Abstract
The Apaf-1 apoptosome is a multi-subunit caspase-activating scaffold that is assembled in response to diverse forms of cellular stress that culminate in apoptosis. To date, most studies on apoptosome composition and function have used apoptosomes reassembled from recombinant or purified proteins. Thus, the precise composition of native apoptosomes remains unresolved. Here, we have used a one-step immunopurification approach to isolate catalytically active Apaf-1/caspase-9 apoptosomes, and have identified the major constituents of these complexes using mass spectrometry methods. Using this approach, we have also assessed the ability of putative apoptosome regulatory proteins, such as Smac/DIABLO and PHAPI, to regulate the activity of native apoptosomes. We show that Apaf-1, caspase-9, caspase-3 and XIAP are the major constituents of native apoptosomes and that cytochrome c is not stably associated with the active complex. We also demonstrate that the IAP-neutralizing protein Smac/DIABLO and the tumor-suppressor protein PHAPI can enhance the catalytic activity of apoptosome complexes in distinct ways. Surprisingly, PHAPI also enhanced the activity of purified caspase-3, suggesting that it may act as a co-factor for this protease.
Keywords: apoptosis, apoptosome, caspase-9, PHAPI, XIAP
Introduction
The caspase family of cysteine proteases plays a critical role in apoptosis by coordinating the controlled destruction of the cell from within (reviewed in Martin and Green, 1995; Cohen, 1997). Caspases are initially synthesized as inactive zymogens that require further post-translational processing for full activity (Stennicke and Salvesen, 1999). During apoptosis, caspase activation appears to be achieved in two main ways: through recruitment of apical caspases to activation scaffolds or through direct proteolytic processing of downstream caspases by other caspases (or non-caspase proteases such as granzyme B).
The Apaf-1 apoptosome is a multi-subunit caspase-activating scaffold that is assembled in response to diverse forms of cellular stress (Li et al, 1997; Cain et al, 1999, 2000; Zou et al, 1999). Assembly of the apoptosome is provoked by the release of cytochrome c from the mitochondrial intermembrane space, an event that is both positively and negatively regulated by members of the Bcl-2 family (Kluck et al, 1997; Yang et al, 1997; Kuwana et al, 2002). Upon entry into the cytosol, cytochrome c associates with monomeric Apaf-1 and promotes a conformational change that permits oligomerization of the latter and recruitment of caspase-9 to the complex (Adrain et al, 1999; Zou et al, 1999; Jiang and Wang, 2000). Electron cryomicroscopy studies have established that apoptosomes assembled from recombinant components adopt a wheel-like configuration and are composed of approximately seven Apaf-1 molecules in complex with an unknown number of caspase-9 dimers (Acehan et al, 2002).
To date, the majority of studies on apoptosome activity, structure and composition have utilized reconstituted apoptosomes prepared from recombinant or purified proteins. Partially purified native apoptosomes have been successfully isolated by gel filtration approaches, but further purification of these complexes has been hampered by instability in high-salt buffers (Cain et al, 1999, 2000; Rodriguez and Lazebnik, 1999). For these reasons, while it is clear that Apaf-1 and caspase-9 form the core components of the apoptosome, the precise composition of native apoptosome complexes remains unclear. Using overexpression analysis, several additional proteins have been implicated as apoptosome constitutents, but the presence of these proteins in native apoptosomes remains to be confirmed. Thus, in addition to Apaf-1/caspase-9, proteins that have been implicated as constitutents of the apoptosome include caspase-3, caspase-7, XIAP, Aven, NAC, Bcl-2 and Bcl-XL (Pan et al, 1998; Cain et al, 1999; Chau et al, 2000; Bratton et al, 2001; Chu et al, 2001).
To explore the protein composition of native apoptosomes, we have used a one-step immunoprecipitation approach to isolate active caspase-9/Apaf-1 apoptosomes. To identify their constituents, these complexes were analyzed by analytical as well as preparative two-dimensional (2D)-PAGE coupled with MALDI mass spectrometry. Using this approach, we have also assessed the ability of putative apoptosome regulatory proteins, such as Smac/Diablo and PHAPI, to regulate the catalytic activity of the apoptosome. Here we describe the major constituents of native apoptosomes and show that the PHAPI tumor-suppressor protein enhances the catalytic activity of these complexes, possibly by acting directly on caspase-3.
Results
To explore the composition of native Apaf-1/caspase-9 apoptosomes, we initially attempted to immunoprecipitate Apaf-1 from cell-free extracts derived from Jurkat T cells. To do this, we used a panel of Apaf-1 antibodies, several of which successfully immunoprecipitated Apaf-1 in the absence of triggers for apoptosome assembly (data not shown). However, when apoptosome assembly was initiated by addition of cytochrome c/dATP to the cell extracts, all Apaf-1 antibodies tested failed to immunoprecipitate Apaf-1/caspase-9 complexes (data not shown). This approach was unsuccessful, presumably because the epitope(s) recognized by these antibodies became inaccessible upon oligomerization of Apaf-1 and recruitment of caspase-9 into the apoptosome.
Differential immunoprecipitation of Apaf-1 via caspase-9
As an alternative approach, we explored whether a monoclonal anti-caspase-9 antibody could differentially co-immunoprecipitate Apaf-1 in the presence or absence of cytochrome c/dATP. As Figure 1A illustrates, this approach was successful as Apaf-1 was readily co-precipitated with caspase-9 in the presence of cytochrome c/dATP, but not in the absence of these co-factors for apoptosome assembly. To explore whether Apaf-1/caspase-9 complexes retained catalytic activity after the extensive washing steps associated with the immunoprecipitation procedure, we assessed the ability of these complexes to cleave synthetic tetrapeptide caspase substrates (Figure 1B). Apoptosomes prepared in this way displayed LEHDase (caspase-9-like), as well as DEVDase (caspase-3-like) activities, but failed to hydrolyze the caspase-1-selective substrate YVAD, as expected (Figure 1B). Importantly, caspase-9 immunoprecipitated in the absence of cytochrome c/dATP failed to display any proteolytic activity (Figure 1B). We also asked whether purified apoptosomes were capable of cleaving natural caspase substrates such as BID, Vimentin and caspase-3 (Figure 1C). As a specificity control, pro-IL-1β was chosen as this cytokine is an established substrate for caspase-1 but not for caspases that are activated during apoptosis. These experiments revealed that apoptosomes purified in this manner retained robust and specific catalytic activity towards natural substrate proteins (Figure 1C).
Figure 1.
