Abstract
Background
Stochastic optical reconstruction microscopy (STORM) and related methods achieves sub-diffraction-limit image resolution through sequential activation and localization of individual fluorophores. The analysis of image data from these methods has typically been confined to the sparse activation regime where the density of activated fluorophores is sufficiently low such that there is minimal overlap between the images of adjacent emitters. Recently several methods have been reported for analyzing higher density data, allowing partial overlap between adjacent emitters. However, these methods have so far been limited to two-dimensional imaging, in which the point spread function (PSF) of each emitter is assumed to be identical.
Methods
In this work, we present a method to analyze high-density super-resolution data in three dimensions, where the images of individual fluorophores not only overlap, but also have varying PSFs that depend on the z positions of the fluorophores.
Results and Conclusion
This approach can accurately analyze data sets with an emitter density five times higher than previously possible with sparse emitter analysis algorithms. We applied this algorithm to the analysis of data sets taken from membrane-labeled retina and brain tissues which contain a high-density of labels, and obtained substantially improved super-resolution image quality.
Background
In recent years, super-resolution optical imaging techniques have been developed to overcome the diffraction limit in fluorescence microscopy (Hell 2007; Huang et al. 2010). Among these techniques, methods based on stochastic switching and localization of individual molecules, such as stochastic optical reconstruction microscopy (STORM) (Rust et al. 2006) and (fluorescent) photoactivated localization microscopy ((F)PALM) (Betzig et al. 2006; Hess et al. 2006), achieve sub-diffraction-limit resolutions by sequentially imaging and localizing fluorescent emitters in the sample. To ensure that fluorophores can be precisely localized, only a sparse subset of fluorophores with non-overlapping images are activated and imaged at any given instant. Iteration of this process allows stochastically different subsets of fluorophores to be switched on and localized. A super-resolution image is then reconstructed from a large number of molecular localizations.
Initially only emitters with well-isolated images were considered in the analysis, with simple fitting algorithms. Recently, several methods have been developed to localize densely distributed emitters, whose images partially overlap with one or more neighbors (Holden et al. 2011; Huang et al. 2011; Cox et al. 2012). These methods are capable of analyzing images with a substantially higher emitter density with only a marginal loss in localization accuracy. This development allows super-resolution images to be acquired with a larger number of localizations per frame, and hence at a higher speed, which is particularly useful for imaging living specimens and specimens with ultra-high labeling density. To date, these methods have been limited to analyzing two-dimensional (2D) images, in which the point spread function (PSF) of all the emitters is assumed to be identical. It is unclear whether such high-density analysis algorithms could be extended to 3D super-resolution imaging which relies on PSFs with variable shapes to determine the z-coordinates of the emitters (Huang et al. 2008; Juette et al. 2008; Pavani et al. 2009).
In this work we demonstrate an extension of the DAOPHOT analysis method (Stetson 1987; Holden et al. 2011), which we term 3D-DAOSTORM, to the analysis of astigmatism-based 3D super-resolution data where the z position of the emitter is encoded in the x and y width of its PSF. 3DDAOSTORM simultaneously fits multiple overlapping images of adjacent emitters with different PSF shapes. To validate the method experimentally, we applied this algorithm to analyze tissue samples labeled with a lectin, Concanavalin-A (ConA). We demonstrate that this lectin, which selectively labels plasma and nuclear membranes, is useful for super-resolution imaging of cellular morphology. Compared to sparse emitter analysis algorithms based on single-emitter fitting, 3D-DAOSTORM substantially increases the localization density and improves the quality of the super-resolution images.
Methods
3D-DAOSTORM algorithm
This algorithm attempts to fit overlapping images of emitters simultaneously with multiple Gaussian peaks using an approach similar to that employed by DAOPHOT, an algorithm previously developed for analyzing images of stars (Stetson 1987). DAOPHOT has been recently used to analyze 2D super-resolution data where the PSFs of all emitters are assumed to have the same shape, an approach termed DAOSTORM (Holden et al. 2011). Here we extend this approach to analyze astigmatism-based 3D super-resolution data, where the PSFs of emitters can be approximated by elliptical Gaussians whose ellipticity depends on the z position of the emitter.
