Abstract
Estimates of genome size using flow cytometry can be biased by the presence of cytosolic compounds, leading to pseudo‐intraspecific variation in genome size. Two important compounds present in coffee trees—caffeine and chlorogenic acid—modify accessibility of the dye propidium iodide to Petunia DNA, a species used as internal standard in our genome size evaluation. These compounds could be responsible for intraspecific variation in genome size since their contents vary between trees. They could also be implicated in environmental variations in genome size, such as those revealed when comparing the results of evaluations carried out on different dates on several genotypes.
Key words: Coffea, nuclear DNA content, flow cytometry, dye accessibility, caffeine, chlorogenic acids
INTRODUCTION
Cytosolic compounds can bias genome size estimates obtained by flow cytometry (Noirot et al., 2000, 2002; Price et al., 2000) and therefore it is important to identify them. Caffeine and chlorogenic acids (CGA), two well‐known compounds in Coffea, could be involved for two reasons: (1) caffeine is an intercalating drug (Tornaletti et al., 1989); and (2) CGA are precursors of polyphenols, which were found to account for a stoichiometric error in genome size evaluation using Feulgen microdensitometry (Greilhuber, 1988). It is necessary to know the main sources of caffeine and CGA content variation in order to predict stoichiometric errors induced by these compounds.
In green coffee beans, three CGA classes, caffeoylquinic acids (CQA), dicaffeoylquinic acids (diCQA) and feruloylquinic acids (FQA), account for approx. 98 % of the CGA content (Clifford and Staniforth, 1977). Note that chlorogenic acids sensus stricto (CGAs.s.) include CQA and diCQA only. Each class includes three isomers, but one isomer, 5‐caffeoylquinic acid (5‐CQA), constitutes 85 % of the CGA content in coffee beans (Clifford et al., 1989; Ky et al., 1999). CGA content varies between and within coffee species. In C. canephora, CGA content is 11·3 % dmb (dry matter basis), ranging from 7·9 to 14·4 % dmb (Ky et al., 2001). Between‐year variations explain 20 (CQA) to 50 % (diCQA) of the content variance (Ky et al., 1999).
Caffeine content in green beans (CAF) varies among African coffee species, ranging from 0 % in C. pseudo‐zanguebariae (PSE) to 2·5 % dmb in C. canephora (Charrier and Berthaud, 1975; Anthony et al., 1993; Mazzafera et al., 1997; Ky et al., 2001). High within‐species variations are noted, e.g. in C. canephora, where CAF varies from 1·5 to 3·3 % dmb between accessions (Ky et al., 2001). Most of this variation is under genetic control (94 % of the total variation), and these effects are mainly additive (Charrier and Berthaud, 1975; Barre et al., 1998; Montagnon et al., 1998). CAF probably results from an accumulation process in green beans, and varies according to the fructification time (FT)—from flowering to ripening—and the daily amount of accumulated caffeine (Ky, 2000). Caffeine binds with 5‐CQA to give rise to caffeine chlorogenate (Payen, 1846, 1851; Baumann et al., 1993) and is accumulated as this complex (Payen, 1851; Rabéchault, 1954).
Four trends could thus be expected, based on the assumed involvement of caffeine and CGA in stoichiometric error: (1) genome size should vary between genotypes within species; (2) genome size should vary over time (this could be verified by evaluating the genome size of a genotype at different dates); (3) genome size should change when using an internal and external standard (in the first case, leaves of the standard and target are chopped simultaneously, releasing cytosolic compounds of both species in the buffer; while in the second case, leaves are separately chopped and cytosolic compounds of the target cannot act on standard nuclei and vice versa); and (4) as a genome size difference (ΔDNA) is expected using these two modes of evaluation, this ΔDNA should change between genotypes within species. It could also be correlated with some biochemical characteristics of these genotypes.
The main objectives of the present study were: (1) to test the effects of caffeine and 5‐CQA on propidium iodide (PI) accessibility to DNA in a standard (Petunia hybrida nuclei); (2) to analyse genome size differences using internal and external standardization (ΔDNA) in interspecific hybrids with a broad range of caffeine and CGA contents and genome size; (3) to correlate this ΔDNA with some biochemical characteristics of these hybrids; and (4) to quantify genotypic and between‐day effects.