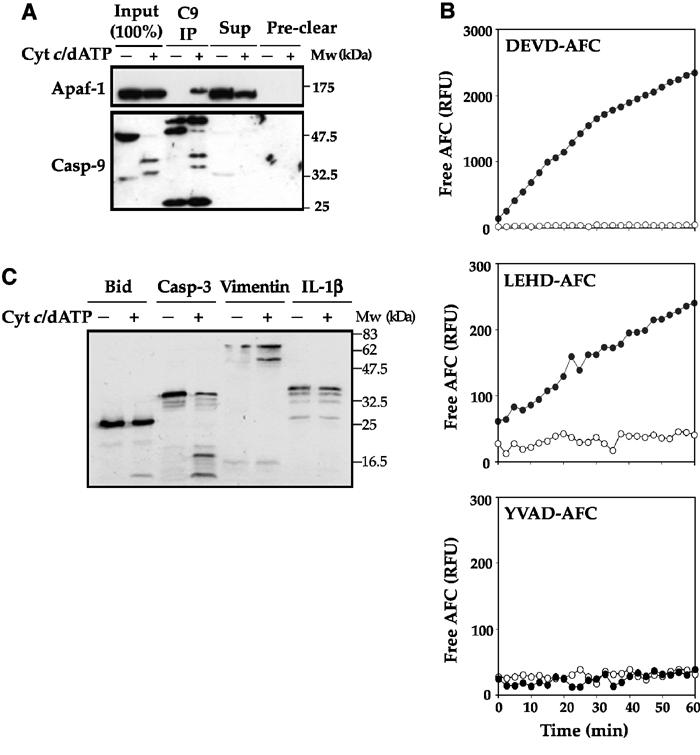
One-step isolation of native apoptosomes using a differential immunoprecipitation strategy. (A) Jurkat cell-free extracts were incubated in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP for 10 min at 37°C. Reactions were adjusted to 50 mM NaCl and 0.3% CHAPS and precleared by incubating with protein A/G agarose beads. Protein complexes immunoprecipitated with a monoclonal anti-caspase-9 antibody (Upstate Biotechnology) were analyzed for the presence of Apaf-1 and caspase-9 by immunoblotting. Equivalent amounts of the input (6.5 μl of the reaction), the IP supernatant (Sup) and protein binding to the preclearing beads (preclear) were loaded to aid comparison. (B) Jurkat cell-free extracts were incubated for 15 min at 37°C in the presence (filled circles) or absence (open circles) of 50 μg/ml cytochrome c/1 mM dATP. Caspase-9 complexes isolated from 100 μl reactions were washed extensively and then incubated for 1 h at 37°C with 50 μM of the indicated fluorogenic peptides. (C) Caspase-9 complexes, isolated from Jurkat cell-free extract incubated for 15 min at 37°C in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP, were incubated with in vitro translated Bid, caspase-3, vimentin or pro-interleukin-1β for 2 h at 37°C. Proteolysis of radiolabeled substrate proteins was analyzed by SDS—PAGE, followed by autoradiography.
Kinetics of apoptosome assembly
We then explored the kinetics of Apaf-1 and caspase-9 association upon addition of cytochrome c/dATP to the cell extracts. As Figure 2A illustrates, in the absence of a trigger for apoptosome assembly, Apaf-1 failed to associate with caspase-9, as expected. However, in the presence of cytochrome c/dATP, Apaf-1 was very rapidly recruited to pro-caspase-9 (within minutes) and this reproducibly occurred during assembly of the reactions at 4°C (Figure 2A). Caspase-9 was observed to undergo rapid proteolytic maturation within the apoptosome and this was routinely complete within 10 min of addition of cytochrome c/dATP to the extracts (Figure 2A). Stable recruitment of caspase-3, but not caspase-6 or -7, to apoptosomes was also observed, providing an explanation for the significant DEVDase activity displayed by these complexes (Figures 2A and 1B).
Figure 2.
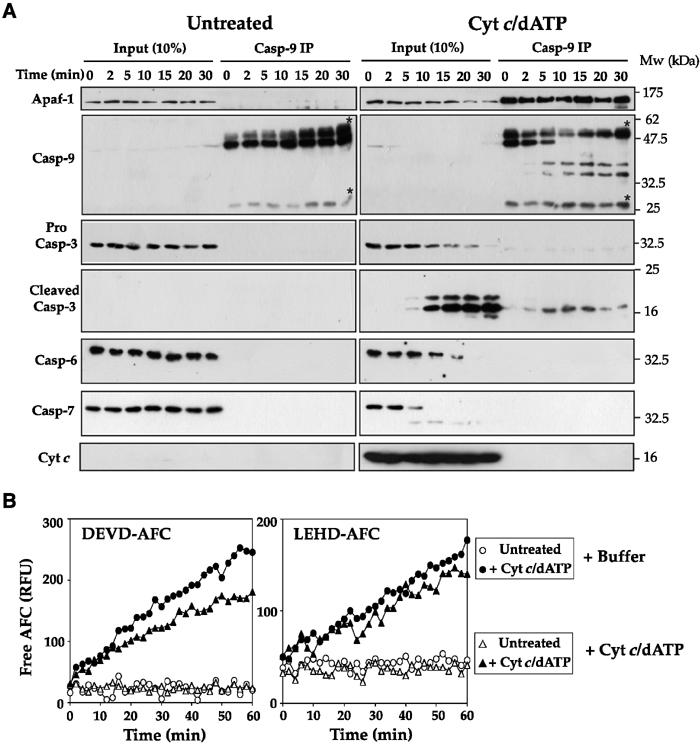
Cytochrome c triggers the rapid and stable association of Apaf-1, caspase-9 and caspase-3. (A) Jurkat cell-free extracts were incubated at 37°C in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP for the times indicated and then brought to 4°C. Caspase-9 was then immunoprecipitated from each reaction as described in the legend to Figure 1A. Caspase-9 immunocomplexes (Casp-9 IP) or 10% of the input reactions (input) were then probed for the presence of Apaf-1, caspase-3, -6, -7 and cytochrome c by immunoblotting. Bands corresponding to immunoglobulin heavy and light chains are indicated by asterisks. (B) Caspase-9 complexes were immunoprecipitated from Jurkat cell-free extracts incubated for 15 min at 37°C in the presence (filled symbols) or absence (open symbols) of 50 μg/ml cytochrome c/1 mM dATP. Caspase-9 immunocomplexes were then washed extensively, as described in Materials and methods, and peptide hydrolysis assays were then performed in the presence (triangles) or absence (circles) of 50 μg/ml cytochrome c/1 mM dATP. Note that for DEVD-AFC hydrolysis reactions complexes were prepared from 50 μl cell-free reactions, whereas for LEHD-AFC hydrolysis assays 150 μl cell-free reactions were required.
Cytochrome c is not stably associated with apoptosomes
Interestingly, we were unable to find stable association of cytochrome c within purified apoptosomes, despite addition of saturating amounts of this protein to cell-free extracts (Figure 2A). This suggests either that cytochrome c acts in a ‘hit-and-run' manner to trigger apoptosome assembly, or that the interaction between cytochrome c and Apaf-1 was of insufficient affinity to remain bound after washing of immunocomplexes. As apoptosomes prepared in this manner displayed robust catalytic activity toward caspase substrates (Figure 1), we favor a model where the continued presence of cytochrome c, after functional apoptosomes have been assembled, is not required. In support of this model, re-addition of cytochrome c and dATP to purified apoptosomes failed to enhance the catalytic activity of these complexes toward tetrapeptide substrates (Figure 2B).
Rapid recruitment of XIAP, but not of other IAPs, to the apoptosome and neutralization by Smac/DIABLO
The inhibitors of apoptosis proteins have been implicated as important regulators of apoptosis through their ability to associate with and repress the catalytic activity of mature caspases (Deveraux et al, 1997). In particular, XIAP has been shown to be capable of associating with active caspase-9 and -3, and has previously been implicated as a constituent of the apoptosome by gel filtration and co-immunoprecipitation analysis (Bratton et al, 2001). To explore whether any of the IAPs were recruited to Apaf-1/caspase-9 apoptosomes, we performed similar immunoprecipitation experiments and probed these complexes for the presence of XIAP, cIAP-1 and cIAP-2. We also explored whether the heat-shock proteins, HSP90 or HSP70, could be detected within the apoptosome, as both have been implicated as negative regulators of apoptosome assembly through binding to Apaf-1 (Beere et al, 2000; Pandey et al, 2000; Saleh et al, 2000). As Figure 3A shows, XIAP was rapidly recruited to apoptosomes and was also observed to undergo proteolytic processing within the complex. Addition of the caspase-3-selective inhibitory peptide Ac-DEVD-CHO to cell-free extracts during preparation of apoptosomes abolished XIAP proteolysis, suggesting that caspase-3 was responsible for this effect (Figure 3B).
