Abstract
Background
Late embryogenesis abundant (LEA) proteins are involved in protecting higher plants from damage caused by environmental stresses. Foxtail millet (Setaria italica) is an important cereal crop for food and feed in semi-arid areas. However, the molecular mechanisms underlying tolerance to these conditions are not well defined.
Results
Here, we characterized a novel atypical LEA gene named SiLEA14 from foxtail millet. It contains two exons separated by one intron. SiLEA14 was expressed in roots, stems, leaves, inflorescences and seeds at different levels under normal growth conditions. In addition, SiLEA14 was dramatically induced by osmotic stress, NaCl and exogenous abscisic acid. The SiLEA14 protein was localized in the nucleus and the cytoplasm. Overexpression of SiLEA14 improved Escherichia coli growth performance compared with the control under salt stress. To further assess the function of SiLEA14 in plants, transgenic Arabidopsis and foxtail millet plants that overexpressed SiLEA14 were obtained. The transgenic Arabidopsis seedlings showed higher tolerance to salt and osmotic stress than the wild type (WT). Similarly, the transgenic foxtail millet showed improved growth under salt and drought stresses compared with the WT. Taken together, our results indicated that SiLEA14 is a novel atypical LEA protein and plays important roles in resistance to abiotic stresses in plants.
Conclusion
We characterized a novel atypical LEA gene SiLEA14 from foxtail millet, which plays important roles in plant abiotic stress resistance. Modification of SiLEA14 expression may improve abiotic stress resistance in agricultural crops.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-014-0290-7) contains supplementary material, which is available to authorized users.
Background
Environmental stresses, such as drought and high salinity, can cause severe damage to plants, leading to considerable reduction in their productivity. To survive under such conditions, plants have developed a series of defense responsive pathways. Among them, Ca2+-dependent signaling leads to the activation of late embryogenesis abundant (LEA)-type genes, which may function in protection and damage repair of plants [1].
LEA proteins were first identified in cotton seeds [2]. The proteins accumulated to high levels in the late stages of seed development [3,4]. Subsequently, they were found to be expressed in vegetative tissues [5,6] and could be induced by abscisic acid (ABA) and various abiotic stresses, such as drought and cold [5-9]. With the development of deep sequencing technology, an increasing number of LEA proteins have been identified. On the basis of their amino acid sequence similarities and conserved motifs, LEA proteins are categorized into different groups [10-13]. In this work, we adopt the classification introduced by Battaglia’s group, in which LEA proteins are categorized into seven distinct families [12]. Groups 1, 2, 3, 4, 6 and 7, which share specific motifs within each group, are considered to be hydrophilic or “typical” LEA proteins. Conversely, group 5 corresponds to atypical LEA proteins. This group includes all LEA proteins with higher content of hydrophobic residues than typical LEA proteins. On the basis of their sequence similarity, group 5 LEA proteins are divided into the subgroups 5A, 5B, and 5C, corresponding to the first described proteins D-34, D-73, and D-95 [3,14] in this group, respectively. Physicochemical properties show that group 5 LEA proteins are not soluble after boiling, suggesting that they may adopt a globular conformation and are not heat stable [3,15,16]. Subsequent reports show that subgroup 5C LEA proteins are natively folded and have more β-sheets than α-helixes [5,17], which is different from subgroups 5A and 5B LEA proteins that are intrinsically unstructured [17-19]. For example, Arabidopsis LEA14 has an αβ-fold consisting of one α-helix and seven β-strands that form two antiparallel β-sheets as determined by nuclear magnetic resonance spectroscopy [16]. Moreover, subgroup 5C LEA proteins have other outstanding characteristics, such as lower instability index, narrower range of GRAVY values, and lower proportion of polar (hydrophilic) and small residues, but higher proportion of non-polar residues than subgroup 5A and 5B members [5]. All of these differences indicate that subgroup 5C LEA proteins may function differently from other LEA proteins in plants.
At present, only a small number of subgroup 5C LEA genes have been characterized. Their transcripts can be upregulated in response to diverse stresses, as reported for cotton LEA14-A [15], Craterostigma plantagineum PcC27-45 [20], soybean D95-4 [21], tomato ER5 [22], hot pepper CaLEA6 [23], Arabidopsis LEA14 [24] and At2g44060 [17], and sweetpotato IbLEA14 [25]. Overexpression of CaLEA6 in tobacco improves tolerance to dehydration and NaCl but not to low temperature [23]. Transgenic sweetpotato non-embryogenic calli that overexpress IbLEA14 show increased tolerance to drought and salt stress by enhancing lignification [25]. Recently, rice OsLEA5 has been reported to enhance resistance against diverse abiotic stresses in recombinant Escherichia coli cells. In vitro analysis showed that OsLEA5 was able to protect lactate dehydrogenase from aggregation under different abiotic stresses [5]. All these results suggest that subgroup 5C LEA proteins are closely associated with resistance to multiple abiotic stresses.
Foxtail millet (Setaria italica (L.) Beauv.), a member of the Poaceae family, has a long history in cultivation of about 7000 years. It has been widely planted in northern China and other Asian countries. Recently, a draft genome sequence for foxtail millet has been completed [26], which enables foxtail millet to be a tractable experimental grass model [27]. As a diploid C4 panicoid crop species, foxtail millet is well known for its remarkable drought resistance. However, the molecular mechanisms underlying this tolerance are not well defined.
In this study, we isolated and functionally characterized a novel member of the atypical subgroup 5C LEA gene, SiLEA14, from foxtail millet. The expression of SiLEA14 was induced by ABA, polyethylene glycol (PEG) and NaCl. Overexpression of SiLEA14 resulted in enhanced resistance to abiotic stresses in E. coli, Arabidopsis and foxtail millet. The SiLEA14 promoter mediated remarkable induction of β-glucuronidase (GUS) expression in transgenic Arabidopsis under various stresses. Cis-acting regulatory elements in the SiLEA14 promoter were also predicted. These data reveal the potential application of SiLEA14 in the genetic engineering of other crops.
Results
SiLEA14 is an atypical LEA protein
The full-length sequence of SiLEA14 was determined by 5′ and 3′ rapid amplification of cDNA ends (RACE) [GenBank: KJ767551]. The sequence is 821 bp in length, with a 100 bp 5′ untranslated region (UTR) and a 208 bp 3′ UTR (with polyA tail) (Additional file 1A). SiLEA14 harbors two exons separated by an intron (Additional file 1B), and encodes an open reading frame of 170 aa with a predicted molecular mass of 18.77 kD and pI of 5.56. It is rich in Ser (10.6%), Lys (8.8%), and Ile (8.2%), but contains low quantities of Trp (1.2%), Asn (1.8%), Cys (1.8%), and Gln (1.8%). Three hydrophobic regions (I, II, and III) were identified in the SiLEA14 protein (Additional file 1C). A motif search of the SiLEA14 protein in InterProScan revealed that it contains a “LEA_2” motif (PF03186, 3.4e-20), which was classified into subgroup 5C (D-95) according to Battaglia’s classification of LEA proteins [12]. Further analysis showed that SiLEA14 contains a lower percentage of polar amino acids (47.2%) and higher percentage of non-polar amino acids (25.8%) than other group LEA proteins (Additional file 1D) [5]. The GRAVY value and the instability index of SiLEA14 is −0.155 and 34.68, respectively (Additional file 1D). All these characteristics were in accordance with those of other members of subgroup 5C [5].
Two additional SiLEA14 homologs (Si003774m and Si003233m, Additional file 2) existed in foxtail millet. They showed 57.7% and 48.24% identity to SiLEA14, respectively. Homologous sequences of SiLEA14 were also present in other plant species, such as rice OsLEA5 (77.06%), maize Lea14-A (50.88%) and Arabidopsis AtLEA14 (47.06%). The homologs shared different levels of sequence similarities, indicating different levels of evolutionary relationship among these proteins (Figure 1A). A phylogenetic tree for SiLEA14 and its homologs was constructed from protein sequences (Additional file 2). The proteins were mainly separated into two clades comprising dicot and monocot species, respectively. The SiLEA14 protein shared the closest relationship with the pearl millet LEA-like protein, followed by OsLEA5 (Figure 1B). Sequence logos were produced to examine the conservation level at each residue position. The sequences lacked significant signature motifs or consensus sequences, except for several conserved residues at specific positions, such as proline at positions 41 and 83 (Additional file 3). This is a common feature of subgroup 5C LEA proteins [5].
Figure 1.
