Abstract
Pancreatic Lgr5 expression has been associated with organoid-forming epithelial progenitor populations but the identity of the organoid-initiating epithelial cell subpopulation has remained elusive. Injury causes the emergence of an Lgr5+ organoid-forming epithelial progenitor population in the adult mouse liver and pancreas. Here, we define the origin of organoid-initiating cells from mouse pancreas and liver prior to Lgr5 activation. This clonogenic population was defined as MIC1-1C3+/CD133+/CD26− in both tissues and the frequency of organoid initiation within this population was approximately 5% in each case. The transcriptomes of these populations overlapped extensively and showed enrichment of epithelial progenitor-associated regulatory genes such as Sox9 and FoxJ1. Surprisingly, pancreatic organoid cells also had the capacity to generate hepatocyte-like cells upon transplantation to Fah-/- mice, indicating a differentiation capacity similar to hepatic organoids. Although spontaneous endocrine differentiation of pancreatic progenitors was not observed in culture, adenoviral delivery of fate-specifying factors Pdx1, Neurog3 and MafA induced insulin expression without glucagon or somatostatin. Pancreatic organoid cultures therefore preserve many key attributes of progenitor cells while allowing unlimited expansion, facilitating the study of fate determination.
Keywords: Organoid, transplantation, reprogramming
Introduction
The potential for progenitor-driven hepatic and/or pancreatic organ regeneration is of significant interest for regenerative medicine. Expression of the Rspondin receptor Lgr5 and activation of the Wnt signaling pathway have been shown to be useful epithelial stem cell markers in these tissues (Barker et al., 2010; Sato et al., 2009). Epithelial Lgr5+ cells are highly clonogenic, initiating self-renewing organoids that can retain differentiation potential despite many rounds of passage/expansion and therefore fulfilling the formal criteria for stem cells. Although Lgr5 is not expressed in uninjured adult liver or pancreas, expression of this marker has been shown to be induced following tissue damage (Huch et al., 2013b). Lgr5+ cells recovered from injured mouse liver form organoids and yield hepatocytes after transplantation and appropriate selection (Huch et al., 2013b). Similarly, Lgr5+ cells from the adult mouse pancreas can establish organoid cultures (Huch et al., 2013a). In both tissues, the precursor cell capable of producing the Lgr5+ population upon injury remains poorly characterized.
Several groups, including ours, have reported the cell surface signature of a biliary duct-resident clonogenic progenitor in t mouse liver (Dorrell et al., 2011; Shin et al., 2011; Suzuki et al., 2008). Our identification employed a clonogenic assay based on two-dimensional growth on conventional tissue culture surfaces, and the self-renewal capacity of progenitors defined by this method was inherently limited. However, given the ability of progenitor-derived cells to express characteristic hepatocyte genes in vitro, we surmised that this population contained (and retained) meaningful differentiation potential.
The pancreas and liver share developmental origins (Kawaguchi, 2013; Zaret and Grompe, 2008) and the ductal epithelial cells in these adult organs have similar functions and morphological appearance. The appearance of hepatocyte-like cells in the pancreata of rats fed a copper-deficient diet (Rao et al., 1986) illustrates the potential for transdifferentiation between cell types from these organs. In mice, the transplantation of pancreatic cells has been shown to yield functional hepatocytes in Fah−/− recipients (Wang et al., 2001). These embryological and functional similarities as well as the existence of an injury induced Lgr5+ population in both tissues raised the possibility that the adult mouse pancreas and liver may harbor a common endodermal progenitor.
In this report we describe the phenotypic definition and FACS-based isolation of a clonal progenitor present at similar frequencies in both the pancreas and liver of adult mice. This hepato-pancreatic MIC1-1C3+/CD133+/CD26− (hereafter M+133+26−) population initiates organoid cultures with unlimited self-renewal potential. The gene expression profiles of these populations revealed that factors such as Sox9 are enriched in both organoid-forming populations in a pattern consistent with the regulation of progenitor function. When single cells from the pancreatic M+133+26− population were examined, heterogeneous expression of Sox9 was observed, suggesting a correlation with organoid-forming capacity. Remarkably, transplantation of organoids derived from sorted pancreatic M+133+26− cells yielded hepatocyte-like cell grafts in the livers of 5/10 of recipient mice, indicating that this population retains important differentiation potential even after massive expansion in culture. Furthermore, M+133+26− organoid cultures yielded insulin-expressing cells after induction of Pdx1/Neurog3/MafA expression, suggesting that a capacity for endocrine differentiation was also retained. The gene expression profiles of the progenitor-enriched populations characterized here reveal new information regarding the nature and potential of adult epithelial progenitors, and may guide future efforts to enhance their activity in situ or to control their fate during ES/iPS cell differentiation.