Figure 3.

XIAP is a constituent of native apoptosomes. (A) Jurkat cell-free extracts were incubated at 37°C in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP for the times indicated and then brought to 4°C. Caspase-9 was then immunoprecipitated from each reaction as described in the legend to Figure 1A. Caspase-9 immunocomplexes (Casp-9 IP) or 10% of the input reactions (Input) were analyzed for the presence of IAPs, HSP90, HSP70, Bcl-2, Bcl-XL and PHAPI by immunoblotting. Bands corresponding to immunoglobulin light chain are indicated by asterisks. (B) Apoptosome assembly was initiated in Jurkat cell-free extracts in the presence or absence of 1 μM Ac-DEVD-CHO, as indicated. Caspase-9 was then immunoprecipitated from each reaction as described in the legend to Figure 1A. Caspase-9 complexes (Casp-9 IP) or 10% of the input reactions (Input) were analyzed for the presence of Apaf-1, caspase-9, -3 and XIAP by immunoblotting. Cleaved XIAP is indicated with an arrow. (C) Caspase-9 complexes were immunoprecipitated from Jurkat cell-free extracts incubated for 15 min at 37°C in the presence (filled symbols) or absence (open symbols) of 50 μg/ml cytochrome c/1 mM dATP. Peptide hydrolysis assays were subsequently performed in the presence of buffer (circles), 0.5 μM (triangles) or 5 μM (squares) recombinant Smac Δ1–55.
In contrast to the recruitment of XIAP to apoptosomes, we failed to find any evidence for recruitment of cIAP-1, cIAP-2, HSP70, HSP90, Bcl-2 or Bcl-xL under the same conditions (Figure 3A). Interestingly, we also failed to find evidence for stable recruitment of PHAPI—recently identified as a regulator of caspase-9 activation within this pathway (Jiang et al, 2003)—within apoptosome complexes (Figure 3A).
Smac/DIABLO is a mitochondrial protein that is released from the mitochondrial intermembrane space during apoptosis and has been shown to neutralize IAPs and potentiate caspase activation (Du et al, 2000; Verhagen et al, 2000). As XIAP was readily detected in association with apoptosome complexes, we explored whether addition of recombinant mature Smac/DIABLO (Δ1–55) to purified apoptosomes would enhance the catalytic activity of these complexes. As Figure 3C illustrates, Smac/DIABLO significantly enhanced the activity of native apoptosome complexes, confirming that XIAP was inhibitory to the complexes and could be neutralized by this IAP-binding protein.
XIAP acts as a tether for caspase-3 within the apoptosome
As XIAP was rapidly and efficiently recruited to apoptosomes, we also explored whether XIAP was required for apoptosome assembly. Thus, we immunodepleted XIAP from Jurkat cell-free extracts and prepared apoptosomes by immunoprecipitating caspase-9 in the presence or absence of cytochrome c/dATP as before. Interestingly, these experiments revealed that caspase-3 failed to be recruited to apoptosome complexes devoid of XIAP (Figure 4A). Consistent with this, apoptosomes prepared under these conditions also failed to exhibit significant DEVDase activity (Figure 4B). In contrast, DEVDase activity in cytochrome c/dATP-treated cell-free extracts depleted of XIAP displayed a net increase relative to mock-depleted extracts, which is consistent with the removal of a caspase inhibitory protein (Figure 4B). Taken together, these data suggest that XIAP is responsible for the stable recruitment of caspase-3 to the apoptosome. Interaction between XIAP and one of the catalytic sites in a mature caspase-3 dimer would be sufficient to tether caspase-3 to the apoptosome, while leaving the other half of the caspase-3 dimer free to act upon substrates. This would explain why significant apoptosome-associated caspase-3 activity is detectable despite interaction between this caspase and its inhibitor within the complex (Figures 1B and 4B).
Figure 4.
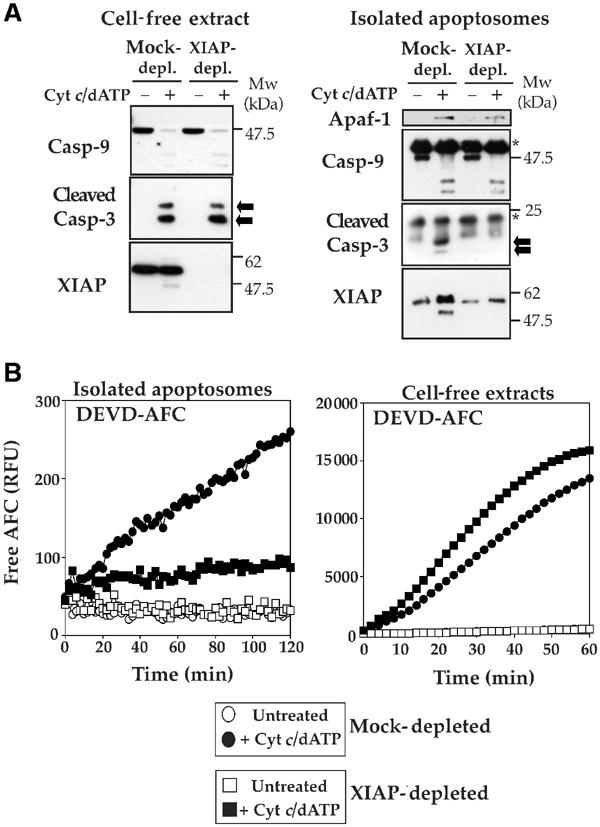
XIAP acts as a tether for recruitment of caspase-3 to the apoptosome. (A) Left panel, mock-depleted or XIAP-immunodepleted cell-free extracts were treated with 50 μg/ml cytochrome c/1 mM dATP for 15 min at 37°C, followed by immunoblotting and probing for the indicated proteins. Right panel, mock-depleted or XIAP-immunodepleted cell-free extracts were treated with 50 μg/ml cytochrome c/1 mM dATP for 15 min at 37°C. Reactions were then brought to 4°C and caspase-9 immunocomplexes were prepared, followed by immunoblotting for the indicated proteins. Arrows indicate mature caspase-3. Asterisks denotes immunoglobulin heavy and light chains. (B) Caspase-9 complexes (left panel) were immunoprecipitated from mock-depleted (circles) or XIAP-depleted (squares) Jurkat cell-free extracts incubated for 15 min at 37°C in the presence (filled symbols) or absence (open symbols) of 50 μg/ml cytochrome c/1 mM dATP. Hydrolysis of DEVD-AFC (50 μM) was monitored for 2 h at 37°C. Cell-free reactions (right panel) from the same extracts were incubated in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP for 30 min at 37°C, and assayed for DEVD-AFC hydrolysis for 60 min at 37°C.