The basic idea behind the DAOPHOT algorithm is to fit the detected emitters in an image, and then examine the residual image after subtracting the fit for evidence of any undetected emitters. These emitters are then fit simultaneously with the emitters identified in the previous cycle, and the process is repeated until there is no further indication of undetected emitters in the residual image. The primary differences between the original DAOPHOT and the 3D-DAOSTORM algorithm developed here are as follows:
(1) DAOPHOT fits the image of every emitter with a fixed-shape PSF. To extend the algorithm to the analysis of astigmatism-based 3D super-resolution data, where the PSFs of emitters vary with their z position and can be modeled by elliptical Gaussians with varying x and y widths, the images of individual emitters are fit with:
| (1) |
where wx and wy are pre-determined functions of z. The dependence of wx and y on was determined by fitting defocusing curves to images of single emitters bound to a coverslip (Huang et al. 2008)
(2) The error in the fit is calculated using the maximum likelihood estimator suitable for a Poisson distribution of error as previously described (Laurence et al. 2010; Mortensen et al. 2010; Smith et al. 2010). This approach gives superior fitting performance for data where the number of detected photons is sufficiently low that the Gaussian distribution of error assumed by least squares fitting is not valid.
(3) The DAOPHOT algorithm groups overlapping images of emitters and simultaneously fits all of them together. This approach involves deciding which emitters overlap sufficiently to be grouped together. It also requires solving a set of coupled linear equations by inverting a matrix with (M×N)2 elements, where M is the number of parameters that describe the PSF of each emitter and N is the number of emitters whose images overlap. As it is computationally expensive to solve these coupled equations due to the poor scaling of matrix inversion with matrix size, we thought to simplify the problem by fitting each emitter independently, but in an iterative fashion. We accomplished this task by performing a single cycle of fit improvement for each emitter independently, then recalculating the overall fit image based on the updated position of every identified emitter (details are given below). This procedure is repeated until the algorithm either converges or reaches a pre-determined maximum number of cycles.
(4) Unlike the DAOPHOT algorithm we do not include a cubic spline term to correct for deviations in the PSF from an idealized Gaussian. Including this term could further improve the fitting accuracy.
The flow of operations of the algorithm is described below:
0. Initialize the algorithm.
0.1. To start by fitting only the peaks from the brightest emitters in the field of view, we first set a threshold value for the peak height to be equal to 4× the user-specified minimum peak height, h0(h0 typically equals to 150 counts).
0.2. Set the residual image equal to the original image.
1. New localization identification.
1.1. Identify pixels in the residual image that are both greater than all neighboring pixels within a user-specified radius (typically 5 pixels) and greater than the current peak height threshold. Mark the center positions of these peak pixels as new localizations, and ignore a pixel if it has already been chosen twice previously as a potential localization. The latter criterion was added to avoid getting trapped in an infinite, futile cycle of first identifying a localization, then removing it due to other screening criteria (such as being too close to a neighbor), and then identifying the same localization again in the next cycle. We found empirically that allowing at most two localizations per pixel was a good compromise, which breaks the repeated, futile localization addition cycle without substantially restricting the addition of valid localizations.
1.2. If no new localizations were identified and the localization height threshold is at its minimum value h0 , then exit the algorithm and return the current list of localizations.
1.3. If the localization threshold value is greater than h0 , decrease the localization height threshold value by h0 in preparation for the next cycle of localization identification.
1.4. Add the newly identified localizations that are at least 1 pixel away from all current localizations into the current list of localizations and flag them as “running”. Even though it is possible that some activated emitters are separated by less than a single pixel, we do not attempt to discriminate them as the signal-to-noise ratio of the images is not high enough. Current localizations that are closer than a user-specified distance (typically 5 pixels) of a newly identified localization are flagged as “running” to indicate that further refinement of their parameters may be necessary.
1.5. Set the parameter-dependent clamp values, Ck , for all the localizations to the default values (1000.0 for h, 1.0 for x and y, 3.0 for wx and wy, 100.0 for bg and 0.1 for z) in preparation for the parameter refinement in the next step.
2. Refining localization parameters.
2.1. For each localization, determine a fitting neighborhood within which fitting to the image is performed to refine localization parameters. We calculate the neighborhood size (defined by X and Y along the x and y directions) based on the current wx and wy values of the localizations. This neighborhood extends to twice wx or wy from the localizations center position.
2.2. Calculate the fit image, f, from the list of localizations by drawing an elliptical Gaussian for each localization using Eq.1 and the current fitting parameters. For reasons of efficiency, f is only calculated within the fitting neighborhood of the localizations as determined in step 2.1.
2.3. Calculate the fit error for each of the “running” localizations using the following equation (Laurence et al. 2010):
Where fi is the value of the fit image at pixel i in the neighborhood of the localization, gi is the actual image intensity at pixel i, and N is the number of pixels in the fitting neighborhood.
If |current error – previous error|/current error is less than a threshold (typically set to 1.0e−6), flag the localization as “converged”.
2.4. For each “running” localization (i.e. those localizations that have not converged as judged by the convergence criteria in step 2.3) perform a single cycle of fit optimization as described below.
