites, both in the same chromosomes as the 45S rDNA sites. In the group with x = 9, P. actinia, P. amethystina, P. edmundoi, P. elegans, P. galbana, P. glandulosa and P. mucronata displayed six 45S rDNA sites, whereas P. alata, P. cincinnata, P. edulis f. flavicarpa, P. edulis var. roxo and P. laurifolia had four sites. In this group, all species were diploid (2n = 18) and had only two 5S rDNA sites. Passiflora foetida, the only species with 2n = 20, had six 45S rDNA sites and four 5S rDNA sites. The species with x = 12 (2n = 24), P. haematostigma and P. pentagona, showed four 45S rDNA sites and two 5S rDNA. In general, the number and location of 5S and 45S rDNA sites were consistent with the hypothesis of x = 6 as the probable ancestral genome for the genus, while the groups of species with x = 9, x = 10 and x = 12 were considered to be of tetraploid origin with descending dysploidy and gene silencing of some redundant gene sites, mainly those of 5S rDNA.
Key words: Passiflora, chromosome base number, FISH, rDNA, karyotype
INTRODUCTION
Passiflora L. is the largest genus of the family Passifloraceae, with approx. 465 species distributed mainly in tropical regions (Vanderplank, 1996; Cervi, 1997). Most of these species are herbaceous, although some representatives are shrubs or trees. Many species of Passiflora are cultivated as ornamentals or for their edible fruits and medicinal properties.
Cytologically, the species of Passiflora for which chromosome numbers are known can be divided into four karyological groups, represented by x = 6, x = 9, x = 10 and x = 12. Most species of Passiflora are diploid, with 2n = 12, 2n = 18 or 2n = 20, although some tetraploids (2n = 24), hexaploids (2n = 36) and octoploids (2n = 72) have been recorded (Snow and MacDougal, 1993; Melo et al., 2001). Different chromosome base numbers (x = 3, 6, 9) have been proposed for the genus, without a clear understanding of this variation and of the phylogenetic relationships among the species (Storey, 1950; Raven, 1975; Morawetz, 1986; Snow and MacDougal, 1993). Recently, Melo et al. (2001), revising the cytotaxonomy of the group, considered x = 6 as the most probable base number for the genus, whereas x = 9, x = 10 and x = 12 were considered secondary base numbers. However, the second‐most probable base number, x = 12, appears to have played an important role in the evolution of the group because it is better represented in other genera of the family. Therefore, with regards to the cytotaxonomy of this group, it is important to know whether x = 6 is derived from x = 12, or vice versa. The mechanisms of chromosome alteration most probably related to these changes are descending dysploidy (x = 12 → 6) or polyploidy (x = 6 → 12) (Guerra, 2000). In this case, any additional indication of polyploidy or dysploidy may be important for understanding the evolution of this group.
In the genus Aloe, Adams et al. (2000) found a correspondence between the number of 5S rDNA sites revealed by FISH (fluorescent in situ hybridization) and the ploidy level of several species. Diploid species displayed two 5S rDNA sites, tetraploids four sites and hexaploids six sites. A similar correspondence was found in Saccharum (D’Hont et al., 1998). However, more often, this relationship has been not observed, mainly because the number of rDNA sites can vary among diploid species. In diploid species of Paeonia, for instance, the number of chromosome pairs carrying 45S rDNA sites varied from three to five (Zhang and Sang, 1999). Exceptionally, the number of 5S and 45S rDNA sites can vary widely within a single species, as in Crocus vernus (Frello and Heslop‐Harrison, 2000). Nevertheless, the number of 5S or 45S rDNA sites is generally larger in tetraploids than in related diploids. In Rhynchospora, the number of 45S rDNA site pairs varied from two to four among diploids, and from four to 15 among polyploids (Vanzela et al., 1998). A similar variation in number of 5S and 45S rDNA sites has been described in diploid and tetraploid species of Hordeum (Taketa et al., 1999). Therefore, in general, polyploids have a higher average number of rDNA sites (mainly 45S rDNA) than diploids.
In the present study, the variability of 5S and 45S rDNA sites was investigated in 20 species of Passiflora, representing different base numbers, to help identify the base number of the genus and relationships among the haploid numbers.
MATERIALS AND METHODS
Material was collected in the field or was obtained from the germplasm bank of Embrapa Mandioca e Fruticultura Tropical—CNPMF (Table 1). Voucher specimens of all materials are deposited at the UFP Herbarium (Federal University of Pernambuco, Brazil), or at TSAH of Embrapa Semi‐Árido (Petrolina‐PE, Brazil).
Table 1.