Analysis of apoptosome composition by preparative 2D-PAGE/mass fingerprinting analysis
To seek additional apoptosome constituents, we used a scaled up approach where apoptosomes were immunoprecipitated from cell-free lysates of 109 Jurkat cells (∼60 mg protein) using anti-caspase-9 mAb (50 μg per IP), in the presence or absence of cytochrome c/dATP. Immunoprecipitates were washed extensively, eluted into urea-based sample buffer and preparative 2D-PAGE gels were run. All protein spots that differentially co-immunoprecipitated with caspase-9 under these conditions were excised from the gels and were analyzed by MALDI-TOF mass spectrometry. As Figure 5A shows, a total of 12 new protein spots were reproducibly detected by silver staining when caspase-9 was immunoprecipitated after apoptosome assembly. By mass fingerprinting analysis, these spots were identified as alternative cleavage products of mature caspase-9 (p35, p37), alternative cleavage products of mature caspase-3 (p20, p17, p12) and XIAP (Table I). These assignments were also confirmed by Western blot analysis on similar 2D gels (Figure 5B).
Figure 5.
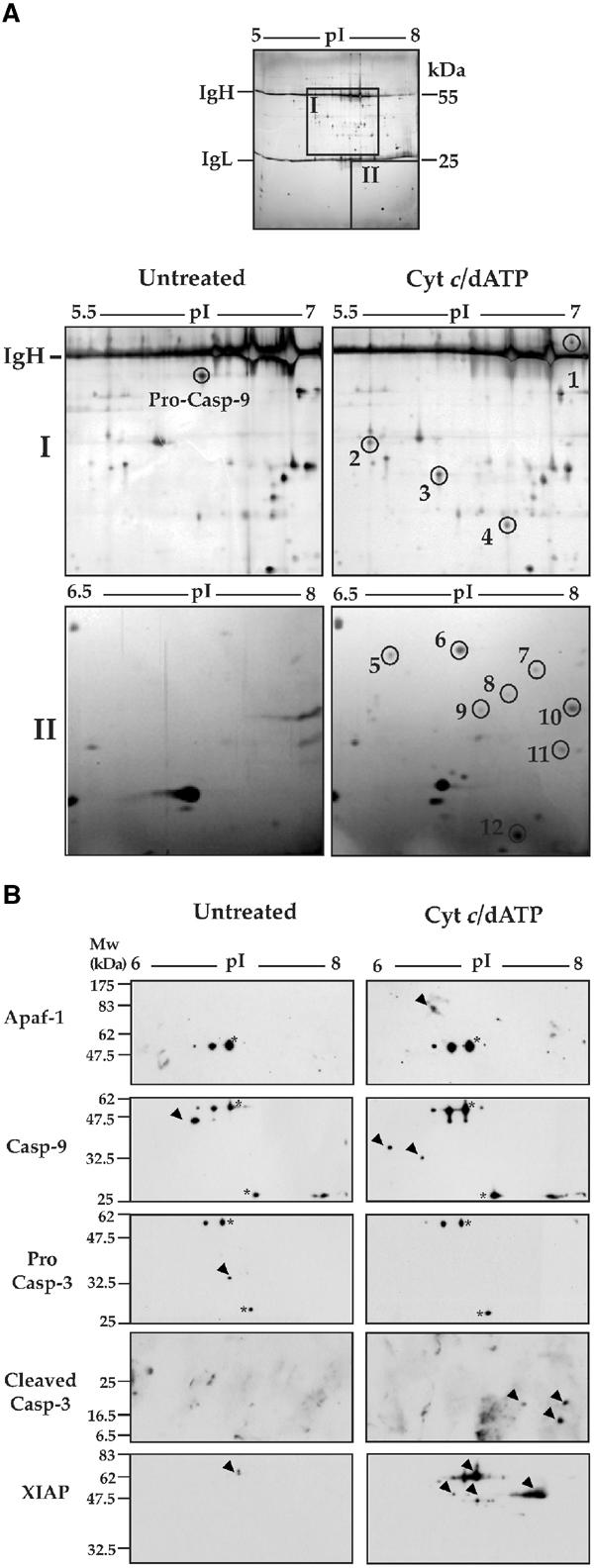
Analysis of native Apaf-1 apoptosomes by 2D-PAGE. (A) Caspase-9 complexes, isolated from 2 ml Jurkat cell-free reactions incubated for 15 min at 37°C in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP, were analyzed by 2D gel electrophoresis (first dimension: pH 5–8, second dimension: 12% SDS–PAGE). Top: A representative silver-stained preparative gel is shown. Heavy and light chains of the immunoprecipitating antibody are indicated. Bottom: enlarged areas containing proteins that differentially immunoprecipitate with caspase-9 (areas I and II in the gel above) are shown. (B) Caspase-9 complexes were isolated from Jurkat cell-free extracts incubated for 15 min at 37°C in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP. Proteins co-precipitating with caspase-9 were analyzed by 2D gel electrophoresis (pH 5–8 first dimension and 12% SDS–PAGE second dimension), followed by immunoblotting with the indicated antibodies. Spots corresponding to immunoglobulin heavy and light chains are indicated by asterisks.
Table 1.
Mass fingerprinting analysis of apoptosome-associated proteins
| Spot no. | Protein ID | Accession no. | No. of matches | Amino-acid coverage |
|---|---|---|---|---|
| Pro-caspase-9 | P55211 | 16 | 14–409/416 (55%) | |
| 1 | XIAP | P98170 | 32 | 11–491/497 (67%) |
| 2 | Caspase-9 (p37) | P55211 | 8 | 16–324/416 (38%) |
| 3 | Caspase-9 (p35) | P55211 | 8 | 57–189/416 (28%) |
| 4 | no ID | |||
| 5 | Rho-GDI2 | P52566 | 3 | 51–164/201 (26%) |
| 6 | Rho-GDI2 | P52566 | 5 | 51–196/201 (56%) |
| 7 | No ID | |||
| 8 | No ID | |||
| 9 | No ID | |||
| 10 | Caspase-3 (p20) | P42574 | 9 | 20–147/277 (34%) |
| 11 | Caspase-3 (p17) | P42574 | 9 | 39–147/277 (30%) |
| 12 | Caspase-3 (p10) | P42574 | 7 | 176–277/277 (26%) |
Interestingly, two of the novel spots were identified as C-terminal cleavage products of Rho-GDI2 (Table I), a member of the Rho family-specific guanine nucleotide dissociation inhibitors and an established caspase substrate (Martin et al, 1996; Na et al, 1996). Despite repeated attempts, four of the novel protein spots could not be identified by mass spectrometry analysis due to the low abundance of these proteins (Figure 5A and Table I). However, one of these spots (Figure 5A, spot 9) was subsequently identified by Western blot analysis as a caspase-3 cleavage product (Figure 5B). Note that Apaf-1 was not identified using mass spectrometry analysis due to poor penetration of high-molecular-weight proteins into the first dimension immobilized pH gradient strips. Apaf-1 was readily detected by direct immunoblot analysis on similar preparations (Figure 5B), but was not present on 2D gels in sufficient quantities to detect by silver staining. These data suggest that, in addition to Apaf-1 and proteolytically processed caspase-9, the major constituents of native apoptosomes are mature caspase-3, XIAP and additional low-molecular-weight proteins that have yet to be identified.
The mass spectrometry-based identification of RhoGDI2 degradation products in apoptosome immunoprecipitates was interesting and suggested that this protein might play a role within the apoptosome. However, using immunoblot analysis, we failed to confirm recruitment of this protein to the apoptosome complex (Figure 6). Moreover, while we readily detected Apaf-1, caspase-9, caspase-3 and XIAP in apoptosomes generated from BJAB and U937 cell-free extracts, we also failed to find evidence for recruitment of RhoGDI2 to these complexes (Figure 6). Thus, it appears likely that RhoGDI2 was nonspecifically co-precipitated with large-scale apoptosome preparations from Jurkat cells.
Figure 6.