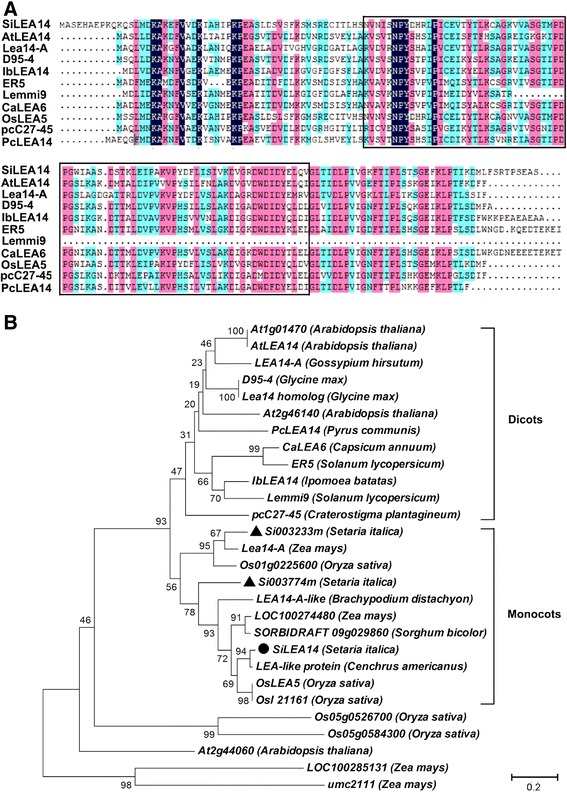
Multiple sequence alignment and phylogenetic relationship for the SiLEA14 protein and its homologs. (A) Multiple sequence alignment of SiLEA14 with its homologs from various plant species. The conserved “LEA_2” motif (PF03168) is boxed. (B) Phylogenetic relationships of SiLEA14 and its homologs. The divergence of the clades between the monocots and dicots is highlighted. SiLEA14 and its two homologs in foxtail millet are highlighted by solid dark circle and triangle, respectively. The GenBank accession numbers are as follows: SiLEA14 (Setaria italic, KJ767551), AtLEA14 (Arabidopsis thaliana, NM_100029), Lea14-A (Zea mays, NM_001159174), D95-4 (Glycine max, U08108), IbLEA14 (Ipomoea batatas, GU369820), ER5 (Solanum lycopersicum, U77719), Lemmi9 (Solanum lycopersicum, Z46654), CaLEA6 (Capsicum annuum, AF168168), OsLEA5 (Oryza sativa, JF776156), pcC27-45 (Craterostigma plantagineum, M62990), pcLEA14 (Pyrus communis, AF386513), At1g01470 (Arabidopsis thaliana, BT015111), LEA14-A (Gossypium hirsutum, M88322), Lea14 homolog (Glycine max, NM_001251780), At2g46140 (Arabidopsis thaliana, NM_130176), Os01g0225600 (Oryza sativa, NM_001048996), LEA14-A-like (Brachypodium distachyon, XM_003567779), LOC100274480 (Zea mays, NM_001148839), SORBIDRAFT 09 g029860 (Sorghum bicolour, XM_002441543), LEA-like protein (Cenchrus americanus, AY823547), OsI21161 (Oryza sativa, CM000130), Os05g0526700 (Oryza sativa, NM_001062639), Os05g0584300 (Oryza sativa, NM_001062985), At2g44060 (Arabidopsis thaliana, BT024723), LOC100285131 (Zea mays, EU970969) and umc2111 (Zea mays, NM_001155750). The sequences used for alignment and phylogenetic tree construction were shown in Additional file 2.
Subcellular localization of SiLEA14
To investigate the potential role of SiLEA14, we examined the subcellular localization of SiLEA14 fused to GFP and GFP alone (as a control) in onion epidermal cells and foxtail millet root protoplasts. The constructs used were shown in Figure 2A. When observed by confocal microscopy, the green fluorescent protein (GFP) fluorescence of SiLEA14-GFP was distributed throughout the cell, consistent with the GFP control, both in the onion cells (Figure 2B) and the foxtail millet root protoplasts (Figure 2C). These results indicated that SiLEA14 was localized in the cytosol.
Figure 2.

Subcellular localization of SiLEA14 protein. (A) Diagram of the constructs of pROK219-GFP and pROK219-SiLEA14-GFP used for subcellular localization. 35S-Pro, cauliflower mosaic virus 35S promoter; SiLEA14, the SiLEA14 coding region without the stop codon; GFP, green fluorescent protein; NOS, nopaline synthase terminator. (B-C) Fluorescent microscopic images of GFP and SiLEA14-GFP fusion protein in the onion epidermal cells (Bar = 200 μm) (B) and in the foxtail millet root protoplasts (Bar = 10 μm) (C).
SiLEA14 expression profiles under normal and stress conditions
Quantitative reverse-transcription PCR (qRT-PCR) was carried out to reveal the temporal and spatial expression of SiLEA14 in foxtail millet (Figure 3A). SiLEA14 was expressed at the highest level in roots, followed by stems and leaves, and at the lowest level in inflorescences. After pollination, SiLEA14 transcription was gradually upregulated with seed maturation, which indicated a potential role for SiLEA14 in the maturation and desiccation phases of foxtail millet seed development.
Figure 3.
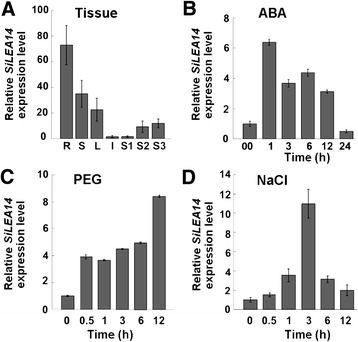
SiLEA14 expression patterns in foxtail millet determined by qRT-PCR. (A) Tissue-specific expression of SiLEA14. Total RNA was extracted from roots (R), stems (S), leaves (L), inflorescences (I) and seeds at 5 (S1), 15 (S2) and 25 (S3) days after pollination, respectively. Foxtail millet Actin7 was used as an internal control. (B-D) SiLEA14 transcripts accumulation in response to various treatments. Two-week-old foxtail millet seedlings were treated with 100 μM ABA (B), 20% (v/v) PEG 6000 (C) and 250 mM NaCl (D) for the indicated times, respectively. Total RNA was extracted from the above-ground parts of ten plants at the indicated times after the treatments. Data represent means and standard errors for three biological replicates.
The expression level of SiLEA14 was also examined by qRT-PCR under various stresses. Under ABA treatment, SiLEA14 transcription abruptly reached its highest level (six-fold) after 1 h treatment, and then decreased gradually and reverted almost to the control level at 24 h (Figure 3B). Under PEG stress, SiLEA14 transcription was rapidly induced within 0.5 h and increased to a peak level (8-fold) at 12 h (Figure 3C). Salt treatment resulted in increased accumulation of SiLEA14 to a maximum level (11-fold) only after 3 h (Figure 3D). These results indicated that SiLEA14 might play important roles in the responses to salt and drought stresses.
SiLEA14 enhances salt tolerance in transformed E. coli
In both prokaryotes and eukaryotes LEA proteins might have similar protective mechanisms [28]. Therefore, the up-regulation of SiLEA14 in response to NaCl prompted us to evaluate the salt stress tolerance of recombinant E. coli overexpressing SiLEA14 (Figure 4). There are no significant differences in colony number between transformed E. coli harboring SiLEA14 and the control under normal conditions, indicating that overexpression of SiLEA14 did not affect the growth of E. coli recombinants in non-stress conditions. However, when grown on Luria–Bertani (LB) plates supplemented with 600 mM NaCl, the number of transformed cells was much higher than that of the control. A similar result was obtained on LB medium supplemented with 600 mM KCl. These results demonstrated that overexpression of SiLEA14 in E. coli significantly enhanced tolerance to salt stress.
Figure 4.
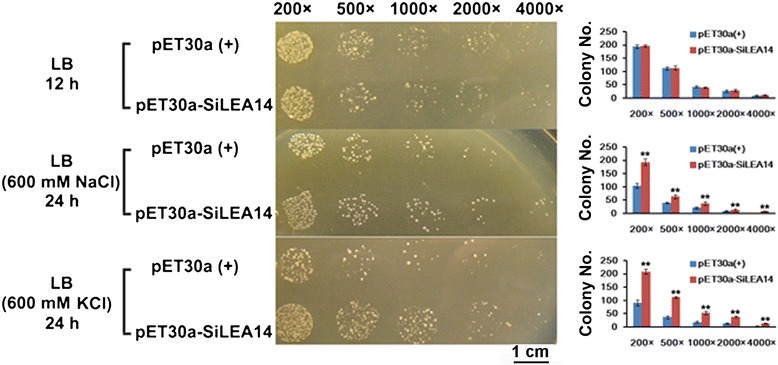
Assay for salt stress tolerance of SiLEA14 transformed E. coli . Transformant E. coli (BL21) with pET30a-SiLEA14 were grown on LB plates with or without 600 mM NaCl and KCl for 12 and 24 h, respectively. Transformant with empty vector pET30a(+) was used as a control. The number of colonies on the plate was calculated. The 200×, 500×, 1000×, 2000× and 4000× represent the dilution fold. Bar = 1 cm. Data represent means and standard errors for three biological replicates. Statistical significance was determined by Student’s t-test. **P < 0.01.