2.4.1. Calculate the Jacobian (J) vector using the following equation (Laurence et al. 2010)
where J is a vector containing the first derivatives of with respect to each parameter in the Gaussian that is fit to the localization, and ak are the parameters describing the Gaussian, which include the background value, bg, the peak height h, the centroid position of the peak in x and y, (x0, y0), and the peak widths in x and y, (wx, wy).
2.4.2. Calculate the Hessian matrix (H) using the following equation (Laurence et al. 2010)
Note that we ignore the second derivative terms in H as suggested in (Laurence et al. 2010).
2.4.3. Calculate the parameter update vector U by solving UH = J using the LAPACK function dposv() (Anderson et al. 1999). The vector U describes how to best adjust each of the parameters ak of the localization to reduce the error in the fit.
2.4.4. Subtract the Gaussian peak calculated using the current localization parameters from the fit image f calculated in 2.2. As more cycles of optimization are performed, more of the localizations will have converged. To avoid having to recalculate f for all of the localizations since many of them will not have changed, we subtract the localizations with the “current parameters” from f in this step and then add the localizations with the “updated parameters” back to f in a later step (step 2.4.8).
2.4.5. Update individual localization parameters (ak) based on the parameter update vector U and the parameter specific clamp value Ck using the formula
If the sign of Uk has changed since the previous iteration, then Ck is first reduced by a factor of 2. The Ck value suppresses oscillations in the optimization as well as damping excessively large corrections (Stetson 1987). Initial values for each Ck of the localization are set when the localization is created (step 1.5).
2.4.6. Flag localizations that have a negative background value, bg, peak height h, or peak widths (wx, wy) as “bad”. These localizations are ignored in subsequent iterations of the fit, and removed from the current list of localizations in step 3.1. 2.4.7. Adjust the size of the localization's neighborhood, X and Y, based on the updated wx and wx parameters.
2.4.8. If the localization is not “bad” add it back to the fit image calculated in 2.2 with the updated parameters.
2.5. If there are still “running” localizations and the maximum number of iterations (typically set to 200) has not been reached, go to step 2.3.
3. Localization cleanup.
3.1. Construct a new localization list containing only those localizations that are “converged” or “running”, have a height (h) greater than h0 and have widths (wx, wy), greater than a user-specified value (typically 0.5 pixels).
3.2. Remove all the localizations in this list whose height is less than that of any neighboring localizations within a user-specified distance (typically 1 pixel). Such localizations tend to be false localizations due to the limited signal-to-noise ratio of our images. Nearby localizations to the ones that are removed are flagged as “running”.
3.3. Repeat step 2 (parameter refinement) with the new list of localizations, then go to step 4. This additional step is performed even if no localizations are removed in step 3.2. It gives localizations that may still be “running” additional cycles to converge. In the event that all the localizations have “converged”, this repetition of step 2 will finish almost immediately.
4. Update the residual image.
4.1. Estimate the background by subtracting the fit image from the original image, then smoothing the result with a 2D Gaussian with a sigma of 8 pixels. The smoothing helps to suppress noise in the background image, and is justified under the assumption that the actual background varies smoothly across the image.
4.2. Calculate the new residual image.
4.2.1. Set the residual image equal to the original image minus the fit image.
4.2.2. Compute the mean value of the residual image.
4.2.3. Subtract the estimated background from the residual image. This step flattens the residual image in situations where the background is not uniform across the image. Flattening the residual image in turn makes it easier to identify to new localizations in subsequent iterations of the algorithm.
4.2.4. Add the mean value from 4.2.2. back to the residual image.
5. Termination of the algorithm.
Go to step 1 if the total number of iterations has not been exceeded (typically 20). If the residual image is such that no new localizations will be found, then the algorithm will exit at step 1.2. If new localizations can still be found in the residual image even after 20 iterations we terminate anyway as we are most likely caught in an infinite loop. For most of the images that we have analyzed the total number of iterations performed is less than 7.
The algorithm was implemented in a combination of the C and Python languages. It is available for download at http://zhuang.harvard.edu/software.html.
Generation of Simulated STORM Images
Simulated STORM images were generated with the following parameters, 20 photons/pixel background, a constant 2000 photons per emitter, an overall camera gain of 3, and a camera read noise of 2. These parameter values are close to real experimental values. The emitters were placed on the image with a uniform random distribution in x and y. The z location of the localization was randomly distributed in a range of 800 nm. Localization widths (wx, wy) were calculated based on the z location using the defocusing curve with w0 = 2 pixels, cx = 150 nm, cy = -150 nm, d = 400 nm and z = -400 nm to 400 nm. These values are again close to real experimental values. The overall image was generated as the sum of the elliptical Gaussian functions associated with individual localizations based on the above-described parameters. The noise due to the EMCCD gain of the acquisition camera was modeled with an exponential distribution.