Analysis of apoptosome constituents from Jurkat, BJAB and U937 cells. Cell-free extracts derived from Jurkat, BJAB or U937 cells were incubated at 37°C in the presence or absence of 50 μg/ml cytochrome c/1 mM dATP for the times indicated and then brought to 4°C. Caspase-9 was then immunoprecipitated from each reaction as described in the legend to Figure 1A. Caspase-9 complexes (CASP-9 IP) or 10% of the input reactions (input) were then immunoblotted for Apaf-1, caspase-3, caspase-9, RhoGDI2 and XIAP.
PHAPI as an enhancer of caspase activity within apoptosomes
Recently, Wang and colleagues have identified the tumor-suppressor protein PHAPI (also known as I1PP2A, Mapmodulin and PP32) as an enhancer of caspase-9 activation in the apoptosome pathway (Jiang et al, 2003). However, it is unclear whether PHAPI acts directly on the apoptosome to stimulate maturation of the caspases within, or whether PHAPI can enhance the catalytic activity of the mature complex (Jiang et al, 2003). As shown in Figure 3A, we failed to detect stable recruitment of PHAPI to the apoptosome, suggesting that this protein is not an integral part of the complex. However, it remained possible that PHAPI could act to enhance apoptosome activity after assembly of the complex. To explore whether this was the case, we added recombinant full-length PHAPI, or a deletion mutant lacking 84 amino acids from the C-terminus (aa 1–163; PHAPI-Δtail), to purified apoptosomes to assess their impact on apoptosome activity (Figure 7). PHAPI had a very significant enhancement effect on the proteolytic activity of apoptosomes, whereas the PHAPI-Δtail deletion mutant failed to display any activity in this assay (Figure 7A, left panel). PHAPI also profoundly enhanced caspase catalytic activity when added to total cell-free extracts in the presence of cytochrome c/dATP, whereas PHAPI-Δtail had no activity in this regard (Figure 7A, right panel). This suggests that the C-terminal region of PHAPI is critical for the activity of this protein.
Figure 7.

PHAPI enhances the catalytic activity of apoptosomes. (A) Caspase-9 apoptosome complexes (left) and Jurkat cell-free extracts (right), incubated for 15 min at 37°C in the presence (filled symbols) or absence (open symbols) of 50 μg/ml cytochrome c/1 mM dATP, were assayed for DEVDase activity in the presence of 0.5 μM recombinant PHAPI (triangles), PHAPI-ΔTail (squares) or buffer alone (circles). (B) Caspase-9 apoptosome complexes (left) and Jurkat cell-free extracts (right), incubated for 15 min at 37°C in the presence (filled symbols) or absence (open symbols) of cytochrome c/dATP, were assayed for DEVDase activity in the presence of 0.5 μM recombinant PHAPI (small circles), GST (large triangles), GST-PHAPI-Tail (small triangles), 15–50 kDa polyglutamine polymers (squares) or buffer alone (large circles). The results shown are representative of three separate experiments. (C) Aliquots (750 ng) of purified bacterially expressed PHAPI, PHAPI ΔTail, GST and GST-PHAP-Tail proteins were separated by SDS–PAGE, and visualized by coomassie blue staining. A degradation product of full-length PHAPI is indicated by an asterisk. (D) Jurkat cell-free extracts, incubated for 30 min at 37°C in the presence (filled symbols) or absence (open symbols) of 2 μg/ml cytochrome c/1 mM dATP, were assayed for DEVDase activity in the presence of 1 μM recombinant PHAPI (small circles), 5 μM okadaic acid (triangles), 1 μM calyculin A (squares) or buffer alone (large circles).
As the C-terminus of PHAPI is highly acidic, we also generated a GST-PHAPI deletion mutant (aa 164–249; GST-PHAPI-Tail) containing this acidic region to ask whether the acidic C-terminal tail was sufficient to enhance caspase activity in isolated apoptosomes or cell-free extracts. To control for nonspecific effects of acidic polypeptides on caspase activity, we also tested polyglutamic acid in the same assays. However, as shown in Figure 7B, neither the acidic PHAP tail nor polyglutamic acid was sufficient to mimic the stimulatory effects of PHAPI on caspase activity.
PHAPI has previously been characterized as a potent inhibitor of protein phosphatase 2A (PP2A; Li et al, 1996). To explore the possibility that PHAPI may enhance caspase activity through inhibition of PP2A, we assessed the effects of two potent inhibitors of this phosphatase (calyculin A and okadaic acid) on Apaf-1-driven caspase activation. However, as Figure 7D illustrates, neither inhibitor had any effect on caspase activation in the apoptosome pathway, arguing that PHAPI does not act via PP2A inhibition in this context.
PHAPI and Smac/DIABLO potentiate caspase activity in distinct ways
To determine whether PHAPI may be acting in a manner similar to Smac/DIABLO, by antagonizing the function of XIAP, we added PHAPI to apoptosomes in the presence of saturating levels of recombinant SmacΔ1-55. As Figure 3C demonstrates, the effect of SmacΔ1-55 on apoptosome activity was already saturated at 0.5 μM. To determine whether PHAPI could further enhance apoptosome activity in the presence of saturating levels of Smac/DIABLO, we combined PHAPI with Smac and monitored DEVDase activity within the apoptosome. This experiment revealed that PHAPI and Smac had an additive effect on the caspase activity of apoptosome complexes, suggesting that these proteins act to enhance apoptosome activity in distinct ways (Figure 8A). Moreover, we also found that PHAPI still enhanced caspase activity in cell-free extracts immunodepleted of XIAP, which strongly indicates that PHAPI acts in an XIAP-independent manner (Figure 8B).
Figure 8.

PHAPI and Smac enhance apoptosome activity in distinct ways. (A) Caspase-9 complexes, prepared from Jurkat cell-free extracts incubated for 15 min at 37°C in the presence (filled symbols) or absence (open symbols) of 50 μg/ml cytochrome c/1 mM dATP, were assayed for DEVDase activity in the presence of 1 μM recombinant PHAPI (triangles), 1 μM recombinant Smac (squares), 1 μM recombinant PHAPI plus 1 μM recombinant Smac (small circles) or buffer alone (large circles). (B) Jurkat cell-free extracts, either mock-depleted (small and large circles) or depleted of XIAP using anti-XIAP antibody (triangles and squares), were incubated for 30 min at 37°C in the presence (filled symbols) or absence (open symbols) of 50 μg/ml cytochrome c/1 mM dATP and then assayed for DEVDase activity in the presence or absence of 1 μM recombinant PHAPI, as indicated. Depletion of XIAP from extracts was confirmed by immunoblotting (inset).
PHAPI enhances the catalytic activity of purified caspase-3
The effects of PHAPI on the proteolytic activity of purified apoptosomes were intriguing and did not appear to be mediated through neutralization of XIAP. Thus, we wondered whether PHAPI could act to directly enhance the catalytic activity of purified recombinant caspases. Rather surprisingly, full-length PHAPI was found to enhance the activity of purified recombinant caspase-3, whereas PHAPI-Δtail, GST-PHAPI-Tail, polyglutamic acid, Smac or BSA had no effect in these assays (Figure 9A and B). These effects of PHAPI on caspase activity appear specific to caspase-3, because no enhancement of the catalytic activity of recombinant caspase-7 or -9 was detected in similar assays (Figure 9C). Moreover, because the recombinant caspase preparations used in these experiments were fully processed enzymes (Figure 9D), this suggests that PHAPI augments caspase activity rather than caspase activation.
Figure 9.