Passiflora species analysed with respective chromosome numbers, number and position of 45S and 5S rDNA sites, herbarium vouchers and provenances
| Taxon | 2n | Number and position of 5S rDNA sites | Number and position of 45S rDNA sites | Voucher number | Provenance* |
| Subgenus Astrophea (DC.) Mast. | |||||
| Section Pseudoastrophea (Harms) Killip | |||||
| P. haematostigma Mart. ex Mast. | 24 | 2 (st) | 4 (st) | PAS‐1641 | Caeté, MG |
| P. pentagona Mast. | 24 | 2 (st) | 4 (st) | TSAH‐1736 | Barra da Estiva, BA |
| Subgenus Plectostemma Mast. | |||||
| Section Cieca (Medic.) Mast. | |||||
| P. morifolia Mast. | 12 | 2 (st) | 2 (st) | UFP‐31195 | Águas de Lindóia, SP |
| P. suberosa L. | 24 | 4 (st) | 10 (st; p) | TSAH‐1742 | Porto Alegre, RS |
| Section Decaloba (DC.) Mast. | |||||
| P. misera Kunth | 12 | 2 (st) | 4 (p) | TSAH‐1737 | Foz do Iguaçu, PR |
| 12 | 2 (st) | 4 (p) | TSAH‐1746 | Cachoeira do Sul, RS | |
| 36 | 6 (st) | 12 (st; p) | TSAH‐1741 | Recife, PE | |
| 36 | 6 (st) | 12 (st; p) | UFP‐31197 | Caruaru, PE | |
| P. tricuspis Mast. | 12 | 2 (st) | 4 (p) | UFP‐31201 | São José do Rio Preto, SP |
| Section Xerogona (Raf.) Killip | |||||
| P. capsularis L. | 12 | 2 (st) | 2 (st) | TSAH‐1745 | Águas de Lindóia, SP |
| P. rubra L. | 12 | 2 (st) | 2 (st) | TSAH‐1744 | Mata do Saltinho, Rio Formoso, PE |
| Subgenus Distephana (Juss.) Killip | |||||
| P. glandulosa Cav. | 18 | 2 (st) | 6 (st) | UFP‐31196/1 | Cultivated: Embrapa CNPMF, Cruz das Almas, BA |
| 18 | 1 (st) | 6 (st) | UFP‐31196/2 | Cultivated: Embrapa CNPMF, Cruz das Almas, BA | |
| Subgenus Passiflora | |||||
| Series Quadrangulares (Harms) Killip | |||||
| P. alata Curtis | 18 | 2 (st) | 4 (st) | – | Cultivated: Embrapa CNPMF, Cruz das Almas, BA |
| Series Laurifoliae Killip ex Cervi | |||||
| P. laurifolia L. | 18 | 2 (st) | 4 (st) | – | Cultivated: Embrapa CNPMF, Cruz das Almas, BA |
| Series Passiflora | |||||
| P. cincinnata Mast. | 18 | 2 (st) | 4 (st) | UFP‐17402 | Buíque, PE |
| P. edulis Sims. f. flavicarpa Deg. | 18 | 2 (st) | 4 (st) | – | Cultivated: Embrapa CNPMF, Cruz das Almas, BA |
| P. edulis Sims var. roxo | 18 | 2 (st) | 4 (st) | – | Cultivated: Embrapa CNPMF, Cruz das Almas, BA |
| Series Kermesinae Killip ex Cervi | |||||
| P. edmundoi Sacco | 18 | 2 (st) | 6 (st) | UFP‐31199 | Palmeiras, BA |
| Series Simplicifoliae (Harms) Killip | |||||
| P. actinia Hook. | 18 | 2 (st) | 6 (st) | PAS‐1592 | Porto Alegre, RS |
| P. galbana Mast. | 18 | 2 (st) | 6 (st) | UFP‐31200 | Mamanguape, PB |
| P. mucronata Lam. | 18 | 2 (st) | 6 (st) | TSAH‐1740 | Igarassu, PE |
| Series Lobatae (Harms) Killip | |||||
| P. amethystina Mikan var. amethystina | 18 | 2 (st) | 6 (st) | UFP‐31202 | Águas de Lindóia, SP |
| 18 | 2 (st) | 6 (st) | – | Domingos Martins, ES | |
| P. elegans Mast. | 18 | 2 (st) | 6 (st) | PAS‐1591 | Porto Alegre, RS |
| Subgenus Dysosmia (DC.) Killip | |||||
| P. foetida L. | 20 | 4 (st) | 6 (p) | TSAH‐1735 | Petrolina, PE |
Species are organized according to the infrageneric division proposed by Killip (1938).
St, Subterminal; p, proximal.
* BA, Bahia; ES, Espírito Santo; MG, Minas Gerais; PB, Paraíba; PE, Pernambuco; PR, Paraná; SP, São Paulo, RS, Rio Grande do Sul.