SiLEA14 improves the abiotic stress resistance of transgenic Arabidopsis
To assess the function of SiLEA14 in plants, it was overexpressed in Arabidopsis under the control of Super promoter (Figure 5A). At least 30 transgenic plants were obtained, and two independent homozygous T3 transgenic lines (L6 than L9) with high expression levels of SiLEA14 (Figure 5B) were chosen for further investigation.
Figure 5.
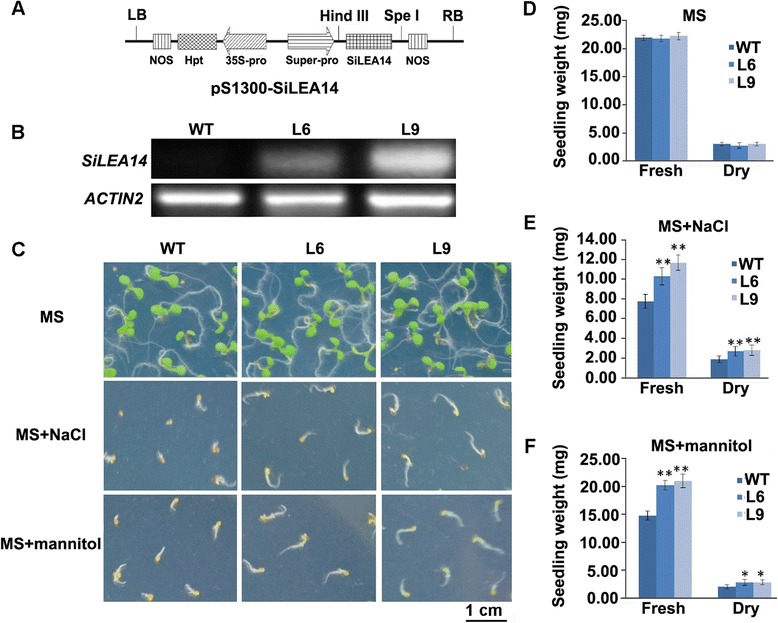
Assay for salinity and mannitol stress tolerance of SiLEA14 overexpression Arabidopsis. (A) Diagram of the T-DNA region of the binary vector pS1300-SiLEA14. LB, left border; RB, right border; 35S-Pro, cauliflower mosaic virus 35S promoter; Hpt, hygromycin B phosphotransferase gene; Super-pro, Super promoter; SiLEA14, SiLEA14 gene; NOS, nopaline synthase terminator. (B) RT-PCR analysis of SiLEA14 in transgenic Arabidopsis lines (L6 and L9). WT was used as a control. Actin2 was a loading control. (C) The phenotype of transgenic and WT Arabidopsis germinated on MS, MS + 125 mM NaCl and MS + 250 mM mannitol, respectively, for 7 days. This experiment had three replicates, and each experiment comprised 30–40 plants. Bar = 1 cm. (D-F) Fresh/dry weights of seedlings germinated on MS, MS + 125 mM NaCl and MS + 250 mM mannitol, respectively. Data represent means and standard errors for three biological replications. Statistical significance was determined by Student’s t-test. *P < 0.05; **P < 0.01.
Under non-stress conditions, no significant differences in the fresh and dry weights were observed between the WT and transgenic plants (Figure 5C and D). However, when grown on Murashige and Skoog (MS) medium supplemented with 125 mM NaCl, the transgenic seedlings showed significantly larger cotyledons and longer roots than those of the WT, even though growth of these organs of the transgenic and WT plants was inhibited compared with normal conditions (Figure 5C). Consistently, the fresh and dry weights of transgenic seedlings were significantly higher than that of the WT (Figure 5E). Similar results were obtained when seedlings were treated with 250 mM mannitol (Figure 5C and F). These results suggest that SiLEA14 may enhance the osmotic stress resistance of transgenic Arabidopsis.
Overexpression of SiLEA14 increases the salt tolerance of transgenic foxtail millet
To further analyze the function of SiLEA14, it was transformed into foxtail millet under the control of ubiquitin promoter (Figure 6A). The SiLEA14 integration was confirmed by genomic PCR using two pairs of primer sets specific to Hpt and 35S promoter, respectively (Additional file 4). QRT-PCR analysis showed higher expression level of SiLEA14 in transgenic lines L68, L76, and L78, which showed a 3:1 segregation ratio at T1 generation, than that of the WT (Figure 6B). The T2 generation of these three lines were used for further analysis.
Figure 6.
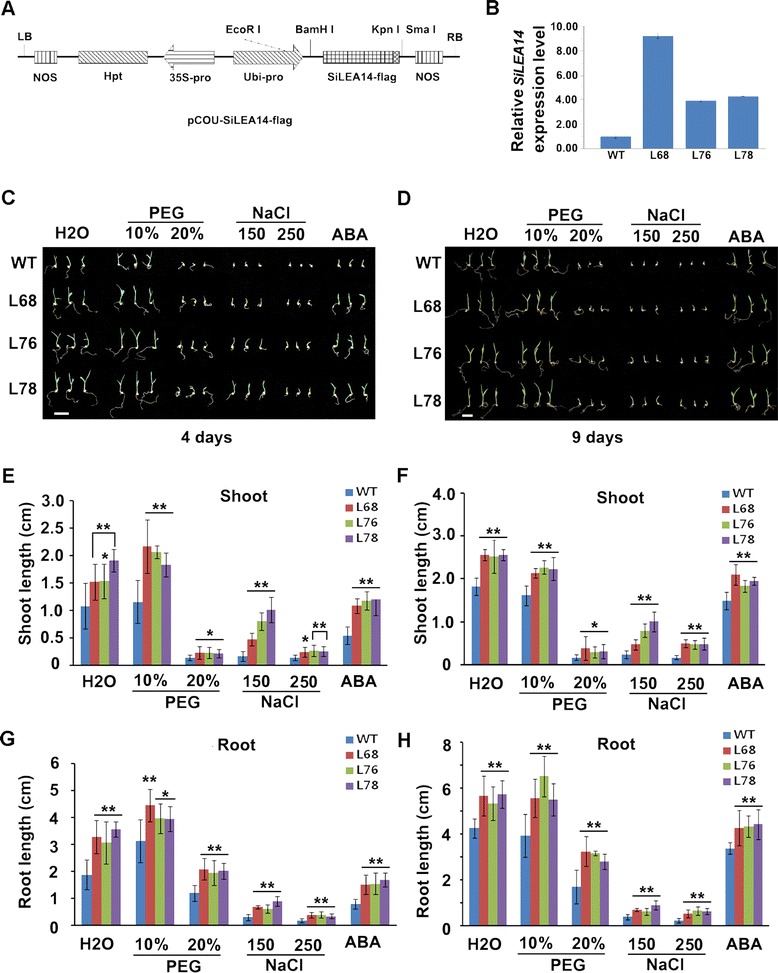
Assay for NaCl, PEG and ABA stress on SiLEA14 overexpression foxtail millet germination. (A) Diagram of the T-DNA region of the binary vector pCOU-SiLEA14-flag. LB, left border; RB, right border; 35S-Pro, cauliflower mosaic virus 35S promoter; Hpt, hygromycin B phosphotransferase gene; Ubi-pro, maize Ubiquitin promoter; SiLEA14-flag, SiLEA14 gene fused with a flag tag; NOS, nopaline synthase terminator. (B) QRT-PCR analysis of SiLEA14 in transgenic foxtail millet lines (L68, L76 and L78). Actin7 was used as an internal control. (C-D) The phenotype of transgenic and WT foxtail millet under various abiotic stress treatment during the germination stage. The T2 seeds soaked in water (as control) or in water containing 150 mM NaCl, 250 mM NaCl, 10% PEG, 20% PEG and 10 μM ABA for 1 day at 30°C and then placed on the filter paper in a Petri dish wet with the same solutions mentioned above for 4 and 9 days, respectively. This experiment had three replicates, and each experiment comprised 30–40 plants. Bar = 1 cm. (E-F) The shoot length of transgenic and WT foxtail millet germinated under above conditions for 4 and 9 days, respectively. (G-H) The root length of transgenic and WT foxtail millet germinated under above conditions for 4 and 9 days, respectively. Data in B, E-H represent means and standard errors for three biological replicates. Statistical significance was determined by Student’s t-test. *P < 0.05; **P < 0.01.