PHAPI specifically enhances the proteolytic activity of caspase-3. (A) The catalytic activity of purified recombinant caspase-3 (5 U/μl) was assayed in the presence or absence of 0.5 μM of the indicated recombinant proteins or BSA. Caspase-3 was allowed to pre-equilibrate with the proteins for 10 min at 4°C prior to addition of Ac-DEVD-AFC substrate. The result shown is representative of four separate experiments. (B) The catalytic activity of purified recombinant caspase-3 (5 U/μl) was assayed in the presence or absence of 0.5 μM of the indicated recombinant proteins. Caspase-3 was allowed to pre-equilibrate with the proteins for 10 min at 4°C prior to addition of Ac-DEVD-AFC substrate. The result shown is representative of three independent experiments. (C) The effect of PHAPI, Smac and control proteins on the catalytic activity of recombinant caspase-7 (0.02 U/μl, left panel) or recombinant caspase-9 (0.05 U/μl; right panel) was assayed as described above. The caspases were allowed to pre-equilibrate with the indicated proteins for 10 min at 4°C prior to addition of Ac-DEVD-AFC or Ac-LEHD substrates. The results shown are representative of three independent experiments. (D) The recombinant caspases used in the above assays were confirmed to be fully processed by immunoblot analysis. In all, 200 U of recombinant caspase-3, 1 U of recombinant caspase-7 and 0.0165 U of recombinant caspase-9 were analyzed by Western blotting, as indicated. As controls, 20 μg of control or activated (treated with 50 μg/ml cytochrome c, 1 mM dATP for 2 h at 37°C) Jurkat cell-free extracts were also run. Blots were probed with a mixture of antibodies that detect the pro-forms as well as the processed forms of caspase-3, -7 and -9. The asterisks denote nonspecific bands recognized by the antibodies.
Discussion
Here we have shown that native apoptosomes derived from Jurkat T cells are largely composed of Apaf-1, mature (proteolytically processed) caspase-9, mature caspase-3 and XIAP (as well as degradation products of this IAP). Mass spectrometry analysis also suggested that proteolytic fragments of RhoGDI2 may be associated with apoptosome preparations from Jurkat cells. However, Western blot analysis of the same complexes failed to confirm the latter interaction. Additional low-molecular-weight proteins may also become incorporated into apoptosomes but due to their low relative abundance within the complex, remain to be identified.
Cytochrome c, while essential for triggering assembly of the apoptosome, did not remain stably associated with the complex and was not required for catalytic activity post-assembly. Assembly of apoptosome complexes in the presence of cytochrome c and dATP was rapid (within minutes) and occurred during incubation at 4°C. The caspase inhibitory protein XIAP was also rapidly recruited to the apoptosome and was found to act as a tether for the stable recruitment of mature caspase-3 to the complex. We have also shown that while the tumor suppressor PHAPI was not stably recruited to apoptosome complexes, it did enhance the activity of the apoptosome very significantly. PHAPI acted in a manner distinct from Smac/DIABLO and these proteins were found to have additive effects on apoptosome activity. We also report that PHAPI was capable of directly enhancing the catalytic activity of free caspase-3. The latter result argues that PHAPI may act as a co-factor for caspase-3 that is capable of sustaining or enhancing the activity of this protease.
Previous studies have ascribed multiple roles to PHAPI, including inhibition of protein phosphatase 2A activity, suppression of oncogene-dependent transformation, inhibition of histone acetyltransferase activity and regulation of microtubule-associated protein binding to microtubules (Chen et al, 1996; Li et al, 1996; Ulitzur et al, 1997; Seo et al, 2002). While it is clearly difficult to reconcile all of these possible functions, the reported tumor-suppressor function of PHAPI is certainly compatible with the observation that this protein can potentiate apoptosis-associated caspase activity. A series of studies have shown that PHAPI displays potent tumor-suppressor activity against a variety of oncogene combinations in co-transformation assays using rodent fibroblast (Chen et al, 1996; Brody et al, 1999). Interestingly, deletion of ∼100 amino acids from the C-terminus of PHAPI completely abolished its inhibitory activity in Ras-Myc transformation assays in rat fibroblasts (Brody et al, 1999). A similar PHAPI-Δtail mutant was used in the present study and this mutant was also found to be devoid of activity towards the apoptosome or purified caspase-3. This suggests that the ability of PHAPI to enhance caspase activity may correlate with its tumor-suppressor activities.
It remains unclear as to precisely how PHAPI influences the catalytic activity of the apoptosome, but our data suggest that PHAPI may enhance the catalytic activity of caspase-3 within the complex. PHAPI-mediated enhancement of caspase-3 catalytic activity would be expected to increase the rate of caspase-9 maturation within the complex through a well-established feedback loop between these caspases (Slee et al, 1999, 2001). Indeed, this is consistent with observations made by Wang and colleagues that caspase-9 maturation was enhanced when PHAPI was added to Hela cell-free extracts (Jiang et al, 2003). Unlike Smac/DIABLO, PHAPI does not appear to mediate its effects through displacement or neutralization of XIAP. Moreover, PHAPI and Smac/DIABLO exerted additive effects on apoptosome activity under conditions where saturating amounts of the latter were employed. Smac/DIABLO had no effect on the activity of purified recombinant caspase-3 as expected, whereas PHAPI enhanced the activity of this caspase. PHAPI did not enhance caspase-3 activation in this context, as the fully processed mature enzyme was used in this assay. One way in which PHAPI might function is through stabilization of caspase-3 dimers. We consistently noted a decline in the activity of recombinant caspase-3 during kinetic peptide hydrolysis assays, suggesting that the enzyme becomes inactivated during incubation at 37°C (Figure 9). The latter effect may be due to dissociation of caspase-3 dimers, but this remains speculative. However, PHAPI sustained the activity of caspase-3 (but not of caspase-7 or caspase-9) in these assays, which leads us to suggest that this protein may stabilize the active caspase-3 zymogen.
Using co-immunoprecipitation analysis, we failed to find evidence for a stable association between PHAPI and the apoptosome, or between PHAPI and active caspase-3 (data not shown). It is possible that the interaction between these proteins is of low affinity and therefore unable to withstand the immunoprecipitation conditions used in this study. Further studies are clearly required to probe the mechanistic details of PHAPI-mediated enhancement of caspase activity. Experiments with a PHAPI deletion mutant lacking the acidic C-terminus of this protein argue that this region of PHAPI is critical for its effects on caspase activity as this mutant had no effect in the assays used. Further studies are clearly required to explore whether PHAPI plays a significant role in setting a threshold for apoptosis in the apoptosome pathway or in other cell death contexts. We suggest that the reported tumor-suppressive effects of PHAPI may be related to the ability of this protein to modulate apoptosis sensitivity by enhancing caspase activity.