Results
Identification and comparison of duct cell subpopulations in the adult mouse pancreas and liver
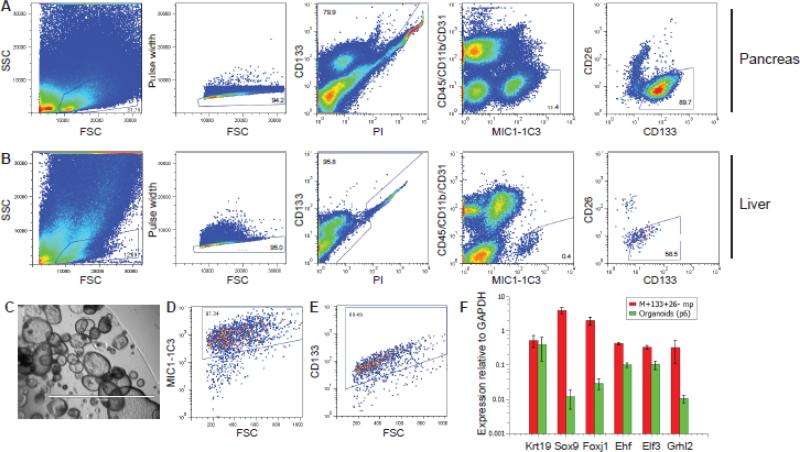
To study adult mouse pancreatic progenitors and compare their characteristics to their hepatic counterparts, cells were obtained by sequential enzymatic tissue dispersal and labeled with combinations of antibodies recognizing cell surface antigens. Figure 1 illustrates the sequential gating strategy used to define subpopulations of mouse pancreatic (A) or hepatic (B) cells. These gates allowed the exclusion of pancreatic acinar cells or hepatocytes (high FSC/SSC), erythrocytes (low FSC/SSC), leukocytes (CD45+/CD11b+) and endothelial cells (CD31+). The percentage of cells labeled by duct cell surface marker MIC1-1C3 (Dorrell et al., 2008) was substantially higher in pancreatic than in liver tissue, as anticipated; the pancreas is substantially more ductal than the liver. Sub-fractionation of the MIC1-1C3+ population by CD133 and CD26 antigenicity revealed that most cells were CD133+, but a smaller (~10% ) population of CD133−CD26+ cells was consistently observed. qRT-PCR expression analysis (Figure 2A) indicated that each population consisted of KRT19+ duct cells, but that these were heterogeneous for progenitor and mature gene expression markers as previously observed in the liver (Dorrell et al., 2011). Both the pancreatic and hepatic M+133+26− subpopulations share a differentially high expression of progenitor associated genes (Sox9/Foxj1), and a collection of transcriptional regulators recently implicated in epithelial cell fate regulation (Ehf, Elf3, Grhl2) (Albino et al., 2012; Senga et al., 2012; Yamaguchi et al., 2010). Thus, the gene expression phenotype of the organoid-initiating cell population was consistent with that of a primitive duct cell type.
Figure 1.
Isolation and cultivation of organoid-forming cell subpopulations from pancreas and liver. Cells isolated from pancreas (A) or liver (B) were enzymatically dispersed, antibody labeled and sorted by FACS. Successive gating shows sequential selection of cell-sized events (FSC vs. SSC), non-doublets (FSC vs. pulse width), live cells (PI-), non-hematopoietic/endothelial events (CD45/CD11b/CD31-) and duct cells (MIC1-1C3 subpopulations). (C) A representative image of a mouse pancreatic organoid culture initiated by M+133+26− cells (original magnification 40x). (D,E) Flow cytometric measurement of duct-associated cell surface markers on cells dispersed from an established organoid culture. (F) qRTPCR analyses of the levels of duct and progenitor-associated genes (relative to GAPDH) in pancreatic M+133+26− cells compared to those from an organoid culture (at passage 6) they were used to establish. These values are the averages of three replicate amplifications.
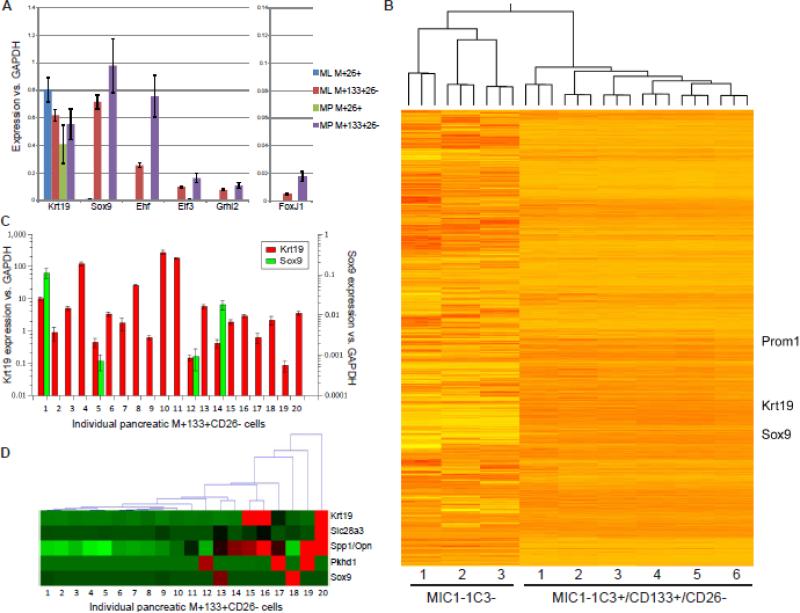
Figure 2.
Expression analysis of progenitor-enriched cell populations. (A) qRT-PCR was used to compare the expression of several genes in MIC1-1C3+ subpopulations of mouse liver and pancreas. Values shown are the averages of values from three pancreatic and two liver samples, each analyzed three times by qRT-PCR. (B) Hierarchical clustering of RNA-seq derived expression values from progenitor-enriched and depleted mouse pancreatic populations. (C) qRT-PCR analyses of twenty individual pancreatic M+133+26− cells captured and processed using the Fluidigm C1 Single-Cell Auto Prep IFC. The levels of Sox9, Krt19 and Actb for each cell were determined in three replicates, and amplified products were validated by electrophoresis. Sox9 and Krt19 expression levels are as delta-Ct relative to Actb. (D) Hierarchical clustering of these same twenty cells on the basis of qRT-PCR-determined expression of duct and associated genes. Expression is normalized per-gene, with red indicating the highest level(s).