For cytological analysis, young root tips were pre‐treated with 0·002 m 8‐hydroxyquinoline at 8 °C for 24 h, and fixed overnight at room temperature in Carnoy 3 : 1 (ethanol : acetic acid). Floral buds of P. pentagona Mast. were collected in the field and fixed directly in Carnoy. Root tips were then stored at –20 °C. Root tips were digested in 2 % cellulase (Sigma, St Louis, Mo, USA) and 20 % pectinase (Sigma) solution for 2 h at 37 °C, and squashed in 45 % acetic acid. Coverslips were removed by freezing in liquid nitrogen and the slides were air‐dried. Slides were stained briefly with DAPI (4′,6‐diamidino‐2‐phenylindol)/glycerol and the best ones were re‐fixed in Carnoy 3 : 1 for 30 min, dehydrated in 100 % ethanol and stored at –20 °C until required for in situ hybridization.
To locate the 45S rDNA sites, probes SK18S and SK25S were used, containing 18S and 25S rDNA of Arabidopsis thaliana L. (Unfried et al., 1989; Unfried and Gruendler, 1990), kindly supplied by Professor Dieter Schweizer (University of Vienna, Austria). The 5S rDNA was obtained from total genomic DNA of P. edulis Sims by PCR using the primers 5′‐GTGCGATCATACCAGC(AG)(CT)TAATGCACCGG‐3′ and 5′‐GAGGTGCAACACGAGGACTTCCCAGGAGG‐3′. The in situ hybridization procedure followed that of Moscone et al. (1996). Probes SK18S and SK25S were labelled with biotin‐11‐dUTP and detected with TRITC (tetramethyl rhodamine isothiocyanate), while the 5S rDNA probe was labelled with digoxigenin‐11‐dUTP and detected with FITC (fluorescein isothiocyanate). Chromosomes were counterstained with DAPI and the slides mounted in Vectashield H‐1000.
Cells were photographed with a DMLB Leica epifluorescence microscope, using Kodak Ultra colour film ASA 400, or their images captured with a Cohu‐CCD video camera using Leica QFish software.
RESULTS
Karyologically, the species were distributed into four groups, in agreement with the base number: six species with x = 6 (2n = 12, 24, 36), 11 with x = 9 (2n = 18), one with x = 10 (2n = 20) and two with x = 12 (2n = 24). Chromosome morphology varied from metacentric to submetacentric. Size chromosome asymmetry was more evident in the group of species with x = 6, especially in P. suberosa, P. capsularis and P. morifolia. Most of the populations had not been analysed previously by Melo et al. (2001), but they exhibited the same number of chromosomes and chromosome morphology as previous samples. Only P. haematostigma, with 2n = 24 (Fig. 1M), was investigated for the first time.
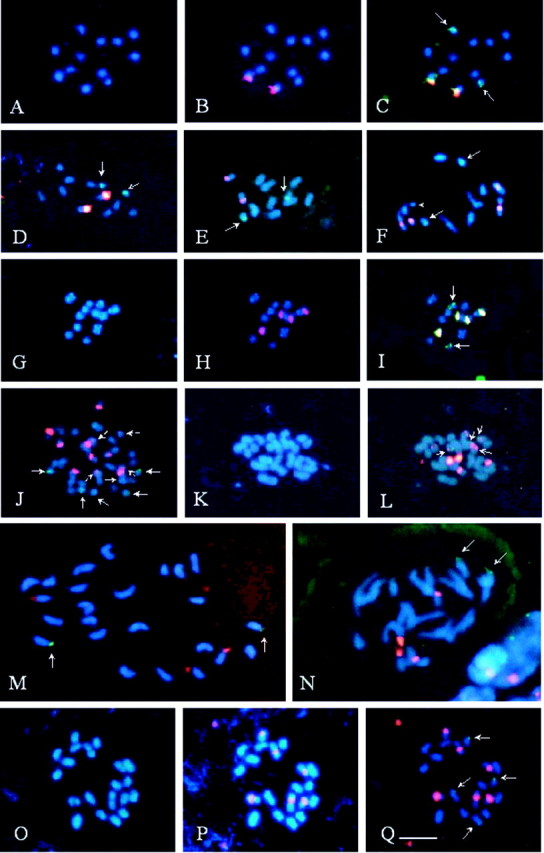
Fig. 1. Fluorescent in situ hybridization with 5S (green) and 45S (red or yellow) rDNA probes, in Passiflora species with x = 6, x = 10 or x = 12. A–C, P. capsularis. Note chromosomes stained with DAPI in A, 45S rDNA sites in B and a pair of 5S rDNA sites in C (arrows). D, P. rubra. Arrows indicate 5S rDNA. E, P. morifolia. Arrows indicate 5S rDNA. F, P. tricuspis. Note a longer pair of chromosomes with terminal regions characteristically less condensed, two pairs with proximal blocks of 45S rDNA, with one of the chromosomes being distended (arrowhead), and a pair with sub‐terminal 5S rDNA (arrows). G–I, P. misera 2x. Note the terminal DAPI+ bands in almost all chromosomes in G, 45S rDNA sites apparently sub‐terminal in one pair of chromosomes and proximal in the other one in H, and 5S rDNA sites in I (arrows). J, P. misera 6x. Broken arrows and arrowhead indicate small 45S rDNA sites. K and L, P. suberosa. Note that three of the five pairs of 45S rDNA sites are located in the smallest chromosomes, two of which also have 5S rDNA (arrows). M, P. haematostigma. Arrows indicate terminal 5S rDNA sites. N, P. pentagona. Arrows indicate 5S rDNA sites. O–Q, P. foetida. Note the secondary proximal constrictions in three chromosome pairs in O, three pairs of 45S rDNA proximal sites in P, and two pairs of 5S rDNA sub‐terminal sites in Q (arrows). All chromosomes were counterstained with DAPI (blue). Bar in Q represents 5 µm.