First, we examined the salt tolerance of transgenic foxtail millet during germination, and the results were shown in Figure 6. Compared with the WT plants, transgenic lines showed better growth performance (Figure 6C) with enhanced shoot and root growth (Figure 6E and G) when germinated in water for 4 days. Under salt stress for 4 days, the transgenic lines showed significantly longer shoot and root than the WT (Figure 6E and G), even though the growth of both WT and transgenic lines were seriously suppressed (Figure 6C). Similar results were obtained for 9 days salt stress treatment (Figure 6D, F and H).
Further, we examined the salt tolerance of transgenic foxtail millet seedlings in soil. No striking differences in the height were observed between the transgenic and the WT plants under non-stress conditions, but wider leaves were observed in the transgenic foxtail millet compared with those of the WT (Figure 7A). This finding is in accordance with the result that transgenic lines showed longer shoot and root lengths when germinated in water compared with the WT (Figure 6C–H). Under 150 mM NaCl stress for 6 days, the first and second leaves of WT plants became curled, whereas there was no obvious effect on the transgenic lines. The WT plants were shorter than those of the transgenic lines. Under 250 mM NaCl stress for 6 days, the growth of WT plants was severely inhibited and leaves were curled, bleached, and senescent. However, the transgenic lines showed healthy growth except that only a few old leaves became bleached (Figure 7A).
Figure 7.
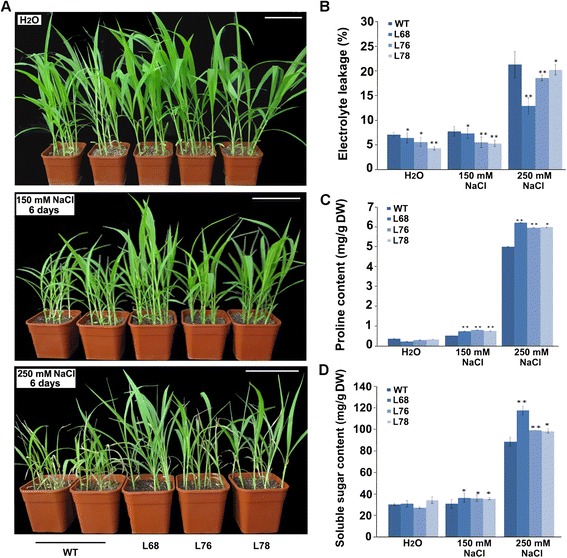
Salt tolerance of SiLEA14 overexpressing foxtail millet seedling. (A) The phenotype of transgenic and WT foxtail millet under normal and salt stress condition. Two-week-old seedlings in soil were irrigated with water, 150 and 250 mM NaCl solution every 3 days. After 6 days, the phenotypes of the transgenic lines and WT were investigated. This experiment had three replicates; and six to eight plants grown in one plot were used in each experiment. Bar = 10 cm. (B) Relative electrolyte leakage in WT and transgenic lines after salt stress. (C) Proline content in WT and transgenic foxtail millet after salt stress. (D) Soluble sugar content in WT and transgenic foxtail millet after salt stress. Data in B-D represent means and standard errors for three biological replicates. Statistical significance was determined by Student’s t-test. *P < 0.05; **P < 0.01.
Electrolyte leakage always occurs following membrane damage under salinity stress [29]. Furthermore, plants accumulate several metabolites, such as amino acids (e.g., proline), sugars, and sugar alcohols (e.g., mannitol and trehalose), to prevent detrimental changes caused by severe osmotic stress [30,31]. Therefore, we measured the electrolyte leakage and change in free proline and soluble sugar contents in control and SiLEA14 transgenic lines with or without salinity stress, respectively. Under optimal or salt-stress conditions, the amount of electrolyte leakage in the transgenic lines was significantly lower than that of the WT plants (Figure 7B). This finding is in agreement with the result that the SiLEA14 transgenic lines showed enhanced germination and growth compared with the WT under both normal and salt-stress conditions (Figures 6C–H, 7A). No significant differences in free proline and soluble sugar contents were observed between the WT and the transgenic lines under normal conditions. Under 150 and 250 mM NaCl stress, the free proline and soluble sugar contents increased in all plants. However, this increase was significantly more pronounced in the transgenic lines than in the WT plants (Figure 7C, D). All of these increased osmotic protectants are beneficial for protection of the plants against salt stress. Moreover, we noted that the transgenic line L68 demonstrated better salt tolerance than the other two transgenic lines (L76 and L78), which correlated with the higher accumulation of SiLEA14 transcripts in L68. All of these results indicated that SiLEA14 overexpressing foxtail millet is more tolerant to salt stress compared with the WT.
Overexpression of SiLEA14 increases drought resistance of transgenic foxtail millet
We also examined the drought resistance of transgenic foxtail millet during germination. When germinated in water for 4 days, the transgenic lines showed better growth performance than WT as described above (Figure 6C, E and G). Compared with in water, no obvious suppression under 10% PEG stress but serious suppression under 20% PEG stress for 4 days were observed (Figure 6C). The shoot and root length of transgenic lines was longer than that of the WT no matter germinated in water, 10% or 20% PEG (Figure 6E and G). When germinated in water or PEG for 9 days, similar results were obtained (Figure 6D, F and H).
The observation that SiLEA14 improved osmotic stress resistance during germination prompted us to investigate the drought resistance of transgenic foxtail millet seedlings in soil. As it is shown in Figure 8A, no significant differences in the height were observed between the transgenic lines and the WT when watered well, except that the leaves of the transgenic lines were wider. However, under drought stress for 7 days, more than 90% of WT leaves, but less than 20% of transgenic plant lines leaves, became curled and wilted. After rewatering, 74–100% plants of trangenic lines survived in contrast to only 14% of WT (Figure 8B).
Figure 8.
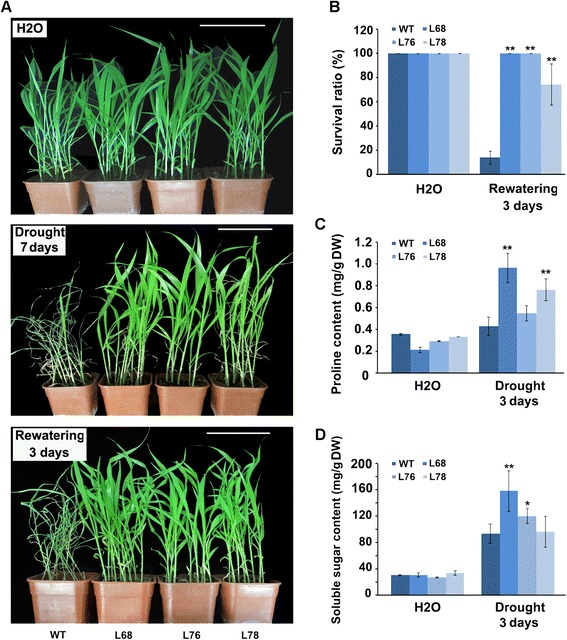
Drought tolerance of SiLEA14 overexpression foxtail millet in soil. (A) The phenotypes of WT and transgenic foxtail millet under normal condition, drought stress for 7 days and after rewatering for 3 days. Two-week-old seedlings were used. Six to ten plants grown in one plot were used in each experiment. This experiment had three replicates. Bar = 10 cm. (B) The survival rate of WT and transgenic foxtail millet plants in (A) under drought stress. (C) Free proline content in WT and transgenic plants after drought stress. (D) Soluble sugar content in WT and transgenic plants after drought stress. Data in B-D represent means and standard errors for three biological replicates. Statistical significance was determined by Student’s t-test. *P < 0.05; **P < 0.01.
The change in free proline and soluble sugar contents in control and SiLEA14 transgenic lines with or without drought stress was examined. No significant differences in free proline and soluble sugar contents were observed between the WT and the transgenic lines under non-stress conditions. Under drought stress for 3 days, the free proline and soluble sugar contents increased in all WT and transgenic plants. However, a significantly higher increase in free proline content was observed in the transgenic lines L68 and L78 when compared to the WT. In addition, compared with the WT, the soluble sugar content increased significantly in the transgenic lines L68 and L76 (Figure 8C, D). Taken together these results indicate that SiLEA14 overexpressing foxtail millet showed improved drought resistance.