Prospective isolation of pancreatic and hepatic organoid-forming cells
We previously reported that among mouse hepatic duct cells, only the MIC1-1C3+/CD133+/CD26− (M+133+26−) epithelial subset contained 2D colony-forming progenitors (Dorrell et al., 2011). Although useful, this assay suffered from a limited capacity for self-renewal and was unsuitable for the expansion of progenitor-initiated cells. Consequently, we adopted the epithelial organoid assay developed by the Clevers lab (Barker et al., 2010; Huch et al., 2013b) and used it to assess the progenitor content of phenotypically-defined pancreatic duct subpopulations.
The frequencies of organoid-forming cells detected in FACS-sorted populations are listed in Table 1. For consistency of enumeration, only organoids estimated to contain ≥500 cells at day 12 were scored as such. As had been observed in the 2D colony assay, the M+133+26− fraction contained by far the highest frequency of colony-forming cells; M+26+ duct cells did not form organoids at a significant frequency. These same patterns were observed with non-parenchymal liver cells, and at similar frequencies. Thus, the mouse organoid forming cells of liver (Huch et al., 2013b) and pancreas (Huch et al., 2013a) can be prospectively isolated and studied using the M+133+26− labeling combination. The self-renewal capacity of organoid-forming cells was assessed by secondary organoid formation. Organoids derived from individually deposited and microscopically confirmed FACS-sorted M+133+26− cells were harvested on d12, dispersed to single cells by trypsinization and re-seeded. Primary organoids which met the aforementioned size criteria yielded multiple daughter organoids >95% of the time (after five independent sorts), indicating that self-renewal potential was consistently maintained. Cell cultures incapable of 10+ passages were excluded from analysis. Culture initiation frequencies were comparable in single-cell (96w plated) and 1000-cell (24w plated) environments (Table 1), indicating environmental independence and clonality. Pancreatic organoids were phenotypically very ductal (Fig. 1C) as previously described (Huch et al., 2013a). Although the surface phenotype of cultured organoid cells remained primarily M+133+26− (Fig. 1D,E) and Krt19 expression was similar to that of the parent population, the expression of progenitor markers such as Sox9 was >10 fold lower (Fig. 1F).
Table 1.
Quantification of organoid-forming progenitors in phenotypically defined pancreatic cell subpopulations.
| Organ | Population | Organoids per 1000 input cells | SD | Single-cell deposition | |
|---|---|---|---|---|---|
| Pancreas | CD45/11b/31+ | Blood/endothelium | <1 | na | |
| CD45−11b−31− | Non-blood/endothelium | 4 | 2 | ||
| CD45−11b−31−MIC1-1C3+ | Duct cells | 31 | 13 | ||
| CD45−11b−31−MIC1-1C3− | Non-duct cells | 2.7 | 2 | ||
| CD45−11b−31−MIC1-1C3+CD26+ | Progenitor-depleted duct cells | <1 | na | ||
| CD45−11b−31−MIC1-1C3+133+26− | Progenitor-enriched duct cells | 44.6 | 14 | 1 in 19.7 | |
| Liver | CD45/11b/31+ | Blood/endothelium | <1 | na | |
| CD45−11b−31− | Non-blood/endothelium | 3.3 | 3 | ||
| CD45−11b−31−MIC1-1C3+CD26+ | Progenitor-depleted duct cells | 1.1 | 2 | ||
| CD45−11b−31−MIC1-1C3+133+26− | Progenitor-enriched duct cells | 38.5 | 11 | 1 in 17.4 |
MIC1-1C3 binds to Integrin alpha 3
MIC1-1C3 reactivity has been invaluable for the FACS isolation of hepato-pancreactic duct cell subsets from mouse tissues. The identification of the associated antigen and investigation of its potential involvement in epithelial cell regulation was therefore of potential interest. Co-IP of protein lysate prepared from a mouse cell line (H2.35) known to exhibit high reactivity with MIC1-1C3 yielded a band of approximately 120 kDa (Fig. S1A). The list of possible targets revealed after band excision and MALDI-TOF mass spectrometric identification is shown in Figure S1B. The two most strongly indicated proteins were integrin beta 1 (ITGB1) and integrin alpha 3 (ITGA3), the two components of the VLA3/CD49c complex. Of these, ITGA3 (molecular weight 117 kDa) matched most closely. RNA expression levels supported this interpretation: ITGA3 mRNA in MIC1-1C3+ cells from liver and pancreas was found to be significantly higher than in MIC1-1C3− cells (Fig. S1C), whereas ITGB1 mRNA levels were not significantly different (Fig. S1D). A comparison of the labeling of MIC1-1C3 and a polyclonal goat anti-CD49c antibody on consecutive sections of pancreatic tissue (Fig. S1E,F) suggested that the same cells were being marked, albeit with different efficiencies.