The positions of 5S and 45S rDNA sites are summarized in Table 1. Figures 1 and 2 illustrate the main results. Note that filter Leica I3 used to excite FITC (green) also excites TRITC (red), changing the colour of the signals captured with filter N2·1 for TRITC. Consequently, the 45S rDNA sites are shown twice and in different colours.
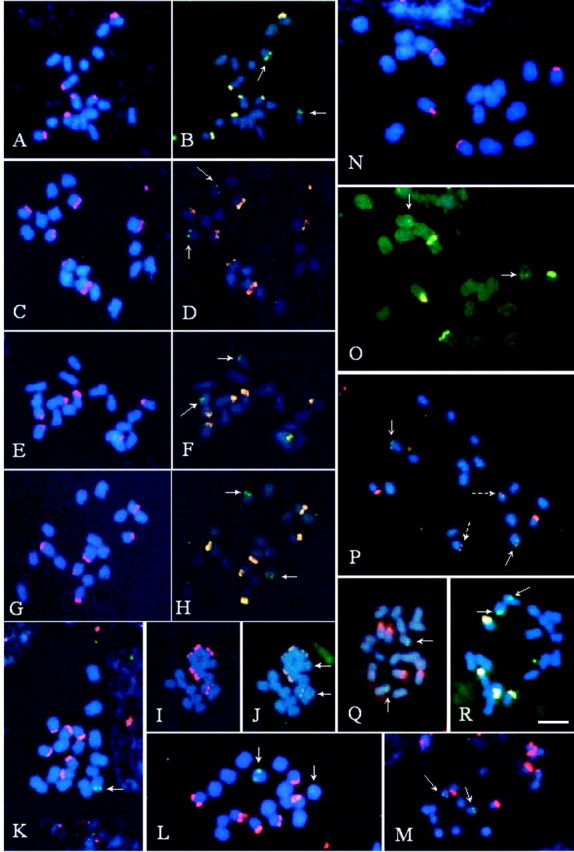
Fig. 2. Fluorescent in situ hybridization with 5S (green) and 45S (red or yellow) rDNA probes, in Passiflora species with x = 9. P. edmundoi (A and B); P. elegans (C and D); P. mucronata (E and F); P. galbana (G and H); P. actinia (I and J); P. glandulosa (K and L); P. amethystina (M); P. alata (N, O); P. edulis (P); P. laurifolia (Q); and P. cincinnata (R). Arrows indicate 5S rDNA sites while broken arrows indicate weak signals of 45S rDNA. All chromosomes were counterstained with DAPI (blue). Bar in R represents 5 µm.
The number of 5S rDNA sites was almost constant, usually a pair for each species, except in P. foetida (2n = 20) and in the tetraploid P. suberosa L., with four sites, and P. misera Kunth (6x), with six sites (Table 1). The position of these sites often seemed to be subterminal and, at least in the species with x = 6, it was always observed on chromosome VI (the smallest pair) or V. In several species, visualization of the 5S rDNA site was limited by its small size. For instance, in P. alata Curtis, the 5S rDNA sites were observed as minuscule points inside the chromosome mass (Fig. 2O). In a single individual of P. glandulosa Cav., the 5S rDNA site of one homologue was not detected, possibly owing to its very small size (Fig. 2K). The interphase nuclei of this sample also displayed only one fluorescent spot. In another individual of this same species, the two 5S rDNA sites showed size heteromorphism (Fig. 2L).
The small chromosome size and the tendency of fluorescence to expand beyond the region marked by the probe did not allow us to distinguish clearly between terminal and subterminal rDNA sites. Since, in some cases, the 45S rDNA sites were located within a subterminal secondary constriction, all sites are referred to as subterminal in Table 1, although some of these may be terminal. In general, 45S rDNA signals were larger, more numerous and more variable than 5S rDNA signals.