GUS activity is induced by various stresses in proSiLEA14::GUS transgenic Arabidopsis
The putative SiLEA14 promoter region was isolated from the foxtail millet genome using PCR. This fragment consisted in nt −1273 to +96 (transcription start site is +1) upstream of SiLEA14 coding sequence. The SiLEA14 promoter was fused to the GUS gene and transformed into Arabidopsis. Then, qRT-PCR was carried out to examine the GUS expression level under various stresses. As it is shown in Figure 9, GUS transcription was up-regulated to its highest level (about 3- and 8-fold increases) after 6 h treatment with ABA and NaCl, respectively. For PEG stress, GUS expression gradually increased and peaked (an almost 12-fold increase) after 18 h treatment. To further confirm these results, a histochemical GUS-staining assay was performed. No visible GUS activity was noted for the control. However, induction of GUS activity was observed with ABA, PEG or NaCl application for 3 to 18 h, although the GUS signals were not particularly strong (Additional file 5). Nevertheless, collectively these results suggest that the selected SiLEA14 promoter sequence is enough to regulate ABA, PEG and NaCl induction of the gene.
Figure 9.
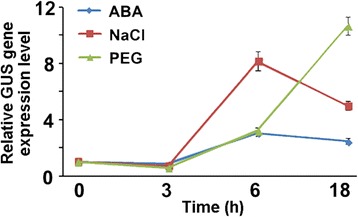
Expression of GUS gene in pro SiLEA14 ::GUS transgenic Arabidopsis upon various stresses determined by qRT-PCR. One-week-old seedlings were treated with 100 μM ABA, 250 mM NaCl and 20% (m/v) PEG 6000 solution for 0, 3, 6 and 18 h, respectively. Total RNA was extracted from at least 30 whole seedlings per treatment. The expression of GUS in proSiLEA14::GUS transgenic Arabidopsis was determined by qRT-PCR using GUS-specific primers (Additional file 4). Actin2 was used to normalize the gene expression. Data represent means and standard errors for three biological replicates.
SiLEA14 promoter contains stress-associated cis-elements
To characterize the mechanism of SiLEA14 function, a 1273 bp SiLEA14 promoter fragment (without the 96 bp 5′UTR) was subjected to a putative cis-acting regulatory element search using the Plant Cis-acting Regulatory DNA Elements (PLACE) database [32]. Cis-acting elements reported to be involved in ABA- and dehydration-mediated gene expression were identified (Table 1). Abscisic acid-responsive elements (ABREs) are common motifs for ABA- and dehydration-mediated regulation of transcription [33-35]. In the SiLEA14 promoter, six ABRE-like sequences and four ABA- and dehydration-responsive ACGT-box motifs [34,36] were identified. A dehydration-responsive element (DRE)-like site, as a cis-acting element involved in ABA induction, drought, salinity, or cold stresses [37,38], was also identified at position −158. MYC and MYB binding sites have been identified as regulatory elements associated with ABA, drought, or cold responsiveness in plants [34,39-43]. In the SiLEA14 promoter, as many as 10 and three MYC- and MYB-like sequences, respectively, were observed. These results suggest that SiLEA14 may be under the regulation of DREB-, DBF-, CBF-, MYC- or MYB-like transcription factors as in Arabidopsis and rice. In addition, other regulatory cis-acting elements were identified in the SiLEA14 promoter, such as low temperature-responsive, pathogen-, defense- and wounding-associated elements. However, whether SiLEA14 participates in these abiotic or biotic stresses awaits further investigation.
Table 1.
Putative cis- acting regulatory elements associated with ABA and various stresses in SiLEA14 promoter region
| Cis -element name | Sequence | Position | Function | Ref. |
|---|---|---|---|---|
| ABRE | ACGTG | −793, −792, −133, −78, −77 | ABA-mediated regulation of transcription; Required for etiolation-induced expression of erd1 in Arabidopsis | [34,35] |
| RYACGTGGY | −79 | ABRE element involved in Arabidopsis dehydration-responsive gene rd22 | [33] | |
| ACGT | −792, −382, −132, −77 | Involved in ABA response; Required for etiolation-induced expression of erd1 in Arabidopsis | [34,36] | |
| DRE | ACCGAC | −158 | DBF1 and DBF2 binding site in the maize rab17 gene promoter involved in ABA induction | [38] |
| RCCGAC | −158 | DRE/CRT regulatory element involved in dehydration, cold or sanility responsiveness | [38] | |
| RYCGAC | −158 | Binding site of barley CBF1 and CBF2 involved in cold acclimation | [37] | |
| MYC | CACATG | −1034, −485 | MYC binding site in rd22 gene of Arabidopsis; ABA-induction | [55] |
| CANNTG | −1034, −867, −793, −666, −588, −553, −485, −468, −433, −78 | MYC recognition site in rd22, CBF3 and many other genes in Arabidopsis; Binding site of ICE1 that regulates the transcription of CBF/DREB1 genes in the cold in Arabidopsis | [41,42] | |
| CATGTG | −1034, −485 | MYC recognition sequence necessary for expression of erd1 in dehydrated Arabidopsis; Binding site of NAC which is stress-inducible | [34,43] | |
| MYB | CNGTTR | −468, −465, −360 | MYB binding site involved in regulation of genes that are responsive to water stress in Arabidopsis | [39,40] |
| YAACKG | −468 | MYB recognition sequence found in the promoters of rd22 and many other genes in Arabidopsis | [41] |
M = C/A; Y = T/C; B = T/C/G; N = A/T/G/C; R = A/G; W = A/T; V = A/C/G; K = G/T.
Discussion
Typical LEA proteins can retain water molecules and protect other proteins from aggregation or desiccation because of their highly hydrophilic properties [44]. Conversely, atypical LEA proteins have higher content of hydrophobic residues than typical LEAs. The latter have been speculated to be involved in diverse stress tolerances, although few studies have been carried out to characterize their functions [5,23,25]. Here, we reported the identification and characterization of SiLEA14, a novel atypical LEA member, in foxtail millet.
As a key phytohormone, ABA plays an important role in plant stress responses. Osmotic stress-regulated genes can be activated through both ABA-dependent and ABA-independent pathways [45]. However, it is considered that stress-signalling pathways for the activation of LEA-like genes completely independent of ABA may not exist [1]. In our study, SiLEA14 accumulation was remarkably induced by ABA and peaked rapidly after 1 h treatment (Figure 3). In addition, SiLEA14 promoter-driven GUS activity was distinctly stimulated by ABA (Figure 9). Compared with the WT, transgenic foxtail millet seeds showed better germination in ABA solution (Figure 6C–H). All of these results indicate that activation of SiLEA14 under salt and drought stresses may be dependent on ABA.
Yeast and E. coli heterologous systems have been widely used to investigate LEA gene functions [5,6,46]. In the present study, overexpression of SiLEA14 protected E. coli cells from damage caused by salt stress (Figure 4). Overexpression of SiLEA14 in Arabidopsis imparted increased tolerance to salt and mannitol stresses (Figure 5). This result suggests that SiLEA14 from foxtail millet, a monocot, can function properly in the dicot Arabidopsis. Interestingly, transgenic foxtail millet overexpressing SiLEA14 exhibited superior germination and subsequent growth in soil compared with the WT even under normal conditions. Under salt and drought stresses, these differences were more remarkable, which were indicative of the key roles of SiLEA14 in foxtail millet (Figures 6, 7 and 8). Consistent with these findings, both SiLEA14 transcription and SiLEA14 promoter-driven GUS activity were remarkably induced by NaCl and PEG (Figures 3, 9 and Additional file 5). However, it should be noted that some discrepancies were observed between the expression pattern of the endogenous SiLEA14 upon abiotic stress and ABA treatment (Figure 3) and the SiLEA14 promoter-driven GUS transcript accumulation responsive to abiotic stress and ABA driven by SiLEA14 promoter region in transgenic Arabidopsis (Figure 9). For example, the SiLEA14 transcripts accumulated to the highest level after 250 mM NaCl treatment for 3 h (Figure 3), whereas the GUS transcripts reached the peak after 6 h treatment (Figure 9). Additionally, the histochemical analysis showed strong GUS signals in the petioles of transgenic Arabidopsis (Additional file 5), whereas the endogenous SiLEA14 was highly expressed in roots (Figure 3). This is probably due to that SiLEA14 is from the monocot foxtail millet and the GUS expression driven by SiLEA14 promoter is performed in the dicot Arabidopsis.
So far, there is only one report on the functional mechanism of the atypical LEA protein in plants [25]. Overexpression of IbLEA14, a homologous gene of SiLEA14, enhanced tolerance to drought and salt stress in the sweetpotato calli. The contents of lignin in the IbLEA14-overexpressing calli were increased under normal conditions. The authors inferred that IbLEA14 may be involved in these functions as a consequence of regulating increased lignin production. In our study, we found that overexpression of SiLEA14 in Arabidopsis and foxtail millet obviously improved the osmotic stress resistance of transgenic plants. Meanwhile, the proline and sugar accumulated at a higher level in transgenic lines, especially in L68 which accumulated higher SiLEA14 transcripts, than WT after osmotic and NaCl stress. These results implied that overexpression of SiLEA14 might up-regulate these metabolites. However, further study is needed.