Lectin labeling of retina and brain tissue samples
3-6 month old C57 mice were euthanized by asphyxiation with CO2 following procedures approved by the Harvard University Animal Care and Use Committees. The eyes or brains were then dissected, fixed by immersion in 4% paraformaldehyde (PFA), and select areas of interest, such as retina or regions of the cerebral cortex were further dissected. The fixed tissue was then washed with phosphate buffered saline (PBS) and stored in PBS at 4°C until use. Fixed tissue was incubated with the Alexa-647 dye labeled lectin at a concentration of 0.25 mg/ml for 3-5 days at 4°C in a labeling buffer containing PBS supplemented with 0.49 mM Mg2+ and 0.90 mM Ca2+. The tissue was then washed extensively with the labeling buffer and fixed overnight with 2% PFA, 0.2% Glutaraldehyde in the labeling buffer. The tissue was sectioned at 50nm (for 2D STORM imaging) or 100nm (for 3D STORM imaging) thickness with a Leica UC6 ultra-microtome. The tissue sections were transferred to cleaned coverslips and stored on coverslips at room temperature prior to use. The following Alexa-647 labeled lectins were used in this study: Concanavalin A (ConA, #C21421), Wheat Germ Agglutinin (WGA, #W32466), Peanut lectin (PNA, #L32460) and Red Kidney Bean lectin (PHA-L, #L32457), all purchased from Invitrogen.
STORM imaging of the lectin-labeled brain and retina tissue
Flow channels containing the tissue samples were constructed by sandwiching two pieces of double stick tape (3M) between the coverslip with the tissue sections and a microscope slide. The following imaging buffer was added to the flow channel: 10mM Tris, 50mM NaCl, 0.1% Triton-X100, pH8.0 supplemented with 0.5 mg/ml Glucose Oxidase (Sigma, G2133), 40 ug/ml Catalase (Sigma, C100), 5% Glucose and 100mM cysteamine (Sigma 30070). For 3D STORM imaging, 1% (v/v) beta-mercaptoethanol (Sigma, 63689) was used instead of 100mM cysteamine. After the addition of the imaging buffer, the flow channel was sealed with 5 minute epoxy and placed on a custom microscope setup built for STORM imaging. Epoxy sealed samples were imaged within a few hours of preparation.
Low resolution conventional fluorescence images were taken with a 2× air objective (Nikon, Plan Apo λ, 0.1NA) first to locate the tissue sections on the coverslip and to find the areas of interest. Once an area of interest was identified, a high resolution conventional fluorescence picture was taken with a 100x oil immersion objective (Nikon, Plan Apo λ, 1.45NA), followed by a STORM image.
STORM imaging was performed on a custom setup built around a Nikon TiU inverted microscope (Bates et al. 2007; Huang et al. 2008). Illumination of the Alexa-647 dye was provided by a 300 mW 656 nm solid state laser (Crystalaser, CL656-300). The 656 nm laser light excites fluorescence from Alexa 647 and switches the dye off rapidly. The same light also reactivates Alexa 647 back to the fluorescent state, but at a very low rate such that only a small fraction of the dye molecules (~0.1%) emit fluorescence at any given instant. When necessary, a 50 mW 405 nm diode laser (Coherent, Cube-405) were used to increase the dye activation rate (Dempsey et al. 2011). The output of the lasers was combined and coupled into a single mode photonic fiber (NKT Photonics, LMA-8) for transmission to the STORM microscope. Light from the fiber was collimated and focused on the back-focal plane of the microscope objective. The illumination was adjusted from epi-flourescence to total internal reflection by translating the illumination beam across the back-focal plane of the objective. Imaging was performed with a 100× oil immersion objective (Nikon, Plan Apo λ , 1.45NA). The laser intensities at the sample were ~1kW/cm2 for the 656nm laser light and ~20W/cm2 for the 405nm laser light. The fluorescence signal was recorded with an EMCCD camera (Andor, DU-897). For 3D STORM images, a 1m focal length cylindrical lens was added to the optical path to provide astigmatism, such that the PSF of individual emitters appear elliptical with ellipticity depending on the z position of the emitter (Huang et al. 2008). In addition the setup had an infrared focus lock system that was used to stabilize the distance between the microscope objective and the sample (Huang et al. 2008).