MATERIALS AND METHODS
Plant material
The plant material consisted of 62 backcross hybrids between C. pseudozanguebariae (PSE) and C. liberica var. dewevrei (DEW): (PSE × DEW) × DEW, and called BCDEW. Beans of PSE ripen in 10 weeks, are caffeine‐free and their CGA content is about 1·5 % dmb (Barre et al., 1998; Ky et al., 1999). The PSE genome size, evaluated using PI and Petunia as internal standard, is 2C = 1·14 pg (Barre et al., 1996). Beans of DEW ripen in 10 months, contain about 0·9 % dmb of caffeine and 8 % dmb of CGA on average (Barre et al., 1998; Ky et al., 1999). DEW genome size, evaluated as for PSE, is 2C = 1·43 pg (Barre et al., 1996).
Hybrids and Petunia hybrida were grown in a glasshouse in Montpellier (France) under tropical climatic conditions (24 °C during the day, 18 °C at night, 70 % relative humidity). The sample size (number of hybrids) varied according to the experiment (see below).
Flow cytometric measurements
Genome size evaluations in backcross hybrids were carried out according to Barre et al. (1996). The main characteristics of the method were: (1) use of a slightly modified version (0·5 % Triton X‐100 and pH = 9·2) of Dolezel’s buffer (Dolezel et al., 1989); (2) nucleus extraction by leaf chopping (Galbraith et al., 1983); (3) use of Petunia as standard (2C = 2·85 pg; Marie and Brown, 1993); (4) nucleus staining with a PI solution at 333 µg ml–1 [95–98 % (TLC)]; (5) use of a FACScan cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) with an argon laser (15 mW) at 488 nm with a pulse area of emissions >590 nm; and (6) use of an amplifier system at constant voltage (557 V) and constant gain. Further details on the method are given in Barre et al. (1996).
Experimental design and statistical analysis
Effect of caffeine.
The aim of this experiment was to test the impact of caffeine (Sigma no. C‐0750; St Quentin, France) on the fluorescence of Petunia nuclei. A caffeine treatment (CAF2: 0·5 %) was thus compared with a caffeine‐free control (CAF1: 0 %). Each treatment (CAF1 and CAF2) was replicated five‐fold.
In practice, a filtrate of Petunia nuclei (4 ml) was obtained and divided into two subfiltrates (2 ml each). The CAF2 treatment consisted of adding 0·5 ml of lysis buffer containing 25 mg ml–1 of caffeine into the subfiltrate, leading to a final caffeine concentration of 5 mg ml–1 (0·5 %). For the CAF1 treatment, the subfiltrate was supplemented with 0·5 ml of lysis buffer without caffeine. Thirty minutes later, three 0·5‐ml replicates were taken from each subfiltrate and stained with PI. A Fisher’s block design (P = 2 blocks; n = 3 replicates per block for each treatment) was applied. The measurement order for the FACScan was randomized within blocks. A two‐way ANOVA with fixed effects (blocks, treatment and interaction) was carried out for the statistical analysis.
Effect of chlorogenic acids.
The aim was to determine the effects of 5‐CQA (Sigma C‐3878) on fluorescence of Petunia nuclei. A 5‐CQA treatment at 0·5 % (CGA2) was thus compared with a 5‐CQA‐free control (CGA1). All steps, including experimental design and statistical analysis, were similar to the caffeine experiment.
Genome‐size differences between evaluations using internal and external standards.
The aim of the experiment was to analyse the relationship between genome size evaluations obtained using internal standardization and external standardization. Thirty‐five BCDEW hybrids were evaluated using Petunia as internal and external standard. In the external standardization conditions, four nuclei extractions were obtained per hybrid for both target and standard, with each coffee–Petunia pair constituting a replicate. In the internal standardization conditions, four coffee–Petunia nuclei extractions were carried out per hybrid. The measurement order for the eight replicates was fully randomized. Each set of eight replicates was preceded by three controls: a DEW control, a PSE control and a fluorescent bead control. The DEW (or PSE) control was obtained by pooling several DEW (or PSE) extracts to obtain a 50 ml solution. This allowed us to verify the stability of nuclei fluorescence over a time course in the two species.
Differences in genome size evaluations (ΔDNA) were also computed and analysed in terms of range and mean.
Relationships between ΔDNA and some biochemical characteristics of hybrids.