Materials and methods
Materials
Mouse anti-caspase-9 antibody used for immunoprecipitation of native apoptosomes was purchased from Upstate Biotechnology (clone 96-2-2). The following antibodies were used for immunoblotting: mouse anti-caspase-9 (Ab-2, Oncogene Research Products), rabbit anti-caspase-9 (Cell Signaling Technology), rabbit anti-caspase-9 D330 (Cell Signaling Technology), rat anti-Apaf-1 (clone 2E12, R&D Systems Inc.), mouse anti-caspase-3 (BD Biosciences), mouse anti-caspase-7 (BD Biosciences), rabbit anti-caspase-7 D198 (Cell signaling Technology), mouse anti-XIAP (clone 48, BD Biosciences), mouse anti-cytochrome c (BD Biosciences), rabbit anti-caspase-3 D175 (Cell Signaling Technology), rat anti-HSP90 (Stressgen), mouse anti-HSP70 (Stressgen), mouse anti-vimentin (Roche), rabbit anti-cIAP2 (R&D Systems Inc.), rabbit anti-RhoGDI (BD Biosciences), goat anti-PHAPI (Santa Cruz Biotechnology), rabbit anti-PHAPI (Biotrend, Germany), mouse anti-Bcl-2 (BD Biosciences), mouse anti-Bcl-XL (BD Biosciences) and rabbit anti-RhoGDI-2 (BD Biosciences). The generation and affinity purification of rabbit polyclonal antibodies against His6-Smac Δ1-55 have been described previously (Adrain et al, 2001) and a similar strategy was used to generate c-IAP1 anti-sera. Briefly, an N-terminal truncated form of c-IAP1 (amino acids 351–618) containing the CARD and RING domains was cloned into pET15b. Recombinant c-IAP1351-618 was expressed in Escherichia coli (BL-21/DE3/pLysS strain), purified over Ni2+-NTA agarose, followed by immunization of rabbits with the purified protein. AFC-coupled synthetic caspase substrate peptides (DEVD, LEHD, YVAD) were purchased from Bachem (UK). Recombinant human caspase-3, -7 and -9 were purchased from Calbiochem (UK). Polyglutamine polymers (15–50 kDa Mr range) were obtained from Sigma (UK).
Assembly and one-step isolation of native apoptosomes from Jurkat cell-free extracts
Cell-free cytosolic S-15 extracts were generated from Jurkat, BJAB or U937 cells as previously described (Slee et al, 2001). Briefly, 5 × 108 cells were packed into a 2 ml Dounce homogenizer and an equal volume of ice-cold cell extract buffer was added (CEB: 20 mM HEPES-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 250 μM PMSF, 1 μg/ml leupeptin, 2 μg/ml aprotinin). Cells were allowed to swell in CEB for 20–30 min on ice and were then lysed by homogenization with ∼15–20 strokes of a B-type pestle. Crude extracts were then centrifuged for 30 min at 15 000 g to remove nuclei, unbroken cells and other debris. Extracts were stored in aliquots at −70°C until required.
To assemble apoptosomes, extracts were incubated at 37°C in the presence of 50 μg/ml bovine heart cytochrome c and 1 mM dATP (the final protein concentration of reactions was 10–12 μg/μl). After 15 min, reactions were placed on ice and insoluble protein complexes were pelleted by centrifugation for 5 min at 20 000 g. The supernatants were collected, diluted in immunoprecipitation (IP) buffer (20 mM HEPES-KOH (pH 7.5), 50 mM NaCl, 0.3% CHAPS, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, I mM DTT, 2 μg/ml aprotinin, 1 μg/ml leupeptin and 250 μM PMSF) and precleared over washed protein A/G agarose beads (Santa Cruz Biotechnology) at 4°C for 1.5 h. A second preclearing step was used in the large-scale immunoprecipitation experiments in order to reduce background binding. Precleared cell-free extracts were incubated with fresh protein A/G agarose beads and anti-caspase-9 antibody (Upstate Biotechnology Inc.) overnight at 4°C. Captured apoptosomes were washed 3 times in IP buffer, and eluted into 1 × SDS-PAGE sample buffer or 2D sample buffer (see below) for analysis by 1D or 2D gels, respectively.
Coupled in vitro transcription/translation
[35S]methionine-labeled proteins were generated using the TNT kit (Promega) as described previously (Martin et al, 1996). Typically, 50 μl reactions were assembled containing 1 μg of template plasmid DNA and 4 μl of translation grade [35S]methionine (1000 μCi/ml, MP Biomedicals, UK).
Caspase substrate hydrolysis assays
For synthetic peptide hydrolysis assays, protein A/G-agarose-immobilized apoptosomes were washed twice in IP buffer and once in CEB, and then resuspended in 45 μl of CEB and transferred to black 96-well fluotrac 200 plates. AFC-coupled peptides (DEVD, LEHD, YVAD) were then added to each well (to a final concentration of 50 μM) and liberation of free AFC was monitored for 1 h at 37°C at excitation and emission wavelengths of 430 and 535 nm, respectively. Recombinant caspase-3, -7 and -9 were assayed in a buffer containing 50 mM HEPES (pH 7.4), 75 mM NaCl, 0.1% CHAPS, 10% glycerol, 1 mM DTT, 250 μM PMSF, 1 μg/ml leupeptin and 2 μg/ml aprotinin, with 50 μM Ac-DEVD-AFC or Ac-LEHD-AFC as a substrate.
Radiolabeled caspase substrate hydrolysis assays were performed as follows. Immunoprecipitated apoptosome complexes were washed as above and brought up in CEB containing in vitro transcribed and translated substrates (in a final reaction volume of 10 μl). Reactions were allowed to proceed for 2 h at 37°C and were stopped by addition of SDS–PAGE sample buffer, followed by analysis by SDS–PAGE/autoradiography.
2D gel electrophoresis
For 2D gel electrophoresis, isolated apoptosomes were eluted in 2D sample buffer (8 M urea, 4% CHAPS, 100 mM DTT, 0.05% SDS, 0.5% ampholyte 3–10 and a trace of bromophenol blue). Samples were rehydrated into IPG strips (BioRad), in 350 or 120 μl of 2D sample buffer for 18 or 7 cm strips, respectively. Passive sample rehydration into IPG strips was performed at room temperature for 6 h or overnight; mineral oil was used to overlay the strips for overnight rehydrations. Isoelectric point focusing (IEF) was performed in a BioRad Protean IEF Cell under the following conditions for 18 cm strips: (1) linear voltage ramp to 500 V over 1 h, (2) 5 h at 500 V, (3) linear voltage ramp to 3500 V over 5 h and (4) 12 h at 3500 V. For 7 cm strips the program used was (1) linear voltage ramp to 250 V over 30 min, (2) linear ramp to 3000 V over 1.5 h and (3) 4 h at 3000 V. Following IEF, the IPG strips were reduced and alkylated with 2% DTT and 2.5% IAA, respectively, in 5 min incubations in an equilibration buffer that contained 6 M urea, 375 mM Tris–HCl (pH 8.8), 2% SDS and 20% glycerol. Strips were then mounted on 12% SDS–PAGE gels using easymelt agarose (Biorad, UK) and electrophoresed at 37.5 mA per gel in a Biorad Protean II xi electrophoresis cell (Biorad, UK). 2D gels were stained using a mass spectrometry-compatible silver-staining protocol that is based on a modification of the EMBL silver-staining protocol (Mortz et al, 2001) or were transferred onto nitrocellulose membranes (Protran, Schleicher & Schuell) for immunoblotting.
In-gel protein digestion and protein identification by MALDI-TOF mass spectrometry
Protein spots were manually excised from 2D gels and were washed extensively in distilled water. Gel pieces were then incubated at room temperature on a shaking platform in oxidation buffer (15 mM K3Fe(CN)6, 50 mM Na2S2O3) until the spots were completely destained. Gel pieces were then washed five times (5–10 min per wash) in 50% methanol/10% acetic acid. Samples were then incubated in 50 mM NH4HCO3 for 5 min, prior to dehydrating in 100% acetonitrile. To further dehydrate the pellets, the acetonitrile was aspirated off and the tubes were spun in a speed-vac (ThermoSavant) for 5 min at room temperature. For trypsin digestion, a 100 μg/ml aliquot of Sequencing grade trypsin (Roche) dissolved in 1 mM HCl was diluted 1:10 in digestion buffer (25 mM NH4HCO3, 0.1 n-octyl β-D-glucopyranoside). Typically, for low-abundance silver-stained spots, 2 μl of trypsin solution (20 ng) was pipetted directly onto the desiccated gel piece. After allowing the gel piece to rehydrate for 5 min, a further 10 μl of digestion buffer was added and samples were incubated overnight at 37°C. Following trypsin digestion, peptides were extracted twice into 40 μl 66% acetonitrile/0.1% trifluoroacetic acid in a sonicating water bath, followed by lyophilization in a speed-vac at room temperature.