Gene expression in pancreatic organoid-forming cells
The transcriptomes of FACS-isolated pancreatic cell populations from six different mice were assessed by RNA-seq and compared to MIC1-1C3− (CD45−CD31−) pancreatic populations from three mice. As shown in Figure 2B, hierarchal clustering of these samples reveals large blocks of genes with distinctive expression in the organoid-forming populations compared to non-ductal cells. Comparative analyses of the M+133+26− and M− population pathways were performed using GSEA; KEGG output is listed in Figure S2A. Among the categories enriched in M+133+26− cells are those known to be associated with duct function (adherens, gap, and tight junctions) and cell proliferation (cell cycle, DNA replication). DNA replication (Fig. S2B), Mismatch repair (Fig. S2C) and RIG1-like receptor signaling (Fig. S2D) had nominal p values <0.05, and are broken out in greater detail.
Although the M+133+26− fractions of mouse pancreas and liver contain all of the organoid-forming activity, only a small percentage of these cells actually initiate organoid cultures (as previously described). This could be a stochastic process in which the cells are essentially homogeneous but form organoids by chance or due to some combination of environmental factors, or because a more primitive subset exists within the M+133+26− subpopulation. To examine this issue, we performed single-cell analyses on individual M+133+26− cell using a Fluidigm C1 apparatus and examined the expression levels of Krt19, Amy2, Sox9, and Actb. In cells with readily detectable beta actin (indicating efficient amplification of the original mRNA) and keratin 19 (confirming the duct identity of the examined cell) and with negligible or absent amylase (i.e. non-acinar), Sox9 levels were highly variable (Figure 2C). In 16/20 cells Sox9 was undetectable, and in the remaining cells the expression of Sox9 varied over a 100-fold range. Thus, the expression of this gene appears to vary from cell to cell within the pancreatic M+133+26− subpopulation. Hierarchical clustering of these results plus those of duct markers Spp1, Slc28a3 and Pkhd1 (Figure 2D) reveals considerable variability, with a tendency for cells with high Sox9 expression to have lower expression of other duct-associated genes. These results may indicate true heterogeneity within this duct population or reflect dynamic transcription within a relatively homogeneous set of cells.
Hepatic differentiation potential of pancreatic organoid cells
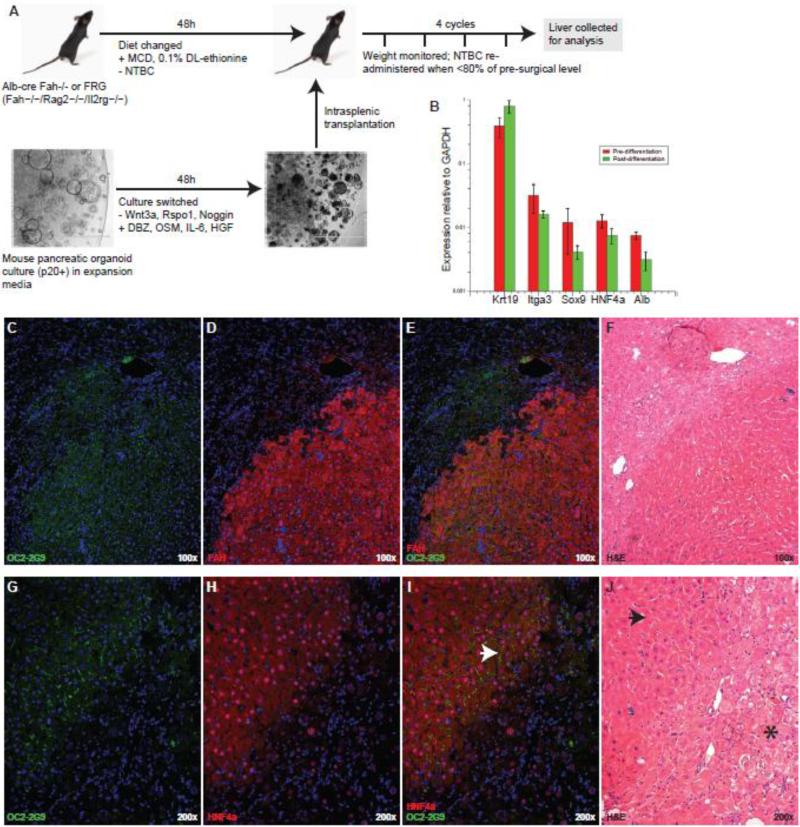
One goal of epithelial progenitor expansion cultures is the derivation of useful numbers of transplantable cells for the treatment of human pathologies. We have previously shown that hepatic organoids can produce hepatocytes upon transplantation to Fah−/− mice (Huch et al., 2013b). Considering the phenotypic and transcriptional similarities between the organoid initiating cells from liver and pancreas, the similar nature of the resulting organoid cultures, and evidence showing the potential for pancreatic/hepatic lineage conversion (Rao et al., 1986; Wang et al., 2001) we tested whether pancreatic organoids could also give rise to hepatocytes after transplantation. Figure 3A illustrates the cell culture and transplantation scheme. Pancreatic organoid cultures were expanded to >100,000 cells per intended recipient mouse then switched to a hepatocyte differentiation medium in which Wnt3, Rspo1 and Noggin were withdrawn and DBZ, Oncostatin M, IL-6, and HGF were administered for a 48h period. Recipient Alb-cre/Fah−/− and Fah−/−/Rag2−/−/Il2rg−/− (FRG) animals were conditioned with a methionine/choline-deficient and ethionine-supplemented (MCD+E) diet over the same 48h interval, with the intention of temporarily creating a “progenitor-friendly” in vivo environment for transplanted cells. This brief period in hepatocyte differentiation culture was not sufficient to produce hepatocyte-associated gene expression. As shown in in Figure 3B, cells remained very ductal and do not express meaningful levels of albumin or HNF4a. The intention of this strategy was to initiate the very earliest steps of differentiation in vitro but allow most of the process to occur in vivo. Pancreatic organoid transplants yielded multiple FAH+ nodular grafts in half (5/10) of the recipient mice. Five additional mice which received MCD+E conditioning but were transplanted with organoids that were not exposed to differentiation culture failed to produce grafts, suggesting that this was an important part of the process. Representative examples of grafts in two different animals are illustrated in Figure 3C-J, Engrafted nodules expressed markers of hepatocyte identity (canalicular OC2-2G9, nuclear HNF4a, and nuclear+cytoplasmic FAH) and exhibited characteristic hepatocytic morphology. H&E staining of consecutive sections of FAH+ nodules revealed healthy tissue, lacking the pale, vacuolar features of tyrosinemia found in surrounding FAH- host liver. Engraftment levels were low (~1% of liver mass), falling below the threshold for therapeutic rescue, so it is unclear whether the engrafting cells had hepatocyte function. The suitability of a R26R-LacZ donor cells and Alb-cre Fah−/− recipients to detect fusion-derived hepatocytes was confirmed by control experiments where R26R-mTmG hepatocytes or bone marrow to an Alb-cre/Fah−/− recipient (Fig. S3). As we have previously described (Willenbring et al., 2004), bone marrow injection results in macrophage-hepatocyte fusion, and viable fusion products with hepatocyte function are selected by NTBC withdrawal in this model. Hepatocyte transplantation (Fig. S3B) does not result in fusion as a detectable frequency, but bone marrow/hepatocyte fusion products are readily detectable. The absence of signal in Fah+ liver nodules after pancreatic R26R-LacZ organoid transplantation is therefore strong evidence for the absence of fusion.
Figure 3.
Pancreatic organoid cultures give rise to hepatocyte-like cells after transplantation to the livers of Fah−/− mice. (A) The experimental design of pancreatic organoid culture and recipient mouse treatment to determine the potential for hepatocytic differentiation. (B) Expression of duct and hepatocyte-associated genes measured by qRT-PCR. No significant differences in the pre- and post-differentiation (pre-transplantation) expression of these genes were observed in two experiments (each sample examined three times by qRT-PCR). (C-F) and (G-J) Representative examples of engrafted nodules identified in two different animals.Immunofluorescent (OC2-2G9, FAH, HNF4a) detection and H&E staining were performed on adjacent sections of FFPE liver. Arrowheads mark examples of properly organized bile canaliculi (I) and healthy hepatocyte-like cells (J), and an asterisk marks a region of unhealthy, highly vacuolated hepatocytes highly invaded by inflammatory cells.
Induction of insulin expression with endocrine programming factors
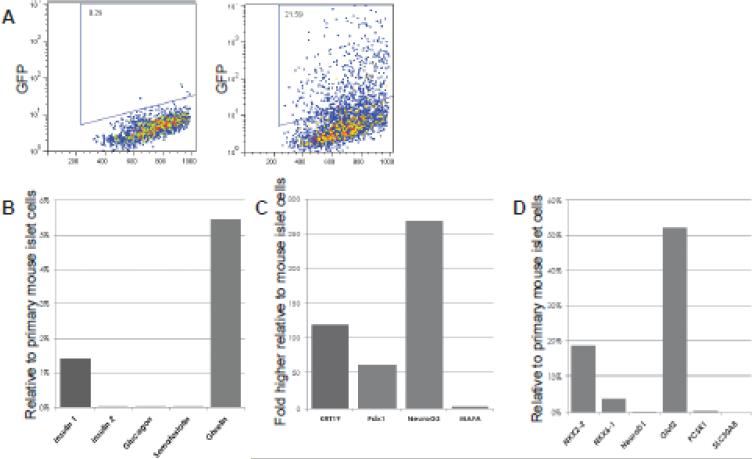
The most obvious objective of pancreatic tissue cultivation is the derivation of glucose-responsive insulin-expressing cells for cell replacement therapy of diabetes. To test whether this highly expandable culture system could yield insulin-expressing cells, we employed a tricistronic adenovector co-expressing MafA, Pdx1, and Neurog3 (Akinci et al.) and organoid cultures derived from the pancreata of MIP-GFP transgenic mice (Hara et al., 2003). As shown in Figure 4A, insulin (GFP) expression was induced in late-passage MIP-GFP pancreatic organoid cells (at a frequency of 5-22%) following tri-cistronic AdV administration. These GFP+ (insulin promoter active) cells showed transcriptional identity partially overlapping that of murine beta cells (Fig. 4B), but with the retained expression of many off-target (non-beta) genes. Nevertheless, the capacity of organoid cultures for dramatic cell number expansion suggests that a more refined reprogramming/differentiation methodology could produce useful cells in unlimited numbers.
Figure 4.
Delivery of endocrine fate-specifying factors to expanded pancreatic organoid cultures yields insulin-expressing cells in vitro. (A) GFP expression in MIP-GFP mouse pancreatic organoid cells 72h after mock transduction or transduction with adenovirus expressing mouse Pdx1, Neurog3 and MafA, measured by flow cytometry. (B-D) Expression of endocrine- and duct-associated genes in Ad-PNM (Pdx1-NeuroG3-MafA) treated pancreatic organoid cells relative to mouse islet cDNA, measured by qRTPCR. Organoid cell cDNA was recovered from the entire culture (GFP+ and GFP-).