In the group of diploid species with x = 6, the 45S rDNA sites varied from one pair in P. capsularis, P. morifolia and P. rubra, to two pairs in P. misera 2x and P. tricuspis (Fig. 1A–I; Table 1). In polyploids, this number was higher. In P. misera 6x, the number and position of rDNA sites were the expected multiple of the diploid form, with 12 45S rDNA sites and six 5S sites. However, while all 45S rDNA sites in P. misera 2x were the same size (Fig. 1H), P. misera 6x showed six large and six small blocks of 45S rDNA (Fig. 1J), and six 5S rDNA sites distributed in three very small chromosome pairs. DAPI bands observed in the diploid cytotype were also observed in the hexaploid one (Fig. 1G).
The tetraploid P. suberosa displayed an asymmetrical karyotype, with eight to 12 larger chromosomes and 12 to 16 smaller ones. It had ten 45S and four 5S rDNA sites. Six 45S rDNA sites were located on the smallest chromosomes: two in the proximal region and four in the subterminal region. These latter four chromosomes also showed the 5S rDNA sites (Fig. 1K and L). The largest 45S rDNA site was located on the second largest chromosome pair. Passiflora tricuspis, although showing a karyotype very similar to that of P. misera 2x, displayed 45S rDNA in chromosome pairs II and III, and 5S rDNA in pair V (Fig. 1F), whereas in P. misera these sites were located in pairs II, IV or V (45S) and VI (5S). Passiflora capsularis, P. rubra and P. morifolia Mast. had only two 45S rDNA sites, located on the short arm of chromosome pair I in the first two species (Fig. 1B and D), and on the metacentric pair V in P. morifolia (Fig. 1E).
In the two species of the group with x = 12, P. haematostigma and P. pentagona, both with 2n = 24, four 45S rDNA sites and two 5S rDNA sites were observed, all terminally located (Fig. 1M and N). They had larger chromosomes than the species with x = 6.
Chromosomes of Passiflora foetida (x = 10; 2n = 20) were larger than those of species with x = 6 and smaller than those of species with x = 9. Six proximal 45S rDNA sites and four terminal or subterminal 5S rDNA sites were observed (Fig. 1P and Q).
In the group of x = 9, the number of 45S rDNA sites varied from two to three pairs (Table 1), all apparently subterminals (Fig. 2). Of the species with three pairs of sites, P. edmundoi, P. elegans and P. mucronata were similar in having 45S rDNA sites in chromosome pairs I and III, whereas the third pair was located on chromosome VII, IX or VIII, respectively (Fig. 2A, C and E). The 5S rDNA was located on the short arm of pair II in P. edmundoi (Fig. 2B), on the long arm of pair VI in P. elegans (Fig. 2D) and on the long arm of pair VII in P. mucronata (Fig. 2F).
In P. galbana two larger blocks of 45S rDNA were observed, located on pairs II and IV, and a smaller one on the pair V (Fig. 2G and H). Passiflora actinia had a symmetrical karyotype with 5S rDNA signals observed as very weak dots (Fig. 2I and J). In Passiflora glandulosa, 45S rDNA sites (Fig. 2K and L) were located on chromosomes V, VII and VIII, and 5S rDNA sites on one or two chromosomes of pair VI. In P. amethystina (Fig. 2M), the 45S rDNA sites were located on pairs V, VI and IX, whereas the 5S rDNA sites were observed in the sub‐terminal region of pair VII.
Passiflora alata had the largest chromosomes of the group with x = 9, with 45S rDNA located on the terminal region of pairs V and VIII, and 5S rDNA subterminally located on the short arm of pairs III or IV (Fig. 2N and O). In Passiflora edulis f. flavicarpa there was a large and a small 45S rDNA site, located on the long arm of pairs VII and IX, respectively, and 5S rDNA was located on the long arm of pair V (Fig. 2P). In P. edulis var. roxo, the number and position of the rDNA sites were the same as those in P. edulis f. flavicarpa although it was impossible to define the chromosome morphology. Passiflora cincinnata and P. laurifolia had 45S rDNA sites of similar size; these were located in pairs IV and VI in the former species, and in pairs V and VIII in the latter (Fig. 2Q and R).
DISCUSSION
In general, both the number and position of 45S rDNA sites detected by FISH coincided with the CMA+ bands and secondary constrictions observed in the karyotypes of Passiflora (Melo et al., 2001). However, some 45S rDNA regions not detected as secondary constrictions could be identified as CMA+ bands. In P. tricuspis, for example, there were two chromosome pairs with CMA+ blocks and 45S rDNA sites but only one secondary constriction (Melo et al., 2001). On the other hand, the number of 5S rDNA sites did not correlate with any other cytological parameter.
Among the species that are of agronomic importance (all concentrated in the group with x = 9), variation in the number and position of rDNA sites was very limited and did not constitute an important cytological marker to characterize species. Nevertheless, inter‐specific hybrids among representatives of the series Quandrangulares (Harms) Killip, Laurifolia Killip ex Cervi or Passiflora, and species of the series Kermesinae Killip ex Cervi, Simplicifoliae (Harms) Killip or Lobatae (Harms) Killip can be easily identified by the difference in the number of 45S rDNA sites.