Results
To increase the imaging speed of 3D super-resolution fluorescence microscopy, we developed a data analysis algorithm, 3D-DAOSTORM, which can determine the position of densely distributed emitters with overlapping images and varying PSFs. Similar to the previously developed DAOSTORM approach (Holden et al. 2011), this algorithm uses multiple rounds of peak identification, fitting and subtraction to determine the positions of emitters with partially overlapping images. However, unlike previous dense-emitter analysis algorithms (Holden et al. 2011; Huang et al. 2011; Cox et al. 2012), which analyze 2D images where the PSF of individual emitters are assumed to have an identical shape, 3D-DAOSTORM can analyze emitters with varying PSF shapes. In particular, we applied this algorithm to the analysis of astigmatism-based 3D STORM data where the PSFs of emitters can be modeled by an elliptical Gaussian shape, and the x and y widths of the Gaussian function, (wx, wy), vary with in z position of the emitter. We compare the performance of this algorithm on both simulated and experimental data to two variations of a sparse emitter analysis algorithm, which does not fit adjacent emitters when their images overlap (Huang et al. 2008). In the first variation of the sparse emitter algorithm (SEA.1) we applied a filter criteria based on the image shape to select only those localizations which had wx and wy values that were within a specified distance of a previously determined defocusing curve. In the second variation of the sparse emitter algorithm (SEA.2) no image-shape-based filtering was applied.
We first compared the performance of the three algorithms on simulated images, where the ground truth of the emitter locations is known. In the simulated images, the emitters were chosen to be randomly distributed in x and y across the entire image and in z over a range of 800 nm. The emitter PSF was modeled as an elliptical Gaussian. The various simulation parameters such as the number of photons detected per emitter, the Gaussian widths as a function of z, the image background, and the camera noise were chosen to match what we typically measure in real STORM images. We quantitatively compared the recall efficiency and localization accuracy of the algorithms as a function of emitter density (Figure 1).
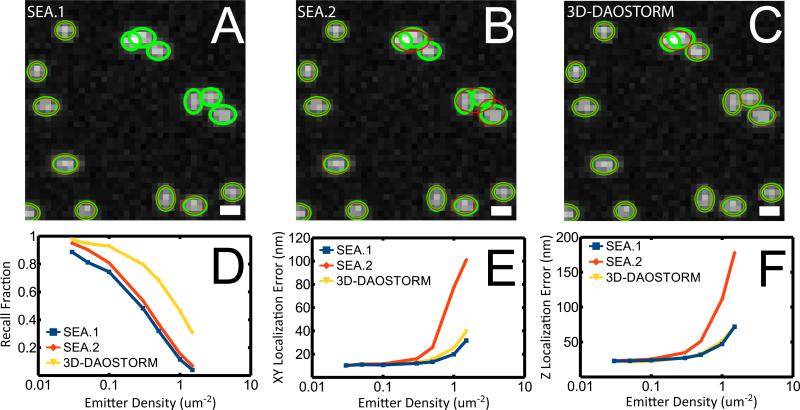
Figure 1.
Comparison of the 3D-DAOSTORM and sparse emitter analysis (SEA) algorithms on simulated data. (A) Analysis of simulated 3D data using the sparse emitter analysis algorithm with image-shape-based filtering (SEA.1). The simulated images of emitters are shown in grey scale and their actual locations are indicated by green ovals. The localizations identified by the analysis are marked by red ovals. The widths of the ovals in x and y are drawn proportional to the simulated PSF widths of the emitters. The molecule density is 0.3 molecules / um2. (B, C) Same as A, except that the analysis was performed using the sparse emitter analysis algorithm without any image-shape-based filtering (SEA.2) (B) and the 3D-DAOSTORM algorithm (C). Scale bars: 4 pixels or 668 nm. (D) A comparison of the recall fraction for the three different analysis methods on the simulated 3D STORM data. The recall fraction is defined as the fraction of the emitters that were identified by the algorithm. (E) A comparison of the localization error in the xy plane for the three different analysis methods on the simulated 3D STORM data. (F) A comparison of the z localization error for the three analysis methods. Data in (D-F) are extracted from ten 256 × 256 pixel images as shown in A-C.