The identification of cis-acting elements in the SiLEA14 promoter may help to provide insight into the molecular mechanism of SiLEA14 function. As a major cis-acting element, ABRE has been identified in the promoters of many ABA-inducible genes of plants such as the cotton LEA gene D-113 [47]. The bZIP transcription factors AREB/ABF can bind to ABRE and activate ABA-inducible gene expression [48-51]. In the present study, ten cis-elements containing the ABRE ACGT-core were identified in the −793 to −77 bp region of the SiLEA14 promoter. The DRE element was first identified in the promoter of rd29A, a gene responsive to dehydration stress in Arabidopsis [52]. In plants, the AP2 transcription factors DREB/CBF specifically bind to the DRE element to regulate the expression of the downstream stress-responsive genes [53]. There are interactions between ABRE and DRE in Arabidopsis rd29A gene expression in response to dehydration and high-salinity stresses [54]. One DRE-like element was predicted in the SiLEA14 promoter. In Arabidopsis, an MYC transcription factor, AtMYC2 and an MYB transcription factor, AtMYB2, have been shown to bind the cis-elements CANNTG and C/TAACNA/G, respectively, to regulate expression of the dehydration-responsive gene RD22 [41,55]. Ten MYC and three MYB-like sequences were identified in the SiLEA14 promoter. Proteins that bind to these elements remain to be isolated. It is necessary to further elucidate the detailed functions of these putative regulatory cis-elements. In addition, pathogen-related elements were also identified. However, whether SiLEA14 participates in biotic stress responses needs to be investigated.
Conclusions
In conclusion, this study characterizes a novel atypical LEA gene SiLEA14 from foxtail millet. SiLEA14 is responsive to ABA, PEG and NaCl and the SiLEA14 is localized in the cytosol. SiLEA14 improves the salt tolerance of E. coli transformant and transgenic Arabidopsis. Furthermore, overexpression of SiLEA14 significantly enhances the salt and drought tolerances of transgenic foxtail millet. SiLEA14 plays important roles in plant abiotic stress resistance and could be used in crops genetic engineering with the aim of improving stress tolerance.
Methods
Plant materials and growth conditions
Seeds of foxtail millet (Setaria italica cv. Jigu11), kindly provided by Prof. Xianmin Diao of the Institute of Crop Science, Chinese Academy of Agricultural Sciences, China, were geminated on moist gauze for 24 h at 30°C, and then grown in pots filled with nutrient soil and vermiculite mixed at 1:1 (v/v) in the controlled chamber (25–26°C, humidity 60-70%, under 16-h light/8-h darkness). Foxtail millet leaves, stems, roots, inflorescences and seeds at 5, 15 and 25 days after pollination were harvested and stored at −80°C after frozen in liquid nitrogen.
Seeds of Arabidopsis thaliana (Col-0), after stratification, were plated on ½MS medium with 2% sucrose and 0.7% agar for three weeks at 21–22°C with a 16 h/8 h (day/night) photoperiod and 60-70% relative humidity. Then, the young seedlings were planted on fertilized soil and grown in the same conditions.
Stress treatments
For SiLEA14 expression profile in response to ABA, NaCl and PEG, two-week-old foxtail millet seedlings were carefully removed from soil and washed. The cleaned plants were fixed in plastic foam, and grown hydroponically in water for 1 d. Then, the seedling roots were immersed separately in water containing 100 μM ABA, 250 mM NaCl and 20% (m/v) PEG 6000 for the indicated time in Figure 3, respectively. Six seedlings were used in each treatment. After drying on the filter paper, the seedlings were harvested, and then immediately frozen in liquid nitrogen and stored at −80°C until use.
For GUS expression in response to ABA, NaCl and PEG, about 20 one-week-old proSiLEA14::GUS transgenic Arabidopsis plants were carefully removed from the plates and immersed in water containing 100 μM ABA, 250 mM NaCl and 20% (m/v) PEG 6000 for 0, 3, 6 and 18 h, respectively. For histochemical GUS staining, three-week-old proSiLEA14::GUS transgenic Arabidopsis plants were used.
Cloning and sequence analysis of SiLEA14
The full-length SiLEA14 was amplified by RACE in accordance with the manufacturer’s protocol (GeneRacer™ Kit, Invitrogen, Carlsbad, CA, USA). The products were cloned into the pMD19-T vector (Takara, Shiga, Japan) and sequenced. Primer sets used are listed in Additional file 4. The isoelectric point and molecular mass predictions were estimated using the compute pI/Mw tool (http://expasy.org/tools/pi_tool.html). Analysis of protein hydropathy was done by constructing hydropathy plots with the Kyte and Doolittle algorithm (http://ipsort.hgc.jp/) [56]. Motif analysis was performed using the Pfam program (http://www.ebi.ac.uk/Tools/InterProScan/). The grand average of hydropathy (GRAVY) and instability index of deduced proteins were predicted using the ProtParam program (http://au.expasy.org/tools/protparam.html). Sequence similarities were determined using the BLAST program and the GenBank database on the NCBI web server. The complete amino acid sequences of subgroup 5C LEA proteins were used to construct a phylogenetic tree. Sequence alignment was performed with ClustalW and adjusted manually. A phylogenetic tree was constructed with the neighbor-joining method using the MEGA4.0 program [57]. Sequence logos for subgroup 5C LEA14 were obtained with the WebLogo website http://weblogo.berkeley.edu/logo.cgi [58].
RNA extraction, semi-quantitative PCR and qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and first-strand cDNA was prepared with SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) after digestion with RNase-free DNase I (Takara, Shiga, Japan). The semi-quantitative RT-PCR were conducted as follows: 95°C for 3 min, then 25 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 30 s for both SiLEA14 and Actin2. For qRT-PCR, 100 ng of cDNAs were used as template in a 20 μL reaction system, containing 10 μL 2× SYBR Premix Ex Taq II (TaKaRa, Shiga, Japan), and 0.5 μM each specific forward and reverse primer (Additional file 4). Amplification was performed using the Bio-Rad CFX96 Real-Time PCR System C1000 Thermal Cycler (Bio-Rad, USA) as follows: 95°C for 30 s, 35 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s. Arabidopsis Actin2 (accession number: NM_180280) and foxtail millet actin7 (accession number: NM_001280818) were used as the endogenous references. Primers used were listed in Additional file 4.
Subcellular localization of SiLEA14
The coding sequence of SiLEA14 without the terminating codon was amplified and inserted into the XbaI/SmaI sites of pROK219-GFP to generate the construct pROK219-SiLEA14-GFP. Onion epidermal cells were bombarded with the constructs pROK219-GFP and pROK219-SiLEA14-GFP, which were validated by sequencing, using a particle gun-mediated system PDS-1000/He (Bio-Rad, Hercules, CA, USA). Foxtail millet protoplast isolation and transfection were carried out according to the procedure described by Zhai et al. [59]. Root tissues from 7-day-old seedlings were sliced and then incubated in a solution containing 1.5% Cellulase RS, 0.75% Macerozyme R10, 0.6 M mannitol, 10 mM MES, 0.1% BSA and 1 mM CaCl2 for 4–5 h at 28°C in the dark with gentle swirling (50 rpm). The constructs pROK219-SiLEA14-GFP and pROK219-GFP were incubated with protoplasts and 40% PEG 4000 for 20 min at room temperature for transient transformation, respectively. GFP signals were observed with a confocal laser scanning microscopy (LSM 510, Carl Zeiss MicroImaging GmbH, Jena, Germany).
Assay for salt-stress tolerance of E. coli transformants
The coding sequence of SiLEA14 without the stop codon was amplified and cloned into the EcoRV/XhoI sites of pET30a(+) to construct the expression vector pET30a-SiLEA14, which was then transformed into E. coli host strain BL21. The pET30a(+) empty vector was used as the control. The expression of SiLEA14 in the recombinant cells was confirmed by SDS-PAGE analysis (Additional file 6). Transformed E. coli BL21 cells carrying pET30a-SiLEA14 or pET-30a (+) were grown in LB liquid medium supplemented with 100 μg/ml ampicillin overnight at 37°C, respectively. The bacterial cultures were diluted 100-fold using fresh liquid LB, and allowed to incubate for 2–3 h at 37°C until OD600 = 0.5–0.6. Isopropylthio-β-D-galactoside was then added to the cultures to a final concentration of 1 mM, and the bacteria were cultured for a further 4 h at 30°C to induce expression of the inserted gene. All induced cultures were adjusted to OD600 = 0.6 using fresh liquid LB medium with 100 μg/ml ampicillin. To measure responses to salt stress, the samples were diluted by 200-, 500-, 1000-, 2000- and 4000-fold with fresh LB medium supplemented with 100 μg/ml ampicillin. Five microliters of each diluted sample were plated on LB agar plates, LB agar plates supplemented with 600 mM KCl and 600 mM NaCl, respectively. After incubation for 12 h on LB agar plates or 24 h on LB agar plates supplemented with 600 mM KCl and 600 mM NaCl at 37°C respectively, the numbers of colonies were calculated. Growth was measured at least three times.