The aim of this analysis was to correlate ΔDNA with some biochemical characteristics of the hybrids. The biochemical evaluation methods are described in Barre et al. (1998) and Ky et al. (1999). The CAF : CGAs.s. ratio was computed since CAF is known to form a complex with CQA and, possibly, also with diCQA. The relationship between CAF/CGAs.s. and ΔDNA from 33 hybrids was analysed using a quadratic regression. The amount of caffeine accumulated daily was also estimated using the CAF : FT ratio, where FT represents fructification time. The relationship between CAF : FT and ΔDNA from 25 hybrids was determined using linear regression.
Between date differences.
The aim of the experiment was to detect differences between genome size evaluations carried out at different dates. Eight hybrids were analysed at dates D1 and D18 (17 d after D1) and five hybrids at dates D1 and D19. At each date, three extracts were prepared per hybrid (replicates). Two comparisons were made, D1 vs. D18 and D1 vs. D19, using a two‐way ANOVA with fixed effects (hybrids, dates and interaction).
RESULTS
Effect of caffeine and chlorogenic acids
Significant differences were observed between CAF1 and CAF2 (F1,8 = 37·3, P = 0·0002), whereas no block effects (F1,8 = 0·14, P = 0·72) or block × treatment interactions (F1,8 = 0·95, P = 0·36) were recorded. Fluorescence peak values (in channel units) were 621 without caffeine and 664 with caffeine, showing that caffeine increased propidium iodide accessibility to Petunia DNA. The gain was about 7 %.
The presence of 5‐CQA at 0·5 % in buffer significantly decreased Petunia nuclei fluorescence (F1,8 = 31·8, P = 0·0005) from 757·3 to 715·7, i.e. a drop of 5 %. As for caffeine, no difference was recorded between blocks and no block × treatment interactions were noted.
Genome size differences between evaluations using internal and external standards
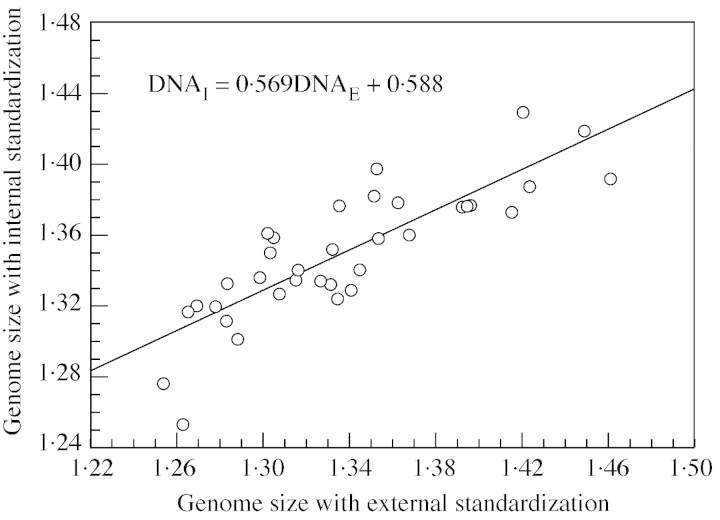
Differences in genome size (ΔDNA) were observed when using internal and external standards, and ranged from –0·059 pg to +0·068 pg (mean –0·012 pg). There was a linear relationship (r = 0·839) between the two modes of evaluation (Fig. 1). The slope of the straight line and intercept were 0·569 and 0·588, respectively, whereas 1 and 0 were expected, respectively.
Fig. 1. Relationship between genome size evaluations using internal and external standardization. Petunia constituted the standard. Each point represents a backcross hybrid (target) between two species differing in their genome size. Segregation of the parental genome size in the second generation explains the range from F1 hybrids to the recurrent parent C. liberica var. dewevrei.