To visually compare the performance of the different analysis algorithms, we overlaid the localizations returned by the algorithms, represented by the red ovals whose widths are proportional to those of the elliptical Gaussian widths, (wx, wy), on the simulated PSFs of the emitters (grey scale image), whose positions and widths are marked by the green ovals (Figure 1A- C). Clearly, all three algorithms can localize the well-isolated emitters precisely, but the 3D-DAOSTORM algorithm performs much better than the sparse emitter analysis algorithm when it comes to identifying and localizing nearby emitters with overlapping images. The sparse emitter algorithm with image-shape-based filtering (SEA.1) identifies substantially fewer emitters (Figure 1A), while the sparse emitter algorithm without any image-shape-based filtering (SEA.2) yields more incorrect localizations (Fig. 1B)
Next, we quantitatively compared the performance of the algorithms as a function of emitter density (Figure 1D- F). We quantify the performance by two parameters: the recall fraction and the localization accuracy. An emitter is considered recalled if the algorithm returns a localization within 30 nm of the true position of the emitter in x and y. Even at a relatively low density of 0.1 emitters per um2, there is already a substantial difference in the recall fraction between the 3D-DAOSTORM algorithm and the sparse emitter analysis algorithms (Figure 1D). While the former identified 95% of the emitters, the latter only found 80% of the existing emitters. This difference increases rapidly as the density of the emitter increases (Figure 1D). At a density of 1.0 emitters per um2, the 3D-DAOSTORM algorithm recalls approximately four times as many emitters as either of the sparse emitter analysis algorithms. Compared to SEA.1, relaxing the image-shaped-based filtering criterion in SEA.2 did not substantially improve the recall fraction, but instead primarily led to higher localization errors. The reason is that fitting images whose shapes did not conform to the predicted shapes of single molecules resulted in mislocalizations that were sufficiently far away from the true emitter positions that they were not considered recalled.
In addition to the recall fraction, we also compared the localization accuracy of all three algorithms, defined as the median XY distance between each localization and the nearest true emitter position in the xy plane (Figure 1E) or the median distance along the z direction (Figure 1F). All the algorithms perform similarly up to a density of 0.3 emitters per um2, at which point the SEA.2 algorithm diverges substantially faster than the SEA.1 and 3D-DAOSTORM algorithms, due to the large number of incorrect localizations that SEA.2 generated in regions where images of adjacent molecule overlap (Figure 1B). At an emitter density of 1.0 per um2, the localization error for the 3D-DAOSTORM algorithm is only about 25% and 45% of that for the SEA.2 algorithm in the xy plane and z direction, respectively. The localization accuracies of the 3D-DAOSTORM and SEA.1 algorithms remain similar. Hence, in the following experimental tests, we compared 3D-DAOSTORM with SEA.1.
To further test the performance of the 3D-DAOSTORM algorithms on experimental samples, we explored tissue samples labeled with fluorescent lectin conjugates. Lectins bind to specific sugar groups that are primarily found on glycosolated proteins and lipids (Sharon et al. 1972; Goldstein et al. 1978). As glycosolated moieties are more commonly found on cellular membranes or in the extracellular matrix between cells, we reasoned that lectins could be valuable super-resolution labels for outlining cells and determining cellular morphology in tissues. The ability to visualize cell morphology with super-resolution fluorescence microscopy is of interest since this approach could be used to determine the connectivity between neurons, as well as to determine the locations of cellular proteins in the context of cellular membranes. Here, we tested four different lectins: concanavalin-A (ConA), wheat germ agglutinin (WGA), peanut lectin (PNA) and red kidney bean lectin (PHA-L), all of which were conjugated with the Alexa-647 fluorescent dye.
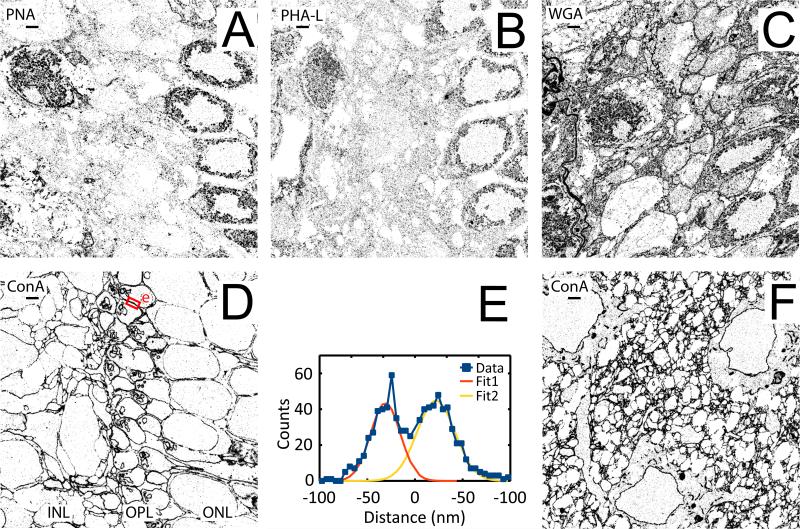
We primarily focused on regions near the outer plexiform layer (OPL) of the retina, where the rod and cone photo-detector cells synapse with the bipolar and horizontal cells (Dowling et al. 1966; Kolb 1970). Figure 2A-D shows the 2D STORM images of the OPL, and nearby regions, from 50 nm thick retina tissue sections labeled with PNA, PHA-L, WGA, and ConA. Among the four lectin labels, ConA appeared to be the best in terms of giving a clean outline of the cell, as it primarily labels the nuclear and cytoplasmic membranes of the cell (Figure 2D). Two plasma membranes that were separated by ~53 nm can easily be resolved in the STORM images (Figure 2E). A similar quality of cell membrane labeling can be achieved in the cortical regions of the brain with ConA (Figure 2F). The WGA lectin performed almost as well, but this lectin appeared to be somewhat less specific, labeling many different structures inside the cell in addition to the plasma membrane (Figure 2C). PNA and PHA-L lectins did a poor job of outlining the cell membranes except for the nuclei (Figure 2A-B). We thus used ConA for the following experiments.