Generation of transgenic plants
For SiLEA14 overexpression in Arabidopsis, the coding region of SiLEA14 was amplified and ligated into the HindIII/SpeI sites of pSB1300, which was kindly provided by Prof. Shuhua Yang of the College of Biological Sciences, China Agricultural University, China, to generate the construct pSB1300-SiLEA14 in which SiLEA14 was controlled by Super promoter. For SiLEA14 promoter assay, the putative SiLEA14 promoter was isolated from the foxtail millet genome using PCR and cloned into the SalI/EcoRI sites of pCAMBIA1391-GUS to generate the construct pCAMBIA1391-proSiLEA14-GUS. After validation by sequencing, the constructs were introduced into Agrobacterium tumefaciens strain GV1301, and transformed into Arabidopsis by the floral dip method [60]. Seeds were obtained following self-pollination.
For SiLEA14 overexpression in foxtail millet, the coding region of SiLEA14 fused with a flag tag at the 3′end was amplified and ligated into the SacI/KpnI sites of the binary vector pCOU [61], which includes the ubiquitin promoter to drive transgene expression. After confirmation by sequencing, the recombinant plasmid pCOU-SiLEA14-flag was introduced into A. tumefaciens strain LBA4404. Transgenic foxtail millet plants were obtained by Agrobacterium-mediated transformation as described previously [62,63]. Seeds were obtained following self-pollination.
Abiotic stress-tolerance assay of SiLEA14 transgenic Arabidopsis
Seeds of WT and T3 transgenic Arabidopsis plants were surface sterilized by the vapor-phase method [60], and sown on MS, MS + 125 mM NaCl and MS + 250 mM mannitol media, respectively. After 3 days vernalization at 4°C, they were cultured in a controlled chamber (22°C, humidity 40-50%, 120–150 μmol/m−2 s−1 under 16-h light/8-h darkness). Photographs were taken after 7 days culture. Fresh and dry weights of each sample were calculated based on the average weight of 20 individual plants.
Abiotic stress tolerance assay of SiLEA14 transgenic foxtail millet
For analysis in the germination stage, 30–40 seeds of both WT and T2 transgenic millet were geminated on the filter paper in a Petri dish wet with water (as control) or water containing 150 mM NaCl, 250 mM NaCl, 10% PEG, 20% PEG and10 μM ABA for 1 day at 30°C, respectively. Photographs were taken after 4 and 9 days, respectively. Shoot and root lengths were measured.
For salt tolerance assay in soil, two-week-old seedlings were irrigated with water, 150 and 250 mM NaCl solution every 3 days, respectively. After 6 days, the phenotypes of the transgenic lines and WT were investigated. Six to eight plants grown in one plot were used in each experiment. Two fully expanded young leaves from each foxtail millet plant per plot were harvested and cut into 1 cm segment for electrolyte leakage measurement.
For drought tolerance assay in soil, two-week-old seedlings were deprived of water for 7 days. Subsequently, the plants were irrigated with water and grown for 3 days. Six to ten plants grown in one plot were used in each experiment. The survived plants were counted. Above-ground parts of treated seedlings were collected and used to measure proline and soluble sugar contents.
Electrolyte leakage assay
Leaf tissue (0.1 g) from each sample was washed and immersed in 20 mL deionized water with 150 rpm shaking for 16 h. The initial electrical conductivity (L1) of the sample was detected using a FE-30 conductivity meter (Mettler-Toledo, Columbus, OH, USA). Then, the samples were autoclaved at 121°C for 10 min and cooled to room temperature. The ultimate conductivity (L2, maximum conductivity of tissues) was measured. Relative electrical conductivity (L) was calculated as the ratio of L1/L2.
Proline content measurement
Free proline content of foxtail millet plants was measured using the method described by Bates et al. [64]. Leaf tissue (0.1 g, dry weight) was used to extract free proline in 3% sulphosalicylic acid at 95°C for 15 min. Then, 2 mL of supernatant was transferred to a new tube and reacted with 2 mL acetic acid and 2 mL acidified ninhydrin reagent for 30 min at 95°C. Next, 5 ml of toluene was added to the tube with full shaking. The absorbance of the toluene layer was determined at 520 nm.
Soluble sugar content measurement in foxtail millet leaves
Soluble sugar content of foxtail millet plants was examined using the method of Yemm and Willis [65]. Leaf tissue (0.1 g, dry weight) was used to extract soluble sugar in 7 ml of 80% ethanol with constant stirring at 80°C for 2 h. Ethanol was evaporated in the boiled water bath. Then, 1 ml water and 5 ml of 0.15% anthrone solution was added. After incubation at 95°C for 15 min and cooling to room temperature, the absorbance of the reaction solution was determined at 620 nm. Glucose was used as a standard.
Histochemical GUS staining
Histochemical GUS staining was performed as described by Jefferson et al. [66]. After the GUS staining, Arabidopsis seedlings were treated with 70% ethanol to remove chlorophyll from the GUS-stained tissue.
Statistical analysis
The survival rate, fresh/dry weights, relative electrolyte leakage rate, proline content and soluble sugar content data were subjected to Student’s t-test analyses using GraphPad Prism 5. All of the experiments were repeated three times.
Availability of supporting data
The SiLEA14 sequence was deposited in GenBank with an accession number of KJ767551. The data supporting the results of this article are included within the article and its additional files.
Acknowledgements
This work was supported by the National Basic Research Program of China (Grant No. 2012CB215301 to YJ).
Additional files
Sequence analyses of SiLEA14. (A) The nucleotide and deduced amino acid sequences of SiLEA14. The transcription site was indicated in large red case. The start codon and stop codon of SiLEA14 are marked in normal red case. The regular distribution of polar amino acid residues in SiLEA14 protein was underlined in black for acidic and amide residuces (N, D, Q and E) and red lines for basic residues (R, K and H). (B) The gene structure of SiLEA14. The position and length of the exons and intron of the SiLEA14 gene are displayed schematically. The green rectangles indicate the exons, and the black line indicates the intron. (C) Hydropathy analysis of SiLEA14 protein sequence using Kyte-Doolittle algorithm. Amino acid position is plotted on the x axis beginning with the N-terminus. Hydrophobic regions (I, II, and III) in the conserved motif were marked. (D) Amino acid content (mol %), GRAVY and instability index of SiLEA14.
Sequence information of SiLEA14 homologs.
The sequence logo for the conserved domain of SiLEA14 protein and its homologs.The overall height of each stack represents the conservation of the protein sequences at that amino acid position, whereas the height of letters within each stack indicates the relative frequency of the corresponding amino acid.
Primer sets used in this study.
Histochemical GUS staining of transgenic Arabidopsis containing pro SiLEA14 -GUS fusion upon various stresses. Three-week-old seedlings subjected to 100 μM ABA, 20% PEG or 250 mM NaCl for 0, 3, 6 and 18 hours, respectively, were used.
SiLEA14 expression in the recombinant E. coli cells by SDS-PAGE analysis. M. Protein marker. Lane 1, pET30a (+) uninduced. Lane 2, pET30a (+) induced. Lane 3, pET30a-SiLEA14 uninduced. Lane 4, pET30a-SiLEA14 induced. Lane 5, Supernate of pET30a-SiLEA14 induced extracts. Lane 6, Pellet of pET30a-SiLEA14 induced extracts. Red triangle represents the HIS tag protein (7 KD). Green triangles represent the SiLEA14 fusion protein.
Footnotes
Meizhen Wang and Ping Li contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MW, PL and JY conceived and designed the experiments. MW, PL, CL, YP and XJ performed the experiments. QZ and DZ participated in the coordination of the experiments. MW and JY wrote the original manuscript. JY thoroughly revised the manuscript and finalized the manuscript. All authors read and approved the final manuscript.
Contributor Information
Meizhen Wang, Email: mzwang1981@163.com.
Ping Li, Email: liping198920062006@126.com.
Cong Li, Email: lnlicong_123@163.com.
Yanlin Pan, Email: panyanlinll@163.com.
Xiyuan Jiang, Email: xiyuan.jiang@gmail.com.
Dengyun Zhu, Email: zhudy@cau.edu.cn.
Qian Zhao, Email: zhaoqian@cau.edu.cn.
Jingjuan Yu, Email: yujj@cau.edu.cn.