Relationships between ΔDNA and some biochemical characteristics of hybrids
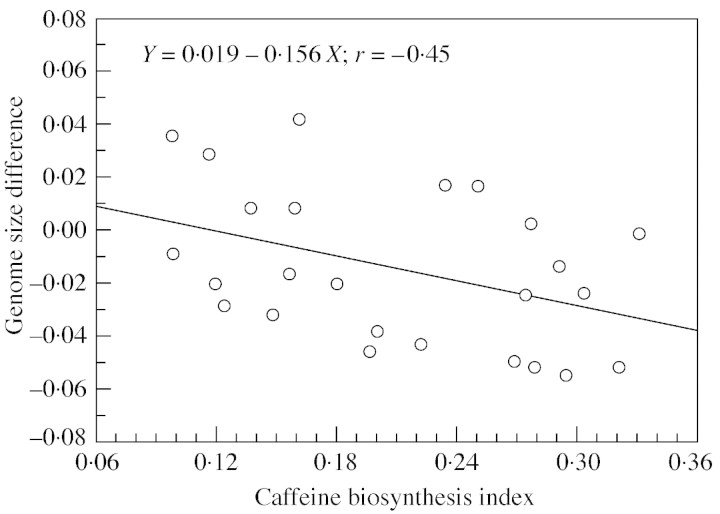
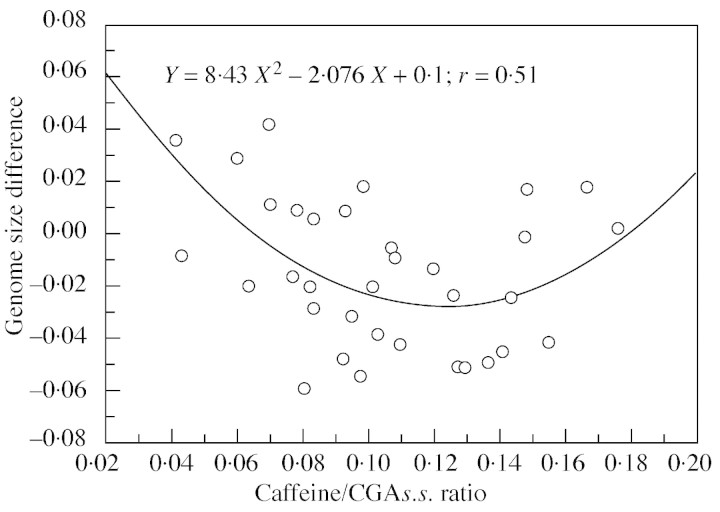
ΔDNA was negatively and linearly related to CAF : FT (Fig. 2): Y = 0·019 – 0·156X. As ΔDNA is estimated to be 0·019 when CAF : FT = 0, the genome size using an external standard would thus be slightly higher in caffeine‐free hybrids. In these hybrids, ΔDNA represents about 1·5 % of the genome size. Conversely, in hybrids with a CAF : FT value of 0·36, characteristic of DEW (see Ky et al., 1999), the ΔDNA would be higher (0·137 pg), representing about 11 % of the genome size. In the latter case, the internal standard would give higher values. ΔDNA was curvilinearly related to CAF : CGAs.s. (Fig. 3): Y = 8·43X2 – 2·08X + 0·0998, indicating that ΔDNA is minimal when CAF : CGAs.s. = 0·123.
Fig. 2. Differences between two genome size evaluations (external vs. internal) in backcross hybrids is linearly related to their CAF : FT ratio (caffeine content per 100 d of fructification time) in mature beans.
Fig. 3. Differences between two genome size evaluations (external vs. internal) in backcross hybrids are curvilinearly related to CAF : CGAs.s. (caffeine content : chlorogenic acids sensus stricto) in mature beans.
Between‐date differences in genome size evaluations
The difference in genome size was 0·011 pg between dates D1 and D18, which was highly significant (F1,32 = 8·63, P = 0·006). The absence of ‘hybrid × date’ interactions (F7,32 = 0·81, P = 0·58) indicated that the between‐date difference was similar for all hybrids. Differences represented about 1·1 % of the PSE genome size.
Five other BC1 hybrids were studied at D1 and D19. A date effect (F1,20 = 18·6, P = 0·0003) with no ‘hybrid × date’ interaction (F4,20 = 0·80, P = 0·54) was noted again. Dates differed by 0·021 pg, i.e. 1·9 % of the PSE genome size.
DISCUSSION
Four trends were highlighted in this study: (1) caffeine and CGA were found to influence Petunia nuclei fluorescence in the presence of propidium iodide; (2) genome size evaluations differed between internal and external standardizations. In addition, the slope and intercept of the regression line between these two evaluations differed from 1 and 0, respectively, indicating a bias. (3) The difference in genome size (ΔDNA) was correlated with some biochemical characteristics of coffee beans; and (4) genome size varied between evaluation dates.