For mass-spectrometric analysis, peptides were solubilized by sonication in 5 μl of 5% formic acid. Digested samples (0.5–1 μl) were applied to a Teflon-coated 96-well MALDI target plate (Applied Biosciences, UK), followed by the addition of 0.5–1 μl of a 10 mg/ml matrix solution of α-cyano-4-hydroxy-cinnamic acid in 60% acetonitrile/0.1% trifluoroacetic acid. Samples were allowed to air-dry at room temperature before analysis in positive reflectron mode in a Voyager DE Pro MALDI mass spectrometer (Applied Biosciences, UK).
Expression of recombinant proteins
Recombinant His6-Smac (Δ1-55), His6-PHAPI, His6-PHAPI-Δtail (aa 1–163), GST and GST-PHAPI-Tail (aa 164–249) were prepared by transforming pET15b-Smac, pQE30-PHAPI plasmids or pGEX4T2-PHAPI-Tail into BL21 E. coli. GST-tagged and polyhistidine-tagged proteins were expressed and purified as described previously (Adrain et al, 1999, 2001).
Acknowledgments
We thank Dr Xiaodong Wang (HHMI, University of Texas Southwestern Medical Center) and Dr Suzanne Pfeffer (Department of Biochemistry, Stanford) for provision of plasmids. We are grateful to the Science Foundation Ireland (PI1/B038) for its generous support to this work and to the European Community (QLG1-1999-00739). SJM is a Science Foundation Ireland Fellow.
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell 9: 423–432 [DOI] [PubMed] [Google Scholar]
- Adrain C, Creagh EM, Martin SJ (2001) Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J 20: 6627–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrain C, Slee EA, Harte MT, Martin SJ (1999) Regulation of apoptotic protease activating factor-1 oligomerization and apoptosis by the WD-40 repeat region. J Biol Chem 274: 20855–20860 [DOI] [PubMed] [Google Scholar]
- Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR (2000) Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol 2: 469–475 [DOI] [PubMed] [Google Scholar]
- Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM (2001) Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J 20: 998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody JR, Kadkol SS, Mahmoud MA, Rebel JM, Pasternack GR (1999) Identification of sequences required for inhibition of oncogene-mediated transformation by pp32. J Biol Chem 274: 20053–20055 [DOI] [PubMed] [Google Scholar]
- Cain K, Bratton SB, Langlais C, Walker G, Brown DG, Sun XM, Cohen GM (2000) Apaf-1 oligomerizes into biologically active approximately 700-kDa and inactive approximately 1.4-MDa apoptosome complexes. J Biol Chem 275: 6067–6070 [DOI] [PubMed] [Google Scholar]
- Cain K, Brown DG, Langlais C, Cohen GM (1999) Caspase activation involves the formation of the aposome, a large (approximately 700 kDa) caspase-activating complex. J Biol Chem 274: 22686–22692 [DOI] [PubMed] [Google Scholar]
- Chau BN, Cheng EH, Kerr DA, Hardwick JM (2000) Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol Cell 6: 31–40 [PubMed] [Google Scholar]
- Chen TH, Brody JR, Romantsev FE, Yu JG, Kayler AE, Voneiff E, Kuhajda FP, Pasternack GR (1996) Structure of pp32, an acidic nuclear protein which inhibits oncogene-induced formation of transformed foci. Mol Biol Cell 7: 2045–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZL, Pio F, Xie Z, Welsh K, Krajewska M, Krajewski S, Godzik A, Reed JC (2001) A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis. J Biol Chem 276: 9239–9245 [DOI] [PubMed] [Google Scholar]
- Cohen GM (1997) Caspases: the executioners of apoptosis. Biochem J 326: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC (1997) X-linked IAP is a direct inhibitor of cell death proteases. Nature 388: 300–304 [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102: 33–42 [DOI] [PubMed] [Google Scholar]
- Jiang X, Kim HE, Shu H, Zhao Y, Zhang H, Kofron J, Donnelly J, Burns D, Ng SC, Rosenberg S, Wang X (2003) Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299: 223–226 [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X (2000) Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J Biol Chem 275: 31199–31203 [DOI] [PubMed] [Google Scholar]
- Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD (1997) The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science 275: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD (2002) Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111: 331–342 [DOI] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z (1996) Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry 35: 6998–7002 [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91: 479–489 [DOI] [PubMed] [Google Scholar]
- Martin SJ, Amarante-Mendes GP, Shi L, Chuang TH, Casiano CA, O'Brien GA, Fitzgerald P, Tan EM, Bokoch GM, Greenberg AH, Green DR (1996) The cytotoxic cell protease granzyme B initiates apoptosis in a cell-free system by proteolytic processing and activation of the ICE/CED-3 family protease, CPP32, via a novel two-step mechanism. EMBO J 15: 2407–2416 [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Green DR (1995) Protease activation during apoptosis: death by a thousand cuts? Cell 82: 349–352 [DOI] [PubMed] [Google Scholar]
- Mortz E, Krogh TN, Vorum H, Gorg A (2001) Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1: 1359–1363 [DOI] [PubMed] [Google Scholar]
- Na S, Chuang TH, Cunningham A, Turi TG, Hanke JH, Bokoch GM, Danley DE (1996) D4-GDI, a substrate of CPP32, is proteolyzed during Fas-induced apoptosis. J Biol Chem 271: 11209–11213 [DOI] [PubMed] [Google Scholar]
- Pan G, O'Rourke K, Dixit VM (1998) Caspase-9, Bcl-XL, and Apaf-1 form a ternary complex. J Biol Chem 273: 5841–5845 [DOI] [PubMed] [Google Scholar]
- Pandey P, Saleh A, Nakazawa A, Kumar S, Srinivasula SM, Kumar V, Weichselbaum R, Nalin C, Alnemri ES, Kufe D, Kharbanda S (2000) Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. EMBO J 19: 4310–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y (1999) Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev 13: 3179–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES (2000) Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat Cell Biol 2: 476–483 [DOI] [PubMed] [Google Scholar]
- Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D (2002) Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem 277: 14005–14010 [DOI] [PubMed] [Google Scholar]
- Slee EA, Adrain C, Martin SJ (2001) Executioner caspases-3, -6, and -7 perform distinct, non-redundant, roles during the demolition phase of apoptosis. J Biol Chem 276: 7320–7326 [DOI] [PubMed] [Google Scholar]
- Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, Green DR, Martin SJ (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol 144: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS (1999) Catalytic properties of the caspases. Cell Death Differ 6: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Ulitzur N, Humbert M, Pfeffer SR (1997) Mapmodulin: a possible modulator of the interaction of microtubule-associated proteins with microtubules. Proc Natl Acad Sci USA 94: 5084–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53 [DOI] [PubMed] [Google Scholar]
- Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X (1997) Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275: 1129–1132 [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X (1999) An APAF-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem 274: 11549–11556 [DOI] [PubMed] [Google Scholar]