Figure 2.
2D STORM images of mouse retina and cortex labeled with lectin – Alexa-647 conjugates. (A-D) Images are centered on the outer plexiform layer (OPL) of the retina and oriented such that the outer nuclear layer (ONL) are to the right and the inner nuclear layer (INL) is to the left. (A) PNA. (B) PHA-L lectin. (C) WGA lectin. (D) ConA lectin. (E) Cross-section profile of the region indicated by the small red rectangle with the “e” next to it in the upper part of (D). The two membranes are separated by a distance of 53 nm and have a width of ~35 nm as determined by multi-Gaussian fitting. (F) Image of a cortex region stained with ConA. All images were taken from lectin labeled tissue sections that are 50nm thick. Scale bars: 1 μm.
To test the 3D-DAOSTORM analysis algorithm, we performed astigmatism-based 3D STORM imaging of 100 nm thick ConA-labeled retina section by inserting a cylindrical lens in to the imaging path (Huang et al. 2008). We imaged a 42×42 μm area of the retina encompassing the inner and outer nuclear layer as well as the outer plexiform layer (Figure 3). Due to the high labeling density of ConA and spontaneous switching of the Alexa-647 dye from a dark to a fluorescent state under 656 nm illumination, the density of the activated Alexa-647 molecules at any given frame was quite high even without the use of a 405nm activation laser. As a result, the images of adjacent emitters often overlap, and the sparse emitter analysis algorithm could identify and localize only a fraction of the emitters in each frame (Figure 3A). In comparison, the 3D-DAOSTORM algorithm localized substantially more emitters per frame (5× on average) (Figure 3B). On average, the 3D-DAOSTORM algorithm returned five times as many localizations per frame for these images. The STORM image created using the 3DDAOSTORM algorithm therefore appears substantially denser and more contiguous, and sometimes sharper than the image of the same field of view generated using the sparse emitter analysis algorithm (Figure 3C-F). This improvement can be seen in cross-sectional profiles of the membranes (Figure 3G-I) where the profiles derived from 3D-DAOSTORM are substantially smoother due to the greater number of localizations that were included in the histogram. In some cases, this reduction in noise allowed us to resolve two adjacent membranes that could not be resolved using the sparse emitter analysis algorithm (Figure. 3G). A similar improvement can also be seen in XZ cross-sections of the cell membrane (Figure. 3J,K). Again, the images from the 3D-DAOSTORM algorithm are substantially less rugged due to the increased number of localizations.
Figure 3.
3D STORM image of the outer plexiform layer of mouse retina labeled with the ConA – Alexa-647 conjugate. (A) A single frame of the STORM data with localizations identified by the sparse emitter analysis algorithm (SEA.1) overlaid as red ovals. (B) Same as A except that the localization were identified using the 3D-DAOSTORM algorithm. (C) STORM image resulting from analysis with the sparse emitter analysis algorithm. The z-coordinates of the localizations are color-coded according to the colored scale bar. (D) STORM image of the same area resulting from analysis with the 3D-DAOSTORM algorithm. (E) A zoom in of the area indicated by the white box in C. (F) A zoom in of the same area in D. (G) Cross-section profile of the image areas in outlined in E and F by the red box with the letter “g” next to it. (H-I) Same as G for the areas indicated by red lines in C and D with a letter “h” (H) and “i” (I) next to them. (J-K) XZ cross-section images of the area in C and D indicated by the letter “j” (J) and “k” (K), respectively. Scale bars: 1 um in A and B, 4 um in C and D, 500 nm in E and F, 100nm in J and K.
To quantify the image resolution, we determine two parameters experimentally, the localization precision and the localization density. The localization precision of Alexa-647 in retinal tissue, measured as the spread of repetitive localizations of the same dye molecules, was found to be 10 nm in xy and 23 nm in z. These values correspond to image resolutions of 23 nm in xy and 54 nm in z. In addition, we found the average distance between neighboring localizations along the boundary between two cells to be 30 nm, corresponding to a resolution limit of 60 nm according to the Nyquist sample theorem. If we were to achieve the same, 50 nm z resolution through ultra-thin sectioning, the section thickness should be no more than 25 nm according to Nyquist sampling theorem, which is 4 times smaller than the 100 nm thickness used here. To reconstruct a large volume of tissue would require 4 times as many sections, which not only requires substantially longer total imaging time but is also subject to more sectioning loss and section alignment errors.