Effects of phenolics on PI accessibility
The addition of 5‐CQA in medium containing PI‐stained Petunia nuclei decreased nuclei fluorescence, i.e. limiting PI accessibility to DNA. As CGA are phenolics and share common chemical properties with tannins, the effect of 5‐CQA could be compared with the stoichiometric error observed by Greilhuber (1988) using Feulgen microdensitometry. This author reported an increase in genome size in woody species (but not in herbaceous species) when using formaldehyde instead of methanol‐acetic acid as fixative. In these species, the genome size ratio between formaldehyde and methanol–acetic acid evaluations ranged from 0·245 to 0·778, showing the importance of the effect. In fact, use of formaldehyde avoids the release of tannin from vacuoles. According to Greilhuber (1988), tannins act on chromatin condensation, thus limiting access of PI to DNA. They could also act on nuclear membrane proteins, thus hindering stain uptake by the nucleus.
Although the effect was emphasized with the 5‐CQA isomer, other affine compounds, including all other CGA isomers, as well as other phenolics with tannic proprieties, should limit dye accessibility. This could explain why flow cytometry is effective for distinguishing brownish coffee tree calli, which are theoretically polyphenol‐rich, from white calli (unpubl. res.).
Effects of caffeine and affine compounds on PI accessibility
The caffeine effect on PI accessibility was tested because caffeine and other oxypurines are known to bind with DNA, modifying DNA‐supercoiling (Tornaletti et al., 1989), and to form a complex with intercalating dye (Traganos et al., 1991). Competition between PI and caffeine is therefore expected, leading to a drop in PI accessibility to DNA. However, the opposite results were obtained, i.e. caffeine increased PI accessibility to DNA in Petunia. How can this paradox be explained?
In fact, caffeine forms complexes with phenolics such as CGA (Payen, 1851; Rabéchault, 1954; Baumann et al., 1993). When added to the buffer, it could trap CGA and other phenolics present in Petunia cell vacuoles and released into the medium by chopping. In this way, caffeine would reduce the negative effects of phenolics on PI accessibility. Rabéchault (1954) noted, ‘A common process to highlight tannins in coffee tree tissues consists of precipitating them using an alkaloid, and in this case we mainly used caffeine’. This means that other oxypurines and alkaloids could also interfere with the phenolic effect.
Comparison between evaluations using internal and external standardization
Genome size evaluations differed when comparing external and internal standardizations, but there were highly significant correlations (r = 0·84) between methods. A statistical interpretation involved considering that regression lines reflected the genome size, with residuals (distance to the straight line) being due to the between‐method effect. From this viewpoint, residual variations would result from differences in phenolic and caffeine contents between hybrids.
The relative importance of residuals with regard to the genome size (1 – r2 = 29 %) depends on the genome size range recorded within the experiment. Correlations will be better with a higher range, while they should vanish with a lower genome size range. In other words, regression line slopes and intersects depend on this range. To avoid such a range‐dependent effect, the use of a least rectangle estimator instead of a least square one should lead to invariant slopes and intersects. The new estimations were then b = 0·68 and a = 0·43 (instead of b = 0·57 and a = 0·59), confirming the presence of stoichiometric error on both nuclei fluorescence and genome size.
Obviously, internal standardization has the benefit of allowing target and standard nuclei in the same medium, so this approach is preferred. This agrees with former recommendations (Vindelov et al., 1983; Tiersch et al., 1989), but it should be kept in mind that the presence of nuclei of both species in the medium does not mean that there will be no stoichiometric error.
Pseudo‐ and true variations in nuclear DNA content in coffee trees
In internal standardization, it is implicitly assumed that target and standard nuclei will react in a similar way to the medium. If this is not the case, differences in phenolic and alkaloid contents between plants will lead to pseudo‐variations in genome size within species. Stoichiometric error can also result from variations in phenolic and alkaloid contents over time or between locations (environmental effect). This should also occur when phenolic and alkaloid contents change with leaf age. This prompts us to discuss controversial intraspecific variations in genome size.