Variation in the number of rDNA loci among species with different base numbers
The species with x = 6 in this study were restricted to the subgenus Plectostemma Mast. (Table 1), with representatives in three of the seven sections proposed by Killip (1938). In this subgenus, most species are diploid (n = 6), although the frequency of polyploidy is higher than that in the other subgenera of Passiflora (Snow and MacDougal, 1993; Melo et al., 2001). Variation in the location of 45S rDNA seems to correlate with the infrageneric division of Killip (1938). Species of section Decaloba (DC.) Mast. had two chromosome pairs with proximal 45S rDNA sites, whereas species of the sections Cieca (Medic.) and Xerogona (Raf.) Killip had only one subterminal site. In species of Xerogona (P. capsularis and P. rubra), the site was found in the largest chromosome pair, whereas in the only species belonging to Cieca studied (P. morifolia), it was located on one of the smallest chromosome pairs.
The only two species analysed that showed intraspecific polyploidy, both with x = 6, had more 45S rDNA sites than the related diploid species. The hexaploid form of P. misera had 12 sites, whereas the tetraploid P. suberosa had ten sites, six more than expected based on the only diploid of the section investigated. Regarding the 5S rDNA sites, in the group of species with x = 6, the number of sites was clearly related to the ploidy level: all diploids had two marked chromosomes, tetraploids had four and the hexaploids had six.
Passiflora suberosa was the only species that had 5S and 45S rDNA loci in the same chromosome, suggesting that the increase in number of 45S rDNA sites in this species occurred by transposition of 45S rDNA repeats from original chromosomes to others that did not possess these sites. Schubert and Wobus (1985) had previously related the capacity of the 45S rDNA repeats to be transposed to different regions of a chromosome complement. The eventual linking of 5S and 45S rDNA sites in the same chromosome has been observed as a derivative characteristic in Hypochaeris (Cerbah et al., 1998), Allium (Lee et al., 1999), Clivia (Ran et al., 2001) and some other genera.
The finding of 2n = 24 in P. haematostigma and P. pentagona reinforces the separation of these species of the subgenus Astrophea into a different karyological group with x = 12 (Melo et al., 2001). The only other species cytologically known in this subgenus, P. lindeniana Triana & Planch., also has n = 12 (Berry, 1987). Although P. haematostigma and P. pentagona have n = 12, like P. suberosa, they possess a more widely diploidized karyotype, with only one pair of 5S rDNA site and two pairs of 45S rDNA sites.
The 11 species analysed of the group with x = 9 are included in the subgenera Distephana (Juss.) Killip and Passiflora. They are diploids with one pair of 5S rDNA sites and two or three pairs of 45S rDNA sites. The karyotype with three pairs of 45S rDNA was found in representatives of subgenera Passiflora and Distephana, whereas that with two pairs was restricted to subgenus Passiflora. Variation in the number of 45S rDNA sites among related diploid species with the same chromosome number has been observed in several other genera (see, for example, Moscone et al., 1999; Široký et al., 2001).
The only species with n = 10, P. foetida, of the subgenus Dysosmia (DC.) Killip, had two pairs of 5S rDNA sites and three pairs of 45S rDNA, and was therefore more similar to tetraploid than to diploid karyotypes of the genus. Thus, P. foetida might be derived from a tetraploid karyotype with n = 12 (x1 = 6), changing to n = 10 by descending dysploidy. The same could have occurred to species with x = 9, which have, on average, more 45S rDNA sites than species with x = 6. Based on the alignment of ITS‐1 and ITS‐2 sequences from 45S rDNA (18S + 5·8S + 25S), Scherer et al. (2000) positioned P. foetida closer to species of the subgenus Passiflora than to those of the subgenus Plectostemma, which is compatible with the evolutionary route suggested by these data. Using molecular phylogenetic analysis of ITS and trnL‐trnF intergenic spacers, Muschner et al. (2003) also placed P. foetida (subgenus Dysosmia) in the same clade of the subgenus Passiflora.
Base chromosome number of the genus
The number of 45S rDNA sites in the present sample varied from one to three per haploid complement (except in the infraspecific polyploids), with a single pair only being found amongst species with x = 6. Given that diploids generally have fewer rDNA sites than the related polyploids, it is reasonable to suppose that the group of species with x = 6 is the originally diploid, single group whereas those with x = 9, 10 and 12 are of tetraploid origin, with descending dysploidy (12 → 10 → 9), and reduction of redundant sites, mainly the 5S rDNA ones. Polyploids have a tendency to reduce the number of duplicated sites owing to diploidization mechanisms and gene silencing (Leitch and Bennett, 1997). The selection pressure to reduce the rDNA site number to a single pair seems to be smaller in 45S rDNA owing to the existence of more efficient inter‐loci homogenization mechanisms for this sequence than for 5S rDNA (Cronn et al., 1996). Therefore, the probability of finding the duplicated 5S rDNA sites would be larger in infraspecific polyploids or in neopolyploids, than in paleopolyploids. Vogel et al. (1999) observed that silencing isoenzyme markers also is more evident in the paleopolyploids than in the neopolyploids.