Discussion
The roles of endogenous stem/progenitor cells in adult pancreatic tissue homeostasis, acute/chronic injury response and tumor initiation remain uncertain. However, it is now clear that a subset of pancreatic duct cells are capable of initiating long-term self-renewing organoid cultures with effectively unlimited potential for expansion in culture. The M+133+26− subsets of both the pancreas and liver of adult mice contains the only cells with this capability. Lgr5, which is a marker for self-renewing stem cells in the colon, small intestine and stomach only becomes expressed after injury in the adult liver and pancreas (Barker et al., 2010; Barker et al., 2007; Huch et al., 2013a; Huch et al., 2013b). Hence, in these latter tissues Lgr5 is only a marker for activated progenitors and cannot be used to prospectively identify clonogenic cells in normal, uninjured tissue. Our study is the first to report the prospective purification of “inactive” pancreatic and liver mouse organoid forming progenitors. We applied population expression profiling with RNA-seq and single cell analyses to determine the transcriptomes and transcriptional heterogeneity of these important populations. The ability to purify both inactive and injury-activated progenitors will permit the study of the molecular circuitry underlying progenitor activation in the future.
Although heterogeneity of pancreatic ducts has long been known based on morphological criteria, we demonstrate here for the first time that pancreatic duct cells are heterogeneous in terms of clonogenic potential. Thus it is appropriate to distinguish between clonogenic and non-clonogenic ducts. The surface marker combinations used herein do not permit the unambiguous identification of these duct subsets in histological sections. Hence, we currently do not know whether clonogenic and non-clonogenic ducts reside in anatomically distinct locations or whether they are intermingled and lie adjacent to each other. Our RNA-seq analysis represents a list of distinctive markers that could be used to localize the progenitors in tissue sections and generate lineage-tracing tools in the future. The observation of high Sox9 expression in only a small subset of M+133+26− is interesting, and it is tempting to speculate that this property is predictive of their organoid-forming capacity. We note that heterogeneity of SOX9 at the protein level has recently been reported in epithelial tissues. Ramalingam et al. found that intestinal epithelial cells express SOX9 at distinct levels, and that only the highest expressing subpopulation had progenitor activity (Ramalingam et al., 2012).
We have recently shown that gall bladder derived epithelial cells can be reprogrammed to become endocrine-like and express insulin using a combination of Pdx1, Ngn3 and MafA (Hickey et al., 2013). Others have shown that biliary duct cells in the liver can be reprogrammed in vivo and cure diabetes in mice (Akinci et al., 2012). Here we demonstrate that pancreatic duct cells can similarly be reprogrammed even after massive expansion in organoid culture. The expandability of the pancreatic progenitors makes them a potential target for generating transplantable beta cells. Ducts and acinar cells are routine byproducts of islet isolation procedures using in clinical transplantation (Baeyens and Bouwens, 2008). Recently, the laboratory of S. Kim has shown that human ducts can also be reprogrammed to become functional beta-like cells (Lee et al., 2013). Therefore, the progenitor population described here may have potential utility in beta cell replacement therapy for diabetes.
The common phenotype and similar frequencies of the pancreatic and liver organoid initiating cells, and the observation that pancreas-derived progenitors can generate hepatocyte-like cells in an appropriate context provides yet more evidence for the commonality of these tissues. One implication is that pancreatic tissue could be a source (or location) for the restoration of lost liver function and vice versa. Additional study of the regulatory events that specify hepatic or pancreatic differentiation from these progenitors will be needed to test these ideas.
Materials and Methods
Tissue sources and pancreatic cell isolation
Animal care and immunization procedures were performed in accordance with protocol IS00000119 of the institutional review committee at Oregon Health & Science University. C57Bl/6 mice, obtained from the Jackson Laboratory, were fed a Purina 5015 diet. For transplantation studies, Fah−/− C57Bl/6 recipient animals were used (Grompe et al., 1993).
Immunofluorescent imaging
Mouse pancreatic cryosections (5 μm) were prepared using a Reichert 2800 Frigocut (Reichert Scientific Instruments) and preserved in acetone for 10’ at −20°C. Non-specific labeling was blocked by pre-incubation in 2% goat serum (Hyclone) for 10’. Antibodies used included anti-CD49c (R&D Systems) and rabbit anti- mouse Fah ((Overturf et al., 1996)). Primary labeling employed hybridoma supernatants diluted 1:20 in DPBS for 30 minutes and secondary labeling with 1:200 dilutions of Cy3-conjugated anti-rat Ig pre-adsorbed against mouse serum proteins (Jackson ImmunoResearch, West Grove, PA) for 20 minutes. Nuclei were visualized with Hoechst 33342 (Molecular Probes, Eugene, OR). A Zeiss Axioskop 2 plus microscope (Carl Zeiss, Jenna, Germany) was used for imaging.