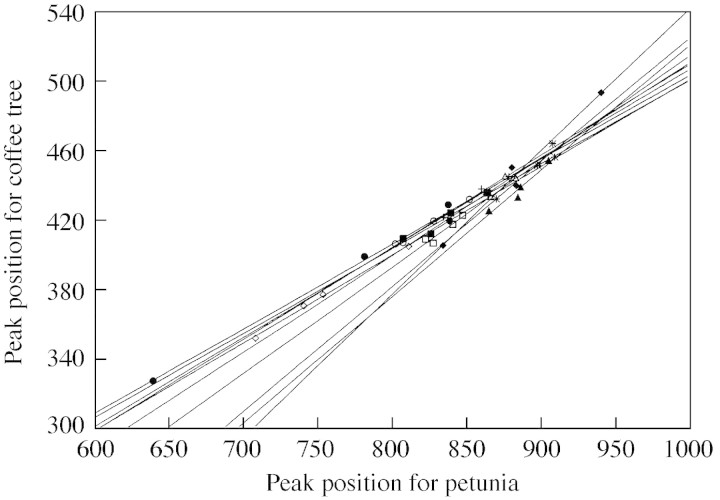
Pseudo‐variations in genome size due to genotypic effects exist in C. liberica var. deweveri (Barre et al., 1996; Noirot et al., 2002). In this species, genome size, estimated using PI and Petunia as internal standard, ranges from 1·408 to 1·439 pg between accessions. In fact, regression lines between target and standard nucleus fluorescence differ between trees; slopes range from 0·486 to 0·824, while intersects range from –282·5 to 15·7 (Fig. 4). Between‐slope variations represent the best evidence that dye accessibility of nuclear DNA changes between standard and target.
Fig. 4. Within‐tree relationships comparing the locations of the Petunia peak and the coffee peak. Each straight line represents a tree (from Noirot et al., 2002) and was estimated from four independent extracts per tree.
Pseudo‐variations in genome size due to environmental effects were highlighted in coffee trees when analysing some genotypes at different dates. Environmental variations in genome size also occur in sunflower in response to light quality and quantity (Johnston et al., 1996; Price et al., 1998, 2000). As CGA are also present in sunflower (Cohen and Ibrahim, 1975; Koeppe et al., 1976) and as CGA are known to be environment‐dependent, this compound could explain environmental variations in genome size in both species.
In summary, stoichiometric error should arise every time phenolic contents vary. As phenolics are common in plants, especially in woody species, stoichiometric error can be expected in all of these cases.
In contrast to CGA, caffeine is (1) not markedly influenced by environmental factors (Barre et al., 1998), and (2) not very widespread in the plant kingdom, at least at high concentration. Caffeine thus could not be a candidate compound to explain between‐date differences in genome size evaluations. Nevertheless, this compound could still account for variations between trees in C. l. dewevrei species (Noirot et al., 2002). Dye accessibility variations in coffee trees are likely to be the result of caffeine–CGA interactions. In practice, caffeine and CGA contents in hybrids would act differently on genome size evaluations under external and internal standardization conditions. Such CGA–caffeine interactions could be extended to other species, such as tea or cacao trees, containing oxypurines. Generally, the simultaneous presence of ‘phenolics–alkaloids’ could lead to interactions and intraspecific variations in genome size.
The existence of a pseudo‐intraspecific variation in genome size does not exclude the possibility of true genome size differences. The latter case concerns subspecies such as Musa acuminata (Dolezel et al., 1994) or Oryza sativa (Ohmido et al., 2000), whose reproductive barriers have prompted questions about their taxonomic classification. In these cases, a change in botanical name would turn intraspecific variations into interspecific variations. Other sources of true DNA content variation include endopolyploidy, as in Brassica oleracea (Kudo and Kimura, 2001), additional chromosomes, as in Quercus petraea (Zoldo et al., 1998), or ploidy level, as in Pennisetum setaceum (Martel et al., 1997). It is also probable that in some species, such as Quercus petrae, both sources of intraspecific variation (true and pseudo) overlap.
Phenotypic relationships between ΔDNA and some biochemical characteristics of hybrids evaluated in green coffee beans
In the present study, a linear relationship was noted between ΔDNA recorded on leaves and the amount of caffeine accumulated per day in green coffee beans. This implicitly assumes that the amount of caffeine accumulated per day in green beans is related to the caffeine content in leaves. This assumption is in line with (1) the fact that caffeine is synthesized in leaves, penetrates all membranes, and is accumulated in beans as a CGA–caffeine complex (Payen, 1851; Baumann et al., 1993), and (2) the absence of direct correlation with caffeine content in green beans.