Conclusions
Here we demonstrated a new algorithm, 3D-DAOSTORM, for analyzing 3D super-resolution data generated by STORM and related methods. 3D-DAOSTORM allows densely distributed emitters with partially overlapping images to be simultaneously localized by repeated cycles of peak identification, fitting and subtraction. We validated the performance of this algorithm both on simulated image data with realistic signal and noise levels as well as on experimental data of retinal tissue sections labeled with lectin-dye conjugates. Compared to sparse analysis algorithms based on single-emitter fitting, 3D-DAOSTORM can analyze super-resolution data with a 4-5 times higher density of emitters per frame with similar localization precision. This improvement will increase the imaging speed of localization-based super-resolution fluorescence microscopy by 4-5 fold.
Acknowledgements
This work is supported in part by a Collaborative Innovation Award from Howard Hughes Medical Institute, a grant from the Gatsby Charitable Foundation, and the National Institutes of Health. X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
Authors’ Contributions
HPB: Designed experiments, conceived and implemented the 3D-DAOSTORM algorithm, prepared the manuscript.
YMS: Contributed to tissue sample preparation.
XZ: Designed experiments and prepared the manuscript.
Competing Interests
The authors declare that they do not have competing interests.
Contributor Information
Hazen Babcock, Center for Brain Science Harvard University Cambridge MA. 02138 hbabcock@fas.harvard.edu.
Yaron M. Sigal, Department of Chemistry and Chemical Biology Harvard University Cambridge, MA. 02138 ysigal@fas.harvard.edu
Xiaowei Zhuang, Center for Brain Science, Harvard University Department of Chemistry and Chemical Biology, Harvard University Department of Physics, Harvard University Howard Hughes Medical Institute Cambridge, MA. 02138.
References
- Anderson E, et al. LAPACK Users Guide, Society for Industrial and Applied Mathematics. 1999 [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor Super-Resolution Imaging with Photo-Switchable Fluorescent Probes. Science. 2007;317(5845):1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Cox S, et al. Bayesian localization microscopy reveal nanoscale podosome dynamics. Nature Methods. 2012;9:195–200. doi: 10.1038/nmeth.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nature Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling JE, Boycott BB. Organization of the Primate Retina: Electron Microscopy. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1966;166(1002):80–111. doi: 10.1098/rspb.1966.0086. [DOI] [PubMed] [Google Scholar]
- Goldstein IJ, Hayes CE. The Lectins: Carbohydrate-Binding Proteins of Plants and Animals. Advances in Carbohydrate Chemistry and Biochemistry. 1978;35:127–340. doi: 10.1016/s0065-2318(08)60220-6. [DOI] [PubMed] [Google Scholar]
- Hell SW. Far-field optical nanoscopy. Science. 2007;316(5828):1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91(11):4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden SJ, Uphoff S, Kapanidis AN. DAOSTORM: an algorithm for high-density super-resolution microscopy. Nature Methods. 2011;8(4):279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- Huang B, Babcock HP, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X. Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science. 2008;319(5864):810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Schwartz SL, Byars JM, Lidke KA. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomedical Optics Express. 2011;2(5):1377–1393. doi: 10.1364/BOE.2.001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nature Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- Kolb H. Organization of the Outer Plexiform Layer of the Primate Retina: Electron Microscopy of Golgi-Impregnated Cells. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1970;258(823):261–283. doi: 10.1098/rstb.1970.0036. [DOI] [PubMed] [Google Scholar]
- Laurence TA, Chromy BA. Efficient maximum likelihood estimator fitting of histograms. Nature Methods. 2010;7(5):338–339. doi: 10.1038/nmeth0510-338. [DOI] [PubMed] [Google Scholar]
- Mortensen KI, Churchman LS, Spudich JA, Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nature Methods. 2010;2010:377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavani SRP, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci U S A. 2009;106(9):2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Lis H. Lectins: Cell-Agglutinating and Sugar-Specific Proteins. Science. 1972;177(4053):949–959. doi: 10.1126/science.177.4053.949. [DOI] [PubMed] [Google Scholar]
- Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieved theoritically minimum uncertainity. Nature Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson PB. DAOPHOT: A Computer Program for Crowded-Field Stellar Photometry. Publications of the Astronomical Society of the Pacific. 1987;99:191–222. [Google Scholar]