Flow cytometry and FACS
Dissociated cells were resuspended at 1×106 cells/ml in DMEM + 2% FBS prior to the addition of MIC1-1C3 hybridoma supernatant (at a 1:20 dilution) or a 1:200 dilution of purified MIC1-1C3 antibody (Novus) and incubation at 4°C (for 30’). After a wash with cold DPBS, cells were resuspended in DMEM + 2% FBS containing a 1:200 dilution of APC-conjugated goat anti-rat secondary antibody adsorbed against mouse serum proteins (Jackson Immunoresearch). After another wash, cells were resuspended in DMEM + 5% rat serum (Serotec) and held on ice (10’) to block the secondary antibody. A final incubation with FITC-conjugated anti-CD26 (BD Biosciences, Franklin Lakes, NJ), PE-conjugated anti-CD133 (eBioscience, San Diego, CA) and PE-Cy7-conjugated anti-CD45 (BD Biosciences) + anti-CD11b/Mac1 (BD Biosciences) + anti-CD31 (Abcam, Cambridge, MA) facilitated cell subfractionation and exclusion gating of hematopoietic (CD45+, CD11b+) and endothelial (CD31+) cells. Propidium iodide staining was used to label dead cells for exclusion. Cells were analyzed and sorted with a Cytopeia inFluxV-GS (Becton-Dickenson, Franklin Lakes, NJ); FSC : Pulse-width gating was used to exclude cell doublets from analysis and collection.
Organoid formation assay
Mouse pancreatic and hepatic organoids were initiated, cultured and cryopreserved as described previously (Huch et al., 2013b) with minor modifications. Gastrin was omitted from the culture media and constitutive administration of ALK5 inhibitor SB431542 (Tocris) was added. For organoid cultures initiated by single FACS-sorted cells, 5% Matrigel was added to chilled liquid media and cells were directly deposited into this mixture. The organoids initiated in these cultures were passaged into “standard” organoid conditions (embedding in >95% Matrigel followed by addition of liquid media) after two weeks of growth.
Adenoviral transduction of organoid cultures
For reprogramming studies, pancreatic organoid cultures initiated from an MIP-GFP transgenic mouse pancreas (Hara et al., 2003). Prior to transduction, organoids were recovered from Matrigel, fragmented by mechanical disruption with a p1000 pipetter (Rainin) and seeded on collagen-coated plasticware in organoid expansion media. Eight hours later, tri-cistronic adenovirus encoding mouse Pdx1, Neurog3 and MafA (ad-PNM) generously provided by the laboratory of Jonathan Slack was added at an MOI of 1:10. The media was changed after over-night transduction, and the emergence of GFP expression was monitored by fluorescence microscopy. After 72h, cells were recovered by exposure to 0.05% Trypsin (Hyclone) and evaluated by flow cytometry and qRT-PCR (described below).
Cell capture and cDNA preparation from individual cells using the Fluidigm C1
Approximately 2,000 MIC1-1C3+CD133+CD26−CD45−CD31−CD11b− cells were isolated from an ActBGFP transgenic mouse pancreas by FACS as described above. These were then loaded onto a 10-17μm C1 Single-Cell Auto Prep IFC chamber (Fluidigm), and cell capture was performed according to the manufacturer's instructions. Capture efficiency was 86% (83/96 wells occupied with a single cell) as determined by fluorescent microscopy. Both the empty wells (8) and doublet-occupied wells (5) were noted and excluded from further analysis. Upon capture, reverse transcription and cDNA pre-amplification were performed in the 10-17μm C1 Single-Cell Auto Prep IFC using the SMARTer Ultra Low RNA and Advantage 2 PCR kits (Clontech). cDNA was then harvested and assessed with an Agilent Bioanalyzer at the OHSU Massively Parallel Sequencing Core. Yielded cDNA concentration was 500-700 pg/μl and the average size of approximately 1.5kb.
Transplantation assay
The injection of sorted cell populations into the spleen and the withdrawal of NTBC to induce hepatocyte selection were performed as previously described (Overturf et al., 1996), with additional procedures listed in Figure 3A Drug withdrawal was performed in cycles of approximately three weeks, terminated when the weight of the recipient mouse dropped below 80% of its pre-surgical level. NTBC was then re-administered until normal weights were restored.
Limiting dilution analysis
For limiting dilution assays of organoid formation, Poisson statistics for the single-hit model were applied. Cells were seeded at densities ranging from 1 to 25 per well and counts were performed at d12.
Antigen identification by mass spectrometry
Whole cell extracts from H2.35 cell lines were fractionated by SDS-PAGE on a 10% Bis-Tris gel under reducing conditions. A unique band was visualized by Bio-Safe Coomasie stain (Bio Rad) and excised. Following trypsin digestion, the fragments were analyzed on a ThermoFinnigan LTQ (Thermo Scientific) tandem mass spectrometer. Spectra files were analyzed with Sequest. Scaffold (Proteome Software Inc., Portland, OR) was used to compile and assign probability scores.
RNA isolation and qRT-PCR
Cell populations were sorted directly into Trizol LS (Invitrogen). First strand cDNA synthesis employed MMLV reverse transcriptase and random oligonucleotide primers (both Invitrogen). RNA levels were assessed by qRT-PCR using a Bio-Rad iCycler and IQ5 detection system. All reactions were performed using 45 cycles of 0:15 at 95°C, 0:20 at 68°C and 0:25 @ 72°C. Reaction mixtures included Platinum Taq DNA polymerase (Invitrogen), 2.5 mM MgCl2, 10 μM 5’ and 3’ primers, 10 mM dNTPs and 0.5x SYBR green. Primer sequences are listed in Table S1. Gene expression levels were reported as the difference between baseline-corrected, curve-fitted cycle thresholds for the gene of interest minus the average cycle thresholds of housekeeping genes GAPDH and/or beta-actin as indicated. Curve-fitting of qRT-PCR cycle threshold results was generated by IQ5 software (Bio-Rad) and statistical mean and standard deviation data were determined with Microsoft Excel.