A curvilinear relationship was also noted between ΔDNA recorded in leaves and the caffeine : CGAs.s. ratio in green coffee beans. As previously discussed, this relationship would involve some constant biochemical and genotypic characteristics in both leaves and beans. The curvilinear relationship is also expected from the antagonistic effects of caffeine and CGA.
Supplementary Material
Received: 30 June 2002;; Returned for revision: 30 January 2003. Accepted: 13 May 2003
References
- AnthonyF, Clifford MN, Noirot M.1993. Biochemical diversity in the genus Coffea L.: chlorogenic acids, caffeine, and mozambioside contents. Genetic Resources and Crop Evolution 40: 61–70. [Google Scholar]
- BarreP, Noirot M, Louarn J, Duperray C, Hamon S.1996. Reliable flow cytometric estimation of nuclear DNA content in coffee trees. Cytometry 24: 32–38. [DOI] [PubMed] [Google Scholar]
- BarreP, Layssac M, D’Hont A, Louarn J, Charrier A, Hamon S, Noirot M.1998. Relationship between parental chromosomic contribution and nuclear DNA content in the coffee interspecific hybrid C. pseudozanguebariae × C. liberica var. dewevrei Theoretical Applied Genetics 96: 301–305. [Google Scholar]
- BaumannTW, Mösli SS, Schulthess BH, Aerts RJ.1993. Interdependence of caffeine and chlorogenic acid (5‐CQA) metabolism in coffee. Proceedings of the International Congress of ASIC 15: 134–140. [Google Scholar]
- CharrierA, Berthaud J.1975. Variation de la teneur en caféine dans le genre Coffea Café Cacao Thé 19: 251–264. [Google Scholar]
- CliffordMN, Staniforth PS.1977. A critical comparison of six spectrophotometric methods for measuring chlorogenic acids in green coffee beans. Proceedings of the International Congress of ASIC 8: 109–113 [Google Scholar]
- CliffordMN, Williams T, Bridson D.1989. Chlorogenic acids and caffeine as possible taxonomic critera in Coffea and Psilanthus Phytochemistry 28: 829–838. [Google Scholar]
- CohenY, Ibrahim RK.1975. Changes in phenolic compounds of sunflower infected by Plamopara hastedii Canadian Journal of Botany 53: 2625–2630. [Google Scholar]
- DolezelJ, Binarova P, Lucretti S 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 31: 113–120. [Google Scholar]
- DolezelJ, Dolezelova M, Novak FJ.1994. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biologia Plantarum 36: 351–357. [Google Scholar]
- GalbraithDW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firozabady E.1983. Rapid flow cytometry analysis of the cell in intact plant tissues. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- GreilhuberJ.1988. ‘Self‐tanning’ – a new and important source of stoichiometric error in cytophotometric determination of nuclear DNA content in plants. Plant Systematics and Evolution 158: 87–96. [Google Scholar]
- JohnstonJS, Jenson A, Czeschin DG Jr, Price HJ.1996. Environmentally induced nuclear 2C DNA content variation in Helia nthus annuus American Journal of Botany 83: 1113–1120. [Google Scholar]
- KoeppeDE, Southwick LM, Bittell JE.1976. The relationship of tissue chlorogenic acid concentration and leaching of phenolics from sunflower grown under varying phosphate nutrient conditions. Canadian Journal of Botany 54: 593–599. [Google Scholar]
- KudoN, Kimura Y.2001. Patterns of endopolyploidy during seedling development in cabbage (Brassica oleracea L.). Annals of Botany 87: 275–281. [DOI] [PubMed] [Google Scholar]
- KyCL.2000.Déterminisme de quelques composés biochimiques de la graine de café vert impliqués dans la qualité à la tasse. Etude d’un croisement interspécifique entre Coffea pseudozanguebariae et C. liberica var. Dewevrei. PhD Thesis, ENSA Montpellier, France. [Google Scholar]
- KyCL, Louarn J, Guyot B, Charrier A, Hamon S, Noirot M.1999. Relations between‐ and inheritance of chlorogenic acid contents in an interspecific cross between Coffea pseudozanguebariae and Coffea liberica var. dewevrei Theoretical Applied Genetics 98: 628–637 [Google Scholar]