The only two samples analysed here that showed site duplication proportional to the ploidy level (P. misera and P. suberosa) were also the only examples of infraspecific polyploidy. Multiplication of the 5S rDNA site number has also been found in other intraspecific polyploids, such as Aloe (Adams et al., 2000), or in neopolyploids, as in common wheat (Mukai et al., 1990). On the other hand, the occurrence of a single 5S rDNA site in P. haematostigma and P. pentagona seems to reflect an oldest origin of polyploidy in these species, as observed in typical paleopolyploids, such as Hevea (Leitch et al., 1998) and Manihot (Carvalho and Guerra, 2002).
Although these data corroborate the hypothesis of x1 = 6 as being the primary base number of Passiflora and Passifloraceae, the hypothesis that the base number is 12 still has in its favour the fact that this is the best represented haploid number among other genera and it is also the base number of Astrophea, a subgenus of shrubby and arboreal species, considered one of the most primitive of Passiflora. However, since polyploidy is a highly recurrent phenom enon in angiosperms, this widespread representation may be due to independent paleopolyploid lines. Guerra (2000) observed that in many taxa, the most primitive representatives are paleopolyploids that conserve ancient characteristics of the group. The polyploid origin of these representatives is suggested by the frequent occurrence of the chromosome number corresponding to the haploid (n = 6, in the case of Passiflora) in close taxa, which would hardly be justified by descending dysploidy (see, for example, Félix and Guerra, 2000).
ACKNOWLEDGEMENTS
The authors are grateful to Dr Armando Carlos Cervi (Federal University of Paraná) for species identification, to Fernando Campos (Zoo‐botanical Garden of Belo Horizonte and Petrobras) for several samples, and to the National Council of Scientific and Technological Development (CNPq) for financial support.
Supplementary Material
Received: 18 December 2002; ; Returned for revision: 12 March 2003. Accepted: 4 May 2003
References
- AdamsSP, Leitch IJ, Bennett MD, Chase MW, Leitch AR.2000. Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae). American Journal of Botany 87: 1578–1583. [PubMed] [Google Scholar]
- BealPR.1973. Cytology of the native Australian and several exotic Passiflora species. 3. Morphology of satellited chromosomes. Queensland Journal of Agricultural and Animal Sciences 30: 19–24. [Google Scholar]
- BennettMD.1995. The development and use of genomic in situ hybridization (GISH) as a new tool in plant cytogenetics. In: Brandham PE, Bennett MD, eds. Kew chromosome conference IV Kew: Royal Botanic Gardens, 167–183. [Google Scholar]
- BerryPE.1987. Chromosome number reports XCV. Taxon 36: 493. [Google Scholar]
- CarvalhoRde, Guerra M.2002. Cytogenetics of Manihot esculenta Crantz (cassava) and eight related species. Hereditas 136: 159–168. [DOI] [PubMed] [Google Scholar]
- CerbahM, Coulaud J, Siljak‐Yakovlev S.1998. rDNA organization and evolutionary relationships in the genus Hypochaeris (Asteraceae). Journal of Heredity 89:312–318. [Google Scholar]
- CerviAC.1997. Passifloraceae do Brasil. Estudo do gênero Passiflora L. subgênero Passiflora Fontqueria 45: 1–92. [Google Scholar]
- CronnRC, Zhao S, Paterson AH, Wendel JF.1996. Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. Journal of Molecular Evolution 42: 685–705. [DOI] [PubMed] [Google Scholar]
- D’HontA, Ison D, Alix K, Roux C, Glaszmann JC.1998. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41: 221–225. [Google Scholar]
- FélixLP, Guerra M.2000. Cytogenetics and cytotaxonomy of some Brazilian species of Cymbidiod orchids. Genetics and Molecular Biology 23: 957–978. [Google Scholar]
- FrelloS, Heslop‐Harrison JS.2000. Chromosomal variation in Crocus vernus Hill (Iridaceae) investigated by in situ hybridisation of rDNA and a tandemly repeated sequence. Annals of Botany 86: 317–322. [Google Scholar]
- GuerraM.2000. Chromosome number variation and evolution in monocots. In: Wilson KL, Morrison DA, eds. Monocots – systematics and evolution, vol. 1 Proceedings of the Second International Conference on the Comparative Biology of the Monocots. Melbourne: CSIRO, 125–134. [Google Scholar]