Highligths.
We report a shared cell surface phenotype for mouse liver and pancreatic progenitors
The organoid-forming cell frequency is very similar in mouse liver and pancreatic ducts
Single cell analysis shows heterogeneity in the pancreatic progenitor population
Pancreatic organoid cells can form hepatocytes after transplantation
Acknowledgments
These studies would not have been possible without the assistance of Meritxell Huch, who introduced the mouse organoid-forming assay to our group. We are grateful for the contributions of Leslie Wakefield, who provided animals and assistance with molecular analyses. John Klimek of the OHSU Shared Proteomics Resource offered guidance with interpretation of mass spectrometry results. The work of Angela Major in embedding, sectioning and staining specimens of mouse liver is greatly appreciated. Thanks also to Ray Hickey, who designed some of the PCR primers employed in these studies.
Grant support: This work was supported by National Institutes of Health grant DK051592 (M.G.) and DK089569 (P.S. and M.G.)
Abbreviations
- FACS
fluorescence-activated cell sorting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: OHSU has commercially licensed a portion of the technology described herein (MIC1-1C3/MPdi1); authors C.D., P.R.S and M.G. are inventors of this antibody. This potential conflict of interest has been reviewed and managed by OHSU. The other authors disclose no conflicts.
References
- Akinci E, Banga A, Greder LV, Dutton JR, Slack JM. Reprogramming of pancreatic exocrine cells towards a beta (beta) cell character using Pdx1, Ngn3 and MafA. Biochem J. 2012;442:539–550. doi: 10.1042/BJ20111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albino D, Longoni N, Curti L, Mello-Grand M, Pinton S, Civenni G, Thalmann G, D'Ambrosio G, Sarti M, Sessa F, et al. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res. 2012;72:2889–2900. doi: 10.1158/0008-5472.CAN-12-0212. [DOI] [PubMed] [Google Scholar]
- Baeyens L, Bouwens L. Can beta-cells be derived from exocrine pancreas? Diabetes Obes Metab. 2008;10(Suppl 4):170–178. doi: 10.1111/j.1463-1326.2008.00949.x. [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Erker L, Lanxon-Cookson KM, Abraham SL, Victoroff T, Ro S, Canaday PS, Streeter PR, Grompe M. Surface markers for the murine oval cell response. Hepatology. 2008;48:1282–1291. doi: 10.1002/hep.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C, Erker L, Schug J, Kopp JL, Canaday PS, Fox AJ, Smirnova O, Duncan AW, Finegold MJ, Sander M, et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011;25:1193–1203. doi: 10.1101/gad.2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG, Soriano P. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- Hara M, Wang X, Kawamura T, Bindokas VP, Dizon RF, Alcoser SY, Magnuson MA, Bell GI. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284:E177–183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- Hickey RD, Galivo F, Schug J, Brehm MA, Haft A, Wang Y, Benedetti E, Gu G, Magnuson MA, Shultz LD, et al. Generation of islet-like cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. 2013;11:503–515. doi: 10.1016/j.scr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013a;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013b;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y. Sox9 and programming of liver and pancreatic progenitors. J Clin Invest. 2013;123:1881–1886. doi: 10.1172/JCI66022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sugiyama T, Liu Y, Wang J, Gu X, Lei J, Markmann JF, Miyazaki S, Miyazaki J, Szot GL, et al. Expansion and conversion of human pancreatic ductal cells into insulin-secreting endocrine cells. Elife. 2013;2:e00940. doi: 10.7554/eLife.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, Daughtridge GW, Johnston MJ, Gracz AD, Magness ST. Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am J Physiol Gastrointest Liver Physiol. 2012;302:G10–20. doi: 10.1152/ajpgi.00277.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Subbarao V, Reddy JK. Induction of hepatocytes in the pancreas of copper-depleted rats following copper repletion. Cell Differ. 1986;18:109–117. doi: 10.1016/0045-6039(86)90005-9. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell. 2012;23:2845–2855. doi: 10.1091/mbc.E12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Walton G, Aoki R, Brondell K, Schug J, Fox A, Smirnova O, Dorrell C, Erker L, Chu AS, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Sekiya S, Onishi M, Oshima N, Kiyonari H, Nakauchi H, Taniguchi H. Flow cytometric isolation and clonal identification of self-renewing bipotent hepatic progenitor cells in adult mouse liver. Hepatology. 2008;48:1964–1978. doi: 10.1002/hep.22558. [DOI] [PubMed] [Google Scholar]
- Wang X, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. Liver repopulation and correction of metabolic liver disease by transplanted adult mouse pancreatic cells. Am J Pathol. 2001;158:571–579. doi: 10.1016/S0002-9440(10)63999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbring H, Bailey AS, Foster M, Akkari Y, Dorrell C, Olson S, Finegold M, Fleming WH, Grompe M. Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nat Med. 2004;10:744–748. doi: 10.1038/nm1062. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Kojima T, Ito T, Kimura Y, Imamura M, Son S, Koizumi J, Murata M, Nagayama M, Nobuoka T, et al. Transcriptional control of tight junction proteins via a protein kinase C signal pathway in human telomerase reverse transcriptase-transfected human pancreatic duct epithelial cells. Am J Pathol. 2010;177:698–712. doi: 10.2353/ajpath.2010.091226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]