References
- 1.Xiong L, Schumaker KS, Zhu J. Cell signaling during cold, drought and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dure L, Galau GA. Developmental biochemistry of cotton seed embryogenesis and germination XIII. Regulation of biosynthesis of principal storage proteins. Plant Physiol. 1981;68:187–194. doi: 10.1104/pp.68.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker J, Steele C, Dure IIIL. Sequence and characterization of 6 LEA proteins and their genes from cotton. Plant Mol Bio. 1988;11:277–291. doi: 10.1007/BF00027385. [DOI] [PubMed] [Google Scholar]
- 4.Bartels D, Singh M, Salamini F. Onset of desiccation tolerance during development of the barley embryo. Planta. 1988;175:485–492. doi: 10.1007/BF00393069. [DOI] [PubMed] [Google Scholar]
- 5.He S, Tan L, Hu Z, Chen G, Wang G, Hu T. Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol Genet Genomics. 2012;287:39–54. doi: 10.1007/s00438-011-0660-x. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Wang L, Xing X, Sun L, Pan J, Kong X, Zhang M, Li D. ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 2013;0:1–16. doi: 10.1093/pcp/pct047. [DOI] [PubMed] [Google Scholar]
- 7.George S, Usha B, Parida A. Isolation and characterization of an atypical LEA protein coding cDNA and its promoter from drought-tolerant plant Prosopis juliflora. Appl Biochem Biotechnol. 2009;157:244–253. doi: 10.1007/s12010-008-8398-6. [DOI] [PubMed] [Google Scholar]
- 8.Dalal M, Tayal D, Chinnusamy V, Bansal KC. Abioticstress and ABA-inducible group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol. 2009;139:137–145. doi: 10.1016/j.jbiotec.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Olvera-Carrillo Y, Campos F, Luis Reyes J, Garciarrubio A, Covarrubias AA. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010;154:373–390. doi: 10.1104/pp.110.158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dure L, III, Crouch M, Harada J, Ho T-HD, Mundy J, Quatrano R, Thomas T, Sung ZR. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12:475–486. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- 11.Bray EA. Molecular responses to water deficit. Plant Physiol. 1993;103:1035–1040. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bies-Ethe’ve N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:107–124. doi: 10.1007/s11103-008-9304-x. [DOI] [PubMed] [Google Scholar]
- 14.Cuming AC. LEA proteins. In: Casey R, Shewry PR, editors. Seed Proteins. The Netherlands: Kluwer Academic Publishers; 1999. pp. 753–780. [Google Scholar]
- 15.Galau GA, Wang HY, Hughes DW. Cotton Lea5 and Lea74 encode atypical Late Embryogenesis-Abundant proteins. Plant Physiol. 1993;101:695–696. doi: 10.1104/pp.101.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh S, Cornilescu CC, Tyler RC, Cornilescu G, Tonelli M, Lee MS, Markley JL. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Sci. 2005;14:2601–2609. doi: 10.1110/ps.051579205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larre C, Satour P, Leprince O. Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol. 2006;140:1418–1436. doi: 10.1104/pp.105.074039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boucher V, Buitink J, Lin X, Boudet J, Hoekstra FA, Hundertmark M, Renard D, Leprince O. MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 2010;33:418–430. doi: 10.1111/j.1365-3040.2009.02093.x. [DOI] [PubMed] [Google Scholar]
- 20.Piatkowski D, Schneider K, Salamini F, Bartels D. Characterization of five abscisic acid-responsive cDNA clones isolated from the desiccation-tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiol. 1990;94:1682–1688. doi: 10.1104/pp.94.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maitra N, Cushman JC. Isolation and characterization of a drought-induced soybean cDNA encoding a D95 family late-embryogenesis-abundant protein. Plant Physiol. 1994;106:805–806. doi: 10.1104/pp.106.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zegzouti H, Jones B, Marty C, Lelievre JM, Latche A, Pech JC, Bouzayen M. ER5, a tomato cDNA encoding an ethylene-responsive LEA-like protein: characterization and expression in response to drought, ABA and wounding. Plant Mol Biol. 1997;35:847–854. doi: 10.1023/A:1005860302313. [DOI] [PubMed] [Google Scholar]
- 23.Kim HS, Lee JH, Kim JJ, Kim CH, Jun SS, Hong YN. Molecular and functional characterization of CaLEA6, the gene for a hydrophobic LEA protein from Capsicum annuum. Gene. 2005;344:115–23. doi: 10.1016/j.gene.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M, Yamamoto YY, Seki M, Sakurai T, Abe T, Yoshida S, Manabe K, Shinozaki K, Matsui M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:ioagrb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Park SC, Kim YH, Jeong JC, Kim CY, Lee HS, Bang JW, Kwak SS. Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta. 2011;233:621–634. doi: 10.1007/s00425-010-1326-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W, Tao Y, Bian C, Han C, Xia Q, Peng X, Cao R, Yang X, Zhan D, Hu J, Zhang Y, Li H, Li H, Li N, Wang J, Wang C, Wang R, Guo T, Cai Y, Liu C, Xiang H, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 2012;2012(30):549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
- 27.Doust AN, Kellogg EA, Devos KM, Bennetzen JL. Foxtail millet: a sequence-driven grass model system. Plant Physiol. 2009;149:137–141. doi: 10.1104/pp.108.129627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robin APS, Reidunn BA. Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis-abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta. 1998;206:476–478. doi: 10.1007/s004250050424. [DOI] [PubMed] [Google Scholar]
- 29.Sreenivasulu N, Grimm B, Wobus U, Weschke W. Differential response of antioxidant compounds to salinity stress in salt-tolerant and salt-sensitive seedlings of foxtail millet (Setaria italica) Physiol Plantarum. 2000;109:435–442. doi: 10.1034/j.1399-3054.2000.100410.x. [DOI] [Google Scholar]
- 30.Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotech. 2005;16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Bhatnagar-Mathur P, Vadez V, Sharma K. Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 2008;27:411–424. doi: 10.1007/s00299-007-0474-9. [DOI] [PubMed] [Google Scholar]
- 32.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (place) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwasaki T, Yamaguchi-Shinozaki K, Shinozaki K. Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet. 1995;247:391–398. doi: 10.1007/BF00293139. [DOI] [PubMed] [Google Scholar]
- 34.Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003;33:259–270. doi: 10.1046/j.1365-313X.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol. 2006;60:51–68. doi: 10.1007/s11103-005-2418-5. [DOI] [PubMed] [Google Scholar]
- 36.Shen Q, Zhang P, Ho TH. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue GP. Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 2002;30:e77. doi: 10.1093/nar/gnf076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L, encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313X.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- 39.Luscher B, Eiseman RN. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990;4:2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- 40.Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinozaki K, Yamaguchi-Shinozaki K. Molecular response to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. doi: 10.1016/S1369-5266(00)80068-0. [DOI] [PubMed] [Google Scholar]
- 46.Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One. 2012;7:e45117. doi: 10.1371/journal.pone.0045117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo K, Zhang G, Deng W, Luo F, Qin K, Pei Y. Functional characterization of a cotton late embryogenesis-abundant D113 gene promoter in transgenic tobacco. Plant Cell Rep. 2008;27:707–717. doi: 10.1007/s00299-007-0482-9. [DOI] [PubMed] [Google Scholar]
- 48.Guiltinan MJ, Marcotte WR, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–71. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 49.Choi H, Hong JH, Ha J, Kang JY, Kim SY. ABFs, a family of ABA-responsive elements binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 50.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcriptional transcription factors involved in an abscisic aciddependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Alonso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–48. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 55.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kyte J, Doolittle RF. A simple method for displaying thehydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 57.Saitu N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic tress. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 58.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhai Z, Jung H, Vatamaniuk OK. Isolation of protoplasts from tissues of 14-day-old Seedlings of Arabidopsis thaliana. J Vis Exp. 2009;30:e1149. doi: 10.3791/1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 61.Zhao L, Liu F, Xu W, Di C, Zhou S, Xue Y, Yu J, Su Z. Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol J. 2009;7:550–561. doi: 10.1111/j.1467-7652.2009.00423.x. [DOI] [PubMed] [Google Scholar]
- 62.Qin F, Zhao Q, Ao G, Yu J. Co-suppression of Si401, a maize pollen specific Zm401 homologous gene, results in aberrant anther development in foxtail millet. Euphytica. 2008;163:103–111. doi: 10.1007/s10681-007-9610-4. [DOI] [Google Scholar]
- 63.Wang M, Pan Y, Li C, Liu C, Zhao Q, Ao G, Yu J. Culturing of immature inflorescences and Agrobacterium-mediated transformation of foxtail millet (Setaria italica) Afr J Biotechnol. 2011;10:16466–16479. [Google Scholar]
- 64.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 65.Yemm EW, Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. Biochem J. 1954;57:508–514. doi: 10.1042/bj0570508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: b-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