- KillipEP.1938. The American species of Passifloraceae. Publications of the Field Museum of Natural History, Botanical Series 19: 1–613. [Google Scholar]
- LeeSH, Do GS, Seo BB.1999. Chromosomal localization of 5S rRNA gene loci and the implications for relationships within the Allium complex. Chromosome Research 7: 89–93. [DOI] [PubMed] [Google Scholar]
- LeitchAR, Lim KY, Leitch IJ, O’Neill M, Chye M, Low F.1998. Molecular cytogenetic studies in rubber, Hevea brasiliensis Muell. Arg. (Euphorbiaceae). Genome 41: 464–467. [Google Scholar]
- LeitchIJ, Bennett MD.1997. Polyploidy in angiosperms. Trends in Plant Science 2: 470–476. [Google Scholar]
- MeloNFde, Cervi AC, Guerra M.2001. Karyology and cytotaxonomy of the genus Passiflora L. (Passifloraceae). Plant Systematics and Evolution 226: 69–84. [Google Scholar]
- MorawetzW.1986. Remarks on karyological differentiation patterns in tropical woody plants. Plant Systematics and Evolution 152: 49–100. [Google Scholar]
- MosconeEA, Matzke MA, Matzke AJM.1996. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105: 231–236. [PubMed] [Google Scholar]
- MosconeEA, Klein F, Lambrou M, Fuchs J, Schweizer, D.1999. Quantitative karyotyping and dual‐color FISH mapping of 5S and 18S‐25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42: 1224–1233. [DOI] [PubMed] [Google Scholar]
- MukaiY, Endo TR, Gill BS.1990. Physical mapping of the 5S rRNA multigene family in common wheat. Journal of Heredity 81: 290–295. [Google Scholar]
- MuschnerVC, Lorenz AP, Cervi AC, Bonatto SL, Souza‐Chies TT, Salzano FM, Freitas LB.2003. A first molecular phylogenetic analysis in Passiflora (Passifloraceae). American Journal of Botany (in press). [DOI] [PubMed] [Google Scholar]
- RanY, Hammett KRW, Murray BG.2001. Phylogenetic analysis and karyotype evolution in the genus Clivia (Amaryllidaceae). Annals of Botany 87: 823–830. [Google Scholar]
- RavenPH.1975. The bases of angiosperm phylogeny: cytology. Annals of the Missouri Botanical Garden 62: 724–764. [Google Scholar]
- SchererNM, Muschner VC, Finkler C, Souza‐Chies TT, Salzano FM, Freitas LB.2000. Aplicação das seqüências dos espaçadores internos transcritos do DNA ribossomal nuclear para estudos filogenéticos com o gênero Passiflora (Passifloraceae). Genetics and Molecular Biology 23: 430. [Google Scholar]
- SchubertI, Wobus U.1985. In situ hybridisation confirms jumping nucleolus organizing regions in Allium Chromosoma 92: 143–148. [Google Scholar]
- ŠirokýJ, Lysák MA, Doležel J, Kejnovský E, Vyskot B.2001. Heterogeneity of rDNA distribution and genome size in Silene spp. Chromosome Research 9: 387–393. [DOI] [PubMed] [Google Scholar]
- SnowN, MacDougal JM.1993. New chromosome reports in Passiflora (Passifloraceae). Systematic Botany 18: 261–273. [Google Scholar]
- StoreyWB.1950. Chromosome numbers of some species of Passiflora occurring in Hawaii. Pacific Science 4: 37–42. [Google Scholar]
- TaketaS, Harrison GE, Heslop‐Harrison JS.1999. Comparative physical mapping of the 5S and 18S‐25S rDNA in nine wild Hordeum species and cytotypes. Theoretical and Applied Genetics 98: 1–9. [Google Scholar]
- UnfriedI, Gruendler P.1990. Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana Nucleic Acids Research 18: 4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UnfriedI, Stocker U, Gruendler P.1989. Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Co10. Nucleic Acids Research 17: 7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderplankJ.1996.Passion flowers. 2nd edn. Cambridge: The MIT. [Google Scholar]
- VanzelaALL, Cuadrado A, Jouve N, Luceño M, Guerra M.1998. Multiple locations of the rDNA sites in holocentric chromosomes of Rhynchospora (Cyperaceae). Chromosome Research 6: 345–349. [DOI] [PubMed] [Google Scholar]
- VogelJC, Barrett JA, Rumsey FJ, Gibby M.1999. Identifying multiple origins in polyploid homosporous pteridophytes. In: Hollingsworth PM, Bateman RM, Gornall RJ, eds. Molecular systematics and plant evolution London: Taylor & Francis, 101–117. [Google Scholar]
- ZhangD, Sang T 1999. Physical mapping of ribosomal RNA genes in peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: implications for phylogeny and concerted evolution. American Journal of Botany 86: 735–740. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