- KyCL, Louarn J, Dussert S, Guyot B, Hamon S, Noirot M.2001. Caffeine, trigonelline, chlorogenic acids and sucrose diversity in wild Coffea arabica L. and C. canephora P. accessions. Food Chemistry 75: 223–230. [Google Scholar]
- LaurieDA, Bennett MD.1985. Nuclear DNA content in the genera Zea and Sorghum Intrageneric, interspecific, and intraspecific variation. Heredity 55: 307–313. [Google Scholar]
- MarieD, Brown SC.1993. A cytometric exercise in plant DNA histogram. Biology of the Cell 78: 41–51. [DOI] [PubMed] [Google Scholar]
- MartelE, De Nay D, Siljak‐Yakovlev S, Brown S, Sarr A.1997. Genome size variation and basic chromosome number in pearl millet and fourteen related Pennisetum species. Journal of Heredity 88: 139–143. [Google Scholar]
- MazzaferaP, Sivarolla MB, Alves de Lima MM, Medina Filho P.1997. Caffeine content of diploid coffee species. Cienca e Cultura 49: 216–218. [Google Scholar]
- MontagnonC, Guyot B, Cilas C, Leroy T.1998. Genetic parameters of several biochemical compounds from green coffee, Coffea cane phora Plant Breeding 117: 576–578. [Google Scholar]
- NoirotM, Barre P, Louarn J, Duperray C, Hamon S.2000. Nucleus‐cytosol interactions – a source of stoichiometric error in flow cytometric estimation of nuclear DNA content in plants. Annals of Botany 86: 309–316. [Google Scholar]
- NoirotM, Barre P, Louarn J, Duperray C, Hamon S.2002. Consequences of stoichiometric error for nuclear DNA content evaluation in Coffea liberica var. dewevrei using DAPI and propidium iodide. Annals of Botany 89: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhmidoN, Kijima K, Akiyama Y, de Jong JH, Fukui K, de Jong JH.2000. Quantification of total genomic DNA and selected repetitive sequences reveals concurrent changes in different DNA families in indica and japonica rice. Molecular and General Genetics 263: 388–394. [DOI] [PubMed] [Google Scholar]
- PayenA.1846. Premier mémoire sur le café. Compte‐Rendus de l’Académie des Sciences (Paris) 22: 724–737. [Google Scholar]
- PayenA.1851.Précis de chimie industrielle. Ed. Hachette, Paris, p. 569.
- PriceHJ, Morgan PW, Johnston JS.1998. Environmentally correlated variation in 2C nuclear DNA content measurements in Helianthus annuus L. Annals of Botany 82: 95–98. [Google Scholar]
- PriceHJ, Hodnett G, Johnston JS.2000. Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propidium iodide fluorescence. Annals of Botany 86: 929–934. [Google Scholar]
- RabéchaultH.1954. Tanins et complexes tanniques chez les caféiers. In Jacques‐Félix H, ed. Contributions à l’études du caféier en Côte‐d’Ivoire Paris: Imprimeri Jouve, 181–219. [Google Scholar]
- TierschTR, Chandler RW, Wachtel SS, Elias S.1989. Reference standards for flow cytometry and application in comparative studies of nuclear DNA content. Cytometry 10: 706–710. [DOI] [PubMed] [Google Scholar]
- TornalettiS, Russo P, Parodi S, Pedrini M.1989. Studies on DNA binding of caffeine and derivatives: evidence of intercalation by DNA‐unwinding experiments. Biochemical and Biophysical Acta 1007: 112–117. [DOI] [PubMed] [Google Scholar]
- TraganosF, Kapuscinski J, Darzynkiewicz Z.1991. Caffeine modulates the effects of DNA‐intercalating drugs in vitro: a flow cytometric and spectrophotometric analysis of caffeine interaction with novantrone, doxorubicine, ellipticine and the doxorubicine analogue AD198. Cancer Research 51: 3682–3689. [PubMed] [Google Scholar]
- VindelovLL, Christensen IJ, Nissen NI.1983. Standardization of high‐resolution flow cytometric DNA analysis by the simultaneous use of chicken and trout red blood cells as internal reference standards. Cytometry 3: 328–331. [DOI] [PubMed] [Google Scholar]
- ZoldoV, Pape D, Brown SC, Panaud O, Siljak‐Yakovlev S.1998. Genome size and base composition of seven Quercus species: inter‐ and intra‐population variation. Genome: 41: 162–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.